Abstract
During infection of Escherichia coli, bacteriophage T4 usurps the host transcriptional machinery, redirecting it to the expression of early, middle, and late phage genes. Middle genes, whose expression begins about 1 min postinfection, are transcribed both from the extension of early RNA into middle genes and by the activation of T4 middle promoters. Middle-promoter activation requires the T4 transcriptional activator MotA and coactivator AsiA, which are known to interact with σ70, the specificity subunit of RNA polymerase. T4 motA amber [motA(Am)] or asiA(Am) phage grows poorly in wild-type E. coli. However, previous work has found that T4 motA(Am)does not grow in the E. coli mutant strain TabG. We show here that the RNA polymerase in TabG contains two mutations within its β-subunit gene: rpoB(E835K) and rpoB(G1249D). We find that the G1249D mutation is responsible for restricting the growth of either T4 motA(Am)or asiA(Am) and for impairing transcription from MotA/AsiA-activated middle promoters in vivo. With one exception, transcription from tested T4 early promoters is either unaffected or, in some cases, even increases, and there is no significant growth phenotype for the rpoB(E835K G1249D) strain in the absence of T4 infection. In reported structures of thermophilic RNA polymerase, the G1249 residue is located immediately adjacent to a hydrophobic pocket, called the switch 3 loop. This loop is thought to aid in the separation of the RNA from the DNA-RNA hybrid as RNA enters the RNA exit channel. Our results suggest that the presence of MotA and AsiA may impair the function of this loop or that this portion of the β subunit may influence interactions among MotA, AsiA, and RNA polymerase.
Bacterial RNA polymerase (RNAP) is a highly conserved enzyme that shares sequence and structural homology with multisubunit polymerases from single-celled archaea to multicellular eukaryotes (19, 55). In bacteria, an RNAP core consisting of five subunits (α1, α2, β, β′, and ω) combines with a specificity factor, σ, to form holoenzyme (13). A primary σ factor, such as σ70 of Escherichia coli, is used for promoter recognition during exponential growth; alternate σ factors are needed under specific growth conditions or times of stress (10, 34). In addition, proteins that bind to RNAP and/or the DNA can influence the activity of polymerase (2, 4, 16, 18, 42).
During infection of E. coli, bacteriophage T4 relies on the host transcriptional machinery for transcription from phage early, middle, and late promoters (reviewed in references 16, 26, and 54). Immediately after infection, synthesis of early RNA is driven by T4 early promoters, which contain strong matches to −10 and −35 DNA elements recognized by σ70. Expression of T4 middle genes is delayed, commencing after 1 to 2 min. Middle RNA is generated both from early promoters, whose transcripts extend into middle genes, and from specific middle promoters. While middle promoters contain the σ70-dependent −10 element, they have a −30 element (MotA box) rather than the −35 element recognized by σ70.
Two T4-encoded early proteins, MotA, a transcription activator, and AsiA, a coactivator, are required for the activation of the middle promoters through a process called σ appropriation (reviewed in references 16 and 18). In this process, AsiA binds tightly to the C-terminal portion of σ70, region 4 (31, 33, 44, 47), structurally remodeling this region (22) and preventing its interaction with the −35 element (3, 9, 27). This remodeling also allows MotA to interact with the C-terminal end of σ70 (6, 36), a portion of σ70 that is normally inaccessibly contained within RNAP (8, 21, 52). In addition, MotA interacts with a DNA sequence in the −30 region of middle promoter DNA (15, 23, 41). Thus, AsiA/MotA serves as a molecular switch: AsiA-bound RNAP is inactive at most host promoters (1, 7, 35, 43, 45, 46) while AsiA-bound RNAP/MotA activates middle promoters (17, 32). Understanding how AsiA and MotA interact with all portions of holoenzyme can provide insight into the intrasubdomain cross talk of RNAP.
Because middle promoters generate much of the prereplicative RNA needed for the synthesis of T4 DNA polymerase and its associated proteins, T4 motA and asiA mutant phage exhibit a strong phenotype. Amber (Am) and temperature sensitive (Ts) mutants exhibit a delay in DNA synthesis and form small plaques (25, 33) while motA and asiA deletions are lethal (5, 37). Analyses of RNA isolated from motA(Am) mutant infections reveal that these mutants are leaky; although transcription from many middle promoters is nearly eliminated, for particular promoters, a low level of RNA can still be detected (11, 24, 49, 51). The mutant E. coli strain TabG was originally identified by its inability to support T4 motA mutant growth (39). Although TabG has no significant growth defect on its own, a T4 motA(Am) infection of TabG yields no detectable plaques. This defect can be complemented by expression of a plasmid-borne motA gene (15). Pulitzer and coworkers reported that the TabG mutation maps to a site within or near the rpoB gene, the gene that encodes the β subunit of core polymerase (39).
In this paper we show that the rpoB gene of TabG contains two mutations, E835K and G1249D, and that it is the G1249D mutation that is responsible for restricting the growth of either a T4 motA(Am) or asiA(Am) mutant. Furthermore, we show that middle-promoter transcription is significantly impaired in a T4 wild-type (wt) infection of the mutant rpoB strain and that the G1249D substitution is responsible for this impairment. In contrast, transcription from early promoters is not compromised by the rpoB mutation. Structures of RNAP of thermophilic bacteria (28, 52) predict that the G1249 residue of the β subunit is located at the entry to the RNA exit channel, immediately adjacent to a hydrophobic pocket (switch 3 loop), which is thought to be responsible for the separation of the RNA from the DNA-RNA hybrid (53). Our results suggest that the presence of MotA and AsiA could impair the function of this loop or that this portion of β may influence interactions among MotA, AsiA, and holoenzyme.
MATERIALS AND METHODS
Phage and strains.
Wild-type T4D+, T4 amG1 (motA) (25), or T4 amS22 (asiA) (33) were used for infections. The E. coli B strains TabG (39), which is restrictive for T4 motA(Am) and motA(Ts) growth, and NapIV (30) have been described previously.
The rpoB mutations present within the TabG strain were transduced into the target strain BL21(DE3) (50) using phage P1vir to minimize gene transfer inefficiency due to the E. coli B-K restriction-modification. Two markers, 50% linked by transduction (29), were first transduced into an E. coli B606 strain from the donor K-12 strain CF2024, giving a B strain CF15179 bearing btuB::Tn10 (Tcr; tetracycline resistant; 20 μg/ml) and rpoB(T563P) (Rifr, rifampin resistant; 100 μg/ml). Phage P1vir grown on the CF15179 B strain was used to transduce the TabG B strain, selecting Tcr and screening for Rifs recombinants to obtain strain CF15192. This increases the likelihood that the rpoBC region beyond RpoB T563 is of TabG origin. Donor P1vir from CF15192 was then used to transduce the target strain BL21(DE3), and Tcr recombinants were screened by infecting with T4 amG1 (motA) to isolate those that could not support T4 motA(Am) growth (as occurs for the TabG strain). The entire rpoBC genes were sequenced from one such transductant, strain B11, which revealed only two codon changes from the wild type. Both occurred within RpoB (E835K and G1249D). Thus, B11 and BL21(DE3) are isogenic strains except for the rpoB mutations and the btuB::Tn10 marker.
DNA.
Plasmids containing either a wild-type rpoB from BL21(DE3) strain or rpoB(E835K G1249D) from TabG were constructed using standard cloning procedures. DNA was isolated from the BL21(DE3) and the TabG strain using a MasterPure DNA Purification Kit (Epicentre). Oligonucleotides directly upstream and downstream of the rpoB gene [4,029 bp in length; E. coli BL21(DE3) chromosome positions 4249149 to 4253177; accession number NC_012947] containing NdeI and BamHI sites, respectively, were used to amplify the rpoB gene. The PCR product was digested with NdeI and BamHI and ligated with pet28a(+) (Novagen) previously digested with those restriction enzymes.
The wild-type and rpoB(E835K G1249D) plasmids described above were used to separate the rpoB(E835K G1249D) mutations into single mutant plasmids. PCR products were obtained from both plasmids starting at rpoB bp position 2756 and ending at bp position 4029 followed by a BamHI site. These PCR products were digested with BsgRI and BamHI, generating inserts from rpoB bp 3689 to 4029. The wt and mutant rpoB plasmids were then digested with the same enzymes. The insert containing wt rpoB DNA from bp 3689 to 4029 was ligated into the mutant rpoB plasmid, generating pE835K/wt1249. The mutant rpoB DNA from bp 3689 to 4029 was ligated into the wt rpoB plasmid, generating pwt835/G1249D. Thus, each plasmid expressed an rpoB gene with only one of the TabG mutations.
rpoB genes and, when indicated, the adjacent rpoC gene were sequence verified in their entirety by the Facility for Biotechnology Resources of the FDA using PCR products that were generated with primers upstream, downstream, and within rpoB. Primer sequences are available upon request.
Complementation.
The ability of various strains to complement T4 motA(Am)or asiA(Am) growth was determined as described previously (6), except cells containing plasmids were grown in the presence of 20 μg/ml kanamycin and 1.7 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After growth to mid-log phase, cells were plated with T4 on LB plates (containing antibiotic, when plasmid was present).
Isolation of in vivo and in vitro RNA and primer extension analyses.
RNA was isolated from T4-infected cells using method II of Hinton (14). Cultures were infected for 5 min; chloramphenicol (to 100 μg/ml) was added immediately before harvesting. When cells with the plasmid-borne mutant rpoB were used, cells were grown in the presence of 1.7 mM IPTG.
Primer extension analyses were performed as described previously (11, 15) using avian myeloblastosis virus (AMV) reverse transcriptase (Life Sciences, Inc.), 5′ 32P-labeled oligonucleotides that annealed approximately 100 nucleotides (nt) downstream of the 5′ end of predicted mRNAs, and the same amount of RNA (from 1 to 10 μg) as measured by absorbance at 260 nm for each analysis (primer sequences are available upon request). RNA was generated in vitro using the plasmid pGEX-5X-3 DNA (Pharmacia Biotech), which contains the Ptac promoter. Transcription reactions were performed as described previously (6) except that the concentration of each ribonucleoside triphosphate was 200 μM, and no labeled ribonucleoside triphosphate was added. In the primer extension analyses shown in Fig. 2B, this Ptac RNA and a 5′ 32P-labeled primer, which annealed 66 nt from the start of the Ptac RNA, was included in the primer extension analyses as a control. Labeled primer extension products were separated on denaturing, polyacrylamide gels. After autoradiography, films were scanned using a Powerlook 100XL densitometer, and various species were quantified using Quantity One software from Bio-Rad, Inc.
For quality control purposes, we performed eight independent primer extension analyses for the gene 46 middle promoter (Pm46) using two independently isolated RNA preparations of the wt T4 and T4 motA(Am) infections of BL21(DE3) and B11; for Pe35.3, the gene 46 early promoter, we performed four independent primer extension analyses using the two different RNA preparations. B11 and BL21(DE3) are the appropriate strains for this analysis since they represent the P1 transductant/parent set. The values and errors for Pm46 and Pe35.3 shown in Fig. 3 are derived from all of these analyses. Furthermore, Pm46 was also analyzed using different primers. With primers that annealed 222 and 419 nt downstream from Pm46, the amount of Pm46 RNA from wt T4 infection of B11 versus BL21(DE3) was 0.53 ± 0.09 and 0.56 ± 0.05, respectively, which agrees well with the value of 0.42 ± 0.14 obtained with the Pm46 primer used in the experiments shown in Fig. 2 and 3 that annealed 98 nt downstream. In addition, the average of values from six primer extension analyses using five independently isolated RNA preparations of wt T4 infections of TabG versus a wt rpoB strain [NapIV or BL21(DE3)] was 0.28 ± 0.11, again using the Pm46 primer that annealed 98 nt downstream. Finally, all of the analyses shown in Fig. 3 were performed using the same RNA preparations that were analyzed for Pm46 and Pe35.3 RNA levels. Thus, the fact that the levels of the early RNAs (except Pe148.6) did not decrease in the B11 infections relative to the BL21(DE3) infections provides an additional control for the differences seen with the middle RNAs.
RESULTS
E. coli TabG contains two mutations within rpoB, the gene encoding the β subunit of RNA polymerase.
TabG is an E. coli B strain, which restricts the growth of the T4 motA(Ts) mutant tsG1 at the nonpermissive temperature and the growth of the T4 motA(Am) mutant amG1; the mutation in TabG needed for this restriction was previously reported to be in or near rpoB (39). We sequenced the rpoBC operon genes from chromosomal DNA of the TabG and wild-type strain BL21(DE3). Only two codon changes were found in the TabG strain, both within rpoB: E835K and G1249D (Fig. 1). These substitutions resulted from rpoB nucleotide changes of a G to A at position 2503 and a G to A at position 3746, respectively.
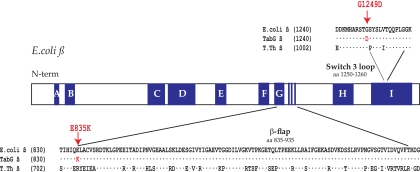
FIG. 1.
Location of TabG rpoB mutations. Schematic representation of the E. coli rpoB gene showing conserved regions (A, B, C, etc.) in dark blue and positions of the TabG substitutions E835K and G1249D within the β flap and adjacent to the switch 3 loop, respectively. Sequences of the residues surrounding the β flap and the switch 3 loop are given for the wt E. coli, for the E. coli TabG and B11, and for the T. thermophilus (T. th.) β proteins.
Previous workers reported that the TabG strain had no growth defects (39). To examine this finding in more detail, we compared the growth properties of the wt rpoB strain, BL21(DE3), with those of B11, a P1 transductant of BL21(DE3) which contains both of the TabG rpoB mutations. We found no significant difference in the abilities of these strains to grow on minimal or LB plates incubated at room temperature, 30°C, 37°C, or 42°C (data not shown).
The G1249D substitution within the β subunit restricts the growth of both T4 motA(Am) and T4 asiA(Am) phage.
We compared the ability of T4 wt, T4 motA(Am), and a T4 asiA(Am), amS22, to plate on the wt rpoB strain BL21(DE3) and on the rpoB mutant strains TabG and B11 (Table 1). wt T4 plaque size and morphology were normal when the phage were plated on each of these strains, and, as expected, both T4 motA(Am) and T4 asiA(Am) produced small plaques on the wt strain. However, the growth of either T4 motA(Am) or asiA(Am) was severely restricted on the rpoB mutant strains, TabG and B11. T4 motA(Am) produced barely perceptible plaques on B11 and no detectable plaques on TabG, while T4 asiA(Am) produced no detectable plaques on either strain. We conclude that the inability of TabG to support T4 motA(Am) or asiA(Am) growth is due primarily to one or both of the rpoB mutations. However, additional differences between TabG and the B11 backgrounds appear to accentuate this effect, allowing for the very tiny plaques seen in the T4 motA(Am) infections of B11.
TABLE 1.
T4 plaque phenotypes on E. coli strains
| Strain | Chromosomal rpoB | Plasmid | Plaque size |
||
|---|---|---|---|---|---|
| T4D+ | T4 motA(Am) | T4 asiA(Am) | |||
| TabG | E835K G1249D | Normal | None | None | |
| BL21(DE3) | wt | Normal | Small | Small | |
| B11 | E835K G1249D | Normal | Nonea | None | |
| TabG | E835K G1249D | pwtrpoB | Normal | Small | NTb |
| BL21(DE3) | wt | pE835K/G1249D | Normal | Smallerc | NT |
| B11 | E835K G1249D | pwtrpoB | Normal | Small | NT |
| B11 | E835K G1249D | pwt835/G1249D | Normal | None | None |
| B11 | E835K G1249D | pE835K/wt1249 | Normal | Small | Small |
Barely perceptible.
NT, not tested.
Smaller plaques than those made by T4 motA(Am) in BL21(DE3) without plasmid.
The presence of wt β, produced from the plasmid pwtrpoB, restored the ability of T4 motA(Am) to form small plaques on either TabG or B11 (Table 1). Conversely, in the wt strain, which produces wt β from its chromosome, synthesis of the plasmid-encoded mutant β from pE835K/G1249D further decreased the plaque size of T4 motA(Am) although it did not eliminate plaque formation. These results are consistent with the previous conclusion (39) that the TabG allele is recessive to wt in its ability to support T4 motA(Am) growth.
To determine whether both β subunit mutations are necessary for TabG restriction of T4 motA(Am) and T4 asiA(Am) phage, we constructed rpoB plasmids containing only one of the substitutions: pE835K/wt1249 or pwt835/G1249D. In each case, expression of the plasmid-borne rpoB was under the control of an IPTG-inducible promoter, and SDS-PAGE demonstrated that each of the mutant β proteins was produced at a high level in the presence of IPTG (data not shown). With the addition of IPTG in the B11 background, the presence of pE835K/wt1249 complemented either T4 motA(Am) or T4 asiA(Am)growth while the presence of pwt835/G1249D did not (Table 1). We conclude that it is the G1249D substitution within rpoB that significantly restricts growth of either T4 motA(Am) or T4 asiA(Am).
rpoB(E835K G1249D) specifically reduces transcription from T4 middle promoters.
Our finding that the TabG mutation is a substitution within the gene for the β subunit of RNAP suggested that this mutation might exert its effect at the level of T4 transcription. To investigate the effect of the TabG rpoB allele on the production of T4 prereplicative transcripts, we isolated RNA from wt or T4 motA(Am) infections of two wt rpoB strains [NapIV and BL21(DE3)] and two rpoB(E835K G1249D) mutant strains (TabG and B11). RNA was isolated 5 min after infection at 37°C, a time point when middle transcripts are the predominant species, but there are residual early transcripts, and late transcripts have started to be synthesized (reviewed in references 12, 40, and 48). The amount of transcription from a number of T4 early and middle promoters was then determined using primer extension analyses.
Previous work has shown that the T4 middle gene 46, which encodes a nuclease important for T4 recombination, is expressed from both the early promoter (Pe35.3), located 678 bp upstream, and the MotA/AsiA-dependent middle promoter (Pm46), located 30 bp upstream of the gene; in addition, transcription from promoters farther upstream, including Pm47, also extend into the gene (11, 20, 24, 26) (Fig. 2 A). As expected, primer extension analyses performed with RNA from wt T4 infection of wt rpoB (wt T4/wt rpoB) revealed products corresponding to transcription from both Pe35.3 and Pm46 (Fig. 2B, lanes 1 and 6), and transcription from Pm46 was dependent on MotA (Fig. 2B, lanes 3 and 8).
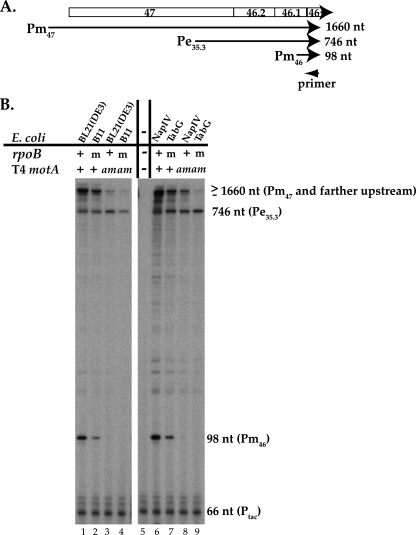
FIG. 2.
The level of T4 gene 46 RNA generated from the MotA-dependent middle promoter Pm46 is reduced by the TabG rpoB allele. (A) Schematic of gene 46 region from T4 map units 36576 (middle promoter Pm47) to 34916 (position of primer) (map units are from reference 26). The relative positions of genes and identified promoters in this region are shown. Transcription from Pm47, Pe35.3, and Pm46 yields primer extension products of 1,660, 746, and 98 nt, respectively. (B) Gels showing the primer extension products of gene 46 RNA. RNA was isolated from wt T4 (+) or T4 motA amG1 (am) infections of the indicated wt rpoB (+) strains, BL21(DE3), NapIV, or rpoB(E835K G1249D) mutant (m) strains, B11 or TabG. Ptac RNA and a 32P-labeled primer that anneals 66 nt from the start of the Ptac RNA were added to the primer extension analyses of the T4 RNA (lanes 1 to 4 and 6 to 9) as a control. The analysis in lane 5 contained the Ptac RNA and Ptac primer alone.
The primer extension analyses of gene 46 RNA isolated from wt T4 infections of either of the mutant β strains (T4/B11 or T4/TabG) revealed patterns that were qualitatively similar to those seen with the wt T4/wt rpoB infections (Fig. 2B, lanes 2 and 7 versus lanes 1 and 6). Transcription was observed from promoters far upstream, from the early promoter Pe35.3, and from the MotA/AsiA-dependent middle promoter Pm46. However, while the amount of transcription from Pe35.3 was similar in these lanes, the amount of Pm46 was significantly reduced relative to that seen in infections of the wt strains (Fig. 2B, compare lane 1 versus 2 and lane 6 versus 7).
To ensure that the differences in RNA levels were not due to experimental error, we checked our analyses in a number of ways. First, for the primer extension analyses shown in Fig. 2B, we included RNA made in vitro from a plasmid that contains the Ptac promoter and an additional 32P-labeled primer that anneals 66 nt downstream of the predicted transcription start. Primer extension using the Ptac RNA and the primer alone revealed a major product of 66 nt as well as slightly larger minor products (Fig. 2B, lane 5). Addition of the Ptac RNA and primer to the in vivo RNA yielded similar levels of the Ptac primer extension products in all the lanes (Fig. 2B, lanes 1 to 4 and 6 to 9). Second, we performed multiple primer extension analyses using independently isolated RNA preparations of wt T4 and T4 motA(Am) infections (see Materials and Methods for details). The values and errors for Pm46 and Pe35.3 that are shown in Fig. 3 are derived from all of these analyses. Thus, we conclude that in a wt T4 infection of the rpoB mutant strain B11, there is a specific decrease in the level of Pm46 RNA relative to that observed in an infection of the wt rpoB parent strain BL21(DE3) while the level of Pe35.3 RNA does not decrease.
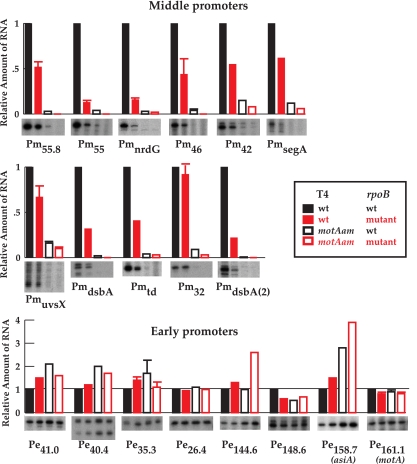
FIG. 3.
Transcription from MotA-dependent T4 middle promoters is specifically reduced in the rpoB(E835K G1249D) mutant strain B11 versus the wt rpoB strain BL21(DE3). Histograms show the levels of RNA from various middle promoters (top two panels) or early promoters (bottom panel) made in the indicated infections: solid black, wt T4/BL21(DE3); solid red, wt T4/B11; open black, T4 motA(Am)/BL21(DE3); open red, T4 motA(Am)/B11. A portion of the gel displaying the indicated primer extension products for each promoter is also shown. [Values for T4 motA(Am) infections were obtained using longer exposures.] For cases in which the primer extension analyses were performed three or more times, an error bar is shown. In the other cases, values from two analyses were averaged. PmuvsX RNA has multiple ends due to the iterative addition of As at the start of the transcript (15).
To investigate whether the findings with gene 46 RNA were general for T4 prereplicative transcription, we performed primer extension analyses of RNA isolated from wt T4 infections of B11 or its parent strain BL21(DE3) using primers that annealed downstream of various T4 middle and early promoters (Fig. 3). As indicated in the legend of Fig. 3, values were obtained from two or more independent primer extension experiments; those analyses performed three or more times are shown with error bars. We observed the same general trend as was seen for gene 46 transcription (Fig. 3, first and second lanes of each panel). Transcription from all of the tested middle promoters, except Pm32, was reduced 2- to 8-fold while transcription from early promoters, except Pe148.6, was the same or enhanced. The fact that the levels of the early transcripts (except Pe148.6) did not decrease in the B11 infections relative to the BL21(DE3) infections indicates that the overall level of T4 RNA is not generally suppressed in the B11 infections. Furthermore, the levels of RNA from the early promoters for MotA (P161.1) and AsiA (P158.7) did not decrease, indicating that the mutant β protein in B11 did not simply lower the level of motA or asiA RNA. Taken together, our results indicate that the rpoB allele in TabG specifically impairs transcription from T4 middle promoters.
As with Pm46 (Fig. 2B, lane 3), we observed a small amount of RNA from many middle promoters in the T4 motA(Am) infections of the wt rpoB strain (Fig. 3, third lane of each middle promoter panel). This low level of middle promoter RNA has been observed by other investigators in T4 motA(Am) infections of wt E. coli (11, 24, 49, 51). Previous work has demonstrated that deletion of motA is lethal (5), suggesting that the motA(Am) mutation is leaky. Thus, this mutant is expected to generate a small amount of full-length MotA protein, allowing a low level of middle-promoter transcription. We find that a T4 motA(Am) infection of B11 (rpoB mutant) further reduces this residual middle promoter transcription (Fig. 3, compare the third and fourth lanes of each middle promoter panel). These results suggest that it is this further impairment of middle-promoter activation by the TabG mutant β protein that is responsible for preventing the growth of T4 motA(Am) and indicates that middle promoter activity is essential for T4 development.
The G1249D mutation within rpoB is responsible for the defect in T4 middle-promoter transcription.
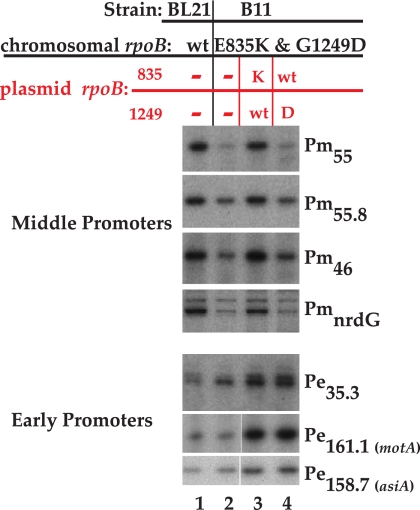
We repeated the primer extension analyses, isolating RNA from wt T4 infections of B11 grown in the presence of IPTG and a plasmid with only one of the rpoB substitutions: either pE835K/wt1249 or pwt835/G1249D. Each of these plasmids produced similar levels of the singly mutant β protein, as judged by SDS-PAGE (data not shown). Using the same RNA preparations, we analyzed RNA from both T4 early and middle promoters (Fig. 4). In the absence of a plasmid, middle-promoter transcripts were again reduced in B11 relative to BL21(DE3) (Fig. 4, lanes 1 and 2). The rpoB plasmid with the wt glycine at 1249 (lane 3) restored the level of middle RNA to that observed in a wt T4 infection of the wt rpoB strain (lane 1), whereas the plasmid with the mutant G1249D did not (lane 4). Thus, we conclude that it is the G1249D mutation that impairs T4 MotA/AsiA-dependent transcription.
FIG. 4.
The rpoB mutation G1249D is responsible for the defect in MotA/AsiA-activated transcription in a wt T4 infection. Portions of gels showing primer extension products were obtained after wt T4 infections of the following: wt rpoB strain BL21(DE3) (lane 1), the double mutant rpoB(E835K G1249D) strain B11 (lane 2), B11 containing pE835K/wt1249 (lane 3), and B11 containing pwt835/G1249D.
As seen before (Fig. 3), the levels of transcription from the early promoters Pe35.3, P158.7, and P161.1, which are immediately upstream of the early genes 46.2, asiA, and motA, respectively, were similar or higher in the T4/B11 than in the T4/wt rpoB infections (Fig. 4, lanes 1 versus 2). Furthermore, the presence of either singly mutant plasmid affected these early RNAs similarly, raising the levels even further over those seen in the T4/wt rpoB infection (lanes 3 and 4). These results indicate that simply increasing motA and asiA transcription cannot compensate for the impairment of middle promoter transcription imparted by the mutant β protein.
DISCUSSION
Bacteriophage T4 gene expression is controlled primarily at the level of transcription through the activity of phage early, middle, and late promoters. Middle transcription, which generates the prereplicative RNAs needed for the synthesis of phage replication proteins, begins approximately 1 to 2 min after infection at 37°C (reviewed in references 26 and 54). Two T4 early proteins, the transcriptional activator MotA and the coactivator AsiA, are required for transcription from nearly 60 known T4 middle promoters (reviewed in references 16 and 18).
The E. coli mutant TabG was originally isolated as a strain that significantly restricts the growth of the T4 motA amber mutant, amG1, and the motA temperature sensitive mutant, tsG1 (39). TabG also restricts T4 asiA(Am), amS22, as we have demonstrated here. Pulitzer and coworkers mapped the TabG mutation to a site in or near rpoB. However, the nature and exact location of the lesions as well as the reason why TabG restricts T4 motA(Am) were unclear. We have shown that TabG rpoB contains two substitutions, E835K and G1249D. Though these mutant residues are separated by over 1,200 bp in the rpoB DNA sequence (Fig. 1), they are relatively close in the structures of RNAP from the thermophilic bacteria, Thermus thermophilus and Thermus aquaticus (28, 52) (Fig. 5). The residue corresponding to E835 in E. coli (R707 of T. thermophilus) is located near the DNA-RNA hybrid at the base of a loop within the β subunit called the β flap, which forms one wall of the RNA exit channel. Our results suggest that the E835K mutation has no significant effect on the growth of T4 or the host, at least under our conditions. G1249 (G1011 of T. thermophilus) is a highly conserved residue, located on the opposite side of the RNA exit channel. Although this mutation does not appear to be deleterious for the host or for transcription from T4 early promoters, our analyses reveal that G1249D confers a defect in MotA/AsiA-dependent transcription. Based on the structure of a T. thermophilus RNAP elongating complex (53), G1249 is immediately adjacent to a structural feature called the switch 3 loop (T. thermophilus β residues 1012 to 1022, corresponding to E. coli residues 1250 to 1260) (Fig. 1 and 5). This portion of β is located at the upstream end of the RNA-DNA hybrid. It has been suggested that switch 3 loop interacts with the RNA, guiding it away from its interaction with the DNA and leading it into the RNA exit channel (53). Thus, this loop is thought to perform an extremely important function for RNAP. However, since the G1249D change is not yet associated with a phenotype other than its effect on MotA/AsiA transcription, this change appears to be an alteration that is relatively benign for the host.
FIG. 5.
Structures of T. thermophilus holoenzyme (top) containing σ (yellow) and β and β′ (teal) (accession number 1IW7) (52) and T. thermophilus elongating complex (bottom) containing β and β′ (teal), RNA (black), and DNA (magenta) (accession number 2O5I) (53), with the positions of the β residues corresponding to E. coli E835 and G1249 shown as red spheres. In the top structure, σ region 4 through the C terminus is boxed in yellow; this region of σ normally interacts with the β flap, with residues at the C-terminal tail of σ in contact with the β-flap tip, as shown here. In σ appropriation, region 4 is dramatically remodeled upon binding to AsiA (22). This remodeling prevents the σ-β-flap interaction, allowing MotA to now interact with the C terminus of σ. In the bottom structure, the residues within the switch 3 loop of β (E. coli residues 1250 to 1260), which are immediately adjacent to G1249, are shown as orange spheres.
Activation of T4 middle promoters by MotA and coactivator AsiA occurs through a process called σ appropriation (reviewed in references 16 and 18). In this process, AsiA binds tightly to free σ70, structurally remodeling region 4, located in the C-terminal portion of the σ70 protein. When the AsiA/σ70 complex associates with core, this remodeling prevents σ70 from interacting with the −35 region of promoter DNA as well as the β flap. Consequently, MotA can interact with its binding site, the MotA box, centered at −30 of middle promoter DNA, as well as with residues at the very end of the C terminus of σ70, which would normally be inaccessible due to their interaction with the β-flap tip. Recent work has indicated that AsiA also binds the β flap, replacing the normal interaction between σ70 region 4 and this portion of the β protein (57). A direct interaction between AsiA and MotA has also been suggested (56) although other work indicates that they do not directly interact (6, 38). Thus, the RNAP/MotA/AsiA complex consists of multiple protein-protein interactions as well as a major conformational change surrounding σ70 region 4. A thorough understanding of the roles of the β flap and σ70 region 4 and how T4 employs these regions to its advantage may provide insight into how the activity of RNAP is regulated.
We can think of three simple models to explain how the G1249D substitution, which will introduce both a negative charge and a decrease in flexibility, might impair MotA/AsiA activation. First, since this substitution lies on a face of β that is across from the β flap (Fig. 5), it seems possible that it might impair the association of the AsiA/σ70 complex with the core, thus affecting the first step in σ appropriation, or impair an association of AsiA with the β flap. Alternately, the binding of AsiA and MotA to RNAP could impose subtle conformational alterations that extend to the switch 3 loop. In particular, the presence of the additional negative charge from the G1249D substitution might now affect the exit of the negatively charged RNA if MotA-induced or AsiA-induced changes in RNAP extend to the RNA exit channel. Finally, the G1249D substitution may identify a region that directly interacts with either AsiA or MotA. In vitro experiments with purified mutant RNAP can test each of these hypotheses. Understanding how the TabG mutation affects MotA/AsiA activation will help elucidate the global arrangement of protein-protein interactions needed for T4 middle gene transcription.
Acknowledgments
We thank C. Jones, K. Decker, L. Knipling, M. Hsieh, R. Bonocora, and T. Cardozo for helpful discussions.
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (T.D.J. and D.M.H.) and by the NIH, Eunice Kennedy Schriver National Institute of Child Health and Human Development (M.C.). T.D.J. was also supported by a Ford Foundation predoctoral fellowship.
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Adelman, K., G. Orsini, A. Kolb, L. Graziani, and E. N. Brody. 1997. The interaction between the AsiA protein of bacteriophage T4 and the σ70 subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 272:27435-27443. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, K., J. Lee, L. Minakhin, K. Severinov, and D. M. Hinton. 2006. Mutational analysis of σ70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA. J. Mol. Biol. 363:931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, L. L., T. G. Smith, and T. R. Hoover. 2007. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 15:530-537. [DOI] [PubMed] [Google Scholar]
- 5.Benson, K. H., and K. N. Kreuzer. 1992. Role of MotA transcription factor in bacteriophage T4 DNA replication. J. Mol. Biol. 228:88-100. [DOI] [PubMed] [Google Scholar]
- 6.Bonocora, R. P., G. Caignan, C. Woodrell, M. H. Werner, and D. M. Hinton. 2008. A basic/hydrophobic cleft of the T4 activator MotA interacts with the C terminus of E. coli σ70 to activate middle gene transcription. Mol. Microbiol. 69:331-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colland, F., G. Orsini, E. N. Brody, H. Buc, and A. Kolb. 1998. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol. Microbiol. 27:819-829. [DOI] [PubMed] [Google Scholar]
- 8.Geszvain, K., T. M. Gruber, R. A. Mooney, C. A. Gross, and R. Landick. 2004. A hydrophobic patch on the flap-tip helix of E. coli RNA polymerase mediates σ70 region 4 function. J. Mol. Biol. 343:569-587. [DOI] [PubMed] [Google Scholar]
- 9.Gregory, B. D., B. E. Nickels, S. J. Garrity, E. Severinova, L. Minakhin, R. J. Urbauer, J. L. Urbauer, T. Heyduk, K. Severinov, and A. Hochschild. 2004. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 101:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 11.Guild, N., M. Gayle, R. Sweeney, T. Hollingsworth, T. Modeer, and L. Gold. 1988. Transcriptional activation of bacteriophage T4 middle promoters by the MotA protein. J. Mol. Biol. 199:241-258. [DOI] [PubMed] [Google Scholar]
- 12.Guttman, B., and E. Kutter. 1983. Overview, p. 8-10. In C. K. Mathews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, DC.
- 13.Helmann, J. D. 2009. RNA polymerase: a nexus of gene regulation. Methods 47:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinton, D. M. 1989. Transcript analyses of the uvsX-40-41 region of bacteriophage T4. Changes in the RNA as infection proceeds. J. Biol. Chem. 264:14432-14439. [PubMed] [Google Scholar]
- 15.Hinton, D. M. 1991. Transcription from a bacteriophage T4 middle promoter using T4 MotA protein and phage-modified RNA polymerase. J. Biol. Chem. 266:18034-18044. [PubMed] [Google Scholar]
- 16.Hinton, D. M. Transcriptional control in the prereplicative phase of T4 development. Virol. J., in press. [DOI] [PMC free article] [PubMed]
- 17.Hinton, D. M., R. March-Amegadzie, J. S. Gerber, and M. Sharma. 1996. Bacteriophage T4 middle transcription system: T4-modified RNA polymerase; AsiA, a sigma 70 binding protein; and transcriptional activator MotA. Methods Enzymol. 274:43-57. [DOI] [PubMed] [Google Scholar]
- 18.Hinton, D. M., S. Pande, N. Wais, X. B. Johnson, M. Vuthoori, A. Makela, and I. Hook-Barnard. 2005. Transcriptional takeover by sigma appropriation: remodelling of the σ70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiology 151:1729-1740. [DOI] [PubMed] [Google Scholar]
- 19.Hirata, A., B. J. Klein, and K. S. Murakami. 2008. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451:851-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, T., and J. D. Karam. 1990. Transcriptional mapping of a DNA replication gene cluster in bacteriophage T4. Sites for initiation, termination, and mRNA processing. J. Biol. Chem. 265:5303-5316. [PubMed] [Google Scholar]
- 21.Kuznedelov, K., L. Minakhin, A. Niedziela-Majka, S. L. Dove, D. Rogulja, B. E. Nickels, A. Hochschild, T. Heyduk, and K. Severinov. 2002. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295:855-857. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, L. J., Y. Wei, V. Schirf, B. Demeler, and M. H. Werner. 2004. T4 AsiA blocks DNA recognition by remodeling σ70 region 4. EMBO J. 23:2952-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.March-Amegadzie, R., and D. M. Hinton. 1995. The bacteriophage T4 middle promoter PuvsX: analysis of regions important for binding of the T4 transcriptional activator MotA and for activation of transcription. Mol. Microbiol. 15:649-660. [DOI] [PubMed] [Google Scholar]
- 24.Marshall, P., M. Sharma, and D. M. Hinton. 1999. The bacteriophage T4 transcriptional activator MotA accepts various base-pair changes within its binding sequence. J. Mol. Biol. 285:931-944. [DOI] [PubMed] [Google Scholar]
- 25.Mattson, T., G. Van Houwe, and R. H. Epstein. 1978. Isolation and characterization of conditional lethal mutations in the mot gene of bacteriophage T4. J. Mol. Biol. 126:551-570. [DOI] [PubMed] [Google Scholar]
- 26.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minakhin, L., J. A. Camarero, M. Holford, C. Parker, T. W. Muir, and K. Severinov. 2001. Mapping the molecular interface between the σ70 subunit of E. coli RNA polymerase and T4 AsiA. J. Mol. Biol. 306:631-642. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, H., and M. Cashel. 2003. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 371:596-601. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, M. A., M. Ericson, L. Gold, and J. F. Pulitzer. 1982. The isolation and characterization of TabR bacteria: hosts that restrict bacteriophage T4 rII mutants. Mol. Gen. Genet. 188:60-68. [Google Scholar]
- 31.Orsini, G., M. Ouhammouch, J. P. Le Caer, and E. N. Brody. 1993. The asiA gene of bacteriophage T4 codes for the anti-sigma 70 protein. J. Bacteriol. 175:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouhammouch, M., K. Adelman, S. R. Harvey, G. Orsini, and E. N. Brody. 1995. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc. Natl. Acad. Sci. U. S. A. 92:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouhammouch, M., G. Orsini, and E. N. Brody. 1994. The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J. Bacteriol. 176:3956-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paget, M. S., and J. D. Helmann. 2003. The sigma70 family of sigma factors. Genome Biol. 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pahari, S., and D. Chatterji. 1997. Interaction of bacteriophage T4 AsiA protein with Escherichia coli sigma70 and its variant. FEBS Lett. 411:60-62. [DOI] [PubMed] [Google Scholar]
- 36.Pande, S., A. Makela, S. L. Dove, B. E. Nickels, A. Hochschild, and D. M. Hinton. 2002. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the σ70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 184:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pene, C., and M. Uzan. 2000. The bacteriophage T4 anti-sigma factor AsiA is not necessary for the inhibition of early promoters in vivo. Mol. Microbiol. 35:1180-1191. [DOI] [PubMed] [Google Scholar]
- 38.Pineda, M., B. D. Gregory, B. Szczypinski, K. R. Baxter, A. Hochschild, E. S. Miller, and D. M. Hinton. 2004. A family of anti-σ70 proteins in T4-type phages and bacteria that are similar to AsiA, a transcription inhibitor and co-activator of bacteriophage T4. J. Mol. Biol. 344:1183-1197. [DOI] [PubMed] [Google Scholar]
- 39.Pulitzer, J. F., A. Coppo, and M. Caruso. 1979. Host-virus interactions in the control of T4 prereplicative transcription. II. Interaction between tabC (rho) mutants and T4 mot mutants. J. Mol. Biol. 135:979-997. [DOI] [PubMed] [Google Scholar]
- 40.Rabussay, D. 1983. Phage-evoked changes in RNA polymerase, p. 167-173. In C. K. Mathews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, DC.
- 41.Schmidt, R. P., and K. N. Kreuzer. 1992. Purified MotA protein binds the −30 region of a bacteriophage T4 middle-mode promoter and activates transcription in vitro. J. Biol. Chem. 267:11399-11407. [PubMed] [Google Scholar]
- 42.Schumacher, J., N. Joly, M. Rappas, X. Zhang, and M. Buck. 2006. Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation. J. Struct. Biol. 156:190-199. [DOI] [PubMed] [Google Scholar]
- 43.Severinova, E., K. Severinov, and S. A. Darst. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 279:9-18. [DOI] [PubMed] [Google Scholar]
- 44.Severinova, E., K. Severinov, D. Fenyo, M. Marr, E. N. Brody, J. W. Roberts, B. T. Chait, and S. A. Darst. 1996. Domain organization of the Escherichia coli RNA polymerase sigma 70 subunit. J. Mol. Biol. 263:637-647. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, A. 1973. An inhibitor of host sigma-stimulated core enzyme activity that purifies with DNA-dependent RNA polymerase of E. coli following T4 phage infection. Biochem. Biophys. Res. Commun. 54:488-493. [DOI] [PubMed] [Google Scholar]
- 46.Stevens, A. 1972. New small polypeptides associated with DNA-dependent RNA polymerase of Escherichia coli after infection with bacteriophage T4. Proc. Natl. Acad. Sci. U. S. A. 69:603-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens, A., and J. C. Rhoton. 1975. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry 14:5074-5079. [DOI] [PubMed] [Google Scholar]
- 48.Stitt, B., and D. M. Hinton. 1994. Regulation of middle-mode transcription, p. 142-160. In J. D. Karam, J. Drake, K. N. Kreuzer, G. Mosig, D. Hall, F. Eiserling, L. Black, E. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, DC.
- 49.Stoskiene, G., L. Truncaite, A. Zajanckauskaite, and R. Nivinskas. 2007. Middle promoters constitute the most abundant and diverse class of promoters in bacteriophage T4. Mol. Microbiol. 64:421-434. [DOI] [PubMed] [Google Scholar]
- 50.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 51.Truncaite, L., L. Piesiniene, G. Kolesinskiene, A. Zajanckauskaite, A. Driukas, V. Klausa, and R. Nivinskas. 2003. Twelve new MotA-dependent middle promoters of bacteriophage T4: consensus sequence revised. J. Mol. Biol. 327:335-346. [DOI] [PubMed] [Google Scholar]
- 52.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 53.Vassylyev, D. G., M. N. Vassylyeva, A. Perederina, T. H. Tahirov, and I. Artsimovitch. 2007. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448:157-162. [DOI] [PubMed] [Google Scholar]
- 54.Weisberg, R., D. M. Hinton, and S. Adhya. 2008. Transcriptional regulation in bacteriophage, p. 174-186. In B. W. J. Mahy and M. H. V. van Regenmortel (ed.), Encyclopedia of virology, 3rd ed. Elsevier, Oxford, United Kingdom.
- 55.Werner, F. 2008. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 16:247-250. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, A. H., and A. Hochschild. 2009. Direct activator/co-activator interaction is essential for bacteriophage T4 middle gene expression. Mol. Microbiol. 74:1018-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, A. H., B. E. Nickels, and A. Hochschild. 2009. The bacteriophage T4 AsiA protein contacts the beta-flap domain of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 106:6597-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]