Abstract
The SasG surface protein of Staphylococcus aureus has been shown to promote the formation of biofilm. SasG comprises an N-terminal A domain and repeated B domains. Here we demonstrate that SasG is involved in the accumulation phase of biofilm, a process that requires a physiological concentration of Zn2+. The B domains, but not the A domain, are required. Purified recombinant B domain protein can form dimers in vitro in a Zn2+-dependent fashion. Furthermore, the protein can bind to cells that have B domains anchored to their surface and block biofilm formation. The full-length SasG protein exposed on the cell surface is processed within the B domains to a limited degree, resulting in cleaved proteins of various lengths being released into the supernatant. Some of the released molecules associate with the surface-exposed B domains that remain attached to the cell. Studies using inhibitors and mutants failed to identify any protease that could cause the observed cleavage within the B domains. Extensively purified recombinant B domain protein is very labile, and we propose that cleavage occurs spontaneously at labile peptide bonds and that this is necessary for biofilm formation.
Staphylococcus aureus is a commensal bacterium that is carried persistently in the anterior nares of about 20% of the human population. The organism can cause superficial skin infections, such as abscesses and impetigo, and more dangerous and potentially life-threatening invasive infections, such as endocarditis, osteomyelitis, and septic arthritis (26). Staphylococcus epidermidis and S. aureus are the major causes of infections associated with indwelling medical devices, such as central venous catheters, cardiovascular devices, and artificial joints (34, 54). The ability to form a biofilm is crucial to the microbes' success in device-related infections. Bacteria in the biofilm matrix are in a semidormant state, are difficult to inhibit with antibiotics, and are impervious to host neutrophils and macrophages (36, 43, 44, 51). Until recently biofilm formation by staphylococci was attributed to the ability to synthesize an extracellular polysaccharide called polysaccharide intercellular adhesin (PIA), which is composed of partially deacetylated poly-N-acetylglucosamine (15, 28, 50). Attachment of bacteria to biomedical devices is mediated by adhesion to the naked plastic or metal surface by a surface component such as the major autolysin Atl (2, 14). Alternatively, adhesion to surfaces that have been conditioned by fibronectin and fibrinogen from host plasma is mediated by surface proteins such as clumping factor A (ClfA) and fibronectin binding proteins (FnBPA/B) of S. aureus or SdrG/Fbe of S. epidermidis (17, 46, 47).
Several surface proteins of staphylococci can also promote the accumulation phase of biofilm: (i) the biofilm-associated protein Bap, which is only expressed by bovine strains of S. aureus (8); (ii) the SasC surface protein of S. aureus (41); (iii) fibronectin binding proteins FnBPA and FnBPB, which are particularly associated with biofilm formation by some types of methicillin-resistant S. aureus (MRSA) (35, 48); (iv) the multifactorial virulence factor protein A, which promotes cell accumulation when expressed at high levels, for example,in mutants defective in the accessory gene regulator Agr (31); (v) the extracellular matrix binding protein (Embp) of S. epidermidis (4); (vi) the accumulation-associated protein (Aap) of S. epidermidis and the related protein SasG from S. aureus (7, 19, 40).
Aap and SasG are typical LPXTG-anchored multidomain cell wall-associated proteins (see Fig. 1A, below). A signal sequence is removed from the N terminus during secretion across the cytoplasmic membrane. The C-terminal domains comprise a sorting signal (LPXTG) and hydrophobic membrane-spanning domain and positively charged residues that are required for covalent attachment of the proteins to cell wall peptidoglycan by sortase A. The N termini of the mature proteins (A domains) comprise related amino acid sequences that have been implicated in adhesion of bacteria to desquamated epithelial cells and could be involved in colonization of the nares and skin (7, 27, 39). The archetypal Aap protein of S. epidermidis RP62a has 12 repeats of almost identical sequences of 128 residues followed by a partial repeat of 68 residues (region B), while SasG from S. aureus strain 8325-4 and strain Newman has seven 128-residue repeats and one partial repeat. The B subunits of Aap and SasG are 64% identical.
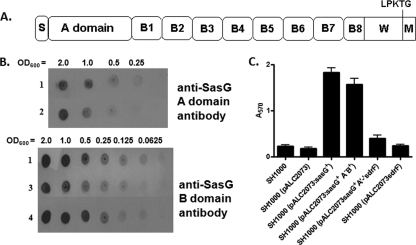
FIG. 1.
(A) Schematic representation of SasG domain organization. The positions of the signal sequence (S), A domain, B region (B1 to -8), and the wall/membrane-spanning regions (W/M) are indicated. The LPKTG motif is recognized by the sortase A enzyme, which covalently anchors the protein to the cell wall peptidoglycan. (B) Whole-cell immunoblot validating expression of A domain and B regions of SasG variants. Serial dilutions of SH1000(pALC2073:sasG+) (row 1); SH1000(pALC2073sasG+ A+B−) (row 2); SH1000(pALC2073sasG+ A−B+) (row 3), and SH1000(pALC2073sasG+ A−B+) induced with tetracycline (90 ng/ml) (row 4) were applied to a nitrocellulose membrane and probed with anti-SasG A domain and anti-SasG B domain antibodies. (C) Biofilm formation by SH1000 constructs expressing SasG variants. Biofilm was allowed to form for 24 h at 37°C under static conditions in microtiter dishes. Biofilm was stained with crystal violet, and the absorbance was measured at 570 nm.
The formation of biofilm by Aap in S. epidermidis is promoted by the removal of the A domain by cleavage by an as-yet-unidentified bacterial protease, an event that can also be precipitated by host proteases (40). The ability of the exposed Aap B domains of different bacterial cells to form homophilic interactions through a Zn2+-dependent zipper mechanism was proposed when it was shown that purified B domains formed dimers in vitro that were dependent on the presence of Zn2+ (6). Purified recombinant B domain protein, but not the A domain, inhibited biofilm formation, as did antibodies that specifically bound to the B domains (40). The Zn2+ chelator diethylenetriaminepentaacetic acid (DTPA) inhibited biofilm formation both by S. epidermidis RP62a (presumed to be due to Aap) and by community-associated MRSA (presumed to be due to SasG) (6).
This study set out to investigate the molecular basis of biofilm accumulation promoted by the SasG protein of S. aureus. We demonstrate that processing of SasG occurs during growth and biofilm formation in a manner that is different from that reported for Aap, and we have investigated the mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this study are listed in Table 1. Escherichia coli was grown in Luria broth at 37°C. Lactococcus lactis was grown at 30°C on M17 agar or broth (Difco) containing 0.5% (wt/vol) glucose. S. aureus and S. epidermidis were grown in tryptic soy broth (TSB) or brain heart infusion broth (BHI; Oxoid) at 37°C. Media were supplemented with glucose (1%, wt/vol), ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), tetracycline (90 ng/ml), E-64 (2 mM; Sigma), dichloroisocoumarin (1 mM; Sigma), 1,10-phenylthroline (10 μM; Sigma), NIFKPST peptide (2 mM; synthesized by Genscript), α2-macroglobulin (2 U/ml; Roche), inhibitor cocktail 1 (Complete protease inhibitor cocktail; Roche), inhibitor cocktail 2 (protease inhibitor cocktail for bacterial extracts; Sigma), or inhibitor cocktail 3 (protease inhibitor cocktail for tissue culture media; Sigma) where appropriate.
TABLE 1.
Bacterial strains
| Strain | Description | Reference or source |
|---|---|---|
| S. aureus strains | ||
| 8325-4 | NCTC8325 cured of prophages | 32 |
| RN4220 | Restriction-deficient mutant of 8325-4 | 23 |
| SH1000 | Strain 8325-4 with repaired defect in rsbU | 18 |
| SH1000 sspA | sspA::pAZ106 Emr | 18 |
| 8325-4 sspB | ΔsspB::Emr | 42 |
| SH1000 sspB | sspB::Emr transduced from 8325-4 | This study |
| RN6390 | Δspl::Emr | 37 |
| SH1000 spl | Δspl::Emr transduced from RN6390 | This study |
| SH1000 scpA | ΔscpA::Emr | 42 |
| SH1000 aur | aur::pAZ106 Emr | 30 |
| SH1000 aur/spl | Δaur spl::Emr | 3 |
| SH1000 htrA1 htrA2 | htrA1::EmrhtrA2::Tcr | Unpublished |
| 8325-4 atl | Δatl::Emr | 11 |
| SH1000 atl | atl:Emr transduced from 8325-4 | This study |
| SA113 | Restriction deficient | 20 |
| SA113 aaa | aaa::Emr | 13 |
| 125 | Clinical isolate st8 | 9 |
| 207 | Clinical isolate st15 | 9 |
| 410 | Clinical isolate st15 | 9 |
| 3093 | Clinical isolate st15 | 9 |
| L. lactis MG1363 | Plasmid-free derivative of strain NCDO 712 | 52 |
| S. epidermidis strains | ||
| TU3298 | Transformable strain | 1 |
| CSF41498 | Cerebrospinal fluid isolate, biofilm positive | 5 |
Plasmid constructions.
Plasmid pALC2073sasG+A−B+ was generated by inverse PCR using plasmid pALC2073sasG+ (7) as template DNA and the primers 5′-GGGAGATCTGCGCCAAAAACAATAACAGAATTAG-3′ and 5′-GGGAGATCTAGCTGCTTCTGCCTCTTGTTGAG-3′, incorporating BglII sites.
The sdrF gene was cloned between KpnI and SacI sites in pALC2073 to give pALC2073sdrF+. An HpaI site exists toward the end of the region encoding the SdrF A domain. Plasmid pALC2073sdrF+ was digested with KpnI and HpaI to release DNA encoding the signal sequence, A domain, and ribosome binding site of SdrF. DNA encoding the A domain, signal sequence, and the ribosome binding site of sasG was amplified from pALC2073sasG+ by using the primers 5′-CGGGGTACCGTAAGTAAAGTGGAAAATATGG-3′ and 5′-GGGGTTAACCATTCTAATTCTGTTATTGTTTTTG-3′. The PCR product was digested with KpnI and HpaI and ligated to KpnI/HpaI-digested pALC2073sdrF+ to create pALC2073sasG+A+B−. Plasmids were introduced into S. aureus RN4220 by electroporation and then transduced into SH1000 using phage 85 (12). Plasmids were introduced into S. epidermidis by electroporation (25).
Biofilm assay.
Bacteria were grown for 18 h in TSB and diluted 1:200 in BHI for S. epidermidis or in BHI with glucose (1%, wt/vol) for S. aureus. Diluted bacteria (200 μl) were added to sterile tissue culture-treated, 96-well polystyrene plates (Nunclon Delta) and incubated statically at 37°C for 24 h. Wells were washed three times with phosphate-buffered saline (PBS) and dried by inversion for 30 min. Adherent cells were stained with 0.5% (wt/vol) crystal violet, and the A570 was measured.
For inhibition assays, DTPA, ZnCl2, HCl, or increasing concentrations of recombinant proteins were added to inoculated wells at the beginning of a biofilm assay, incubated, and treated as described above.
Primary attachment assay.
Attachment assays were based on the method of Lim et al. (24). Bacteria were grown overnight in BHI medium supplemented with 1% (wt/vol) glucose, diluted in the same medium, and approximately 300 CFU in 100 μl was spread on the base of empty petri dishes. Dishes were incubated upright at 37°C for 30 min, washed three times with 5 ml of sterile PBS, and covered with BHI agar. Bacterial plate counts were run in parallel, and the percent attachment was calculated. Each experiment was repeated three times. Statistical significance was determined with Student's t test, using GraphPad software.
Aggregation assay.
Bacteria were grown overnight in TSB and diluted to an optical density at 600 nm (OD600) of 1 in BHI supplemented with 1% (wt/vol) glucose. Tubes were incubated statically at 37°C for 24 h. One milliliter of broth was removed from the top of the tube, and the OD600 was measured. The remaining culture was vortexed to resuspend the cells, and the OD600 was measured again. The percent aggregation was calculated using the following formula: 100 × [(OD600 of vortexed sample − OD600 before vortexing)/(OD600 of vortexed sample)]. Statistical significance was determined with Student's t test, using GraphPad software.
Western immunoblotting.
Cell wall-associated proteins of S. aureus were prepared as previously described (38). Stationary-phase cultures were harvested, washed in PBS, and resuspended to an OD600 of 40 in lysis buffer (50 mM Tris-HCl, 20 mM MgCl2; pH 7.5) supplemented with 30% (wt/vol) raffinose and Complete protease inhibitors (40 μl/ml; Roche). Cell wall proteins were solubilized by incubation with lysostaphin (200 μg/ml; AMBI, NY) for 10 min at 37°C. Protoplasts were removed by centrifugation at 12,000 × g for 10 min, and the supernatant containing solubilized cell wall proteins was aspirated and boiled for 5 min in Laemmli sample buffer (Sigma). To release noncovalently bound proteins, cells were heated to 70°C for 10 min prior to lysostaphin digestion. For supernatant fractions, bacteria were removed from an overnight culture by centrifugation, and the supernatant was passed through a 0.2-μm filter. Where necessary, protein was concentrated by trichloracetic acid precipitation.
Proteins were separated on 7.5% (wt/vol) polyacrylamide gels, transferred onto polyvinylidene difluoride membranes (Roche), and blocked in 10% (wt/vol) skimmed milk proteins. Blots were probed with polyclonal anti-SasG A domain (1:20,000), anti-SasG B domain (1:3,000), and anti-ClfA A domain (1:5,000) antibodies. Bound antibodies were detected using horseradish peroxidase (HRP)-conjugated protein A (1:500; Sigma). Reactive bands were visualized using the LumiGLO reagent and peroxide detection system (Cell Signaling Technology).
Whole-cell immunoblotting.
SH1000(pALC2073) or SH1000(pALC2073::sasG+ A−B+) cells were grown statically overnight and diluted 1:200 in BHI supplemented with 1% (wt/vol) glucose, rB2.5-His (5 μM), and ZnCl2 (5 mM). Bacteria in suspension were removed, washed twice with PBS, and resuspended to an OD600 of 2 in PBS. Doubling dilutions (5 μl) were spotted on a nitrocellulose membrane (Protran). The membrane was blocked in 10% (wt/vol) skimmed milk proteins and probed with anti-His6 monoclonal antibody 7E8 (49) followed by goat anti-mouse peroxidase-conjugated F(ab)2 fragments (Abcam).
To estimate the concentration of SasG fragments in culture supernatants, bacteria were removed from an overnight culture by centrifugation and the supernatant was passed through a 0.2-μm filter. Doubling dilutions of supernatant (5 μl) or rA-His (1 to 20 nM; 5 μl) were spotted on a nitrocellulose membrane (Protran). The membrane was blocked in 10% (wt/vol) skimmed milk proteins and probed with anti-SasG A domain antibodies (1:20,000) followed by HRP-conjugated protein A (1:500; Sigma).
Expression and purification of recombinant proteins.
Plasmid pQE30sasGA+ (38) expresses N-terminal hexahistidine-tagged SasG A domain protein. Plasmid pQE30sasGBrep+ (39) expresses His-tagged SasG B protein (two full and one partial B domain). Plasmid pQE30sasGBrep+ was digested with BamHI and HindIII to release DNA specifying the B domains, and this was cloned into the pGEX-KG vector to give pGEX-KGsasGBrep+, expressing glutathione S-transferase (GST)-tagged B domains (rB2.5-GST). Plasmid pCF41 expresses His-tagged ClfA N2N3 domains (33). E. coli strain XL-1 Blue (Stratagene) was used as the host for selecting recombinant plasmids following cloning, and E. coli strain TOPP 3 (Stratagene) was used for expression of recombinant proteins. The codon-optimized sequence of a single SasG B domain was synthesized (GenScript Corporation) and subcloned into the pSKB2 vector, employing NdeI and BamHI restriction sites, to give pSKB2-B. A single SasG B domain was expressed with an N-terminal hexahistidine tag in E. coli BL21-Gold(DE3) (Stratagene).
Recombinant His-tagged proteins were expressed and purified by nickel affinity chromatography as described previously (33). To obtain untagged single B domain protein, the N-terminal hexahistidine tag was removed using human rhinovirus (HRV) 3C protease (Promega). GST-tagged protein was purified as described previously (33). Highly purified protein was obtaining by passing the protein through an anion exchange column following affinity chromatography. Untagged SasG B domain protein was obtained by treating rB2.5-GST with thrombin to remove the GST tag and passing this through a GS-Trap column (GE Healthcare), followed by a benzamidine column (GE Healthcare) to remove GST and thrombin.
Surface plasmon resonance.
Surface plasmon resonance (SPR) was performed using the BIAcore X100 system (GE Healthcare). Goat anti-GST IgG (30 μg/ml; GE Healthcare) was diluted in 10 mM sodium acetate buffer at pH 5.0 and immobilized on CM5 sensor chips using amine coupling. This was performed using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride, followed by N-hydroxysuccinimide and ethanolamine hydrochloride, as described by the manufacturer. rB-GST (100 μg/ml) in PBS was passed over the anti-GST surface of one flow cell while recombinant GST (100 μg/ml) was passed over the other flow cell to provide a reference surface. rB-His (4 μM) in PBS or in PBS with ZnCl2 (10 mM) was flowed over the surface at a rate of 5 μl/min. All sensorgram data presented were subtracted from the corresponding data from the reference flow cell. The response generated from injection of buffer over the chip was also subtracted from all sensorgrams.
SEC-MALLS.
The size exclusion chromatography-multiangle laser light scattering (SEC-MALLS) experiment was performed using a Superdex 75 HR10/30 column (GE Healthcare) and Shimadzu high-performance liquid chromatography system. Data for a single recombinant B domain were collected in the presence of EDTA (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) and zinc acetate (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM ZnAc2). Samples (100 μl) of protein at a concentration of 100 μM were loaded onto a gel filtration column and eluted with one column volume (24 ml) of appropriate running buffer at a flow rate 0.5 ml/min. The eluting fractions were monitored using a Dawn HELEOS-II 18-angle light-scattering detector (Wyatt Technologies), an SPD20A UV/Vis detector (Shimadzu), and an Optilab rEX refractive index monitor (Wyatt Technologies). Recorded data were analyzed using Astra (Wyatt Technologies).
RESULTS
Role of SasG in the accumulation phase of biofilm formation.
Expression of SasG by laboratory strains such as SH1000 and Newman could not be detected by Western blotting. However, SasG is expressed at high levels by many clinical isolates (7). Expression of SasG by S. aureus SH1000 has been achieved by placing the sasG gene under the control of the Pxyl/tetO tetracycline-inducible promoter of the pALC2073 vector (7). Boles and Horswill (3) previously reported that S. aureus strain SH1000 forms biofilms independently of PIA, and our previous work showed that biofilm formation by SH1000(pALC2073sasG+) did not require PIA. SasG-mediated biofilm formation occurred in a manner that was dependent on the inducer concentration and also on the length of the SasG protein (7). Here, the role(s) of SasG in the primary attachment and the cell accumulation phases of biofilm formation was examined. Expression of SasG did not increase the adherence of bacteria to polystyrene [67% for SH1000(pALC2073) and 64% for SH1000(pALC2073::sasG+)], suggesting that it is not involved in primary attachment. To examine the effect of SasG expression on cell accumulation, bacterial suspensions were allowed to settle for 24 h and the percent aggregration was determined. Strains expressing SasG showed increased aggregation, with 81.9% for SH1000(pALC2073sasG+) compared to 47.7% for SH1000(pALC2073) (P = 0.0234). This suggests that SasG plays a role in the accumulation phase but not the primary attachment phase of biofilm formation.
Identification of the region of SasG responsible for biofilm formation.
Variants of SasG comprising only the B region or the SasG A domain linked to the repeated regions of SdrF (B-repeats and serine-aspartate repeat regions) were expressed in SH1000 to give SH1000(pALC2073sasG+ A−B+) and SH1000(pALC2073sasG+ A+B−), respectively. SdrF, an LPXTG-anchored surface protein of S. epidermidis, has C-terminal repeat regions of a similar length to the B region of SasG, and these were used as spacers between the A domain of SasG and the bacterial cell surface.
Whole-cell immunoblotting with anti-SasG A domain and anti-SasG B repeat antibodies was employed to quantify the level of expression of SasG by SH1000(pALC2073sasG+A−B+) and SH1000(pALC2073sasG+A+B−). Similar expression levels of the SasG A domain were detected from SH1000(pALC2073sasG+ A+B−) and SH1000(pALC2073sasG+) (Fig. 1 B). However, expression of the B domains from SH1000(pALC2073sasG+ A−B+) was approximately 2-fold lower than from SH1000(pALC2073sasG+). To achieve equal levels of expression, tetracycline (90 ng/ml) was added to SH1000(pALC2073sasG+ A−B+) to increase expression from the Pxyl/tetO promoter. The integrity of the constructs was validated by Western immunoblotting analysis of proteins solubilized from the cell wall during stable protoplast formation (cell wall extracts) and detected with anti-SasG A domain and anti-SasG B domain antibodies (data not shown).
Each strain was tested for biofilm formation. SH1000(pALC2073sasG+) and SH1000(pALC2073sasG+ A−B+), strains that express the B region of SasG, formed robust biofilms, whereas SH1000(pALC2073sasG+ A+B−) and SH1000(pALC2073sdrF+) did not (Fig. 1C). This provides evidence that the B repeat region of SasG and not the A domain is responsible for biofilm formation.
Inhibition of biofilm formation by recombinant SasG domains.
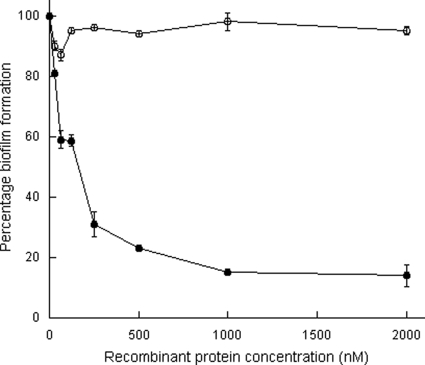
To further investigate a role for the B repeat region in SasG-mediated biofilm formation, increasing concentrations of recombinant hexahistidine-tagged A domain (rA-His) or rB2.5-His protein (comprising two full and one partial B repeat) were added to bacterial cultures at the beginning of the biofilm assay. The rB2.5-His protein inhibited biofilm formation in a dose-dependent manner, whereas rA-His had no effect (Fig. 2). A single B domain (rB-His) also inhibited biofilm formation (data not shown). This provides further evidence that the B repeat region of SasG is responsible for biofilm formation.
FIG. 2.
Inhibition of biofilm formation by recombinant SasG domains. SH1000(pALC2073:sasG+) was incubated with increasing concentrations of rB2.5-His (•) or rA-His (○) at 37°C for 24 h. Biofilm was stained with crystal violet, and the absorbance was measured at 570 nm. Values are expressed as the percentage of wells lacking recombinant protein, and background readings from SH1000 wells were subtracted in each case. Results shown are the mean values of triplicate samples. This experiment was performed three times with similar results.
Several clinical isolates that express SasG (7) can form biofilms. In the case of strains 207 and 3093, biofilms could be inhibited by the addition of rB2.5-His (data not shown). This demonstrated that SasG is likely to promote biofilms in these strains. Strains 125 and 410 formed a biofilm that was not inhibited by rB2.5-His (data not shown). It is possible that SasG does not mediate biofilm formation in these strains and that PIA or another surface protein is responsible. The addition of rB2.5-His had no effect on biofilm formation by the Aap-expressing S. epidermidis isolate CSF41498. This strain reportedly forms protein-dependent biofilms (5, 16). Either a surface protein other than Aap is responsible or the SasG B domains are not sufficiently similar to Aap B domains to bind and cause inhibition.
The effect of zinc chelation on SasG-mediated biofilm formation.
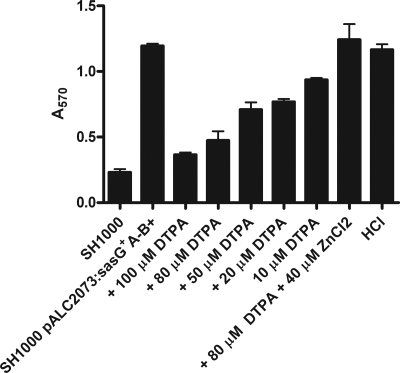
Previous studies demonstrated the necessity of Zn2+ for biofilm formation by staphylococci (6). The zinc chelator DTPA inhibited biofilm formation by S. epidermidis RP62a and community-associated MRSA USA300. Biofilm formation by USA300 isolates is unlikely to be mediated by SasG, as the sasG gene contains a nonsense mutation that precludes expression of the full-length protein (10). To determine if zinc chelation could inhibit SasG-mediated biofilm formation, the effect of DTPA was tested. Biofilm formation by SH1000(pALC2073sasG+ A−B+) was reduced in a manner that was dependent on the concentration of DTPA (Fig. 3). Zinc chelation had no effect on the attachment of SH1000 to polystyrene (data not shown). Biofilm was restored by the addition of ZnCl2. Furthermore, the concentration of Zn2+ present in the BHI broth used for biofilm formation was measured by inductively coupled plasma-mass spectroscopy and found to be 4.8 μM, very close to the concentration in human plasma (10.7 to 18.3 μM) (21). These data illustrate that Zn2+ is required at physiologically relevant concentrations for SasG-mediated biofilm formation.
FIG. 3.
Inhibition of biofilm formation by zinc chelation. SH1000(pALC2073::sasG+ A−B+) was incubated with DTPA, ZnCl2, or HCl (buffer control) for 24 h at 37°C. Biofilm was stained with crystal violet, and the absorbance read at 570 nm.
Recombinant B domain interactions in Zn2+.
The B domains of Aap form dimers in the presence of ZnCl2 (6). The B domains of SasG share 64% amino acid identity with Aap B domains, and SasG-mediated biofilm formation was inhibited by zinc chelation (Fig. 3). Therefore, the ability of SasG B domains to interact in a Zn2+-dependent manner was tested using SPR and SEC-MALLS. A recombinant protein comprising two full and one partial SasG B domain was expressed in E. coli with an N-terminal GST affinity tag (rB2.5-GST). Recombinant B2.5-GST was captured on the surface of a sensor chip that had been coated with a polyclonal anti-GST antibody. A tag-free recombinant protein (rB2.5) was generated by cleaving the GST tag from rB2.5-GST. When rB2.5 was passed over the surface of the rB2.5-GST-coated chip, no interaction could be detected (Fig. 4 A). However, in a solution of ZnCl2, rB2.5 bound to the rB2.5-GST-coated chip (Fig. 4A). Similar behavior was observed for a single recombinant B domain (molecular mass, 14,501.2 g/mol) using SEC-MALLS. Molar masses calculated for the major species eluted from a gel filtration column revealed that in the absence of divalent cations the B domain is monomeric (15,150 ± 1,212 g/mol), while in the presence of ZnAc2 it forms dimers (29,840 ± 2,026 g/mol) (Fig. 4B). Both SPR and SEC-MALLS data demonstrate that SasG B domains are capable of associating and that this interaction is Zn2+ dependent.
FIG. 4.
rB domain interactions in the presence of Zn2+. (A) Surface plasmon resonance analysis of SasG B domain interactions. GST-tagged SasG B domains were captured on the surface of a CM5 sensor chip by using an anti-GST polyclonal antibody. The same concentration (4 μM) of untagged B protein (rB2.5) was passed over the surface either in the presence of 10 mM ZnCl2 (solid line) or without ZnCl2 (dashed line). Injections started at 0 s and ended at 180 s. Results shown are representative of two independent experiments. (B) SEC-MALLS analysis of the oligomeric state of a single B domain in the presence of EDTA (solid line) and ZnAc2 (dashed line). The graph shows an overlay of the calculated molar mass (dotted line) and the 90° light scattering (solid line) as a function of elution volume. The calculated average molecular mass for the single B domain was 15,150 ± 1,212 g/mol in the presence of EDTA and 29,840 ± 2,026 g/mol in the presence of ZnAc2.
Cellular localization of SasG.
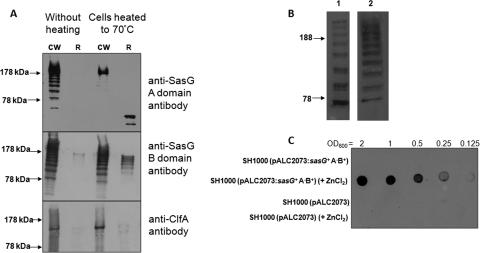
Western immunoblotting of proteins from the cell wall of SH1000(pALC2073sasG+) grown under planktonic culture conditions solubilized by lysostaphin revealed a dominant band of 220 kDa that was presumed to correspond to full-length SasG (predicted molecular mass of 190 kDa) and several smaller bands that were also recognized by anti-SasG A domain antibodies (Fig. 5 A). A similar cleavage pattern was noted for proteins isolated from the cell wall of bacteria that were derived from a biofilm (data not shown). This suggests that SasG undergoes limited cleavage within the B repeat region. The bands differed in molecular mass by 18 to 22 kDa, the approximate size of one B domain, suggesting that there are two cleavage sites in each B domain.
FIG. 5.
Cellular localization of SasG. (A) SH1000(pALC2073sasG+) was either heated to 70°C for 10 min or not heated. Cell wall extracts (CW) and released proteins (R) were probed with anti-SasG A domain, anti-SasG B domain, and anti-ClfA A domain antibodies. Size markers (in kDa) are indicated. (B) Detection of of SasG in culture supernatants. SH1000(pALC2073sasG+) culture supernatants were probed with anti-SasG A domain (row 1) or anti-SasG B domain (row 2) antibodies. Size markers (in kDa) are indicated. (C) Whole-cell immunoblot demonstrating the binding of His-tagged SasG B domain protein to the S. aureus cell surface. SH1000(pALC::2073) or SH1000(pALC2073::sasG+ A−B+) was grown in broth supplemented with ZnCl2 (5 mM) or without added ZnCl2 and rB2.5-His. Serial dilutions of washed bacteria were applied to a nitrocellulose membrane and probed with mouse anti-His6 monoclonal antibody 7E8 followed by goat anti-mouse peroxidase-conjugated F(ab)2 fragments.
The covalent anchoring of SasG to the bacterial cell wall is catalyzed by the sortase A enzyme, which recognizes an LPKTG sorting motif at the C terminus of the protein. If the protein is cleaved within the B region, then only the full-length form of the protein should be linked to the cell wall and be detectable in cell wall extracts when probed with anti-A domain antibodies. All other A domain-containing fragments should be released into the supernatant. However, as shown above, SasG fragments of different sizes were detected in cell wall extracts by anti-SasG A domain antibodies. One explanation is that cleaved SasG fragments have become attached noncovalently to the cell wall.
To investigate this, SH1000(pALC2073sasG+) cells were heated to 70°C in order to promote the release of noncovalently bound proteins. The anti-SasG A domain antibodies recognized proteins of different sizes in the cell wall fraction of control unheated cells (Fig. 5A). In contrast, anti-A domain antibodies only recognized the full-length protein in the cell wall fractions of cells that had been heated to 70°C, indicating that truncated A domain-containing fragments were noncovalently bound and had been released. The fragments appeared to have undergone proteolysis within the SasG A domain upon release from the cell. This may suggest that heating cells to 70°C releases and/or activates a cell surface-associated protease that degrades the SasG A domain.
Anti-SasG B domain antibodies recognized proteins of different sizes in cell wall extracts of heated and nonheated cells (Fig. 5A). This revealed that some of the SasG B domain-containing fragments are covalently associated with the cell wall. Fragments of different sizes were released from cells that had been heated to 70°C, implying that some fragments containing SasG B-repeats are associated noncovalently with the cell wall. These experiments suggest that SasG fragments containing an A domain and various numbers of B domains are associated noncovalently with the cell wall. ClfA served as a control for a typical covalently anchored cell wall protein. No protein was released by heating to 70°C, and no difference was seen between cells that had or had not been heated (Fig. 5A).
SasG fragments of different lengths and showing the characteristic cleavage pattern were also detected in culture supernatants by anti-SasG A domain and anti-SasG B domain antibodies (Fig. 5B). This shows that fragments of SasG are being released from the cell and cleaved during growth of the culture and not during lysostaphin treatment.
Given that recombinant B domains inhibit SasG-mediated biofilm formation, it is possible that released fragments of SasG are inhibitory. The concentration of SasG present in the culture supernatant of SH1000(pALC2073::sasG+) cells was estimated by whole-cell dot immunoblotting with anti-SasG A domain antibodies and compared to that for a known concentration of rA-His. Comparing the intensity of the dots allowed the concentration of SasG in the culture supernatant to be estimated at 10 nM (data not shown). A much higher concentration of rB2.5-His (31.25 nM) was required to inhibit biofilm formation by 20% (Fig. 2). Thus, it is unlikely that released fragments of SasG reach a high enough concentration to inhibit the formation of biofilm.
Binding of SasG B domains to SasG-expressing cells.
Given the ability of SasG rB-repeats to dimerize in the presence of Zn2+, it seemed possible that released SasG fragments were reattaching noncovalently to exposed SasG B-repeats on the cell surface. To test this hypothesis, rB2.5-His was added to BHI broth prior to inoculation with SH1000(pALC2073) or SH1000(pALC2073sasG A−B+). After growth, cells were washed and probed with a monoclonal antibody to the His6 tag in whole-cell immunoblot assays. rB2.5-His did not bind detectably to SH1000(pALC2073). rB2.5-His bound to SH1000(pALC2073sasG A−B+) when ZnCl2 was present in the growth medium (Fig. 5C). This illustrates that SasG B domains can bind to the cell surface-expressed SasG B region. It also strongly suggests that released SasG B domain-containing fragments reassociate with the bacteria surface by attaching to exposed cell wall-anchored B-repeats in a Zn2+-dependent manner. Furthermore, this demonstrates that SasG B domains do not associate with any other cell surface component.
Investigating the role of proteolysis in the cleavage of SasG.
It is possible that SasG is cleaved by a protease during growth. Aap, the SasG homologue from S. epidermidis, must be proteolytically cleaved at a site close to the C terminus of the A domain to allow biofilm formation (40). The cleavage of SasG in S. aureus is different and occurs within the B domains. To try to identify the S. aureus protease responsible, SH1000(pALC2073sasG+) was grown in the presence of various protease inhibitors. A recent study demonstrated that a combination of the protease inhibitors E-64, 1-10-phenylanthroline, and dichloroisocoumarin reduced protease activity of S. aureus strain UAMS-1 (45). However, protease inhibitors used either singly or in combination had no effect on the cleavage of SasG (data not shown). Similarly, commercially available protease inhibitor cocktails did not inhibit the cleavage of SasG. Previously it was reported that the broad-spectrum protease inhibitor α2-macroglobulin prevented SasG-mediated biofilm formation (7). However, cell wall extracts of bacteria grown in α2-macroglobulin had the same SasG cleavage profile as those grown in its absence (data not shown). Interestingly, addition of α2-macroglobulin to a suspension of SH1000(pALC2073sasG+) cells inhibited aggregation (41% compared to 81%), suggesting that its effect is mediated by binding to the cell surface and preventing accumulation rather than inhibiting protease activity.
Expression of SasG by protease-deficient mutants was tested. Strains deficient in each of the known extracellular proteases and in the membrane-bound proteases HtrA1 and HtrA2 were examined after introduction of pALC2073sasG+. Protease-deficient strains showed similar SasG cleavage patterns compared to SH1000(pALC2073sasG+) (data not shown). SH1000 aur spl has very low levels of extracellular protease activity (3), but expression of SasG in this host exhibited the same pattern of cleavage as in the wild type (data not shown). In addition, each of the protease-deficient strains carrying pALC2073sasG+ formed biofilm at levels similar to SH1000(pALC2073sasG+) (data not shown). Strains defective in the autolysins Atl and Aaa were also tested. Once again no difference in the SasG cleavage profile was observed (data not shown). SH1000 atl(pALC2073sasG+) failed to form biofilm, but this was attributed to a reduced level of attachment to the polystyrene plates [67% for SH1000(pALC2073sasG+) and 32% for SH1000 atl(pALC2073sasG+); P = 0.032], in agreement with previous studies demonstrating a role for Atl in the primary attachment phase of biofilm formation (2). Taken together these results indicate that none of the known extracellular proteases is responsible for the cleavage of SasG and that HtrA1/HtrA2 and autolysins Atl and Aaa can also be excluded.
Cleavage during secretion.
SasG must be cleaved within the B region either during or after secretion to account for cleaved fragments being present in the supernatant and on the cell surface. Truncates formed by intracellular cleavage would lack an N-terminal signal sequence and would not pass through the Sec secretion system. The possible role of membrane-bound enzymes was investigated. S. aureus expresses two membrane-bound sortases. Sortase A anchors surface proteins containing LPXTG motifs to cell wall peptidoglycan. Sortase B is only expressed under iron-limiting conditions and is responsible for anchoring a single surface protein, IsdC, via an NPQTN sorting motif. Examination of the amino acid sequence of a SasG B domain revealed the presence of two sequences resembling sortase cleavage motifs (NPETG and NPKTG). It seemed possible that SasG could be cleaved at these sequences to a limited degree by sortase during secretion. To test this hypothesis, bacteria were grown in the presence of the sortase inhibitor E-64, which reduced the amount of cell wall-bound SasG, resulting in elevated levels of SasG being released into the culture supernatant. However, the same pattern of cleavage of SasG occurred, suggesting that sortases do not play a role in the cleavage of the B region of SasG (Fig. 6 A).
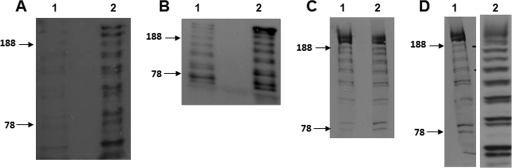
FIG. 6.
Western immunoblotting results demonstrating cleavage of SasG. Cell wall extracts or culture supernatants were separated on 7.5% acrylamide gels and probed with anti-SasG A domain antibodies. Size markers (in kDa) are indicated. (A) Culture supernatants from SH1000(pALC2073:sasG+) grown without (lane 1) or in the presence of sortase inhibitor E-64 (lane 2). (B) Culture supernatants from SH1000(pALC2073:sasG+) (lane 1) and SH1000 srtA(pALC2073::sasG+) (lane 2). (C) Cell wall extracts from SH1000(pALC2073::sasG+) grown without (lane 1) or in the presence of SpsB inhibitor peptide NIFKPST (lane 2). (D) Cell wall extracts from S. epidermidis(pALC2073::sasG+) (lane 1) and L. lactis(pKS80::sasG+) (lane 2).
Next, pALC2073sasG+ was expressed in an srtA mutant. SasG failed to be sorted while cleaved SasG was evident in the culture supernatant, supporting the conclusion that sortase A has no role in the cleavage of SasG B domains (Fig. 6B). An elevated level of SasG protein was detected in the supernatant of the srtA mutant. We conclude that this is due to (i) the failure of SasG to be anchored to the cell wall and (ii) the inability of released SasG fragments to reattach to the cell surface in the absence of surface-bound SasG B domains.
Another possibility was that the SasG protein was being cleaved to a limited degree during secretion by the Sec pathway. The membrane-bound signal peptidase SpsB cleaves AXA consensus motifs in the signal sequence of proteins passing through the Sec secretory system. The spsB gene is essential in S. aureus. SpsB is resistant to all common protease inhibitors. A peptide inhibitor of SpsB (NIFKPST peptide) inhibits SpsB activity at concentrations that cause a small reduction in growth of SH1000 (22). SH1000(pALC2073sasG+) was grown in media containing concentrations of NIFKPST peptide that were subinhibitory to growth. However, no reduction in cleavage of SasG was observed, indicating that the SpsB peptidase is not responsible for the processing of SasG within the B region (Fig. 6C).
Next, SasG was expressed in L. lactis(pKS80::sasG+) and S. epidermidis(pALC2073sasG+) to investigate whether cleavage occurs in other Gram-positive hosts. The same pattern of SasG cleavage was noted, suggesting that the same mechanism of cleavage occurs in these organisms (Fig. 6D). In addition, L. lactis expressing SasG showed increased aggregation [66% for L. lactis(pKS80::sasG+) compared to 38% for L. lactis(pKS80); P = 0.031], illustrating that the expression of SasG is sufficient to promote accumulation during biofilm formation.
The failure to identify a protease responsible for processing SasG may indicate that SasG undergoes spontaneous cleavage. This phenomenon has been reported for several bacterial proteins, and in some cases the molecular mechanism has been elucidated (53). rB2.5-GST was subjected to extensive purification to remove any contaminating proteases, and its stability was assessed after incubation at 37°C for 16 h (Fig. 7). A control, recombinant ClfA A domain protein remained intact after similar treatment. The rB2.5-GST protein with an apparent molecular mass of 78 kDa, was degraded into a number of smaller fragments during the incubation period. This suggests that the SasG B domains undergo cleavage spontaneously in the absence of a protease.
FIG. 7.

Cleavage of SasG B domains. Highly purified recombinant B2.5-GST was incubated at 37°C for 16 h, separated by SDS-PAGE, and stained using Coomassie blue. Size markers (in kDa) are indicated.
DISCUSSION
We previously reported that the S. aureus surface protein SasG promotes biofilm formation independently of PIA (7). This study set out to investigate the molecular basis of SasG-mediated biofilm formation. SasG was found to play a role in the accumulation phase but not the primary attachment phase of biofilm formation. Variants of SasG comprising only the A or B domains were expressed on the surface of S. aureus. The region of SasG responsible for biofilm formation was localized to the B region, with the A domain playing no role. This was confirmed by the ability of recombinant domain B but not A to inhibit biofilm formation.
Previous studies demonstrated the requirement for Zn2+ in biofilm formation by staphylococci (6). Here, Zn2+ chelation inhibited SasG-mediated biofilm formation, and Zn2+ was required at concentrations found in plasma for biofilm formation to occur. Conrady et al. (6) proposed that Zn2+-dependent Aap B domain dimerization on the staphylococcal surface represents the basis of biofilm formation by S. epidermidis RP62a. This study on SasG further supports this hypothesis. SasG B domains, like Aap B domains, form dimers in ZnCl2. Furthermore, recombinant B domain protein bound to the surface of cells expressing SasG B region, demonstrating that B domain oligomerization occurs on the cell surface. This is the first direct evidence that B domain interactions occur on the cell surface. These interactions likely mediate cell accumulation in biofilm formation.
The interaction between recombinant B domain protein and surface-bound SasG B region was only detected when Zn2+ was added to the growth medium. The reason for this is likely to be due to the addition of a high concentration of recombinant B protein reducing the concentration of Zn2+ in the medium. Thus, more Zn2+ must be added to allow biofilm formation.
Western immunoblotting of proteins solubilized from the cell wall by lysostaphin revealed that SasG is cleaved into several fragments. The cleavage is limited and occurs within the B region. SasG fragments could be released from cells by heating, indicating that some are attached to the cell surface in a noncovalent manner. Attachment most likely occurs following release of cleaved SasG into the medium by B domain dimerization on the cell surface. This was demonstrated directly by the binding of recombinant B domain protein to the surface of cells expressing the B domain of SasG. We postulated that a secreted or cell envelope-associated protease was responsible for the limited cleavage of SasG B domains. However, cleavage of SasG could not be prevented by adding protease inhibitors to the growth medium. Furthermore, strains deficient in extracellular proteases and the membrane-bound HtrA proteases showed the same pattern of SasG cleavage. This implies that proteases may not be responsible for cleavage of SasG. This is in contrast to the S. aureus surface protein Bap, which is degraded by the extracellular metalloprotease aureolysin and the serine protease SspA in a sigB mutant and loses the ability to promote biofilm formation (29). Strains defective in the autolysins Atl and Aaa showed no difference in the SasG cleavage profile. The same pattern of SasG cleavage also occurred in L. lactis and S. epidermidis, indicating that the same mechanism of cleavage is responsible. Failure to identify a protease responsible for SasG processing could indicate that more than one protease can mediate cleavage or that the protease is resistant to protease inhibitors. Alternatively, spontaneous cleavage at labile bonds in the SasG B domain could occur. This is supported by the observation that recombinant B domain protein that had been extensively purified was cleaved during incubation in buffer at 37°C. Spontaneous cleavage of peptide bonds promoted by arginine occurs during processing of the EscU protein of E. coli and the YscU protein of Yersinia enterocolitica at a consensus motif, NPTH (53). A similar mechanism may occur here.
We propose a model for SasG-mediated biofilm formation by S. aureus (Fig. 8). Initially, the full-length SasG protein is covalently attached to the cell wall by sorting. Limited cleavage of SasG within the B region occurs during growth. SasG fragments of different lengths are released into the supernatant, and some fragments reattach to exposed B domains on the cell surface in a noncovalent manner that is dependent on Zn2+. The cleaved and exposed SasG B domains on neighboring cells interact with each other in a Zn2+-dependent manner, leading to cell accumulation and biofilm formation.
FIG. 8.
Model for SasG processing and biofilm formation. The smaller circles represent the B domains of SasG, while the large circle is the N-terminal A domain. (A) The full-length SasG protein is attached to the cell wall peptidoglycan by sortase. (B) Cleavage in the B region results in detachment of the N-terminal A region with some B-repeats. (C) The released fragment can reattach to the cell wall-anchored B domain in a Zn2+-dependent manner. (D) Cell-cell interactions occur by exposed B domains dimerizing in a Zn2+-dependent manner during the accumulation phase of biofilm formation.
We believe that SasG must be cleaved either during or after secretion to account for cleaved fragments being present both in the supernatant and on the cell surface. Truncated forms created by cleavage intracellularly would lack an N-terminal signal sequence and would not pass through the Sec secretion system. If a protease is responsible for the processing of SasG, it was not identified using mutants, and the cleavage of SasG could not be inhibited by any protease inhibitor. The inability to inhibit cleavage of SasG prevents us from determining if this event is required for biofilm formation. In the case of Aap, the SasG homologue from S. epidermidis, proteolysis within the A domain has been shown to be a prerequisite for biofilm formation (40). The identity of the S. epidermidis protease is also unknown. We propose that cleavage of SasG is also necessary for biofilm formation to occur. While the cleavage of Aap and SasG occurs at different sites, the effect is the same: the A domain is removed, leaving B domains exposed on the surface.
S. aureus is a major cause of infections associated with indwelling medical devices. It will be of interest to determine if SasG-promoted biofilm formation contributes to virulence in animal models of foreign body infection. The fibronectin binding proteins FnBPA and FnBPB enhance colonization of catheters in mouse models of foreign body infection, while the absence of the ica operon has no effect (48). This suggests that surface proteins can mediate biofilm formation in vivo.
Acknowledgments
This work was funded by a Science Foundation Ireland Programme Investigator grant (to T.J.F.). J.R.P. and D.T.G. thank the British Heart Foundation for financial support. This work was also supported by grants from the Health Research Board (to J.P.O.) and Fondazione CARIPLO “Vaccines 2009-3546” (to P.S.).
We thank the Centre for Microscopy and Analysis (Trinity College Dublin) for carrying out inductively coupled plasma-mass spectroscopy and A. Loughman and M. Tallant (Trinity College Dublin) for assistance with plasmid construction.
Footnotes
Published ahead of print on 3 September 2010.
REFERENCES
- 1.Allgaier, H., G. Jung, R. G. Werner, U. Schneider, and H. Zahner. 1986. Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur. J. Biochem. 160:9-22. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, R., L. Voggu, U. K. Simon, P. Hentschel, G. Thumm, and F. Gotz. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259:260-268. [DOI] [PubMed] [Google Scholar]
- 3.Boles, B. R., and A. R. Horswill. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christner, M., G. C. Franke, N. N. Schommer, U. Wendt, K. Wegert, P. Pehle, G. Kroll, C. Schulze, F. Buck, D. Mack, M. Aepfelbacher, and H. Rohde. 2010. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75:187-207. [DOI] [PubMed] [Google Scholar]
- 5.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrady, D. G., C. C. Brescia, K. Horii, A. A. Weiss, D. J. Hassett, and A. B. Herr. 2008. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U. S. A. 105:19456-19461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrigan, R. M., D. Rigby, P. Handley, and T. J. Foster. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435-2446. [DOI] [PubMed] [Google Scholar]
- 8.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 10.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 11.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, T. J. 1998. Molecular genetic analysis of staphylococcal virulence, p. 433-454. In P. Williams, J. Ketley, and G. Salmond (ed.), Methods in microbiology. Academic Press Ltd., London, England.
- 13.Heilmann, C., J. Hartleib, M. S. Hussain, and G. Peters. 2005. The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73:4793-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 16.Hennig, S., S. Nyunt Wai, and W. Ziebuhr. 2007. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297:117-122. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann, M., P. E. Vaudaux, D. Pittet, R. Auckenthaler, P. D. Lew, F. Schumacher-Perdreau, G. Peters, and F. A. Waldvogel. 1988. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J. Infect. Dis. 158:693-701. [DOI] [PubMed] [Google Scholar]
- 18.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain, M., M. Herrmann, C. von Eiff, F. Perdreau-Remington, and G. Peters. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65:519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, L. A., A. J. Pesce, and S. C. Kazmierczak. 2003. Clinical chemistry: theory, analysis, correlation, 4th ed. Mosby, St. Louis, MO.
- 22.Kavanaugh, J. S., M. Thoendel, and A. R. Horswill. 2007. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 65:780-798. [DOI] [PubMed] [Google Scholar]
- 23.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lofblom, J., N. Kronqvist, M. Uhlen, S. Stahl, and H. Wernerus. 2007. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 102:736-747. [DOI] [PubMed] [Google Scholar]
- 26.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 27.Macintosh, R. L., J. L. Brittan, R. Bhattacharya, H. F. Jenkinson, J. Derrick, M. Upton, and P. S. Handley. 2009. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 191:7007-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marti, M., M. P. Trotonda, M. A. Tormo-Mas, M. Vergara-Irigaray, A. L. Cheung, I. Lasa, and J. R. Penades. 2010. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12:55-64. [DOI] [PubMed] [Google Scholar]
- 30.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276:29969-29978. [DOI] [PubMed] [Google Scholar]
- 31.Merino, N., A. Toledo-Arana, M. Vergara-Irigaray, J. Valle, C. Solano, E. Calvo, J. A. Lopez, T. J. Foster, J. R. Penades, and I. Lasa. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 33.O'Connell, D. P., T. Nanavaty, D. McDevitt, S. Gurusiddappa, M. Hook, and T. J. Foster. 1998. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J. Biol. Chem. 273:6821-6829. [DOI] [PubMed] [Google Scholar]
- 34.O'Gara, J. P., and H. Humphreys. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50:582-587. [DOI] [PubMed] [Google Scholar]
- 35.O'Neill, E., C. Pozzi, P. Houston, H. Humphreys, D. A. Robinson, A. Loughman, T. J. Foster, and J. P. O'Gara. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190:3835-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto, M. 2006. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306:251-258. [DOI] [PubMed] [Google Scholar]
- 37.Reed, S. B., C. A. Wesson, L. E. Liou, W. R. Trumble, P. M. Schlievert, G. A. Bohach, and K. W. Bayles. 2001. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 69:1521-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 39.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759-2767. [DOI] [PubMed] [Google Scholar]
- 40.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883-1895. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder, K., M. Jularic, S. M. Horsburgh, N. Hirschhausen, C. Neumann, A. Bertling, A. Schulte, S. Foster, B. E. Kehrel, G. Peters, and C. Heilmann. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 45.Tsang, L. H., J. E. Cassat, L. N. Shaw, K. E. Beenken, and M. S. Smeltzer. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One 3:e3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaudaux, P., D. Pittet, A. Haeberli, E. Huggler, U. E. Nydegger, D. P. Lew, and F. A. Waldvogel. 1989. Host factors selectively increase staphylococcal adherence on inserted catheters: a role for fibronectin and fibrinogen or fibrin. J. Infect. Dis. 160:865-875. [DOI] [PubMed] [Google Scholar]
- 47.Vaudaux, P. E., P. Francois, R. A. Proctor, D. McDevitt, T. J. Foster, R. M. Albrecht, D. P. Lew, H. Wabers, and S. L. Cooper. 1995. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 63:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vergara-Irigaray, M., J. Valle, N. Merino, C. Latasa, B. Garcia, I. Ruiz de Los Mozos, C. Solano, A. Toledo-Arana, J. R. Penades, and I. Lasa. 2009. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77:3978-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visai, L., Y. Xu, F. Casolini, S. Rindi, M. Hook, and P. Speziale. 2000. Monoclonal antibodies to CNA, a collagen-binding microbial surface component recognizing adhesive matrix molecules, detach Staphylococcus aureus from a collagen substrate. J. Biol. Chem. 275:39837-39845. [DOI] [PubMed] [Google Scholar]
- 50.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279:54881-54886. [DOI] [PubMed] [Google Scholar]
- 51.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 52.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 53.Zarivach, R., W. Deng, M. Vuckovic, H. B. Felise, H. V. Nguyen, S. I. Miller, B. B. Finlay, and N. C. Strynadka. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124-127. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerli, W., A. Trampuz, and P. E. Ochsner. 2004. Prosthetic-joint infections. N. Engl. J. Med. 351:1645-1654. [DOI] [PubMed] [Google Scholar]