Abstract
A number of bacteriophages have been identified that target the Vi capsular antigen of Salmonella enterica serovar Typhi. Here we show that these Vi phages represent a remarkably diverse set of phages belonging to three phage families, including Podoviridae and Myoviridae. Genome analysis facilitated the further classification of these phages and highlighted aspects of their independent evolution. Significantly, a conserved protein domain carrying an acetyl esterase was found to be associated with at least one tail fiber gene for all Vi phages, and the presence of this domain was confirmed in representative phage particles by mass spectrometric analysis. Thus, we provide a simple explanation and paradigm of how a diverse group of phages target a single key virulence antigen associated with this important human-restricted pathogen.
Bacteriophages are dependent for their survival on the presence of susceptible host bacteria in their environment. The first stage of recognition of the bacterial host normally involves binding of a specific phage attachment protein to a receptor molecule on the bacterial surface. Bacteria can evade phage infection by various mechanisms, including accumulating escape mutations in the receptor, acquiring phage inhibitory proteins, or directly modifying the receptor, for example, lipopolysaccharide (LPS) (43). In addition, phage can adapt to recognize different receptors through a number of genetic mechanisms involving evolution of their attachment proteins (20) or by tropism switching (21, 22).
Phage can exploit capsular exopolysaccharides as receptors, some of which are associated with virulence in pathogens (5, 23, 35). A notable example is the Vi capsule found in Salmonella enterica serovar Typhi (S. Typhi) and some isolates of S. Dublin and Citrobacter freundii (29). The Vi capsule of S. Typhi is an important virulence factor, facilitating the bacteria to escape opsonization and other forms of immune surveillance (14, 30) as well as potentially helping the bacteria to evade phage that would otherwise target the O:9 LPS, which the Vi capsule can, at least in part, mask (27). In the middle of the last century, a set of lytic phages were isolated that utilized the Vi capsule as a receptor (6). These Vi phages were exploited in diagnostic laboratories as a “typing set” to distinguish between different strains of S. Typhi isolated from typhoid patients (8). A secondary typing set was generated from Vi typing phage II by adapting this phage to grow on different S. Typhi hosts (6). At this time, typhoid was still common in many parts of Europe and North America, and clinicians tested some of these Vi phages for their potential in phage therapy experiments with human typhoid patients (11). Although this work showed significant promise, phage therapy gradually disappeared from clinical practice in many countries as antibiotics became readily available.
S. Typhi is a monophyletic serovar of the broad enteric species S. enterica (16, 31). Interestingly, S. Typhi is host restricted to humans and has no known zoonotic source. Unlike many other S. enterica serovars, S. Typhi normally causes a systemic infection and does not persist in the intestine efficiently, where high levels of bacteriophage are present. Although it is rare in developed countries, S. Typhi is still a significant cause of mortality in many developing countries (26). Most current clinical isolates are Vi positive when first isolated (2), but it is noteworthy that the Vi capsule biosynthesis and export genes are carried by an operon within a potentially unstable island called Salmonella pathogenicity island 7 (SPI-7) (29).
Although some phenotypic characterization of the Vi phage has been undertaken (1), very little has been performed at the molecular level. We previously showed that Vi typing phage II-E1 is related to the S. Typhimurium phage ES18 (4, 28), with synteny in many capsid and tail proteins. We have now further characterized the other members of this S. Typhi Vi phage collection, designated types I, III, IV, V, VI, and VII (abbreviated from here on as Vi phages I, III, IV, etc.) (6, 11), by utilizing electron microscopy and genomic analysis. This analysis shows that this collection of Vi phages represents a diverse group of bacteriophages that have adapted to growth on S. Typhi through convergent evolution within their tail spike protein genes and the acquisition of conserved acetyl esterase domains.
MATERIALS AND METHODS
Original sources of S. Typhi Vi bacteriophages, bacterial strains, and growth conditions.
Vi phages I, III, IV, V, VI, and VII were obtained from the Health Protection Agency (HPA), Colindale, London, United Kingdom, but the original sources of these phages date from the 1930s through 1955. Vi phages I to IV were isolated from clinical samples of stools obtained from patients with typhoid fever in Toronto, Canada (7-9), while Vi phages V and VI were originally isolated by Desranleau in the state of Quebec, Canada (11), and Vi phage VII originated from Germany in 1955 (3). Vi phage II-E1 was obtained from Stanford University (28).Vi phage I produces very small plaques of 0.02 cm on S. Typhi Ty2 lawns, and Vi phage II-E1 produces 0.15-cm plaques. Vi phage III plaques are 0.25 cm in diameter, while Vi phage IV plaques are more heterogeneous in size and appearance. Vi phages V, VI, and VII all form larger 0.3- to 0.4-cm plaques, with Vi phage VI plaques being consistently the largest and having strikingly clear centers. In contrast, phage V and VII plaques have turbid centers.
The requirement of S. Typhi Vi phages III, IV, V, VI, and VII to target the Vi capsule for infection was confirmed using S. Typhi strain BA256, in which the tviB gene of the S. Typhi viaB operon (29) had been knocked out by insertion of a kanamycin cassette. None of these phages were able to infect under these conditions.
For propagation of all phage, S. Typhi BRD948 was used as the host. This strain not only is heavily capsulated but also is attenuated and can be used in a containment level 2 environment (38). S. Typhi BRD948 contains deletions in two genes of the aromatic pathway (aroC and aroD) and an additional deletion within the gene for the heat shock protein HtrA (38). All S. Typhi isolates were grown in Luria broth or agar plates at 37°C supplemented with aromatic amino acids as described previously (28). Phage lysates were made as described previously and used 0.35% agar in the top layer and 1.2% agar in the base (33).
Preparation of phage stocks and CsCl-purified DNA and restriction enzyme analysis of phage DNA.
Purified phage DNA was obtained via treatment of CsCl-purified phage particles. These were purified using standard procedures described previously (28) and summarized below.
One liter of phage lysate was obtained by infecting S. Typhi BRD948 at multiplicities of infection ranging from 0.1 to 10 with the various S. Typhi Vi phage isolates at 37°C. It typically took 7 h to generate high-titer stocks. After being centrifugally spinned to remove cellular debris, RNase A and DNase were added to eliminate bacterial nucleases. The phage particles were concentrated by addition of NaCl (to a final concentration of 1%) and polyethylene glycol 8000 (PEG8000) (10%, wt/vol). After overnight incubation at 4°C to precipitate the phage particles from the solution, they were spun down at 11,000 × g for 30 min, and the phage pellet was resuspended in 16 ml of lambda diluent. An equal volume of chloroform was added to remove any remaining cell debris and PEG and spun once more for 15 min at 11,000 × g. Finally, the phage pellet was resuspended in lambda diluent and purified by two-stage CsCl buoyant-density ultracentrifugation. DNA was obtained from the purified phage particles by proteinase K and phenol-chloroform treatments.
Vi phage restriction enzyme analysis.
Preliminary analysis and comparison of the various Vi phage DNA preparations were carried out by restriction analysis in order to confirm the quality of the DNA and any degree of relatedness between the phages. Restriction enzymes were purchased from Roche UK and used as recommended by the manufacturer.
DNA sequencing and annotation.
A combination of Sanger sequencing methods and 454 Illumina sequencing technologies was used to sequence all the phages to ensure complete coverage. PCR and primer walking was used to fill any gaps with low coverage. Artemis (32) was used to facilitate annotation of the Vi phage genomes. Pairwise whole-genome comparisons of Vi phage with the related phages T7 (GenBank accession number V01146), K1E (GenBank accession number AM084415), K1F (GenBank accession number AM084414), and SP6 (GenBank accession number AY370673) were performed using tBLASTx and visualized using the Artemis Comparison Tool (ACT) (36). Circular diagrams were made using DNAPlotter (37), and genome comparison figures were produced using easyFig (M. Sullivan, unpublished data).
Comparisons of the maturation-adhesion genes were carried out using the ClustalW package found within MacVector 7.2 DNA analysis software (Invitrogen). Pfam was used to identify significant domains (http://pfam.sanger.ac.uk/search), and to identify conserved structures in selected proteins, we used the tool Phyre (protein homology/analogy recognition engine search) (http://www.sbg.bio.ic.ac.uk/phyre/html/). The latter program combines primary and secondary structure profile information using optimized profile-profile comparison algorithms and can predict domain functions that may be missed by Pfam.
Electron microscopy studies of the Vi phage.
CsCl-purified phage particles for electron microscopy (EM) analysis were dialyzed against three changes of lambda diluent to remove the heavy metal from the sample. The phage particle suspension was mixed with fresh 0.1 M ammonium acetate buffer diluted until just slightly turbid. A total of 5 μl of suspension was applied for 30 s to freshly glow-discharged carbon/Formvar-coated 200-mesh copper grids. Finally, 5 μl of 5% aqueous ammonium molybdate with 1% trehalose was added for a few seconds and then removed with filter paper. The grid was air dried for 30 min and then imaged on a 120-kV Philips Tecnai Spirit BioTwin transmission electron microscopewith a Tietz F415 charge-coupled-device (CCD) TemCam camera at magnifications ranging from ×10,000 to ×60,000.
Mass spectrometry.
Mass spectrometric analysis of the structural proteins was performed similarly as previously described (28), with some modifications. As previously described, CsCl-purified phage particles were used for this study after extensive dialysis against lambda diluent to remove CsCl and other impurities from the preparations. Polypeptide bands were excised, destained completely in 50% methanol-50% ammonium bicarbonate at 50 mM, and digested with trypsin (sequencing grade; Roche) overnight. Peptides were then extracted with 0.5% formic acid-50% methanol, dried, and resuspended in 0.5% formic acid prior to mass spectrometric analysis.
The mass spectrometric analysis was performed with a Finnigan LTQ FT Ultra mass spectrometer (Thermo Electron), controlled by Xcalibur 2.0 SR2, and coupled with an UltiMate 3000 capillary/nano-high-performance liquid chromatography (HPLC) system (Dionex). Samples were desalted on a trap (PepMap C18, 300 μm [inside diameter] by 5 mm; Dionex) and then separated on a bridged ethyl hybrid (BEH) C18 column (75 μm [inside diameter] by 100 mm; Waters) with a gradient of 4 to 32% acetonitrile-0.1% formic acid in 45 or 60 min. LTQ FT Ultra was operated in standard data-dependent acquisition mode, with a Fourier transform ion cyclotron resonance (FT-ICR) resolution of 100,000 at m/z 400 and a survey scan within m/z 400 to 1,500. The four most abundant multiply charged ions were subject to tandem mass spectrometry in the LTQ ion trap at an isolation width of 2 D, a dynamic exclusion width of ±20 ppm, and a duration of 90 s. Peak lists were generated by using BioWorks 3.3 (Thermo Electron). The data were subjected to a database search with Mascot Server 2.2 (Matrix Science) against an in-house-built phage-translated genomic database using the following parameters: trypsin/P with a maximum of 3 missed cleavages sites, peptide mass tolerance at ±20 ppm, tandem mass spectrometry fragment mass tolerance at ±0.5 Da, and variable modifications for acetyl (N-term), carbamidomethyl (C), deamidated (NQ), dioxidation (M), formyl (N-term), Gln→pyro-Glu (N-term Q), Glu→pyro-Glu (N-term E), methyl (E), and oxidation (M). The matched peptides were manually validated.
Nucleotide sequence accession numbers.
The complete genome annotation for Vi phages I and VI can be found in GenBank or EMBL with accession numbers FQ312032 and FR667955, respectively. Full annotation details and comprehensive homology scores for all the Vi phages can be found at the following website: ftp://ftp.sanger.ac.uk/pub/pathogens/Phage/. Visualization of the EMBL FTP files with ARTEMIS is a suggested option (http://www.sanger.ac.uk/resources/software/artemis/).
RESULTS
Morphology of the Vi phage.
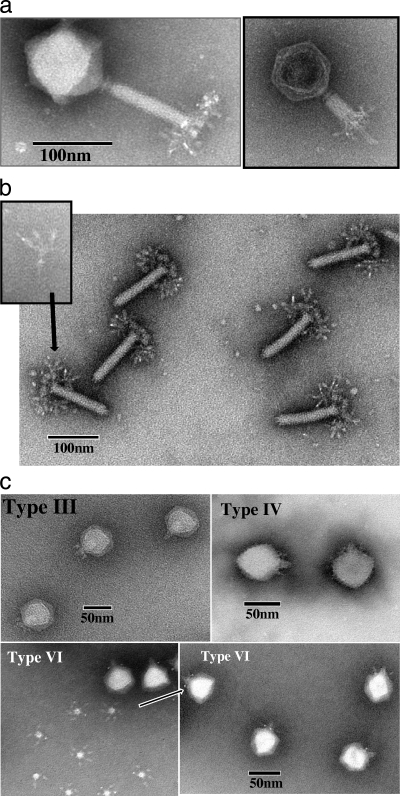
Table 1 provides a general overview of the seven Vi phages that constitute this collection. We have previously shown that Vi phage II is a member of the Siphoviridae, related to S. Typhimurium phage ES18 (28). In order to refine the classification of the remaining different Vi typing phages, we exploited a combined approach involving electron microscopy and DNA sequencing. Initially, each of the different phages was examined under the electron microscope following negative staining of purified phage preparations. This microscopy data allowed the assignment of the phage to Myoviridae morphotype A1 (Vi phage I) and the Podoviridae (Vi phages III, IV, V, VI, and VII). This is in good agreement with a previous study by Ackermann et al. (1) (Fig. 1). Myoviridae Vi phage I shows a very complex overall tail fiber structure reminiscent of a number of recently described phages, and in particular, Salmonella phage Det7 (41). Figure 1a shows that Vi phage I possesses an isometric icosahedron-shaped head, with a dimension of ∼88 nm from apex to tail joint and ∼90-nm width. The tail shows clear evidence of a collar, followed by the major tail structure at ∼115 nm in length and at ∼18 nm in diameter. The smaller inset image shown in Fig. 1a most likely shows the phage after tail contraction. Each corner of the icosahedron-shaped head has a visible small extension (Fig. 1a). An end plate is present, terminating the tail, with a further intricate array of minor tail fibers observable following loss of the capsid (Fig. 1b). These fine tail fibers can be seen as having a four-pronged arrangement, with a further fine fiber attaching this structure to the array surrounding the phage tail base. These unfolded and ramified branched tail fiber structures are very similar to those reported for S. Heidelberg phage 10 (10).
TABLE 1.
General characteristics of S. Typhi phages I to VIIa
| Vi phage | Morphology | Genome size (bp) | G+C ratio (%) | No. of CDSs (no. of introns) |
|---|---|---|---|---|
| I | Myoviridae | 157,061 | 45.37 | 210 (0) |
| II | Siphoviridae | 45,051 | 47.03 | 52 (0) |
| III | Podoviridae | 38,969 | 50.75 | 60 (1) |
| IV | Podoviridae | 44,618 | 47.12 | 62 (0) |
| V | Podoviridae | 38,582 | 48.98 | 52 (1) |
| VI | Podoviridae | 38,367 | 49.47 | 51 (1) |
| VII | Podoviridae | 39,248 | 49.57 | 54 (1) |
This data summarizes the basic information collected for all seven Vi phages, including genome size and number of predicted proteins.
FIG. 1.
(a) Electron micrograph images of S. Typhi Vi phage I. Note the fine tail structure and the presence of a collar and tail end plate. The minor tail fibers surround the bottom of the major phage tail. The contracted tail sheath exposes a long linear structure that extends beyond the collar. (b) Electron micrograph image of the Vi phage I tail. It is possible in these photographs to see the intricate detail of the minor tail fibers—the tail spikes. These surround the major tail fiber in a chandelier-like fashion. They have at least 4 prongs, plus a fifth prong that is attached to the region surrounding the bottom of the major tail fiber (see magnified insert of this structure, which was seen separated from the tail in various electron micrograph images of this phage). These ramified branched structures represent an unfolding of the original closely packed tail fibers shown in panel a. (c) Vi bacteriophages III, IV, and VI. Types V and VII are not shown, but they have morphology similar to that of type IV. All these phages can be characterized as possessing short stumpy tail structures. In a number of these phage images, the tail structure seems to extend into the main body of the capsid in a manner similar to that seen in a number of Podoviridae phages.
The other Vi phages examined, III to VII, are all morphologically members of the Podoviridae (Fig. 1c), which represents an extensive family of bacteriophages, with T7 and SP6 among the best studied examples (12, 36). These Vi phages each have an icosahedral capsid with a diameter of ∼60 nm. All these Vi Podoviridae phages possess short tails and an associated electron-dense structure that extends into the capsid. The tail structure is observed separated from the capsid in Fig. 1c. This central tail-portal structure is frequently very well stained. Based upon the arrangement previously described in structural studies of T7 and K1E phage (20), the arrangement of six minor fine tail fibers seen in the electron micrograph for Vi phage VI (Fig. 1c) is likely to possess the tail spike protein required for receptor recognition.
Comparative sequence analysis of Vi phages I, III, IV, V, VI, and VII.
DNA was prepared from each of the different typing phages, and the DNA sequence was determined for each one, as described in Materials and Methods. Annotation and analysis of each of the Vi phage genomes revealed significant information about the genetic and evolutionary relationships between each of the phages. Analysis of the intron and intein contents of each Vi phage can be found in the supplemental material.
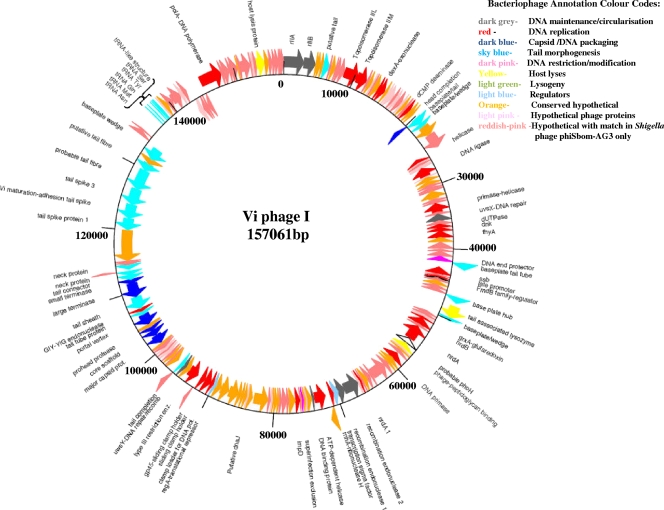
The genome of Vi phage I, shown in Fig. 2, closely resembles that of the large lytic phage of Shigella boydii, phiSboM-AG3 (GenBank accession number NC_013693), displaying remarkable synteny along its entire length. The 157-kbp genome is circular, and it encodes an estimated 209 proteins, of which 170 have homology within this Shigella phage. Seventy-two of these predicted proteins are presently unique to Vi phage I and phiSboM-AG3 only. Twenty-one predicted proteins are hypothetical, with no similarities to any proteins presently found in the databases. A cluster of genes carried by Vi phage I encoding capsid, DNA packaging, and neck proteins (Vi01_152c to Vi01_166c) are also found as a gene cluster with variable homology within a variety of cyanobacterial lytic phages, such as Prochlorococcus phage P-SSM2 and Synechococcus phage S-PM2. A significant number of genes with known prototypes carried by the T4 phage were identified during the annotation (25, 44), and these were added to GenBank (see Materials and Methods). Significantly, the number and type of T4 prototypes found in the Vi phage I genome closely matched those identified in Synechococcus phage S-PM2 (24).
FIG. 2.
Genomic map of Vi phage I. Genes with predicted functions are shown alongside a key to the gene colors used.
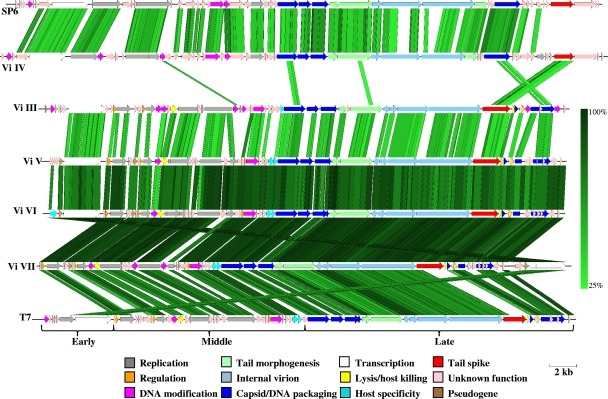
Sequence analysis of Vi phage III shows that it closely resembles enterobacterial phage K1F, while Vi phages V, VI, and VII most closely resemble the T7 phage (Fig. 3). The genomes of Vi phage V, VI, and VII were very similar to each other, as Fig. 3 illustrates. At the DNA level, homology exceeded 95% with respect to Vi phages VI and VII and 90% for Vi phage V with respect to VI and VII. Synteny is well conserved throughout these four genomes, and most gene products have very high similarity scores in excess of 90% at the amino acid level, with the exception of Vi phage III. Vi phage III also has marked similarity to a number of other lytic phages, particularly the enterobacterial phages EcoDS1 and T7.
FIG. 3.
Comparison of the genomes of Vi phage III, IV, V, VI, and VII with those of T7 and SP6. Comparison of the genomes of the S. Typhi Vi phage types III, IV, V, VI, and VII, shown aligned with the genomes of the related phages SP6 (GenBank accession number AY370673) and T7 (GenBank accession number V01146). Regions with significant amino acid similarity between the genomes are linked by shading (percentage identity [tBLASTx] indicated on the right). The phage genes identified are color coded according to their predicted function (see key). The scale bar indicates genome length. Vi phages V, VI, and VII are highly related to phage T7. Vi phage III is more distant from T7 and is in fact more similar to a T7-like phage, K1F (data not shown). Vi phage IV is even more distant from the T7 genus and is most similar to K1E (data not shown) and SP6. In spite of the differences between the genomes of these five Vi phages, their tail spike proteins are highly conserved but unrelated to the tail spike proteins carried by SP6 and T7.
Vi phage IV most clearly resembles the S. Typhimurium lytic phage SP6 (36). The coding sequence (CDS) synteny and identity again are consistently high throughout the genome, with only a small number of genes not found in SP6, K1-5, or K1E (Fig. 3) (20). This includes a putative gene for S-adenosylmethionine hydrolase, which is not present in any of these three phages. At the DNA level, there are particular regions of Vi phage IV that are >80% identical to SP6, while in other regions, there is little or no DNA identity.
Using this DNA sequence data, we were able to classify Vi phages III, V, VI, and VII as belonging to the Autographivirinae subfamily, T7-like genus (19). Vi phage IV has emerged from a different phage lineage from III, V, VI, and VII (13). Vi phage III is significantly more distant from Vi phages V, VI, and VII (Fig. 3).
The tail spike proteins of Vi phages I to VII harbor conserved acetyl esterase domains.
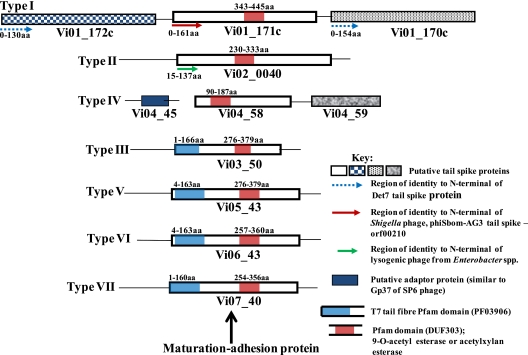
All seven Vi phages encode a maturation-adhesion tail spike protein that recognizes the Vi exopolysaccharide capsule as the receptor. Additionally, Vi phage I harbors two further candidate tail spike proteins, and Vi phage IV harbors one extra tail spike. This is summarized in Fig. 4. A comprehensive analysis of the maturation-adhesion tail spike receptor binding proteins was performed using ClustalW (see Table S2 in the supplemental material) in combination with the Pfam and Phyre programs. The latter two programs search for known protein domains and secondary structures, respectively. Using these approaches, we were able to define clear domains shared within the tail spikes for all Vi phages (Fig. 3). With the exception of Vi phage IV, the Podoviridae Vi phages examined all harbored N-terminal domains with homology to known tail spike proteins that likely carry the region involved in phage tail attachment to the capsid. For example, the first 160 amino acids of the tail spike proteins of Vi phages V, VI, and VII are most similar to those of the corresponding tail spike protein of the T7 phage, which is Gp17, or in phage K1F, Gp36.
FIG. 4.
Tail spike proteins of all the sequenced Vi phage I to VII. Using a combination of BLASTP and Pfam searches, we identified all the likely tail spike proteins of the classic Vi phage set and the likely target receptor. At least one tail spike protein was identified in all of the Vi phages. Significantly, at least one tail spike protein in all had a Pfam domain for an acetyl esterase. A number of phages in the Vi phage set also encoded other tail spikes, but their targets are unknown. aa, amino acids.
Unlike the tail spike protein of the other Vi Podoviridae phage examined, the candidate Vi phage IV tail spike protein does not show any homology at the N terminus to any known phage adaptor gene which is required for tail spike attachment to the capsid. Significantly Vi phage IV and other closely related phages, including SP6, K1E, and K1-5, have been shown to be attached to the phage particle by a different mechanism, a trimeric adaptor protein designated gene product gp37 (20). A candidate gene, Vi04_45, was identified in the Vi phage IV genome, which is over 90% identical to the trimeric adaptor protein gp37 of SP6 phage. As with SP6, a further gene encoding a probable tail spike protein is present in phage IV (12), and this is designated Vi04_59 and highlighted in Fig. 4. This gene has significant similarity to a probable tail fiber gene of Vi phage I, Vi01_0173c, which is adjacent to the three tail spike genes of this phage.
The receptor for all Vi phages has been postulated to include the acetyl groups that decorate the Vi exopolysaccharide capsule, which is a polymer of α-1,4-linked N-acetyl galactosaminuronate. Significantly, a further domain with significant homology to acetyl esterases was identified by BLASTP and Pfam data in all seven maturation-adhesion tail spikes proteins of the Vi phage (Fig. 3 and 4; see also Table S2 in the supplemental material). This conserved domain is likely involved directly in deacetylation of the Vi exopolysaccharide and is annotated as a putative acetyl esterase based upon Pfam identification and Phyre analysis. The acetyl esterase of Vi phages V, VI, and VII can be grouped together in particular due to their very high similarity, as shown in the ClustalW alignment provided in Table S2 and a dot matrix comparison (see Fig. S5 in the supplemental material). A putative adhesion region was identified downstream of the acetyl esterase domain in all seven phages by BLASTP analysis, showing some homology to motifs such as that of yadA (15). This was further confirmed by Phyre analysis (data not shown).
Mass Spectrometric analysis of S. Typhi Vi phages I and VI.
Mass spectrometric analysis was performed on purified preparations of Vi phages I and VI in order to further characterize the phages and their tail spike proteins. Vi phage I contains a number of features that make this a phage of particular special interest. The annotated sequence indicated the presence of three tail spikes, including Vi01_171c associated with recognition of the acetyl modification on the Vi exopolysaccharide.
The intricate tail fiber structure visible in electron microscopy indicated that we might anticipate multiple distinct tail spikes on Vi phage I. The mass spectrometric data, summarized in Table 2, identified 41 proteins, of which 18 are likely to be involved with tail morphogenesis. Other proteins identified included a number of those associated with the capsid. Significantly, the three candidate tail spike proteins designated Vi01_170c, Vi01_171c, and Vi01_172c were also present in this data set. Vi01_171c is the tail spike protein that recognizes the acetyl modification on the Vi capsule. Eighteen genes previously identified in annotation as putative or hypothetical phage proteins were positively identified in the mass spectrometric analysis, and these have therefore been renamed as phage-associated proteins in Table 2. Eight of these are unique to Vi phage I and phiSboM-AG3.
TABLE 2.
Mass spectrometry analysis of Vi phage I
| Gene designationa | Description | Mol wt | No. of peptide sequences identified per protein |
|---|---|---|---|
| Vi01_006 | Putative tail protein product 5503:6312 | 27,801 | 13 |
| Vi01_034 | T4-like baseplate tail tube cap 20271:21239 | 36,118 | 8 |
| Vi01_035 | Gp53 baseplate wedge subunit 21252:21806 | 21,534 | 8 |
| Vi01_036 | Conserved phage associated protein 21806:23194 | 52,590 | 16 |
| Vi01_037 | Phage-associated protein 23205:25145 | 70,019 | 22 |
| Vi01_065c | Gp2 DNA end protector protein 41170:41874 rev | 28,329 | 11 |
| Vi01_066 | Baseplate tail tube initiator 41929:42873 | 35,020 | 5 |
| Vi01_076 | Baseplate hub subunit 48036:49661 | 59,079 | 7 |
| Vi01_077 | Gp25 baseplate wedge protein 49726:50106 | 14,119 | 7 |
| Vi01_081c | Glutaredoxin 51249:51476 rev | 8,472 | 1 |
| Vi01_092c | Conserved phage-associated protein 60113:60466 rev | 13,026 | 3 |
| Vi01_093c | Phage-associated protein 60527:61099 rev | 20,383 | 7 |
| Vi01_094c | Phage-associated protein 61147:63546 rev | 87,994 | 108 |
| Vi01_096c | Phage-associated protein 64004:64642 rev | 22,864 | 4 |
| Vi01_105 | Conserved phage-associated protein 71048:71815 | 28,321 | 15 |
| Vi01_112c | Phage-associated protein 75633:75920 rev | 10,868 | 1 |
| Vi01_117c | Phage-associated protein 78753:79199 rev | 17,304 | 15 |
| Vi01_140c | Tail completion and sheath stabilizer protein 95723:96217 rev | 18,533 | 2 |
| Vi01_142c | Conserved phage-associated protein 96906:97631 rev | 25,368 | 3 |
| Vi01_149c | Phage-associated protein 99867:100310 rev | 16,901 | 2 |
| Vi01_152c | Major capsid protein 100868:102190 rev | 47,971 | 36 |
| Vi01_154c | Gp21 prohead protease 103168:103836 rev | 24,454 | 10 |
| Vi01_155c | Conserved phage-associated protein 103847:104161 rev | 12,349 | 5 |
| Vi01_157c | Gp20 portal vertex protein of the head 104382:106064 rev | 63,067 | 48 |
| Vi01_158c | Gp19 tail tube protein 106135:106665 rev | 19,900 | 10 |
| Vi01_160c | Gp18 tail sheath protein 107211:109106 rev | 68,380 | 32 |
| Vi01_163c | Gp15 proximal tail sheath stabilization 112033:112731 rev | 26,876 | 2 |
| Vi01_164c | Gp14 neck protein 112734:113375 rev | 24,743 | 6 |
| Vi01_166c | Gp13 neck protein 113678:114430 rev | 29,000 | 11 |
| Vi01_169c | Conserved phage-associated protein 115043:119881 rev | 177,452 | 87 |
| Vi01_170c | Tail spike protein 119976:121766 rev | 64,411 | 25 |
| Vi01_171c | Maturation-adhesion tail spike protein 121816:124014 rev | 79,294 | 31 |
| Vi01_172c | Tail spike protein 124070:126610 rev | 91,415 | 26 |
| Vi01_173c | Hemolysin-type calcium-binding protein 126661:129834 rev | 111,484 | 41 |
| Vi01_174c | Putative tail fiber protein 129881:131071 rev | 42,525 | 35 |
| Vi01_175c | Conserved phage-associated protein 131074:131928 rev | 33,009 | 7 |
| Vi01_176c | Gp6 baseplate wedge subunit 131912:133693 rev | 65,582 | 35 |
| Vi01_179 | Phage-associated protein (Vi phage I only) 138943:139176 | 8,054 | 3 |
| Vi01_180 | Conserved phage-associated protein 139186:139701 | 18,551 | 4 |
| Vi01_183 | Phage-associated protein (Vi phage I only) 141852:142301 | 16,820 | 7 |
| Vi01_184 | Phage-associated protein 142337:142792 | 17,332 | 1 |
A significant number of CDSs were identified during this analysis, including those for many hypothetical proteins and a full range of structural proteins. These included the major capsid, collar, baseplate, and all three predicted tail spike proteins.
The mass spectrometric data for purified Vi phage VI, which belongs to the same genus as the T7 phage, are summarized in Table 3. Constituents of the tail were identified (part of gp17 of T7), as were the head-to-tail joining protein (gp8) and a variety of internal virion proteins equivalent to gp14, gp15, and gp16 of T7. We also confirmed the presence of the predicted capsid assembly protein Vi06_34 and the major capsid protein Vi06_35. The tail spike that recognizes the Vi exopolysaccharide, Vi06_43, was also positively identified in the mass spectrometric analysis.
TABLE 3.
Mass spectrometry analysis of Vi phage VIa
| Gene designation | Description | Equivalent gene in phage T7 | Mol wt | No. of peptide sequences identified per protein |
|---|---|---|---|---|
| Vi06_30 | Conserved hypothetical phage protein 16148:16414 | gp6.7 | 9,450 | 13 |
| Vi06_32 | Host specificity protein B 16819:17092 | gp7.3 | 9,862 | 13 |
| Vi06_33 | Predicted head-to-tail joining protein 17107:18717 | gp8 | 59,029 | 47 |
| Vi06_34 | Predicted capsid assembly protein 18760:19692 | gp9 | 34,075 | 7 |
| Vi06_35 | Predicted major capsid protein 19829:20881 | gp10A | 36,993 | 42 |
| Vi06_37 | Predicted tail tubular protein A 21082:21672 | gp11 | 22,278 | 14 |
| Vi06_38 | Predicted tail tubular protein B 21693:24080 | gp12 | 89,481 | 47 |
| Vi06_40 | Predicted internal virion protein B 24589:25179 | gp14 | 20,778 | 26 |
| Vi06_41 | Predicted internal virion protein C 25186:27429 | gp15 | 84,421 | 93 |
| Vi06_42 | Predicted internal virion protein D 27456:31412 | gp16 | 143,700 | 105 |
| Vi06_43 | Maturation-adhesion tail fiber protein 31494: 33467 | gp17 | 71,792 | 43 |
This phage represents the array of Podoviridae phages that are the predominant type found in this Vi phage collection. A array of CDSs similar to those found with mass spectrometry of T7 phage were identified.
DISCUSSION
In this paper, we have described the morphology and genome structure of the classic S. Typhi Vi phage set. This phage collection dates from the early 1930s for Vi phages I to IV (8), 1948 for Vi phages V and VI (11), and 1955 for Vi phage VII (3). The present study was undertaken to definitively classify the phages and to understand how all these phages had become adapted to recognize the Vi capsular antigen as a receptor. Morphologically, these phages are diverse and belong to three phage families, the Myoviridae (Vi phage I), the Siphoviridae (Vi phage II) (28), and the Podoviridae (Vi phages III to VII). The combined data indicate that phages belonging to distinct genus and related to classical phages such as T7 and SP6 have retained their normal gene complement but have exchanged, via recombination, their tail spikes to target the Vi capsule. The preponderance of the T7 and SP6 genera shows that these phage classes allowed diverse genetic exchanges to occur while retaining fitness. Though Vi phages III, IV, V, VI, and VII are all Podoviridae members, sequencing revealed that Vi phages V, VI, and VII are highly related and form a distinctive clade with extensive synteny to T7 phage, whereas Vi phage III is slightly more distant but has significant homology to another member of the T7-like phage genus, K1F. Vi phage IV displays significant synteny to Salmonella phage SP6. We found that Vi phage I is remarkably similar to the recently database-submitted phage phiSboM-AG3 that infects S. boydii (GenBank accession number NC_013693). Vi phage II has been previously shown to share regions of synteny with the lysogenic phage ES18.
Although the Vi phages are genetically and morphologically diverse, they all share a domain linked to the tail spike that is required for recognition and deacetylation of the Vi exopolysaccharide (17, 18, 37). Unlike the endosialidases of bacteriophage that degrade the backbones of Escherichia coli K1 or K5 capsular polysaccharides (20, 36), the acetyl esterases of the seven Vi phages specifically target the acetyl modification on the sugars themselves (17, 39, 40). It is possible that there are phages with the capability to target and degrade the sugar backbone of the Vi capsule in a way similar to that of the K1 phage, but these have not been identified to date. However, this deacetylation enzyme may itself destabilize the long linear Vi fibers due to loss of hydrogen bond cohesion (42). Thus, targeting of the acetyl modification represents a simple but efficient mechanism that allows these phages to infect S. Typhi. Vi phage I encodes three tail spike proteins identified in both the annotation and the mass spectrometric studies. Two of these proteins share significant homology to the N-terminal regions of the well-characterized tail spike protein of Salmonella phage Det7 (41) and to orf00207 of S. boydii phage phiSboM-AG3. The third tail spike protein encodes acetyl esterase and shares significant homology in the first 161 amino acids with orf00210, one of the tail spike proteins from phage phiSboM-AG3. Vi phage IV contains one additional tail spike, and this is similar in arrangement to other members of this SP6-like genus of the Autographivirinae, which tend to recognize two receptors, e.g., phage K1-5, which can recognize E. coli capsular antigens K1 and K5 (34, 41).
While the mechanism by which phages exchange their receptor-recognition tail spikes via homologous recombination is relatively well documented (20), the conditions within the bacterial host that permit these exchanges remain vague but are beginning to be explored (34). Phages possessing multiple tail spikes, such as those reported here, may offer a distinct advantage in a mixed-phage infection by enhancing these possible recombination exchanges, as they will carry not only the receptor used to infect the host bacteria but also other “surplus” receptor-recognition cassettes as well. Thus, such an arrangement would permit a large member of the Myoviridae, such as Vi phage I, to coinfect a Vi-negative bacterial host along with a Podoviridae member, theoretically yielding progeny for the latter that may now carry the receptor for the Vi capsule.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust.
We also thank Hans Ackermann for invaluable advice and comments on all aspects of Vi phage morphology. We thank Simon Harris at the Sanger Institute for constructing the Vi01 figure from the annotated data.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ackermann, H. W., L. Berthiaume, and S. S. Kasatiya. 1970. Ultrastructure of Vi phages I to VII of Salmonella typhi. Can. J. Microbiol. 16:411-413. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S., K. Holt, E. van de Vosse, P. Roumagnac, S. Whitehead, E. King, P. Ewels, A. Keniry, F. X. Weill, D. Lightfoot, J. T. van Dissel, K. E. Sanderson, J. Farrar, M. Achtman, P. Deloukas, and G. Dougan. 2008. High-throughput genotyping of Salmonella enterica serovar Typhi allowing geographical assignment of haplotypes and pathotypes within an urban district of Jakarta, Indonesia. J. Clin. Microbiol. 46:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandis, H., and P. S. Imamura. 1956. Comparative studies of Vi-bacteriophages. Z. Hyg. Infektionskr. 143:50-61. (In German.) [PubMed] [Google Scholar]
- 4.Casjens, S. R., E. B. Gilcrease, D. A. Winn-Stapley, P. Schicklmaier, H. Schmieger, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett, D., and I. S. Roberts. 2008. Capsular polysaccharides in Escherichia coli. Adv. Appl. Microbiol. 65:1-26. [DOI] [PubMed] [Google Scholar]
- 6.Craigie, J., and C. H. Yen. 1938. The demonstration of types of B. typhosus by means of preparations of type II Vi phage: the stability and epidemiological significance of V form types of B. typhosus. Can. Public Health J. 29:484-496. [Google Scholar]
- 7.Craigie, J., and K. F. Brandon. 1936. Bacteriophage specific for the O-resistant V form of B. typhosus. J. Pathol. Bacteriol. 43:233-248. [Google Scholar]
- 8.Craigie, J., and C. H. Yen. 1938. The demonstration of types of B. typhosus by means of preparations of type II Vi phage. I. Principles and technique. Can. Public Health J. 29:448-463. [Google Scholar]
- 9.Craigie, J., and C. H. Yen. 1937. V bacteriophages for B. typhosus. Trans. R. Soc. Can. 5:79-87. [Google Scholar]
- 10.Demczuk, W., R. Ahmed, and H. W. Ackermann. 2004. Morphology of Salmonella enterica serovar Heidelberg typing phages. Can. J. Microbiol. 50:873-875. [DOI] [PubMed] [Google Scholar]
- 11.Desranleau, J. M. 1949. Progress in the treatment of typhoid fever with Vi bacteriophages. Can. J. Public Health 40:473-478. [PubMed] [Google Scholar]
- 12.Dobbins, A. T., M. George, Jr., D. A. Basham, M. E. Ford, J. M. Houtz, M. L. Pedulla, J. G. Lawrence, G. F. Hatfull, and R. W. Hendrix. 2004. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 186:1933-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, R. J., T. Cheasty, and B. Rowe. 1977. Isolation of bacteriophages specific for the K1 polysaccharide antigen of Escherichia coli. J. Clin. Microbiol. 6:548-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirose, K., T. Ezaki, M. Miyake, T. Li, A. Q. Khan, Y. Kawamura, H. Yokoyama, and T. Takami. 1997. Survival of Vi-capsulated and Vi-deleted Salmonella typhi strains in cultured macrophage expressing different levels of CD14 antigen. FEMS Microbiol. Lett. 147:259-265. [DOI] [PubMed] [Google Scholar]
- 15.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt, K. E., J. Parkhill, C. J. Mazzoni, P. Roumagnac, F. X. Weill, I. Goodhead, R. Rance, S. Baker, D. J. Maskell, J. Wain, C. Dolecek, M. Achtman, and G. Dougan. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 40:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwiatkowski, B. 1969. Location of the VI-phage II enzyme causing deacetylation of VI-polysaccharide. Biul. Inst. Med. Morsk. Gdansk. 20:235-242. [PubMed] [Google Scholar]
- 18.Kwiatkowski, B., H. Beilharz, and S. Stirm. 1975. Disruption of Vi bacteriophage III and localization of its deacetylase activity. J. Gen. Virol. 29:267-280. [DOI] [PubMed] [Google Scholar]
- 19.Lavigne, R., D. Seto, P. Mahadevan, H. W. Ackermann, and A. M. Kropinski. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406-414. [DOI] [PubMed] [Google Scholar]
- 20.Leiman, P. G., A. J. Battisti, V. D. Bowman, K. Stummeyer, M. Muhlenhoff, R. Gerardy-Schahn, D. Scholl, and I. J. Molineux. 2007. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 371:836-849. [DOI] [PubMed] [Google Scholar]
- 21.Liu, M., R. Deora, S. R. Doulatov, M. Gingery, F. A. Eiserling, A. Preston, D. J. Maskell, R. W. Simons, P. A. Cotter, J. Parkhill, and J. F. Miller. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091-2094. [DOI] [PubMed] [Google Scholar]
- 22.Liu, M., M. Gingery, S. R. Doulatov, Y. Liu, A. Hodes, S. Baker, P. Davis, M. Simmonds, C. Churcher, K. Mungall, M. A. Quail, A. Preston, E. T. Harvill, D. J. Maskell, F. A. Eiserling, J. Parkhill, and J. F. Miller. 2004. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J. Bacteriol. 186:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, L., M. Conover, H. Lu, M. R. Parsek, K. Bayles, and D. J. Wozniak. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann, N. H., M. R. Clokie, A. Millard, A. Cook, W. H. Wilson, P. J. Wheatley, A. Letarov, and H. M. Krisch. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parry, C. M., T. T. Hien, G. Dougan, N. J. White, and J. J. Farrar. 2002. Typhoid fever. N. Engl. J. Med. 347:1770-1782. [DOI] [PubMed] [Google Scholar]
- 27.Pickard, D., J. Li, M. Roberts, D. Maskell, D. Hone, M. Levine, G. Dougan, and S. Chatfield. 1994. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickard, D., N. R. Thomson, S. Baker, J. Wain, M. Pardo, D. Goulding, N. Hamlin, J. Choudhary, J. Threfall, and G. Dougan. 2008. Molecular characterization of the Salmonella enterica serovar Typhi Vi-typing bacteriophage E1. J. Bacteriol. 190:2580-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raffatellu, M., D. Chessa, R. P. Wilson, C. Tukel, M. Akcelik, and A. J. Baumler. 2006. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect. Immun. 74:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roumagnac, P., F. X. Weill, C. Dolecek, S. Baker, S. Brisse, N. T. Chinh, T. A. Le, C. J. Acosta, J. Farrar, G. Dougan, and M. Achtman. 2006. Evolutionary history of Salmonella typhi. Science 314:1301-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver, R. P., W. Aaronson, and W. F. Vann. 1988. The K1 capsular polysaccharide of Escherichia coli. Rev. Infect. Dis. 10(Suppl. 2):S282-S286. [DOI] [PubMed] [Google Scholar]
- 36.Stummeyer, K., D. Schwarzer, H. Claus, U. Vogel, R. Gerardy-Schahn, and M. Muhlenhoff. 2006. Evolution of bacteriophages infecting encapsulated bacteria: lessons from Escherichia coli K1-specific phages. Mol. Microbiol. 60:1123-1135. [DOI] [PubMed] [Google Scholar]
- 37.Szczeklik, H., and A. Taylor. 1973. The presence of Vi-polysaccharide deacetylase in three morphologically different Vi-bacteriophages. Biul. Inst. Med. Morsk. Gdansk. 24:165-167. [PubMed] [Google Scholar]
- 38.Tacket, C. O., M. B. Sztein, G. A. Losonsky, S. S. Wasserman, J. P. Nataro, R. Edelman, D. Pickard, G. Dougan, S. N. Chatfield, and M. M. Levine. 1997. Safety of live oral Salmonella typhi vaccine strains with deletions in htrA and aroC aroD and immune response in humans. Infect. Immun. 65:452-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, K. 1965. Enzymatic deacetylation of Vi-polysaccharide by Vi-phage. II. Biochem. Biophys. Res. Commun. 20:752-756. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, K. 1965. A study on the loss of the receptor activity of Vi-polysaccharide during incubation with Vi-phage II. Acta Biochim. Pol. 12:157-166. [PubMed] [Google Scholar]
- 41.Walter, M., C. Fiedler, R. Grassl, M. Biebl, R. Rachel, X. L. Hermo-Parrado, A. L. Llamas-Saiz, R. Seckler, S. Miller, and M. J. van Raaij. 2008. Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J. Virol. 82:2265-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West-Nielson, M., P. M. Dominiak, K. Wozniak, and P. E. Hansen. 2006. Strong intramolecular hydrogen bonding involving nitro- and acetyl-groups. J. Mol. Struct. 789:81-91. [Google Scholar]
- 43.Wollin, R., B. A. Stocker, and A. A. Lindberg. 1987. Lysogenic conversion of Salmonella typhimurium bacteriophages A3 and A4 consists of O-acetylation of rhamnose of the repeating unit of the O-antigenic polysaccharide chain. J. Bacteriol. 169:1003-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood, W. B., and H. R. Revel. 1976. The genome of bacteriophage T4. Bacteriol. Rev. 40:847-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.