Abstract
Small noncoding RNAs (sRNAs) regulate gene expression in Escherichia coli by base pairing with mRNAs and modulating translation and mRNA stability. The sRNAs DsrA and RprA stimulate the translation of the stress response transcription factor RpoS by base pairing with the 5′ untranslated region of the rpoS mRNA. In the present study, we found that the rpoS mRNA was unstable in the absence of DsrA and RprA and that expression of these sRNAs increased both the accumulation and the half-life of the rpoS mRNA. Mutations in dsrA, rprA, or rpoS that disrupt the predicted pairing sequences and reduce translation of RpoS also destabilize the rpoS mRNA. We found that the rpoS mRNA accumulates in an RNase E mutant strain in the absence of sRNA expression and, therefore, is degraded by an RNase E-mediated mechanism. DsrA expression is required, however, for maximal translation even when rpoS mRNA is abundant. This suggests that DsrA protects rpoS mRNA from degradation by RNase E and that DsrA has a further activity in stimulating RpoS protein synthesis. rpoS mRNA is subject to degradation by an additional pathway, mediated by RNase III, which, in contrast to the RNase E-mediated pathway, occurs in the presence and absence of DsrA or RprA. rpoS mRNA and RpoS protein levels are increased in an RNase III mutant strain with or without the sRNAs, suggesting that the role of RNase III in this context is to reduce the translation of RpoS even when the sRNAs are acting to stimulate translation.
RpoS is a general stress response sigma factor made in Escherichia coli in response to several types of stresses and in the stationary phase of growth. An increase in the cellular levels of RpoS results in transcription of many genes, whose products help the cell cope with stress. The levels of RpoS in the cell are very tightly controlled and are modulated in response to specific changes in the growth environment (reviewed in reference 19).
Regulation of RpoS levels in response to environmental signals occurs at the levels of transcription, translation, and protein stability. In the absence of stress, when cells are growing rapidly, RpoS levels are very low, synthesis is low, and the protein is rapidly degraded. When cells run out of nutrients or encounter other types of stress, RpoS levels rise rapidly, a result of changes in both synthesis and degradation.
For instance, during exponential growth at 37°C, RpoS is rapidly degraded by the ClpXP ATP-dependent protease (46). This degradation requires the adaptor protein RssB (also called SprE), which binds to RpoS and delivers it to ClpXP (37, 40, 64). In response to specific suboptimal growth conditions, RpoS becomes stable. Stabilization of RpoS occurs in response to specific antiadaptor proteins that bind to RssB, blocking its ability to deliver RpoS to the protease. Antiadaptors made in response to phosphate starvation, magnesium starvation, and DNA damage have been described (3, 4).
The major translational control of RpoS depends upon the 5′ untranslated region (UTR) of rpoS and small noncoding RNAs (sRNAs). This regulation was first demonstrated in experiments showing that translation of rpoS is enhanced during growth at low temperature (20 to 25°C). Translational control under this growth condition is completely dependent on the presence of the sRNA DsrA (48). Transcription of DsrA is activated at low temperature, and upon accumulation, this sRNA base pairs with rpoS mRNA in the 5′ UTR, resulting in increased translation of RpoS (28, 43; reviewed in reference 30).
Regulation of translation by sRNAs is a widely studied phenomenon in E. coli and is rapidly being recognized as an important mechanism in other bacteria (62). A major class of regulatory sRNAs is synthesized in response to many kinds of stresses and base pairs with target mRNAs. Base pairing is stimulated by the RNA chaperone Hfq, which binds to both the sRNA and the target mRNA (22; reviewed in reference 5). The most frequent outcome of pairing is negative regulation of the target mRNAs, including degradation of the mRNA. Degradation of those target mRNAs that have been tested in E. coli is mediated by the degradosome, a protein complex containing the endoribonuclease RNase E, the exoribonuclease polynucleotide phosphorylase (PNPase), and other components (16, 31, 35, 57).
A rarer outcome of pairing is positive regulation by sRNAs, such as has been seen for regulation of rpoS translation by DsrA (28). A second sRNA, RprA, stimulates RpoS synthesis in response to cell envelope stress and base pairs with rpoS mRNA in the same region of the 5′ UTR as DsrA (27, 29). A number of other examples of positive regulation by sRNAs have been described (34, 38, 41, 55, 60). Thus, it seems likely that the positive regulation seen with DsrA and RprA is mimicked by many other sRNAs. Many of these positively acting sRNAs also act negatively on other mRNAs.
A number of studies have examined the interaction of DsrA and RprA and their targets in vitro and in vivo, and crucial base pairs for the action of DsrA and RprA have been identified (25, 26, 28, 30, 33, 50). Furthermore, genetic studies and in vitro analysis have suggested that the rpoS mRNA forms a hairpin that occludes the ribosome binding site in the 5′UTR if Hfq or the sRNAs are absent (6, 26). In vitro, base pairing with the sRNA can relieve this occlusion (26). Hfq binding to multiple sites on the rpoS mRNA significantly enhances binding by the sRNAs (49, 58). However, little is known about the fate of the mRNA upon binding to an sRNA in vivo.
The present study analyzes the fate of the rpoS mRNA in the presence and absence of the sRNAs DsrA and RprA in vivo. We find that rpoS mRNA is unstable and is degraded in a fashion that involves the two major endoribonucleases, RNase E and RNase III. Expression of either DsrA or RprA can stabilize the mRNA, and this requires base pairing; the stabilization overcomes degradation by RNase E but not by RNase III. Blocking degradation of the mRNA is, however, not sufficient to improve RpoS translation; pairing with the sRNA is still necessary to allow productive translation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains were grown on LB (Lennox broth; KD Medical) plates at 37°C with or without the following antibiotics, as appropriate: ampicillin (Ap; 50 μg/ml), kanamycin (Km; 25 μg/ml), chloramphenicol (Cm; 10 μg/ml), or tetracycline (Tc; 25 μg/ml). Strains containing the rne-3071 allele were maintained at 30°C. All E. coli strains used are derivatives of DJ480 and are listed in Table 1. DJ480 is a ΔlacX74 derivative of MG1655 (7). The primers and biotinylated oligonucleotide probes used in this study were purchased from Integrated DNA Technologies and can be found in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| CM1000 | DJ480 ΔdsrA14 | NM308 + P1(NM14) |
| CM1001 | DJ480 ΔdsrA14rpoS* | NM307 + P1(NM14) |
| CM1010 | DJ480 ΔdsrA14rne-3071zce-726::Tn10 | CM1000 + P1 (EM1277) |
| CM1050 | DJ480 ΔdsrA14 Δrnc-1223::cat | CM1000 + P1(CM1223) |
| CM1052 | DJ480 ΔdsrA14rne-3071zce-726::Tn10 Δrnc-1223::cat | CM1010 + P1(CM1223) |
| CM1060 | DJ480 ΔdsrA14 Δara714 | CM1000 + P1(LMG194), P1(EM1448) |
| CM1061 | DJ480 ΔdsrA14 Δara714 rpoS* | CM1001 + P1(LMG194), P1(EM1448) |
| CM1062 | DJ480 ΔdsrA14 rprA1::kan Δara714 | CM1060 + P1(NM22521) |
| CM1063 | DJ480 ΔdsrA14 rprA1::kan rpoS* Δara714 | CM1061 + P1(NM22521) |
| CM1064 | DJ480 ΔdsrA14rne-3071zce-726::Tn10 Δara714 | CM1060 + P1(EM1277) |
| CM1080 | DJ480 ΔdsrA14 Δara714 Δrnc-1223::cat | CM1060 + P1(CM1223) |
| CM1082 | DJ480 ΔdsrA14 rprA1::kan Δara714 Δrnc-1223::cat | CM1062 + P1(CM1223) |
| CM1083 | DJ480 ΔdsrA14 rprA1::kan rpoS* Δara714 Δrnc-1223::cat | CM1063 + P1(CM1223) |
| CM1086 | DJ480 ΔdsrA14rne-3071zce-726::Tn10 Δrnc-1223::cat Δara714 | CM1064 + P1(CM1223) |
| CM1223 | NM300 Δrnc-1223::cat | This study |
| DJ480 | MG1655 ΔlacX74 | 7 |
| DY330 | W3110 ΔlacU169 gal-490λcI857 Δ(cro-bioA) | 63 |
| EM1277 | DJ480 rne-3071zce-726::Tn10 | 31 |
| EM1448 | DJ480 Δara714leu+ | Lab strain |
| JNB001 | DJ480 ΔdsrA14rprA1::kan | CM1000 + P1(NM22521) |
| JNB002 | DJ480 ΔdsrA14rprA1::kanrpoS* | CM1001 + P1(NM22521) |
| LMG194 | Δara714 leu::Tn10 | 18 |
| MG1655 | Wild type | Lab strain |
| NC397 | HME45 lacI′<>kan-Ter<>cat-sacB<>lacZYA | 51 |
| NM1 | DY330 ΔdsrA14::cat-sacB | This study |
| NM14 | DY330 ΔdsrA14 | This study |
| NM22521 | C600 rprA1::kan | Lab strain |
| NM300 | DJ480 mini-λ tet | 54 |
| NM301 | NM300 rpoS* | This study |
| NM307 | NM301 rpoS* ΔdsrA::cat-sacB his::Tn10 | NM301 + P1(NM311) |
| NM308 | DJ480 ΔdsrA::cat-sacB his::Tn10 | DJ480 + P1(NM311) |
| NM111 | Hfr H ΔdsrA::cat-sacBhis::Tn10 | SG2204 +P1(NM1) |
| SG2204 | Hfr H his::Tn10 | 56 |
| Plasmids | ||
| pACS21 | pBR322 rnc+ | 53 |
| pNM12 | pBAD24 derivative | 28 |
| pNM13 | pBAD-dsrA+ | 28 |
| pNM33 | pBAD-dsrA* | 28 |
| pNM100 | pBAD-rprA+ | 29 |
| pNM109 | pBAD-rprA* | 29 |
TABLE 2.
Oligonucleotide primers and probe used in this study
| Primer or probe | Sequence |
|---|---|
| Primers | |
| ΔdsrAcat-For | CGTTGAATGCACAATAAAAAAATCCCGACCCTGAGGGGGTCGGGATGAAAATGAGACGTTGATCGGCACGTA |
| ΔdsrAsacB-Rev | CGTTAATCATTCATATGGCGAATATTTTCTTGTCAGCGAAAAAAATTGCGATCAAAGGGAAACTGTCCATATG |
| MtntSeq_RpoS-FOR | ATAGCGACCATGGGTAGCACC |
| Rnccatfor | TCGTGTGCTGAATTGTTGACGCATTTATTTATTGGTATCGCAAAATGAGACGTTGATCGGCACG |
| Rnccatrev | TCCGACGATGGCAATAAATCCGCAGTAACTTTTATCGATGCAACCAGCAATAGACATAAGCG |
| RpoS-NcoI-dpndnt | CACCGGAACCAGTTCAACACGCTTGCATTTTGAAATTCCATGGACAAGGGGAAATCCGTAAACCCGCTGCGTTATTTGCC |
| RpoS_NcoI-REV | CTTCTTCGGCCGTTAACAGTGGTG |
| rpoSRACE235 | CGGCCGTTAACAGTGGTGAAT |
| ΔdsrA replacement oligonucleotide | CGTTGAATGCACAATAAAAAAATCCCGACCCTGAGGGGGTCGGGATGCGCAATTTTTTTCGCTGACAAGAAAATATTCGCCATATGAATGATTAACG |
| Probe RpoS-N3 | CAAATCGTTATCACTGGGTTCCTG |
The rpoS* allele replaces 3 nucleotides (nt) of the rpoS 5′ UTR with 5 nt, generating an NcoI restriction site in the rpoS DNA sequence, and was previously used and referred to as NcoI′ (28, 29). rpoS* was introduced directly into the chromosome by bacteriophage lambda red recombination (63) using the single-stranded mutagenic oligonucleotide RpoS-NcoI-dpndnt. The NM300 strain (DJ480 carrying a mini-bacteriophage λ [54]) was electroporated with 100 ng of the oligonucleotide, resuspended in LB medium, serially diluted, plated on LB medium, and counted. The leftover electroporated cells were diluted the next day into 100 μl each in a 96-well microtiter plate to have about 6 to 10 bacteria per well, and the plate was incubated overnight at 37°C. One microliter from each well was then screened by PCR using the RpoS_NcoI-REV and MtntSeq_RpoS-FOR primers, whose last 3 nucleotides at the 3′ end match the mutated sequence and not the wild-type sequence. Three wells yielded a PCR product, and their contents were serially diluted and plated for single colonies. Thirty-two colonies from the 3 wells were screened further by PCR, and 8 were positive for the mutation. These were purified, their DNA was sequenced to confirm the mutation, and one isolate was saved as NM301.
The unmarked ΔdsrA14 deletion was constructed using the ΔdsrAcat-For and ΔdsrAsacB-Rev primers to amplify the cat-sacB cassette from strain NC397 (51) by PCR. The product was introduced by bacteriophage λ red recombinase-mediated recombination (recombineering) into strain DY330, selecting for Cmr and screening for Sucs, as described previously (63), to yield strain NM1. Recombineering was used again to replace the cassette in NM1 by the single-stranded ΔdsrA replacement oligonucleotide to yield strain NM14.
Transductions with P1vir were performed as described previously (47) to create the strains used here, as indicated in Table 1, generally selecting for antibiotic resistance. To make an unmarked deletion of dsrA, the ΔdsrA::cat-sacB allele from NM1 was first introduced into SG2204 to link the mutation to a his::Tn10 auxotrophy marker (linkage, ∼1%) to yield strain NM111. The linked his and dsrA mutations were then introduced into DJ480 and NM301 (rpoS*), selecting for Tcr and screening for Cmr (linkage, ∼1%), followed by P1 transduction from NM14, containing the ΔdsrA14 allele, selecting for his+ growth on M63 glucose minimal plates (KD Medical), and screening for Cms to yield strains CM1000 (rpoS+) and CM1001 (rpoS*), respectively. Δara derivatives of CM1000 and CM1001 were made by P1 transduction of Δara714 leu::Tn10 from LMG194 (18), selecting for Tcr; Leu+ versions were generated by P1 transduction from EM1448 with selection on M63 glucose minimal medium.
The rne(Ts) mutation, rne-3071 (9), was introduced by P1 transduction from EM1277, selecting for a linked marker (zce-726::Tn10) (31).
The rnc gene was deleted and replaced with a cat cassette by linear recombination (63). The cat cassette from NC397 was amplified by PCR with the primers Rnccatfor and Rnccatrev and electroporated into NM300, selecting for Cmr, to make CM1223. The entire rnc open reading frame from the start codon to the stop codon was deleted in this strain. The start codon of the downstream gene, era, overlaps the stop codon of rnc, and the start codon of era was deleted in this construct. This rnc allele will be referred to as Δrnc-1223::cat. Despite the fact that era is reported to be essential (53), chloramphenicol-resistant colonies were obtained, perhaps through a fusion of the cat open reading frame to era or readthrough from the cat transcript. We confirmed by complementation experiments that the phenotypes studied here were due to the absence of RNase III and not a lack of Era. CM1223 grows slowly at low temperature, similar to other rnc mutants (53), and this slow-growth phenotype was complemented by pACS21, a plasmid expressing rnc but not era (53). Other phenotypes of the Δrnc-1223::cat mutant strain (effects on RpoS levels; see Results) were also complemented by this plasmid (data not shown).
Plasmids pNM12, pNM13, pNM33, pNM100, and pNM109 (28, 29) were used to transform appropriate strains and selected with ampicillin, as described previously (10).
Immunodetection of RpoS.
Overnight cultures were prepared in LB medium with Ap (100 μg/ml) (LB Ap) at 32°C. Strains were diluted 1:1,000 and grown to mid-exponential phase at 30°C in LB Ap. sRNA expression was induced from plasmids with 0.02% arabinose (KD Medical) for 20 min. One-milliliter samples were taken after the induction, and total protein was collected by trichloroacetic acid (TCA) precipitation, as described previously (42). When the effects of the rne-3071 allele were tested, cells were heat shocked for 10 min at 43.5°C following sRNA induction, and samples were collected and TCA precipitated immediately before and after heat shock.
Protein samples from equal cell numbers, as determined by measurement of the optical density, were separated by SDS-PAGE using 10% NuPAGE polyacrylamide gels (Invitrogen) and 1× morpholinepropanesulfonic acid (MOPS) buffer, as recommended by the manufacturer. Protein was transferred to nitrocellulose membranes, and RpoS was detected with anti-RpoS antibody (provided by S. Wickner, NIH), as described previously (42). Sample dilutions were included on every gel, and several film (Kodak) exposures were taken to ensure detection in the linear range. Films were scanned with an Epson flatbed scanner at 600 dpi with a calibrated optical density step wedge (Stouffer). ImageJ software was used to quantify the RpoS levels on different exposures, as follows. Each film exposure was calibrated with the step wedge to allow measurement of the optical density. The mean optical density of each band was measured and used for comparison to the mean optical densities of the other bands from the same gel within the linear range. Membranes were also probed with anti-EF-Tu antibody (provided by M. Maurizi, NIH) and analyzed as described above to ensure equal loading of protein on each gel.
RNA preparation and Northern blotting.
Overnight cultures were prepared in LB Ap at 32°C. Strains were diluted 1:1,000 and grown to mid-exponential phase at 30°C in LB Ap. sRNA expression was induced from plasmids with 0.02% arabinose for 15 or 20 min, as noted in the legends to Fig. 2 to 6. When mRNA accumulation was measured, samples were collected after the induction for RNA preparation using the hot phenol method, as described previously (31). When the effects of the rne-3071 allele were measured following the 20-min sRNA induction, cells were heat shocked for 10 min at 43.5°C. Samples were collected immediately before and after the heat shock. To determine mRNA half-lives, rifampin (250 μg/ml) was added after the arabinose induction and RNA samples were collected immediately following addition (time zero samples) and then every 2.5 or 5 min for 20 min, as noted in the legends to Fig. 4. Samples were treated with hot phenol immediately after collection, as described above.
rpoS mRNA was detected by Northern blotting, as follows. Ten micrograms of RNA from each sample was separated by 1.2% agarose gels in 1× MOPS buffer at 100 V. Serial dilutions of at least one sample were included on each gel. RNA was transferred to nylon Nytran N 0.45-μm-pore-size membranes (Whatman) or nylon Zeta-probe membranes (Bio-Rad) using the capillary transfer method in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (Invitrogen) overnight. After UV cross-linking, membranes were incubated with Ultrahyb solution (Ambion) at 42°C for 30 min and then hybridized to the RpoS-N3 5′-biotinylated probe (Table 2) (100 ng/ml in Ultrahyb) for approximately 15 to 20 h at 42°C. RpoS-N3 anneals to the rpoS mRNA 93 nucleotides downstream of the start codon. The blots were washed, and labeled RNA was detected using a BrightStar BioDetect kit (Ambion), as recommended by the manufacturer. RNA levels in each sample were measured after exposure to film (Kodak) using ImageJ, as described above for the RpoS immunoblots. After exposure to film, the blots were stripped of probe by incubation in boiling 0.5% SDS for 10 min, washed three times in diethyl pyrocarbonate (DEPC)-treated water, and then hybridized as described above with a 5′-biotinylated probe for SsrA (17). Detection and analysis of SsrA levels were as described above. The rpoS mRNA accumulation reported for a given sample was normalized to the SsrA level in the same sample; very little variation in SsrA was observed.
5′ Rapid amplification of cDNA ends (RACE) of rpoS mRNA.
RNA samples were prepared from CM1001/pNM33 following induction with arabinose or from CM1010/pNM13 following induction with arabinose and heat shock at 43.5°C, using hot phenol as described above. Ten micrograms of each RNA sample was treated with 4 U Turbo DNase (Ambion) for 30 min at 37°C. Following phenol-chloroform extraction and ethanol-sodium acetate precipitation, samples were split and half was treated with tobacco acid pyrophosphatase (Epicentre), as described previously (1), while the other half was left untreated. Ligation of the 5′ ends to an RNA adapter was performed as described previously (1). cDNA was obtained by reverse transcription with the SuperScript III enzyme (Invitrogen) and the primer rpoSRACE235, as recommended by the manufacturer. cDNA products were amplified by PCR with an adapter-specific primer and the gene-specific primer rpoSRACE235. PCR products were separated on a Tris-acetate-EDTA agarose gel, purified with a kit (Qiagen), and cloned using the TOPO-TA cloning kit (Invitrogen), as recommended by the manufacturers. Several clones were sequenced with the rpoSRACE235 primer.
RESULTS
One of the best-studied examples of sRNA-mediated positive regulation is provided by the regulation of rpoS by DsrA and RprA. Base pairing between the sRNA and rpoS mRNA (Fig. 1) is absolutely required for efficient RpoS translation. Specific point mutations in the sRNA or in rpoS that disrupt this pairing prevented stimulation of an rpoS-lacZ fusion by DsrA or RprA (28, 29). Restoration of pairing through compensatory mutations restored stimulation of the fusion. We have built on these observations to investigate in more detail the mechanism and consequences of positive regulation by these small RNAs in vivo.
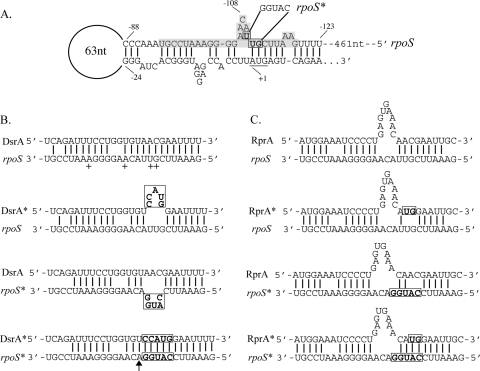
FIG. 1.
(A) Nucleotides in the 5′ UTR of the rpoS mRNA upstream of the start codon form a hairpin structure in the absence of binding to an sRNA. Bases are numbered backward from the start codon. The transcription start site was mapped at 584 bases upstream of the start codon (see text). The region that pairs with the sRNAs is highlighted, and the start codon is underlined. Mutated bases are boxed, and nucleotide changes in the rpoS* allele are indicated. (B) Predicted pairings are shown between the rpoS or rpoS* mRNA with DsrA or DsrA*. The plus sign and the arrow indicate 5′ ends of the cleaved forms of rpoS mRNA as mapped by 5′ RACE (+, high-temperature forms; arrow, DsrA*-rpoS* forms). Boxed nucleotides indicate the bases that were mutated in the dsrA* and rpoS* alleles. (C) Predicted pairings between the rpoS or rpoS* mRNA with RprA or RprA* are shown, and boxes indicate mutated nucleotides.
Previous work examining the mode of action of DsrA and RprA used translational fusions to lacZ. In order to look at the roles of DsrA and RprA in the context of the wild-type rpoS message and still use the required pairing as a way to ensure specificity of action, a mutant derivative of the native rpoS gene with a mutation in the region of the 5′ leader necessary for pairing with DsrA or RprA was created (Fig. 1, rpoS*). The rpoS* chromosomal mutation was sufficient to block activation of translation of rpoS*-lacZ by DsrA and limited activation of translation by RprA. The mutant forms of the sRNAs, dsrA* and rprA*, restored pairing to rpoS* (Fig. 1) and activation of the fusion (28, 29).
RpoS protein levels were measured in these backgrounds by immunoblotting to confirm the effectiveness of sRNA pairing with the wild-type and mutant rpoS (Fig. 2). For this analysis, dsrA rprA double mutant strains with a wild-type copy of rpoS or the rpoS* mutation were transformed with vector or plasmids containing dsrA, rprA, dsrA*, or rprA* under the control of the arabinose-inducible PBAD promoter (see base-pairing combinations in Fig. 1).
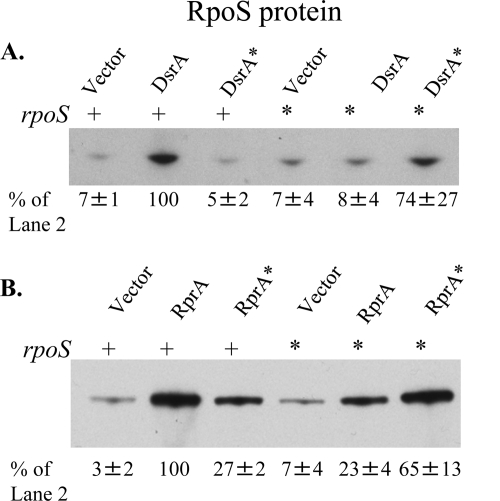
FIG. 2.
RpoS expression in strains with optimal or mismatched pairing between sRNAs and rpoS. CM1062 (ΔdsrA rprA::kan rpoS+ Δara714) and CM1063 (ΔdsrA rprA::kan rpoS* Δara714) with the pNM12 vector or its derivatives expressing dsrA or dsrA* (A) and CM1062 and CM1063 with the vector or its derivatives expressing rprA or rprA* (B) were grown in LB Ap at 30°C to mid-exponential phase, and sRNA expression was induced with 0.02% arabinose for 20 min. Total protein was precipitated and analyzed by Western blotting. RpoS accumulation in each strain is described as a percentage of the accumulation in the rpoS+/pBAD-dsrA+ (A) or rpoS+/pBAD-rprA+ (B) strain. Accumulation is presented as the mean percentage ± the standard deviation (n = 3).
As was observed with the translational fusions, there was little or no detectable RpoS when only the vector was present. However, as with the rpoS-lacZ fusion (28), wild-type DsrA significantly stimulated RpoS accumulation (14-fold in Fig. 2A; in other experiments, even higher fold changes were seen [see Fig. 5B]). Expression of DsrA from the PBAD promoter under these conditions yielded 3-fold more RpoS than the amount from strains expressing DsrA from the chromosomal gene under these growth conditions (data not shown). In contrast, the DsrA* derivative, which is unable to fully pair, had no stimulatory effect (Fig. 2A). Mutating rpoS to reduce pairing (rpoS*) also eliminated most RpoS synthesis in the presence of DsrA. As with the translational fusions (28), expressing the dsrA* plasmid in a host carrying the rpoS* allele restored RpoS accumulation (Fig. 2A), though the RpoS levels were reduced compared with those from the wild-type pairing. The level of RpoS accumulation in the strains with the paired compensatory mutations was generally somewhat lower than that of the strains with the wild-type combination, suggesting that this region of the 5′ UTR may have some direct effects beyond its role in pairing.
The results with expression of RprA are consistent with this picture. RprA expressed from the plasmid stimulated RpoS translation significantly. In the case of the RprA mismatched sets, however, the rprA*-rpoS+ and rprA+-rpoS* combinations gave significant amounts of RpoS, although they were less than those in cells in which rprA and rpoS have either the wild type or rprA*-rpoS* pairing (23 to 27% of the amount for rprA+-rpoS+) (Fig. 2B). This finding is similar to results seen with these mutations when monitored with rpoS-lacZ fusions (29). The level of RpoS accumulation in the strains with the paired compensatory mutations was 65% that of the wild type.
Stability of rpoS mRNA.
The rpoS mRNA was examined in the presence and absence of optimal pairing with the sRNAs. RNA isolation was performed on a dsrA rprA double mutant transformed with the same plasmids as described above, and samples were analyzed by Northern blotting using the oligonucleotide probe RpoS-N3 (Table 2).
When cells expressed neither DsrA nor RprA, rpoS mRNA levels were very low (Fig. 3 A and B). When wild-type DsrA or RprA can pair with wild-type rpoS, the mRNA accumulated to a high level (Fig. 3A and B). The 1.6-kb mRNA detected here is what we expected for a transcript starting at the PrpoS promoter within the nlpD open reading frame (23, 52). 5′ RACE analysis of the full-length rpoS mRNA identified the same transcription start site mapped previously (23, 52). Quantitative analysis of the Northern blots shows that DsrA or RprA expression from the plasmid resulted in a 7-fold higher level of rpoS mRNA compared to that in strains carrying the vector control (Fig. 3A and B). These results suggest that expression of DsrA or RprA stabilized the rpoS mRNA. The effect of the sRNAs on the rpoS mRNA half-life is addressed below.
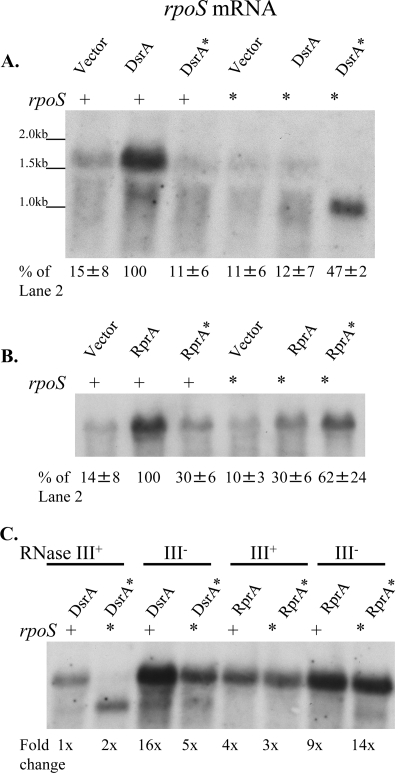
FIG. 3.
Northern blot analysis of rpoS mRNA levels in strains with optimal or mismatched pairing between sRNAs and rpoS. CM1062 (ΔdsrA rprA::kan rpoS+ Δara714) and CM1063 (ΔdsrA rprA::kan rpoS* Δara714) with the pNM12 vector or its derivatives expressing dsrA or dsrA* (A) and CM1062 and CM1063 with the pNM12 vector or its derivatives expressing rprA or rprA* (B) were grown in LB Ap at 30°C to mid-exponential phase, and sRNA expression was induced with 0.02% arabinose for 20 min. RNA was collected by the hot phenol method and analyzed by Northern blotting with the probe RpoS-N3. rpoS mRNA accumulation is described as a percentage of the accumulation in the rpoS+/pBAD-dsrA+ (A) or rpoS+/pBAD-rprA+ (B) strains. Accumulation is presented as the mean percentage ± the standard deviation (n = 3). (C) rpoS mRNA was examined in the same manner as for panels A and B for the RNase III mutant strains (CM1082 and CM1083) containing the same plasmids. rpoS mRNA accumulation was measured relative to that of the rpoS+ rnc+-pBAD-dsrA+ strain.
To explore the role of base pairing with the sRNAs on rpoS mRNA, the mRNA accumulation was examined in strains where sRNA base pairing with the rpoS mRNA was disrupted by mutation. In strains expressing DsrA* in the rpoS+ background or expressing DsrA in the rpoS* background, rpoS and rpoS* mRNA levels remained very low (Fig. 3A). This is consistent with a requirement for productive base pairing (leading to translation) of DsrA to rpoS mRNA for stabilization of the mRNA.
Cells in which pairing was restored (dsrA*-rpoS*) had elevated levels of rpoS mRNA (Fig. 3A). Surprisingly, the mRNA accumulating in the dsrA*-rpoS* strain was 0.5 kb shorter than the full-length rpoS mRNA (Fig. 3A). Northern analysis using a probe that anneals to the 5′ end of rpoS mRNA did not detect this truncated band, suggesting that the shorter RNA is missing sequences at the 5′ end upstream of the pairing region (data not shown). Some full-length rpoS mRNA can be detected in this background, and some of the truncated form can be detected in the wild type with long exposures (data not shown).
5′ RACE was performed to determine the 5′ end of the truncated form of the rpoS mRNA in the dsrA*-rpoS* background. This analysis identified an A at the 5′ end, −109 bases upstream of the AUG start codon (marked with an arrow in Fig. 1B), within the region that pairs with DsrA. This result was obtained both from RNA samples treated with tobacco acid pyrophosphatase and from untreated samples, suggesting that this is a cleavage product.
The endoribonuclease RNase III cleaves double-stranded RNA molecules, including mRNAs, in E. coli and has been shown to cleave rpoS mRNA in vitro and to play a role in the degradation of rpoS mRNA and the accumulation of RpoS in vivo (14, 44). Northern blot analysis of rpoS mRNA accumulation in the dsrA*-rpoS* strain in combination with a deletion of rnc, the gene encoding RNase III, showed that the mRNA is primarily full length in this background (Fig. 3C). This finding suggests that the cleaved form of rpoS* seen in the rnc+ background is generated by RNase III. Additionally, full-length rpoS mRNA levels in the dsrA+-rpoS+ strain were much higher in the Δrnc-1223::cat mutant than in the rnc+ background (Fig. 3C). In a strain background expressing dsrA from its native promoter on the chromosome, the levels of rpoS mRNA were increased 3- to 4-fold in the Δrnc-1223::cat mutant relative to those in an isogenic rnc+ strain (data not shown). Together, these findings show that RNase III plays a complex role in the cleavage and stability of rpoS mRNA, and this role is discussed further in the next section.
In the case of RprA, the effect of disrupting base pairing with the rpoS mRNA by the mutations in Fig. 1 was less drastic that that seen for DsrA and rpoS, although protein levels were the highest when RprA and rpoS had optimal pairing (Fig. 2A and B). Consistent with this, the accumulation of rpoS mRNA was also the highest in the cases where pairing was optimal and was reduced but not abolished when pairing was incomplete (Fig. 3B). As with RpoS protein levels, the rpoS mRNA levels in the rprA*-rpoS* strain were close to, but less than, the levels of rpoS mRNA in the strain with wild-type pairing (62%; Fig. 3B). Unlike the dsrA*-rpoS* case, however, no truncated rpoS mRNA was detected.
Surprisingly, rpoS mRNA accumulated to a 3- to 4-fold higher level, on average, with induction of RprA than with DsrA, when the levels of accumulation were compared directly (Fig. 3C). However, rpoS mRNA is less susceptible to RNase III in the presence of RprA than it is in the presence of DsrA. The levels of rpoS mRNA increased only 2- to 3-fold in the Δrnc background with RprA expression (Fig. 3C). A bulge in the predicted paired sequence of RprA with rpoS mRNA near the likely site of RNase III action may make the RprA-rpoS duplex a poorer substrate for the enzyme (Fig. 1).
Increased accumulation of rpoS mRNA most likely reflects increased stability of the mRNA during the 20-min period of DsrA or RprA expression, since the rpoS mRNA levels were low in their absence. To measure the stability of rpoS mRNA directly, strains with wild-type rpoS or with rpoS* were induced for expression of DsrA, RprA, DsrA*, or RprA* and treated with rifampin, and RNA was collected at regular intervals. In strains with no DsrA or RprA expression, rpoS mRNA levels were too low to measure the half-life (data not shown; note that the chromosomal copy of dsrA is also absent from these strains). When DsrA was expressed from the PBAD promoter in the rpoS+ strain, the rpoS mRNA half-life was 3 min and the truncated form of rpoS in the rpoS*-dsrA* strain had a similar half-life of 2 min (Fig. 4 A). In strains where pairing was disrupted by the mismatch mutations in dsrA or in rpoS, the half-life could not be measured, because the mRNA became undetectable immediately after the chase began (data not shown). This is consistent with the findings presented in a previous report showing some increase in the half-life of rpoS upon overexpression of DsrA, though the authors stated that the half-life was difficult to measure due to the overall instability of rpoS mRNA (24). The rpoS mRNA half-lives in the rprA+-rpoS+ and rprA*-rpoS* strains were always higher than those in the unpaired sets and higher than the half-lives measured with DsrA expression, although significant variation in these values was seen from experiment to experiment (see Fig. 4 legend). In a representative experiment, the half-life measured for the rpoS mRNA in the rprA+-rpoS+ strain was 10 min, and that for the rprA*-rpoS* strain was 15 min (Fig. 4B). In strains where pairing was disrupted (rprA+-rpoS* and rprA*-rpoS+), the half-lives were reduced to 2 min (Fig. 4B). Pairing between RprA and rpoS* or RprA* and rpoS is apparently sufficient to allow some partial stabilization of the mRNA, as well as to partially allow translation, as seen in Fig. 2B.
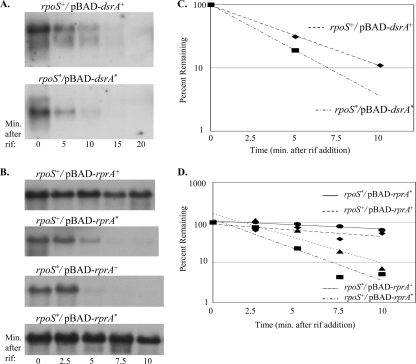
FIG. 4.
rpoS mRNA half-life determination when it is paired with DsrA or RprA. CM1000 (ΔdsrA rpoS+)/pBAD-dsrA and CM1001 (ΔdsrA rpoS*)/pBAD-dsrA* (A) and JNB001 (ΔdsrA rprA::kan rpoS+) and JNB002 (ΔdsrA rprA::kan rpoS*) with pBAD-rprA or pBAD-rprA* (B) were grown in LB Ap at 30°C to mid-exponential phase, and sRNA expression was induced with 0.02% arabinose for 15 min. RNA samples were collected by the hot phenol method at the indicated times after the addition of 250 μg/ml rifampin (rif). rpoS mRNA levels were analyzed by Northern blotting. The experiment whose results are shown in panel A was performed three times, resulting in half-lives measured for rpoS and rpoS* of 2 to 3 min in each case. The experiment whose results are shown in panel B was performed four times, with some variability in half-lives being measured for rpoS and rpoS*. In each experiment, the mRNA half-lives of the mismatched pairs (rpoS+-rprA* and rpoS*-rprA+) are shorter than that of the optimally paired sets. However, the mRNA half-lives in the rpoS+-rprA+ and rpoS*-rprA* strains varied from 4 to 15 min, and the mRNA half-lives in the mismatched pairs varied from 1 to 10 min. (C and D) Graphical representation of rpoS mRNA decay.
When cells with wild-type chromosomal dsrA were grown at 25°C (a permissive temperature for DsrA expression) to mid-exponential phase and treated with rifampin, rpoS mRNA had a half-life of approximately 2 to 4 min (data not shown), comparable to that seen with DsrA overexpression at 30°C (Fig. 4A). In a ΔdsrA strain, rpoS decay was too fast to measure; rpoS could be detected in the time zero sample only.
These results demonstrate that DsrA and RprA pairing to rpoS mRNA not only increases protein translation, as shown above, but also increases mRNA stability and that disruption of base pairing is sufficient to destabilize the mRNA.
Degradation of rpoS mRNA by RNase III- and RNase E-mediated mechanisms.
The results described thus far show that rpoS mRNA is stabilized by base pairing with DsrA and RprA. Thus, in the absence of pairing, the rpoS mRNA must be degraded by one or more RNases. RNase E and RNase III are two major RNA endonucleases in E. coli; both have been implicated in initiating degradation of sRNAs and mRNAs (20, 31, 35, 61). Cleavage of double-stranded mRNA regions by RNase III can lead to mRNA decay but can also lead to activation of mRNA translation (reviewed in reference 13). In fact, RNase III has been shown to play a role in the degradation of rpoS mRNA and the accumulation of RpoS in vivo (14, 44). Above, we showed that RNase III can generate a stable cleavage product in the dsrA*-rpoS* background. RNase E, in contrast, cleaves single-stranded RNA in AU-rich regions and acts in concert with the degradosome proteins, including the exoribonuclease PNPase, to degrade mRNA (reviewed in reference 8).
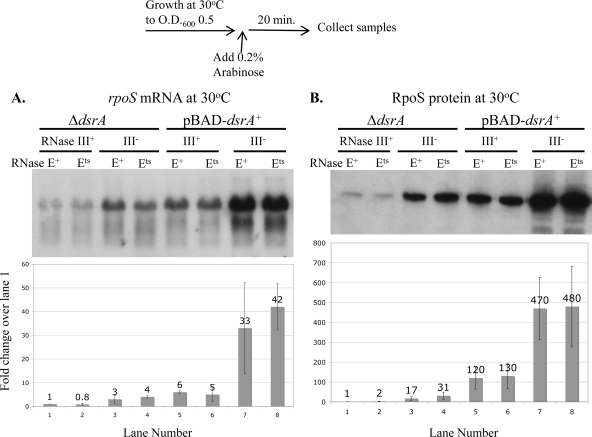
A set of isogenic strains was constructed to evaluate the roles of both endoribonucleases in the turnover of rpoS mRNA in the presence and absence of DsrA, as well as to evaluate the contributions of mRNA instability on the translation of RpoS. The strains used carried a deletion of dsrA, with or without a deletion of the gene encoding RNase III, rnc, and with a wild-type or a temperature-sensitive (Ts) allele of the gene encoding RNase E, rne. All strains carried either the vector or the pBAD-dsrA+ plasmid. After these strains were grown to mid-exponential phase at 30°C, DsrA synthesis was induced with arabinose for 20 min and RNA and protein were isolated after the induction. Cultures were then transferred to 43.5°C for a 10-min heat shock to inactivate RNase E, and RNA and protein samples were again collected. rpoS mRNA was analyzed by Northern blotting, and RpoS protein levels were measured by Western blotting analysis. mRNA and protein levels were normalized to those for the wild-type strain (rnc+ rne+ carrying the vector control).
Figure 5 shows rpoS mRNA and RpoS protein levels after induction of DsrA synthesis at 30°C. In these samples, RNase E should have been active in both the rne+ and rne(Ts) strains. Therefore, these two alleles had similar phenotypes under this condition. Consistent with the results shown above (Fig. 2 and 3), the rpoS mRNA and RpoS protein levels were low in the absence of dsrA (Fig. 5A and B). In the Δrnc and ΔdsrA background, there was an increase in the rpoS mRNA level (3-fold; Fig. 5A) and the RpoS protein level (17-fold; Fig. 5B) compared to the levels in the rnc+ ΔdsrA strain. Therefore, RNase III plays a role in degradation of the rpoS mRNA, as shown by Resch et al. (44), and negatively regulates RpoS translation in the absence of DsrA.
FIG. 5.
rpoS mRNA and RpoS levels in a Δrnc mutant strain in the presence and absence of DsrA. CM1000 (rnc+ rne wild type), CM1010 [rnc+ rne(Ts)], CM1050 (Δrnc-1223::cat rne wild type), and CM1052 [Δrnc-1223::cat rne(Ts)] with the pNM12 vector or pBAD-dsrA were grown at 30°C in LB Ap to mid-exponential phase; sRNA expression was induced with 0.02% arabinose for 20 min. Samples were collected for protein and RNA isolation, as described in Materials and Methods (O.D.600, optical density at 600 nm). (A) Northern blot analysis of RNA samples was performed using the oligonucleotide probe RpoS-N3 to detect rpoS mRNA. (B) Western blot analysis of protein samples was performed using the anti-RpoS antibody to detect RpoS. Graphical analyses show the mean accumulation of the full-length rpoS mRNA or RpoS protein ± the standard deviation (n = 3) relative to that of CM1000 with the vector control. Samples were also tested by Western blotting for EF-Tu, which was close to identical in each sample. Values over the bars indicate the mean fold change.
As seen earlier (Fig. 2 and 3), expression of DsrA led to an increase in rpoS mRNA levels (6-fold; Fig. 5A) and a significant induction of RpoS translation (120-fold; Fig. 5B) in the rnc+ strains. rpoS mRNA levels were higher in Δrnc strains than in rnc+ strains in the presence of DsrA (6-fold), and there is a 4-fold increase in translation (Fig. 5A and B). Notably, expression of DsrA in the Δrnc background increased both the rpoS mRNA level and the level of translation of RpoS significantly, comparable to the increase seen in the rnc+ host (Fig. 5A and B). Therefore, RNase III decreases accumulation of rpoS mRNA and translation of RpoS significantly in both the absence and presence of DsrA. Furthermore, DsrA works to stimulate translation with or without RNase III.
In the Δrnc cells expressing DsrA (and having high levels of rpoS mRNA), a short form of the rpoS mRNA was detected (Fig. 5A), suggesting that there could be cleavage by another enzyme. Truncation of the rpoS mRNA is discussed further below.
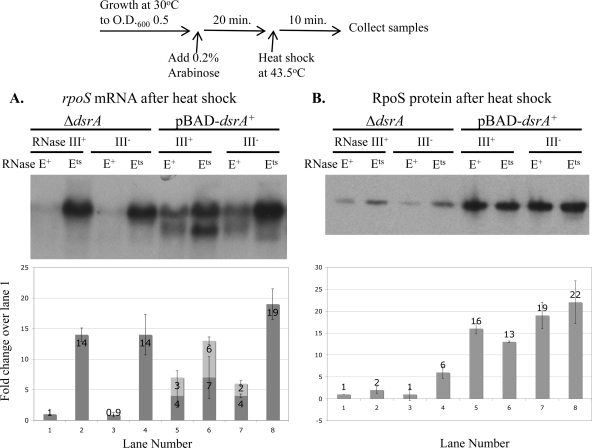
Figure 6 shows rpoS mRNA levels and RpoS protein levels after DsrA induction followed by a 10-min heat shock. The 10-min heat shock treatment itself increased the basal level of expression of RpoS protein in the rne wild-type background in the presence or absence of DsrA by as much as 10- to 20-fold compared to the levels shown in Fig. 5 (data not shown; see Fig. 6 legend for further detail). This is consistent with the findings of previous studies showing increased RpoS after heat shock (21, 36). While Muffler et al. reported that this increase in RpoS was primarily due to stabilization of the protein (36), we also see a 3-fold increase in the levels of rpoS mRNA after the heat shock (data not shown); the effects described here are relative to these increased basal levels.
FIG. 6.
rpoS mRNA and RpoS levels in an rne(Ts) mutant strain in the presence and absence of DsrA. CM1000 (rnc+ rne wild type), CM1010 [rnc+ rne(Ts)], CM1050 (Δrnc rne wild type) and CM1052 [Δrnc rne(Ts)] with the pNM12 vector or pBAD-dsrA were grown at 30°C in LB Ap to mid-exponential phase; sRNA expression was induced with 0.02% arabinose for 20 min. Cultures were heat shocked for 10 min at 43.5°C, and RNA and protein samples were collected, as described in Materials and Methods (O.D.600, optical density at 600 nm). (A) Northern blot analysis of RNA samples was performed using the oligonucleotide probe RpoS-N3 to detect rpoS mRNA. Graphical analysis shows the mean accumulation of full-length rpoS mRNA (dark bars) and the truncated form of the rpoS mRNA (light bars) ± the standard deviation (n = 3) relative to that of CM1000 with the vector control. rpoS mRNA accumulation in lane 1 is 3 times that for the same strain before heat shock (at 30°C). (B) Western blot analysis of protein samples was performed using the anti-RpoS antibody to detect RpoS. Protein samples were diluted 5-fold before electrophoresis. Graphical analysis shows mean accumulation of RpoS protein ± the standard deviation (n = 3) relative to that of CM1000 with the vector control. RpoS protein accumulation in lane 1 is 20 times that of the same strain before heat shock; in lane 5, RpoS accumulation is 10 times that of the same strain before heat shock. As with Fig. 5, equal protein input per lane was confirmed by Western blotting for EF-Tu. Values over the bars indicate the mean fold change.
As described above, in the absence of DsrA, rpoS mRNA levels were low compared to those of other samples in this set, as was the RpoS protein level (Fig. 6A and B). Under these conditions, inactivation of RNase E resulted in a significant accumulation of full-length rpoS mRNA (14-fold; Fig. 6A), suggesting that RNase E plays a major role in degradation of rpoS in the absence of DsrA. However, there was only a 2-fold accumulation of RpoS protein (Fig. 6B, lane 2). Therefore, in the absence of DsrA, inactivation of RNase E is not sufficient for efficient translation of RpoS.
There was little or no effect of the rnc mutation on either rpoS mRNA or RpoS protein after the heat shock (Fig. 6A and B). However, there was a slightly larger increase in RpoS production when RNase E was inactivated in the absence of RNase III (Fig. 6B, lane 4), which may reflect increased accumulation of RpoS in the rnc mutant strain during growth before the heat shock.
DsrA expression resulted in an increase in the total rpoS mRNA level (7-fold) and the level of induction of RpoS translation (16-fold) compared to the levels for the vector controls in a ΔdsrA host (Fig. 6A and B). Interestingly, however, 40% of the rpoS mRNA was truncated in the presence of DsrA (see below). Inactivation of RNase E resulted in only a 2-fold additional increase in the levels of both the full-length and short forms of rpoS mRNA and no increase in the level of RpoS (Fig. 6A and B). These data suggest that DsrA expression overcomes most but not all of the RNase E-mediated degradation of rpoS and can induce maximal translation of RpoS even in the presence of RNase E. While expression of DsrA stimulated translation in the rne(Ts) strain, the level of total rpoS mRNA in the pBAD-dsrA+-rne(Ts) strain was not any more than that in the pBAD vector-rne(Ts) strain, suggesting that DsrA cannot stabilize the mRNA any further in the rne(Ts) strain (Fig. 6A and B, compare lanes 2 and 6).
The truncated form of the rpoS mRNA from the dsrA+ rne(Ts) sample shown in Fig. 6A, lane 6, possesses 5′ ends located in the DsrA pairing region and ranging from −101 to −112 bases upstream of the AUG, as determined by 5′ RACE (Fig. 1B, marked with + signs). Northern blot analysis confirmed that the 5′ end of rpoS, upstream of the DsrA interaction region, is missing in this form (data not shown). The short form seen here is the same size as that described above in the dsrA*-rpoS* strain.
Again, there was no effect of RNase III in the presence of DsrA at this temperature (Fig, 6A and B). However, in the Δrnc rne(Ts) strain there was maximal accumulation of full-length rpoS mRNA but little to no accumulation of truncated form (Fig. 6A, lane 8). Cleavage of rpoS mRNA in the rne(Ts) background requires DsrA and RNase III. Thus, in the Δrnc rne+ strain, some cleavage is seen but must be mediated by an unidentified enzyme.
Taken together, Fig. 6A suggests that RNase E plays an important role in the degradation of rpoS mRNA. rpoS mRNA is degraded by an RNase E-mediated mechanism in the absence of dsrA. DsrA expression stabilizes the rpoS mRNA, and DsrA-bound rpoS is significantly less susceptible to RNase E. RNase III is not required for RNase E-mediated degradation of rpoS mRNA in the absence of dsrA or for the DsrA-mediated protection of the rpoS mRNA.
A striking effect seen here is that stabilizing rpoS mRNA by inactivation of RNase E does not lead to much improvement in translation in the absence of DsrA. DsrA is still required for maximal translation even after the 10-min heat shock and inactivation of RNase E. Thus, although expression of DsrA protects the mRNA from degradation, these data clearly show that protecting from degradation is not sufficient to lead to translation. DsrA plays a separate and critical role by enhancing translation, even under conditions when degradation is significantly reduced.
DISCUSSION
RpoS accumulation is regulated at multiple levels, including the tight regulation of translation dependent upon the RNA chaperone Hfq and the regulatory sRNAs DsrA and RprA. A basic model for stimulation of translation by DsrA and RprA was developed on the basis of predicted base pairing between these RNAs and the upstream leader of rpoS mRNA and was confirmed by the demonstration that this base pairing was necessary for sRNA action, using rpoS-lacZ translational fusions (28, 29), as well as in vitro experiments (26, 49, 58). We looked in more detail at the in vivo characteristics of stimulation of RpoS translation by DsrA and RprA, examining the action of these sRNAs on the native rpoS mRNA and the translation of RpoS from this mRNA. The advantage of this system is that any characteristics of the mRNA and its degradation are studied under the control of the native promoter and without the use of reporters that may introduce or abolish possible sites of action of ribonucleases or other factors. However, we note that the behavior of the fusions is entirely consistent with what we have observed here.
Previous studies on the action of DsrA and RprA have clearly demonstrated an increase in translation that is dependent on sRNA binding. In in vitro experiments, sRNA binding results in remodeling of the rpoS mRNA to uncover the ribosome binding site, the likely explanation for the increased translation (26, 58). Our results are entirely consistent with those findings. Our work also shows that overexpression of both DsrA and RprA significantly increases the rpoS mRNA half-life and, therefore, the level of mRNA accumulation during the short induction period. The levels of mRNA generally correlate well with the levels of translated protein in these experiments.
One interpretation of this increase in mRNA stability is that it is secondary to increased translation, since it is known that translation helps to protect mRNA from degradation (reviewed in reference 11). An alternative interpretation would be that the sRNAs, by increasing the stability of the mRNA, lead to increased translation. If this were the case, stabilizing the mRNA by inactivating the RNases responsible for degradation should be as effective as the sRNAs in increasing translation. Our results using RNase mutants make the second possibility unlikely.
RNase E and the degradation and translation of rpoS mRNA.
RNase E is an important and essential endoribonuclease in E. coli. It cleaves single-stranded RNA and has previously been implicated in the negative regulation of mRNAs by small RNAs (31, 35, 39). The data presented here suggest a central role for RNase E in the degradation of rpoS mRNA. Levels of the mRNA increased significantly when RNase E was inactivated by heat shock in strains with a temperature-sensitive allele of rne, the gene encoding RNase E (Fig. 6A). DsrA expression led to very little additional accumulation of RNA when RNase E was inactivated and DsrA was expressed (compare lanes 6 and 8 to lanes 2 and 4 in Fig. 6A). This suggests that DsrA's major effect on rpoS mRNA accumulation is in overcoming RNase E-dependent degradation of this mRNA.
These experiments also very clearly show that stabilization of the mRNA is not sufficient to lead to RpoS translation and that the effect of DsrA on mRNA stability is not as strong as the effect on translation. Thus, DsrA played an important role in promoting translation even when rpoS mRNA levels were relatively high (Fig. 6B; compare RpoS protein levels in lanes 6 and 8 to those in lanes 2 and 4), and, even more strikingly, protein levels were high when DsrA was expressed, even when not as much mRNA accumulated (Fig. 6B, lanes 5 and 7). Another way to measure the relative effect of DsrA on RpoS translation versus the effect on mRNA stability is by a measure of translation efficiency (a comparison of relative protein levels to relative mRNA levels) (Table 3). If we normalize translation efficiency to that for the wild-type strain in the absence of DsrA at 30°C (set equal to 1), DsrA stimulated translation relative to mRNA levels greater than 20-fold [Table 3, rnc+ rne+ dsrA+ and rnc+ rne(Ts) dsrA+]. At 43.5°C, both protein and mRNA levels were higher, so we separately normalized the translation efficiency to 1 for the wild-type strain in the absence of DsrA under this condition (see below for temperature-specific effects independent of RNase E). At 43.5°C, the stimulation by DsrA was 4-fold in both rne+ strains (Table 3, rnc+ rne+ dsrA+ and Δrnc rne+ dsrA+) but was only 1-fold when RNase E was inactivated, because the level of RNA was already very high and was barely increased by DsrA (compare lanes 2 and 4 to lanes 6 and 8 in Fig. 6A). Because the levels of RpoS protein were similar when DsrA was present, with or without RNase E (Fig. 6B, compare lane 5 to lane 6 and lane 7 to lane 8), this also suggests that the level of mRNA is not limiting for DsrA-dependent translation.
TABLE 3.
Efficiency of RpoS translation
| Strain | Translation efficiencya |
|
|---|---|---|
| 30°C | 43.5°C | |
| rnc+rne+ ΔdsrA | 1 | 1 |
| rnc+rne(Ts) ΔdsrA | 2 | 0.1 |
| Δrnc rne+ ΔdsrA | 6 | 2 |
| Δrnc rne(Ts) ΔdsrA | 8 | 0.4 |
| rnc+rne+dsrA+ | 21 | 4 |
| rnc+rne(Ts)dsrA+ | 24 | 1 |
| Δrnc rne+dsrA+ | 14 | 5 |
| Δrnc rne(Ts)dsrA+ | 12 | 1 |
In a previous study, Urban and Vogel examined the ability of DsrA and RprA to stimulate the translational fusion rpoS::GFP (green fluorescent protein) in cells carrying rne-701, defective in the degradosome portion of RNase E, and saw significant stimulation there as well (59). Our results significantly extend those observations and demonstrate that DsrA efficiently overcomes the ability of RNase E to degrade the rpoS mRNA and allows very efficient translation, whether or not RNase E is functional. We note that Basineni et al. (2) did not see an effect of inactivating RNase E on rpoS turnover. We do not currently have an explanation for this difference, although in their experiments cultures were shifted to 42°C, not 43.5°C, which we find necessary for inactivation of RNase E, and were also treated with hydrogen peroxide before the mRNA half-life was measured. In other work (45), a strain with mutations in rne, pnp, and rnb was examined and reported to have increased levels of RpoS compared to those in wild-type cells, consistent with our findings for rne mutants, but the contribution of each of these genes was not determined.
RNase III and degradation and translation of rpoS mRNA.
In E. coli, RNase III is a nonessential double-stranded endoribonuclease. Evidence of a role for RNase III in decay of rpoS mRNA has been reported (2, 14, 44). The role of RNase III in the degradation of rpoS mRNA is rather different and somewhat more complex than that of RNase E. Consistent with the report by Resch et al. (44), we find that RNase III reduces the level of rpoS mRNA, presumably by degrading it, and that this degradation occurs even when DsrA is present (44). mRNA levels were increased 3- to 4-fold in the absence of RNase III in the ΔdsrA background, and this increase in rpoS mRNA was accompanied by a significant (17-fold) increase in protein levels (Fig. 5A and B). Under the conditions of this experiment (absence of dsrA in the chromosome, growth of cells at 30°C), RNase III clearly contributes to keeping the levels of RpoS low. Freire et al. (14) found a negative effect, rather than a positive effect, of an RNase III mutation on rpoS mRNA stability and protein levels under carbon starvation conditions. We have not tested our mutants under these conditions, but these results suggest additional complexity for the role of RNase III.
While DsrA clearly suppresses the effects of RNase E, it does not overcome the effects of RNase III. Thus, DsrA is additive with the effects of an rnc mutant. The level of mRNA increased 10-fold in the rnc mutants when DsrA was expressed, and the level of protein increased more than that, so that the level of mRNA and the level of RpoS protein were the highest when DsrA was present and RNase III was absent (Fig. 5, lanes 7 and 8). This suggests that the ability of RNase III to negatively affect rpoS is not overcome by translation or by DsrA annealing. This interpretation is somewhat different from that reached by Resch et al. (44), who suggested that translational activation involves RNase III-dependent processing of the rpoS 5′ leader (44). The clear activation that we see by DsrA even in the absence of RNase III demonstrates that RNase III cleavage is not necessary for DsrA to act (compare lanes 7 and 8 to lanes 3 and 4 in Fig. 5B).
How and where does RNase III act on rpoS mRNA? One possibility that we cannot currently rule out are indirect effects of an rnc mutant. However, this discussion is based on the assumption that RNase III directly acts on rpoS mRNA. In vitro, Resch et al. (44) find RNase III cleavage of the rpoS mRNA 5′ UTR in the absence of sRNA at −15 and −94 nucleotides upstream of the start codon, in the paired region of the hairpin. In the presence of chromosomally encoded DsrA, they find cleavage at these two sites as well as at −112 nucleotides upstream of the AUG in the region of pairing with DsrA (44).
In our experimental setup, we are overproducing DsrA, so that pairing with rpoS should be relatively complete. If we assume that essentially all rpoS mRNA is paired with DsrA, the negative effect of RNase III on RNA accumulation when DsrA is present (Fig. 5A) is consistent with a model in which RNase III can cleave the DsrA-rpoS double-stranded structure after hairpin opening. Direct evidence for such a cleavage in vivo was seen in the rpoS*-DsrA* strain (Fig. 3C), where all of the rpoS mRNA was cleaved. The 5′ ends of this cleavage product mapped to the DsrA pairing region of the rpoS mRNA, at position −109, a few nucleotides from the −112 position found by Resch et al. in vitro (Fig. 1) (44). However, we did not see this processed band in the strain with wild-type DsrA and wild-type rpoS; if the cleavage occurs, either it is not very efficient or the resulting cleaved rpoS mRNA is rapidly degraded. We suggest that both are occurring. Part of the mRNA is cut and destroyed, reducing the extent of RpoS accumulation. Results shown in Fig. 6 suggest that RNase E is participating in the rapid degradation of the cleaved rpoS mRNA; there is evidence of accumulation of the truncated message at high temperature, when RNase E was inactivated (Fig. 6, lane 6). Cleavage of rpoS mRNA by RNase III will provide a 5′-monophosphate for the remaining rpoS message, a better substrate for subsequent RNase E cleavage downstream (12). The rest of the mRNA may not be cut, even though it is paired with DsrA. The uncut mRNA would then be an efficient substrate for translation. It is possible that the wild-type DsrA-rpoS duplex may bind to RNase III less effectively than the DsrA*-rpoS* duplex or that the wild-type duplex may bind to some other factor that prevents RNase III action.
When RprA is present and paired with the rpoS mRNA, a loop in RprA is found in the region where RNase III is found to cleave the DsrA-rpoS duplex (Fig. 1C). The RprA*-rpoS* duplex also contains this bulge and is not truncated like the rpoS* mRNA paired with DsrA* (Fig. 3B). While there are RNase III-dependent effects on rpoS in the presence of RprA (Fig. 3C), the RprA-rpoS duplex may be less susceptible to this endonuclease than the DsrA-rpoS duplex. This may explain the increased stability of rpoS mRNA after RprA expression in rnc+ strains compared to that of rpoS mRNA after DsrA expression (Fig. 3C and 4).
Effects of high temperature on RpoS translation and mRNA stability.
In order to test the effects of inactivating the essential endonuclease RNase E, it was necessary to carry out one set of analyses after a 10-min heat shock at 43.5°C. This has provided some insight into the effect of high temperature on the regulation of rpoS. There was an increase in the level of RpoS protein (in the absence of DsrA) of 20-fold, somewhat higher than the increase of 3- to 5-fold after heat shock reported previously (36). In addition, we detected a 3-fold increase in rpoS mRNA levels as well. Thus, in addition to the reported stabilization of RpoS protein at high temperature, it seems likely that either there is increased mRNA stability, possibly from increased translation, or increased transcription.
A striking observation was that RNase III showed little or no effect on rpoS mRNA and protein levels after the heat shock (Fig. 6). One model for both this observation and the increased mRNA levels might be that heat shock may lead to melting of the secondary structure within the hairpin, rendering it resistant to RNase III at these sites. Alternatively, RNase III could be either inactive or unavailable (busy with other substrates) under this condition. The heat shock-related increase in rpoS mRNA and translation may simply be due to loss of RNase III action on the rpoS mRNA.
Acknowledgments
We thank Gisela Storz, Nicholas De Lay, Kyung Moon, Aurelia Battesti, Pierre Mandin, Daniel Schu, and Paola Milanesio for their comments on the manuscript.
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, and by a National Research Service Award to C.A.M. (award F32 GM075392, National Institutes of Health).
Footnotes
Published ahead of print on 27 August 2010.
REFERENCES
- 1.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic region of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 2.Basineni, S. R., R. Madhugiri, T. Kolmsee, R. Hengge, and G. Klug. 2009. The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol. 6:584-594. [DOI] [PubMed] [Google Scholar]
- 3.Bougdour, A., C. Cunning, P. J. Baptiste, T. Elliott, and S. Gottesman. 2008. Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68:298-313. [DOI] [PubMed] [Google Scholar]
- 4.Bougdour, A., S. Wickner, and S. Gottesman. 2006. Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev. 20:884-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125-133. [DOI] [PubMed] [Google Scholar]
- 6.Brown, L., and T. Elliott. 1997. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 179:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera, J. E., and D.-J. Jin. 2001. Growth phase and growth rate regulation of the rapA gene, encoding the RNA polymerase-associated protein RapA in Escherichia coli. J. Bacteriol. 183:6126-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis, A. J. 2007. The RNA degradosome of Escherichia coli: a multiprotein mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61:71-87. [DOI] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., G. van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNAse E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 10.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deana, A., and J. G. Belasco. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19:2526-2533. [DOI] [PubMed] [Google Scholar]
- 12.Deana, A., H. Celesnik, and J. G. Belasco. 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451:355-358. [DOI] [PubMed] [Google Scholar]
- 13.Drider, D., and C. Condon. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8:195-200. [DOI] [PubMed] [Google Scholar]
- 14.Freire, P., J. D. Amaral, J. M. Santos, and C. M. Arraiano. 2006. Adaptation to carbon starvation: RNase III ensures normal expression levels of bolA1p mRNA and σS. Biochimie 88:341-346. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman, S., C. A. McCullen, M. Guillier, C. K. Vanderpool, N. Majdalani, J. Benhammou, K. M. Thompson, P. C. FitzGerald, N. A. Sowa, and D. J. FitzGerald. 2007. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp. Quant. Biol. 71:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guillier, M., and S. Gottesman. 2008. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 36:6781-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillier, M., and S. Gottesman. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59:231-247. [DOI] [PubMed] [Google Scholar]
- 18.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamoto, H., Y. Koide, T. Morita, and H. Aiba. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61:1013-1022. [DOI] [PubMed] [Google Scholar]
- 23.Lange, R., D. Fischer, and R. Hengge-Aronis. 1995. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 177:4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 25.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. U. S. A. 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lease, R. A., and S. A. Woodson. 2004. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 344:1211-1223. [DOI] [PubMed] [Google Scholar]
- 27.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 28.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U. S. A. 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 30.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 40:93-113. [DOI] [PubMed] [Google Scholar]
- 31.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massé, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 33.Mikulecky, P. J., K. Meenakshi, C. C. Brescia, J. C. Takach, D. Sledjeski, and A. L. Feig. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A)RNAs. Nat. Struct. Mol. Biol. 11:1206-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita, T., K. Maki, and H. Aiba. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA stabilization mediated by bacterial noncoding RNAs. Genes Dev. 19:2176-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muffler, A., M. Barth, C. Marschall, and R. Hengge-Aronis. 1997. Heat shock regulation of σS turnover: a role for DnaK and relationship between stress responses mediated by σS and σ32 in Escherichia coli. J. Bacteriol. 179:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the σs subunit of RNA-polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 38.Opdyke, J. A., J.-G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer, V., K. Papenfort, S. Lucchini, J. C. D. Hinton, and J. Vogel. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 16:840-846. [DOI] [PubMed] [Google Scholar]
- 40.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. U. S. A. 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prévost, K., H. Salvail, G. Desnoyers, J. F. Jacques, E. Phaneuf, and E. Massé. 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis Mol. Microbiol. 64:1260-1273. [DOI] [PubMed] [Google Scholar]
- 42.Ranquet, C., and S. Gottesman. 2007. Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189:4872-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Repoila, F., and S. Gottesman. 2003. Temperature sensing by the dsrA promoter. J. Bacteriol. 185:6609-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Resch, A., T. Afonyushkin, T. B. Lombo, K. J. McDowall, U. Blasi, and V. R. Kaberdin. 2008. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 14:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos, J. M., P. Freire, F. S. Mesquita, F. Mika, R. Hengge, and C. M. Arraiano. 2006. Poly(A)-polymerase I links transcription with mRNA degradation via σS proteolysis. Mol. Microbiol. 60:177-188. [DOI] [PubMed] [Google Scholar]
- 46.Schweder, T., K.-H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σs) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 49.Soper, T. J., and S. A. Woodson. 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14:1907-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, X., and R. M. Wartell. 2006. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry 45:4875-4887. [DOI] [PubMed] [Google Scholar]
- 51.Svenningsen, S. L., N. Costantino, D. L. Court, and S. Adhya. 2005. On the role of Cro in λ prophage induction. Proc. Natl. Acad. Sci. U. S. A. 102:4465-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takayanagi, Y., K. Tanaka, and H. Takahashi. 1994. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol. Gen. Genet. 243:525-531. [DOI] [PubMed] [Google Scholar]
- 53.Takiff, H. E., S. M. Chen, and D. L. Court. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171:2581-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 189:4243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tramonti, A., M. De Canio, and D. De Biase. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70:965-982. [DOI] [PubMed] [Google Scholar]
- 56.Trisler, P., and S. Gottesman. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Udekwu, K. I., and E. G. Wagner. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Updegrove, T., N. Wilf, X. Sun, and R. M. Wartell. 2008. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry 47:11184-11195. [DOI] [PubMed] [Google Scholar]
- 59.Urban, J. H., and J. Vogel. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35:1018-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urban, J. H., and J. Vogel. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 6:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel, J., L. Argaman, E. G. Wagner, and S. Altuvia. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14:2271-2276. [DOI] [PubMed] [Google Scholar]
- 62.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets σS for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]