Abstract
Hydrolytic deamination of cytosine to uracil in cellular DNA is a major source of C-to-T transition mutations if uracil is not repaired by the DNA base excision repair (BER) pathway. Since deamination increases rapidly with temperature, hyperthermophiles, in particular, are expected to succumb to such damage. There has been only one report of crenarchaeotic BER showing strong similarities to that in most eukaryotes and bacteria for hyperthermophilic Archaea. Here we report a different type of BER performed by extract prepared from cells of the euryarchaeon Archaeoglobus fulgidus. Although immunodepletion showed that the monofunctional family 4 type of uracil-DNA glycosylase (UDG) is the principal and probably only UDG in this organism, a β-elimination mechanism rather than a hydrolytic mechanism is employed for incision of the abasic site following uracil removal. The resulting 3′ remnant is removed by efficient 3′-phosphodiesterase activity followed by single-nucleotide insertion and ligation. The finding that repair product formation is stimulated similarly by ATP and ADP in vitro raises the question of whether ADP is more important in vivo because of its higher heat stability.
After depurination, hydrolytic deamination of cytosine to uracil is the most frequent event that damages DNA (36), and it results in G·C-to-A·T transition mutations if the damage is not repaired. In addition, some dUTP molecules escape hydrolysis by dUTPase, which results in a certain amount of dUMP introduced into DNA opposite adenine during replication (32). Irrespective of the mode of appearance, all cells contain uracil-DNA glycosylase (UDG) (EC 3.2.2.3) enzymes to remove uracil from DNA (17). The resulting abasic or apurinic/apyrimidinic (AP) site can subsequently be removed, and the integrity of the DNA can be restored by the so-called base excision repair (BER) pathway, which consists in its simplest form of the sequential actions of 5′-acting AP endonuclease, 5′-deoxyribose phosphate (dRP) lyase, DNA polymerase, and DNA ligase. The BER pathway can be initiated by one of several DNA glycosylases with different substrate specificities (17, 36, 57), and quantitatively it is the most important repair mechanism for the removal of spontaneously generated base modifications. Genes encoding bacterial and eukaryotic UDGs exhibiting significant selectivity for uracil have been cloned and sequenced in the last 2 decades, and the results have demonstrated that there is a high degree of conservation between distantly related species. Family 1 UDGs (for a review of UDG families 1 to 3, see reference 44), typified by the Escherichia coli Ung enzyme (37), recognize uracil in an extrahelical or flipped-out conformation in double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA). Several family 1 enzymes have been extensively characterized, both structurally and at the cell and organism levels. Family 2 UDGs, which includes E. coli Mug and mammalian thymine-DNA glycosylase, are mismatch specific and recognize guanine on the complementary strand rather than the lesion itself and thus are inactive with ssDNA. Family 3 UDGs, typified by the SMUG1 enzyme of human cells, have similar substrate requirements but exhibit a stronger preference for uracil in ssDNA than family 1 enzymes (17, 27, 57, 67).
UDG activity in hyperthermophilic microorganisms was first reported in 1996 (33). Three years later, Sandigursky and Franklin (47) cloned and overexpressed an open reading frame (ORF) of the hyperthermophilic bacterium Thermotoga maritima that typifies the family 4 UDGs that are able to remove uracil from U·G and U·A base pairs, as well as from ssDNA. By means of homology searches, these workers found ORFs homologous to the T. maritima UDG gene in several prokaryotic genomes, including that of the hyperthermophilic archaeon Archaeoglobus fulgidus, a strict anaerobe that grows optimally at 83°C (60; for a review of DNA repair in hyperthermophilic archaea, see reference 20). Subsequently, they cloned and overexpressed the A. fulgidus ORF in E. coli by producing a His-tagged fusion protein. As expected, the purified A. fulgidus recombinant Afung (rAfung) protein exhibited UDG activity (48). However, whether Afung is the major UDG of A. fulgidus or is just a minor glycosylase with uracil-releasing ability remained to be determined.
As a continuation of previous biochemical and physicochemical studies (31) of non-His-tagged rAfung protein, here we characterized a family 4 UDG in archaeon cell extract. The abundance of Afung in vivo was determined, and evidence indicates that this enzyme is the principal UDG of A. fulgidus. Here we also describe the mechanism of dUMP repair employed by this euryarchaeon, which differs in important ways from the mechanism reported for the crenarchaeon Pyrobaculum aerophilum (50).
MATERIALS AND METHODS
Cultivation of A. fulgidus and preparation of cell extract.
A. fulgidus type strain VC16 (= DSMZ 4303) (60) was grown anaerobically at 83°C in a 1-liter culture under Ar (31). Cells were harvested in the early stationary phase by centrifugation at 7,000 × g and stored at −20°C. The cell extract used in glycosylase assays was prepared by using a procedure described previously (31). This procedure was also used to prepare extracts for other purposes, except that extraction was performed with 70 mM MOPS [3-(N-morpholino)propanesulfonic acid] (pH 7.5), 1 mM EDTA, 1 mM dithiothreitol (DTT), 30% (vol/vol) glycerol and cells were frozen in a 70% ethanol bath and stored at −70°C. For extraction, 5 ml buffer per g (wet weight) cell material was used, and cell debris was removed by high-speed centrifugation with a microcentrifuge.
Cloning and expression of the afung gene and purification of the rAfung and His-tagged rAfung proteins.
Amplification and overexpression of the afung gene and subsequent purification of the rAfung protein were performed as described previously (31). Prior to expression employing the vector pBAD/HisA (Invitrogen), the gene was amplified from genomic A. fulgidus DNA by PCR using oligonucleotide primers 5′-GCGCTCGAGATGGAGTCTCTGGACGACATA-3′ (forward primer containing an XhoI site [underlined]) and 5′-GCGGGGTACCTTATAGGTAATCAAAGAGCGTGGG-3′ (reverse primer containing a KpnI site [underlined]). The PCR was performed using a mixture (total volume, 50 μl) containing 1.5 U Platinum Taq high-fidelity DNA polymerase (Invitrogen), 100 ng A. fulgidus VC16 DNA as the template, each deoxynucleoside triphosphate (dNTP) at a concentration of 0.32 mM, 2 mM MgSO4, each primer at a concentration of 0.4 μM, and 1× high-fidelity PCR buffer (Invitrogen). The reaction mixture was incubated at 94°C for 2 min for 1 cycle, at 94°C for 30 s, at 53°C for 30 s, and at 68°C for 1 min for 30 cycles, and at 68°C for 10 min for 1 cycle. The PCR product and expression vector were cut with KpnI (Promega) in buffer J (10 mM Tris-HCl [pH 7.5], 7 mM MgCl2, 50 mM KCl, 1 mM DTT), purified by agarose (0.8%) gel electrophoresis (using a gel extraction kit [Stratagene]), and then cut with XhoI (Promega) in buffer D (6 mM Tris-HCl [pH 7.9], 6 mM MgCl2, 150 mM NaCl, 1 mM DTT). After another round of purification using an agarose gel and a gel extraction kit, ligation was performed as described previously (46). The correct nucleotide sequence of the insert was confirmed by DNA sequencing (DNA Sequencing Facility at the High Technology Centre in Bergen, Norway). The expression vector plus the insert produced a recombinant Afung-His fusion protein in which the N terminus of rAfung was extended with the peptide MGGSHHHHHHGMASMTGGQQMGRDLYDDDDLDRWGSG (rAfungH). The protein was purified with TALON Superflow resin (BD Biosciences, Clontech).
Enzymatic assays for DNA glycosylase activities.
Substrate DNA was incubated with a protein extract or enzyme in 50 μl 70 mM MOPS (pH 7.5), 1 mM EDTA, 1 mM DTT, 100 mM KCl, 5% (vol/vol) glycerol (reaction buffer) at 80°C for 10 min, unless otherwise stated, and other details are described elsewhere (31). One unit of UDG was defined as the amount of enzyme that catalyzed the release of 1 pmol of uracil per min under standard conditions at 80°C. [3H]uracil-containing DNA (with a specific activity of 1,110 dpm/pmol) was a gift from G. Slupphaug and B. Kavli.
Assays for incision of uracil-containing DNA fragments.
Single-stranded oligonucleotides (end protected by phosphorothioate, with 4 bonds at each end, to avoid degradation of the substrate by nucleases present in the cell extract; melting temperature, 65.8°C) with a uracil residue inserted at one position were supplied by Eurogentec S.A., labeled with 32P at the 5′ end using T4 polynucleotide kinase and [γ-32P]ATP (Amersham Biosciences) or at the 3′ end by one-nucleotide extension using [α-32P]dCTP and terminal deoxynucleotidyl transferase (Amersham Biosciences), and purified on 20% polyacrylamide gels containing 7 M urea. Double-stranded oligonucleotide substrates were prepared by annealing each 32P-labeled single-stranded oligomer to a complementary strand with an A or G residue inserted opposite U. Reactions with cell extract were performed as described below and were terminated by addition of 20 mM EDTA, 0.5% (wt/vol) sodium dodecyl sulfate (SDS), proteinase K (190 μg/ml) and incubation at 37°C for 30 min, which was followed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and precipitation with ethanol. Separation of incision products was carried out on 20% polyacrylamide gels containing 7 M urea. Incubation with E. coli Ung plus AP endonuclease (Nfo) or AP lyase (Nth or Fpg) was performed in parallel to define the 3′ end of the incised DNA. E. coli Ung and bacterial alkaline phosphatase were supplied by Invitrogen (catalog no. 18054-015 and 18011-015, respectively); the Nth and Fpg proteins were a gift from Serge Boiteux; and Nfo was supplied by Trevigen (catalog no. 4050-100-EB).
BER assay.
BER assays were performed essentially as described previously (30). A double-stranded oligonucleotide substrate with uracil inserted at a specific position was incubated with cell extract (incubation times and temperatures varied as indicated below) in 45 mM HEPES (pH 7.8), 0.4 mM EDTA, 1 mM DTT, 70 mM KCl, 5 mM MgCl2, 2% (vol/vol) glycerol, 1.4 μg/μl bovine serum albumin (BSA), 40 μM dATP, 40 μM dTTP, 40 μM dGTP, [α-32P]dCTP in an 11-μl (final volume) mixture, and 2 mM ATP and/or 1 mM NAD was added as indicated below. The reactions were terminated and the repair intermediates and products were separated as described above for the incision assays.
Polyclonal antiserum against rAfungH.
Polyclonal antiserum was raised in a rabbit as described previously (13). Purified rAfungH protein (200 mg) was mixed with Freund's complete adjuvant (1:1, vol/vol) for the first injection, which was followed by three injections of rAfungH (200 mg) mixed with Freund's incomplete adjuvant (1:1, vol/vol). The injections were performed every third week. Blood serum aliquots were prepared 3 weeks after the last injection and stored at −20°C. Serum was also withdrawn from the rabbit used for immunization before the first injection of rAfungH (control serum). The specificity of the polyclonal antiserum was confirmed by Western blot analysis.
Western blot analysis.
A. fulgidus cell extract and purified rAfung and rAfungH were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (12%) using Precision protein standards obtained from Bio-Rad (catalog no. 161-0372) as molecular weight markers. The separated material was transferred from the SDS-PAGE gel to nitrocellulose membranes, which was followed by incubation in blocking solution consisting of 3% (wt/vol) gelatin in 20 mM Tris, 500 mM NaCl (pH 7.5) (Tris-buffered saline [TBS]) for 1 h at room temperature and then by immunoblotting with rabbit serum containing antibodies against rAfungH diluted in antibody buffer containing TBS, 0.1% (vol/vol) Tween 20, and 1% (wt/vol) gelatin for 1 to 2 h at room temperature. The protein bands recognized by the antibodies were visualized with an Immun-Blot horseradish peroxidase assay kit [goat anti-rabbit IgG(H+L); catalog no. 170-6463; Bio-Rad].
Immunoprecipitation.
Afung in archaeon cell extract or fAfungH (following enzymatic removal of the His tag using 0.85 U enterokinase) was immunoprecipitated by incubating the extract (240 μg) or rAfungH (0.88 μg) with 0.5 to 17.5 μl of antiserum at 4°C for 1 h with gentle mixing. Protein A-Sepharose 4 Fast Flow beads (Amersham Biosciences) were washed and resuspended in the same packed volume of 70 mM MOPS (pH 7.5), 1 mM EDTA, 1 mM DTT, 5% (vol/vol) glycerol. Washed and resuspended protein A beads (50 μl) were added to the extract-antibody mixture, which was followed by agitation at 4°C for 1 h. Supernatant was collected by centrifugation at 13,000 rpm for 30 s at room temperature.
Homology modeling of Afung.
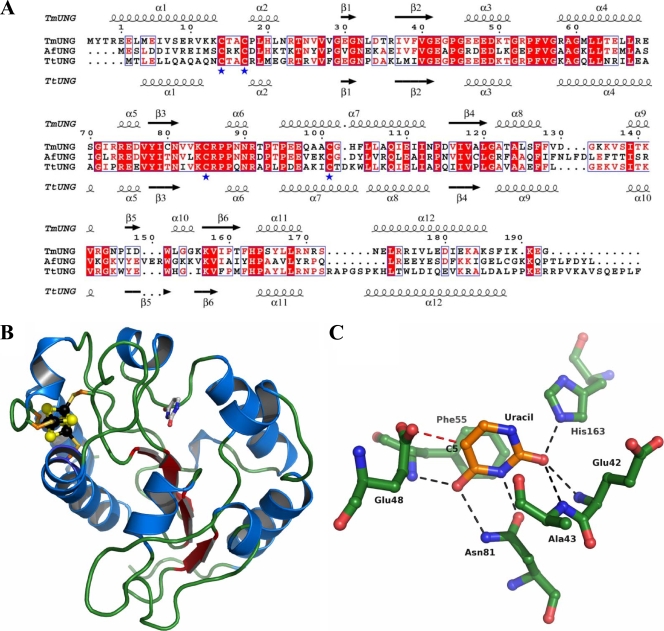
Homology modeling of Afung was based on crystal structures of two other family 4 UDGs, Ttung of Thermus thermophilus (pdb2UI0 and pdb2UI1) (24) and the deposited but unpublished structure of Tmung of T. maritima (pdb1L9G and pdb1VK2), which based on their amino acid sequences exhibit 35.5% and 44.4% identity to Afung, respectively. A sequence alignment was constructed by separate pairwise alignment using ClustalW (68) and was displayed using ESPript (19) (see Fig. 5A). A homology model was constructed by threading the sequence of Afung onto the crystal structure of Ttung using the tools available in SwissPDBViewer (21). Insertions and deletions were constructed and side chain geometries were manually adjusted using “O” (26), with the aim of keeping the overall homology model for Afung as conserved as possible compared to the structures of Ttung and Tmung.
General procedures.
Protein concentrations were determined by the method of Bradford (5) using BSA as the standard.
RESULTS
Immunodepletion of UDG activity present in archaeon cell extract.
After detection of UDG activity in A. fulgidus cell extract (Fig. 1 A), it was of interest to determine whether this activity is a function of one or more gene products. Then, following characterization of the rAfung protein (31, 48), it was crucial to determine whether Afung is indeed the principal DNA glycosylase enzyme that excises uracil from A. fulgidus DNA or instead is a minor UDG enzyme in this organism. Initial comparisons of the abilities of A. fulgidus cell extract and rAfung to release uracil (Fig. 1) indicated that the characteristics of the extract and the protein, including inhibition caused by different agents, were similar (31). To examine this issue, we performed experiments in which cell extract was treated with polyclonal antibodies raised against purified rAfungH protein to perhaps remove all or most of the wild-type Afung (wtAfung) present. The extract was also treated with control serum. When [3H]uracil-labeled DNA was used as the substrate, the results showed that there was ∼60% depletion of the UDG activity in cell extract with the smallest amount of antiserum added. Addition of the largest amount of anti-rAfungH serum (35 times more antiserum) resulted in 98% depletion (2.25% of the UDG activity remained), whereas no loss of UDG activity was detected following treatment with the same amount of control serum (Fig. 2 A). As a positive control, rAfungH with its His tag enzymatically removed (rAfungH′) was treated with anti-rAfungH serum under the same conditions. After addition of the smallest amount of antiserum, a significant increase in UDG activity was observed, and the effect of immunodepletion was more apparent at higher serum concentrations; 3.8% of the UDG activity remained with the highest concentration of antiserum employed (Fig. 2A). We therefore concluded that small amounts of antiserum have a stimulatory effect on rAfung, which was not unexpected because of possible nonspecific stabilizing effects of proteins (31) present in the antiserum. In conclusion, the results demonstrated that Afung is the principal and probably only glycosylase in A. fulgidus that removes uracil incorporated opposite adenine in DNA during replication.
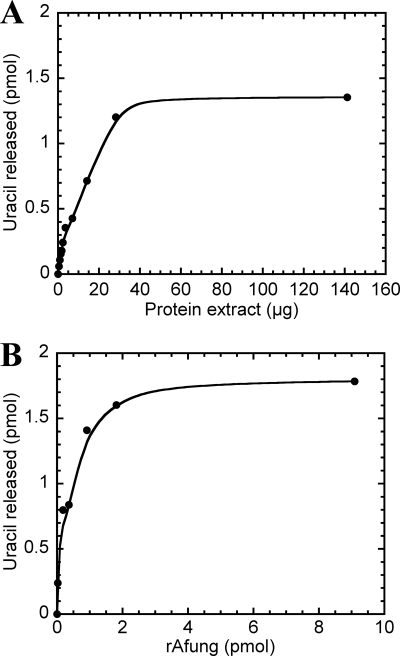
FIG. 1.
Dependence on protein for excision of uracil from DNA by A. fulgidus cell extract (A) and rAfung (B). Different amounts of cell extract (A) or rAfung (B) were incubated with [3H]uracil-containing DNA (2,400 dpm; 2.2 pmol DNA uracil residues) in reaction buffer at 80°C for 10 min. Each value is the average of two independent measurements.
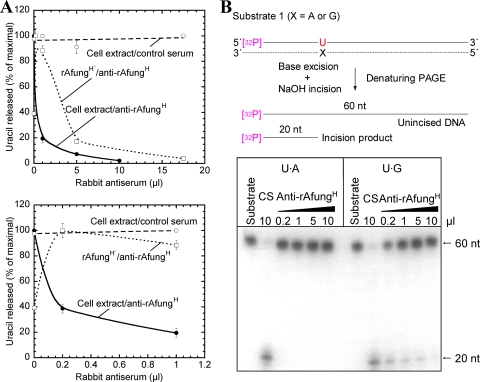
FIG. 2.
Immunodepletion of the uracil-releasing activity present in A. fulgidus cell extract. Protein extract (240 μg) (in panel A also purified enzyme [0.88 μg]) was treated with different volumes of rabbit antiserum plus reaction buffer in a 500-μl (total volume) mixture, which was followed by centrifugation as described in Materials and Methods. (A) Supernatant following the immunodepletion procedure (25 μl) was incubated for 10 min with [3H]uracil-containing DNA (2,000 dpm; 2 pmol DNA uracil residues) in reaction buffer at 80°C. Symbols: •, cell extract treated with anti-rAfungH; ○, cell extract treated with control serum; □, rAfungH′ (rAfungH following enzymatic removal of the His tag) treated with anti-rAfungH. Each value is the mean of three independent measurements. The lower graph is an expansion of the 0-to-1 μl part of the upper graph. (B) Supernatant following immunodepletion (5 μl) was incubated with substrate 1 (32P-labeled 5′-TAGACATTGCCCTCGAGGTAUCATGGATCCGATTTCGACCTCAAACCTAGACGAATTCCG-3′ plus complementary strand; 4 fmol) at 60°C for 10 min in reaction buffer (20 μl). As a result of uracil excision and base-catalyzed phosphodiester bond cleavage (generating a mixture of β- and δ-elimination products; see Fig. 7A), each 32P-end-labeled oligonucleotide (60 nt) was converted into one 32P-labeled 20-nt product and one 40-nt unlabeled product. The strand opposite the damage-containing strand is indicated by a dashed line; the sizes of the 32P-labeled repair intermediates and products are indicated by solid lines. CS, control serum.
Since the [3H]uracil-labeled DNA contained only uracil opposite adenine and removal of premutagenic deaminated cytosines should be the most important UDG function in vivo, Afung-depleted cell extract was also assayed using a defined 5′- 32P-labeled DNA sequence with uracil inserted opposite G, and the same sequence with uracil paired with A was included for comparison (substrate 1). Incised DNA following uracil excision can be observed as a radiolabeled 20-nucleotide (nt) band on a denaturing polyacrylamide gel. The results show that virtually all activity directed toward uracil opposite G, as determined at both 50°C (data not shown) and 60°C (Fig. 2B), disappeared from the cell extract when it was treated with a sufficient amount of anti-rAfungH serum and that there was no observable effect after addition of high concentrations of control serum (Fig. 2B). The fact that more antiserum was needed to deplete “all” activity for excision of U opposite G than to deplete “all” activity for the excision of U opposite A (Fig. 2B) is consistent with our previous results showing that rAfung excises uracil more efficiently when it is opposite G than when it is opposite A (31). Thus, we concluded that most or perhaps all of the UDG activity that initiates BER in A. fulgidus is activity of the Afung protein.
Determination of the molecular weight and cellular abundance of wtAfung by Western blot analysis.
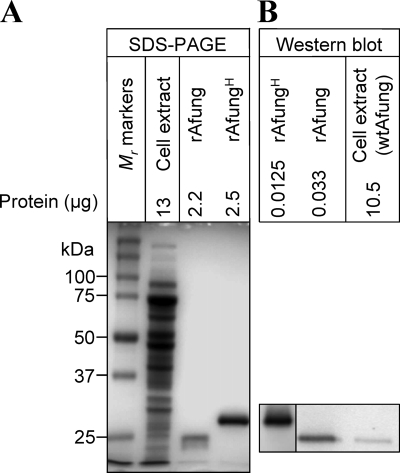
Based on the amino acid sequence, the estimated Mr of Afung is 22,718 (31), which is consistent with the Mr of rAfung (24,600 ± 400) determined by eight measurements (data not shown) obtained by SDS-PAGE (Fig. 3 A). Similarly, the Mr of the rAfungH fusion protein is 26,791 and 28,300 ± 900 as determined from the amino acid sequence and by SDS-PAGE (three measurements) (data not shown), respectively (Fig. 3A). The molecular weight of wtAfung was determined by SDS-PAGE analysis of A. fulgidus cell extract together with rAfung, followed by Western blot analysis using different dilutions of anti-rAfungH serum (Fig. 3). Several experiments demonstrated that wtAfung is virtually the same size (Mr, 24,400 ± 200 based on five measurements) as rAfung (Fig. 3B), indicating that there is no major covalent modification of the translated afung gene transcript in vivo. Taking into account the finding that each A. fulgidus cell contains about 0.033 pg soluble protein (31), the amount of wtAfung per cell was estimated to be ∼500 molecules (Fig. 3B).
FIG. 3.
Western blot analysis of the Afung protein present in A. fulgidus cell extract. Following SDS-PAGE of A. fulgidus cell extract and purified rAfung and rAfungH (the results of one such experiment performed with a 12% [wt/vol] polyacrylamide gel stained with Coomassie blue are shown in panel A), Western blot analysis (B) using antiserum raised against rAfungH (left panel, diluted 1/4,000; right panel, diluted 1/1,000) was performed with purified rAfung and rAfungH and cell extract as described in Materials and Methods.
Kinetics of excision of uracil by wtAfung.
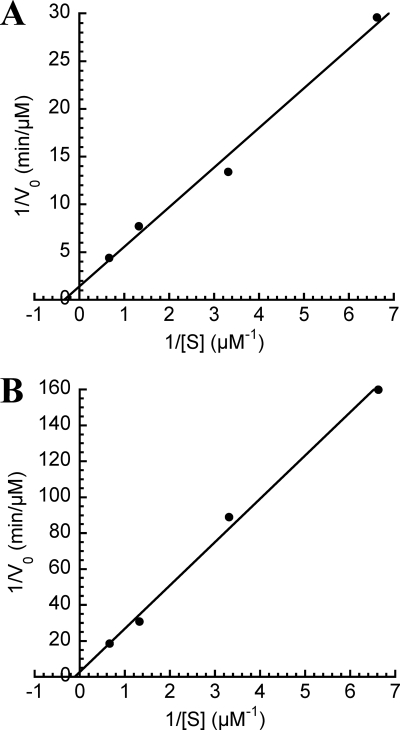
Based on the conclusion that Afung is the principal and probably only UDG in A. fulgidus, cell extract was considered equivalent to wtAfung, and a more detailed comparison with the previously characterized rAfung protein (31) was performed. To specifically examine the efficiency of uracil excision from [3H]uracil-labeled DNA by wtAfung, initial velocities were measured as a function of the substrate concentration. Enzymatic release of uracil was determined over a substrate concentration range of 0.15 to 1.5 μM using 14 μg of protein extract in preparations incubated at 80°C, which is close to the optimal growth temperature of A. fulgidus. In addition, rAfung (6.2 pmol) was reexamined using the same conditions. Analysis of the results by using Lineweaver-Burk plots (Fig. 4), taking into account the cellular abundance of Afung as determined by Western analysis (Fig. 3B), resulted in the kinetic parameters shown in Table 1. The almost 3-fold decrease in the Km and the 100-fold increase in kcat/Km for uracil exhibited by wtAfung compared to rAfung indicate that the enzyme may be covalently modified or is associated with certain cofactors or additional subunits in vivo to facilitate uracil recognition and/or binding. This hypothesis was supported by the turnover numbers (kcat) showing that 1 rAfung molecule excises about 3 uracil residues per min, whereas about 100 uracil residues per min are excised by wtAfung.
FIG. 4.
Lineweaver-Burk plots for excision of uracil from DNA by wtAfung (A) and rAfung (B). Protein extract (14 μg containing 0.33 pmol wtAfung) or rAfung (6.2 pmol) was incubated with different amounts of [3H]uracil-containing DNA (7.55 to 75.5 pmol DNA uracil residues) in reaction buffer at pH 7.5 for 10 min at 80°C. Each value is the average of two to six independent measurements.
TABLE 1.
Kinetic parameters of Afung at 80°C
| Enzyme | Km (μM) | Vmax(μM/min) | kcat (min−1) | kcat/Km (μM−1 min−1) |
|---|---|---|---|---|
| rAfung | 8.7 | 0.36 | 2.9 | 0.33 |
| wtAfunga | 3.0 | 0.72 | 109 | 36 |
Cell extract.
Homology modeling of Afung structure.
As determined by an overall comparison with the sequences of one T. thermophilus UDG (Ttung) and T. maritima UDG (Tmung), Afung is well conserved, and although the levels of sequence identity are relatively low (range, 35 to 45%), there are only a limited number of insertions and deletions (Fig. 5 A). When the homology model was prepared and evaluated (Fig. 5B), it was clear that amino acid residues involved in coordination of the iron-sulfur cluster (Cys18, Cys21, Cys89, and Cys105) are identical and clearly able to play a coordinating role in Afung, similar to previous observations for the crystal structures of Ttung and Tmung. In addition, amino acids that are known to be involved in coordination of an extrahelical (flipped-out) uracil base are ideally positioned to have a similar role in Afung. Therefore, the homology model was also used as a way to understand the substrate specificity of Afung (Fig. 5C) based on observations made in previous biochemical studies of purified rAfung (31).
FIG. 5.
Sequence alignment of Afung, Tmung of T. maritima, and Ttung of T. thermophilus (A), Afung protein structure (B), and structure of the Afung substrate-binding site with a uracil base inserted (C). (A) Secondary structure elements of Tmung (also designated TmUDG [Table 2]) and Ttung (also designated TtUDGa [Table 2]) are indicated above and below the alignment, respectively. α-Helices are indicated by spirals, and β-strands are indicated by arrows. Identical residues in all of the sequences and identical residues in two of the sequences are indicated by white letters and red letters, respectively. Residues indicated by a blue asterisk are the cysteines that coordinate the iron-sulfur cluster present in the family 4 UDGs. The residue numbering is consistent with Afung numbering. The sequences were aligned using ClustalW as described in Materials and Methods. (B) Cartoon showing the overall homology model of Afung. The secondary structure elements are shown; α-helices are indicated by blue spirals, β-strands are indicated by red arrows, and the loop structure is indicated by green tubes. The iron-sulfur cluster was modeled based on the structure of Ttung, and black and yellow spheres represent iron and sulfur, respectively. The coordinating cysteine residues are indicated by sticks. Uracil in the substrate-binding pocket, based on the structure of Ttung, is also indicated by a stick representation. The cartoon was drawn using PyMOL (10). (C) Close-up of the substrate-binding pocket of Afung and the putative interactions with uracil, showing uracil with orange carbon atoms and protein residues with green carbon atoms. Other atoms are also shown (blue, nitrogen; red, oxygen). Hydrogen bonds involving uracil are indicated by black dashed lines. Repulsion between side chain atoms of Glu48 and a substituent in the C-5 position in uracil is indicated by a dashed red line. The cartoon was drawn using PyMOL (10).
Characterization of the incision products following excision of uracil from DNA.
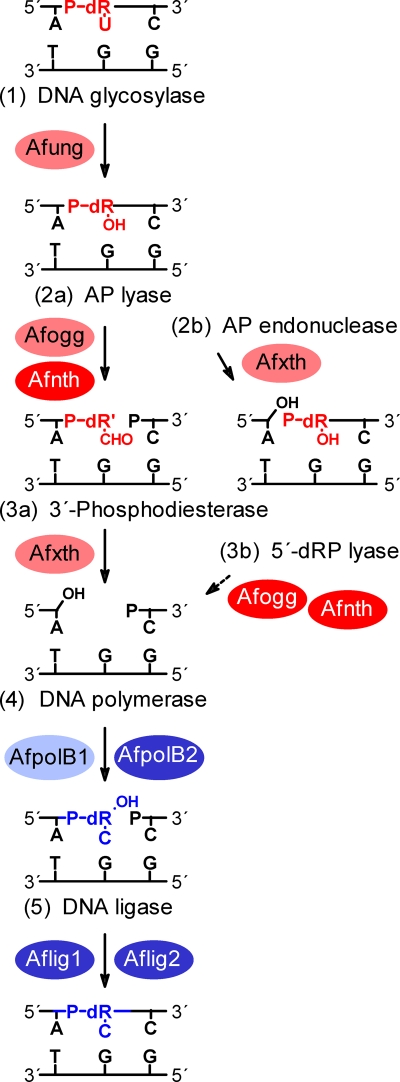
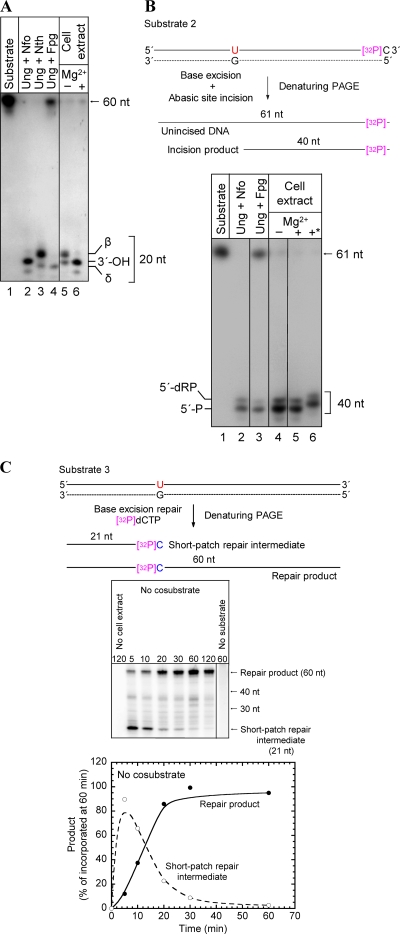
To characterize the 3′ incision products following glycosylase-catalyzed excision of uracil from DNA (Fig. 6, step 1), a defined single-stranded oligonucleotide with uracil inserted at a specific position was labeled with 32P at the 5′ end, annealed to a complementary strand with uracil opposite G, and used as a substrate for A. fulgidus cell extract (substrate 1). Incubation at 60°C for 10 min caused detectable incision of the 32P-labeled strand, resulting in a 3′-α,β-unsaturated aldehyde (β-elimination product), as defined by E. coli Nth (Fig. 7 A, lane 3), as the major product (Fig. 7A, lane 5). No incision was observed when the enzyme was not added (Fig. 7A, lane 1). This result conclusively established that there is a strong AP lyase function in A. fulgidus (Fig. 6, step 2a). When incubation was performed in the presence of MgCl2, only a product with a 3′-OH end was formed (Fig. 7A, lane 6), as defined by the E. coli Nfo protein (Fig. 7A, lane 2). Such 3′-OH products are formed by either an Mg2+-dependent hydrolytic AP endonuclease (45) (Fig. 6, step 2b) or an Mg2+-stimulated 3′-phosphodiesterase (Fig. 6, step 3a). A substantial fraction of a 3′-OH product was also observed when MgCl2 was not added (Fig. 7A, lane 5), while no δ-elimination product as defined by the E. coli Fpg protein (Fig. 7A, lane 4) was detected (Fig. 7A, lanes 5 and 6). A specific AP endonuclease function (Fig. 6, step 2b), as defined by E. coli Nfo (Fig. 7B, lane 2), was observed when a significant fraction of the incised DNA contained a 5′-dRP moiety (Fig. 7B, lane 4), as shown when the uracil-containing strand was labeled with 32P at the 3′ end (substrate 2). However, 5′-phosphates, as defined by E. coli Fpg (Fig. 7B, lane 3) or Nth (data not shown), predominated (Fig. 7B, lane 4). No incision was observed in the control lane, in which no enzyme was added (Fig. 7B, lane 1). The presence of a phosphate group at the 5′ ends was confirmed by alkaline phosphatase treatment (Fig. 7B, lane 6). Importantly, since addition of MgCl2 did not increase the amount of 5′-dRP compared to the 5′-phosphate product (Fig. 7B, lanes 4 and 5), the A. fulgidus AP endonuclease function is not significantly stimulated by Mg2+, demonstrating that the previously observed increase in the amount of the 3′-OH product caused by the presence of Mg2+ (Fig. 7A, lanes 5 and 6) was due solely to 3′-phosphodiesterase-catalyzed removal of the 3′-α,β-unsaturated aldehyde (Fig. 6, step 3a). Thus, excision of the 3′ remnant seems to be much more efficient than excision of the 5′-dRP residue (at least in the presence of MgCl2), since a significant fraction of the latter was still present following the 10-min incubation period despite the fact that much less substrate was employed (Fig. 7B, lanes 4 and 5). It should be recognized that dRP moieties are extensively detached from 5′ ends by the alkaline conditions during electrophoresis, leaving behind 5′-phosphate groups (Fig. 7B, lane 2). The amount of 5′-dRP ends observed should therefore be considered an underestimate. In conclusion, our results demonstrate that incision of the AP site following uracil removal by A. fulgidus cell extract can be carried out both by a preferred AP lyase-catalyzed β-elimination reaction (Fig. 6, step 2a) and by a less favored AP endonuclease-catalyzed hydrolytic mechanism (Fig. 6, step 2b), which is not stimulated by addition of Mg2+. The 3′ remnant is removed by efficient 3′-phosphodiesterase activity stimulated by Mg2+ (Fig. 6, step 3a), while it is still not clear that there is a 5′-dRP lyase function (Fig. 6, step 3b).
FIG. 6.
Diagram of the steps of the BER pathway of A. fulgidus for repair of uracil in DNA. The residues removed and the results of replacement are indicated by red and blue, respectively; proteins with verified biochemical activity (e.g., Afung and Afogg; however, whether Afxth contains 3′-phosphodiesterase activity and whether AfpolB1 is able to fill in one-nucleotide gaps have not been verified [6, 52]) are indicated by black type; putative proteins based on genomic sequences (ORFs) are indicated by white type. The genes encoding the proteins are as follows (Swiss-Prot/TrEMBL accession numbers are indicated in parentheses): Afogg, AF0371 (O29876); Afnth, AF1692 (O28581); Afxth, AF0580 (O29675); AfpolB1, AF0497 (O29753); AfpolB2, AF0693m; Aflig1, AF0623 (O29632); and Aflig2, AF1725 (O28549). dR, deoxyribose residue; dR′, modified deoxyribose residue (i.e., α,β-unsaturated aldehyde); dRP, deoxyribose phosphate; P, phosphate group. A solid arrow indicates a step confirmed experimentally; a dashed arrow indicates a step not demonstrated experimentally yet.
FIG. 7.
Experimental verification of the different steps of the BER pathway of A. fulgidus for repair of uracil in DNA. The 3′ (A) and 5′ (B) incision products and the short-patch repair intermediate and product (C) were determined following excision of uracil from DNA by A. fulgidus cell extract. (A and B) Protein extract (11.4 μg) was incubated with 0.177 pmol substrate 1 (A) (see Fig. 2B) or 2.475 fmol substrate 2 (sequence identical to substrate 1 sequence except for an additional dCMP at the 3′ end as the 32P label; complementary strand identical to that of substrate 1) (B) at 60°C for 10 min in 45 mM HEPES-KOH (pH 7.8), 0.4 mM EDTA, 1 mM DTT, 2% (vol/vol) glycerol, 70 mM KCl with or without 5 mM MgCl2 (total volume, 10 μl) with E. coli enzymes (1 U of Ung; 1 μg of Nfo, Nth, or Fpg) at 37°C for 10 min in buffer without MgCl2. 5′-dRP, 5′-deoxyribose phosphate; 3′-OH, AP endonuclease product; 5′-P, 5′-phosphate; β, β-elimination product (α,β-unsaturated aldehyde); δ, δ-elimination product; *, alkaline phosphatase treatment. (C) Results of a typical experiment showing conversion of repair intermediates to repair product as a function of time. Substrate 3 (0.5 pmol), which is an unlabeled version of substrate 1, was incubated with A. fulgidus protein extract (14 μg) at 60°C in 45 mM HEPES-KOH (pH 7.8), 0.4 mM EDTA, 1 mM DTT, 2% (vol/vol) glycerol, 70 mM KCl, 5 mM MgCl2, 1 μg/μl BSA, 13 μM dATP, 13 μM dTTP, 13 μM dGTP, [α-32P]dCTP in a 15-μl (final volume) mixture. The lower panel shows the relative concentrations of the short-patch repair intermediate and repair product as a function of time based on the average values of two independent measurements.
Complete short-patch BER of uracil in DNA.
Next, we examined whether A. fulgidus is capable of complete BER, using an unlabeled version of the 60-nt DNA substrate employed in previous experiments, in which one uracil residue was inserted opposite guanine at a specific position (substrate 3). Completely repaired DNA could be observed as a radiolabeled 60-nt band, because the dUMP lesion was replaced with [α-32P]dCMP. Since uracil was placed 21 bp downstream from the 5′ end and the final DNA ligase reaction has been reported to be rate limiting (63), the nonligated intermediate was expected to produce a 21-nt band (Fig. 7C, top) if the short-patch mode of BER was operating (Fig. 6, step 4). As anticipated, the presence of 60-nt bands of increasing strength after incubation with cell extract (14 μg) at 60°C for 5 to 60 min demonstrated that A. fulgidus has the enzymes necessary for complete repair of uracil-containing DNA, and the occurrence of 21-nt bands of decreasing strength (Fig. 7C, middle panel) indicated that the short-patch repair mode described previously (30) was present. The concentration of the 21-nt repair intermediate was high after 5 min and decreased as the concentration of the 60-nt repair product increased, after completion of the ligase reaction (Fig. 7C, lower panel). The absence of 22- to 59-nt bands of increasing strength due to incorporation of an increased number of [α-32P]dCMP molecules eliminated the possibility that nonspecific DNA synthesis to the end of the substrate rather than true short-patch repair is the major activity. When cell extract (20 μg protein) was incubated with substrate 3 at 77°C for increasing times (15 to 120 min), addition of 10 U of T4 DNA ligase converted all of the visible 21-nt band into the 60-nt final product, confirming the anticipated ligation of the 21-nt band, as required for conclusion of the BER pathway (data not shown). As a positive control, human cell extract also formed the expected 21-nt repair intermediate, as well as the fully repaired 60-nt product (data not shown). As expected, no band of a relevant size (i.e., a possible derivative of substrate 3) was observed in an analysis of samples after incubation without cell extract or after incubation with cell extract without substrate (Fig. 7C, middle panel). The results obtained with the latter control exclude the possibility that radiolabeled derivatives of, e.g., genomic DNA may falsely be identified as a repair intermediate(s) and/or product.
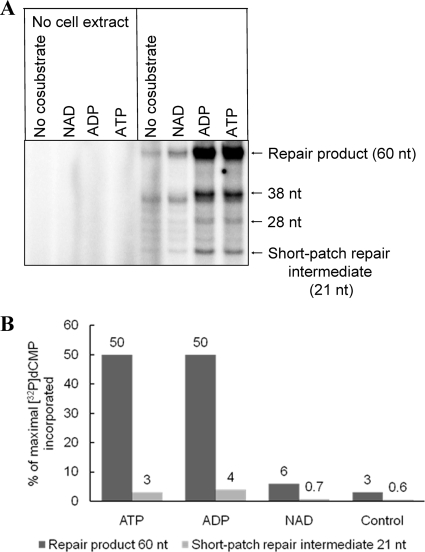
Bacterial DNA ligases use NAD and the mammalian enzymes use ATP for adenylation (32). Although there are exceptions (62, 71), the archaeal ligases are considered ATP dependent and exhibit sequence homology with eukaryotic enzymes (12), and some of them have also been demonstrated to utilize ADP with similar efficiency (25). This is consistent with our results, which showed that addition of NAD increased the level of repaired DNA to a level somewhat greater than the control level, while addition of ATP and ADP resulted in a >1-order-of-magnitude increase in the level of the repair product following incubation for 60 min (Fig. 8). After addition of ATP or ADP, only a low level of the 21-nt BER intermediate accumulated. No clearly visible repair product or intermediate was detected in samples obtained from control incubations without cell extract (Fig. 8).
FIG. 8.
Effect of nucleotides on BER of uracil in DNA by A. fulgidus cell extract. (A) Results of a typical experiment comparing ATP, ADP, and NAD as stimulatory agents for conversion of a repair intermediate to a repair product following 60 min of incubation. (B) Average concentrations of repair intermediate and repair product as determined in three independent experiments, as shown in panel A. Substrate 3 (0.5 pmol) was incubated with A. fulgidus protein extract (17 μg) at 60°C in 45 mM HEPES-KOH (pH 7.8), 0.4 mM EDTA, 1 mM DTT, 70 mM KCl, 5 mM MgCl2, 2% (vol/vol) glycerol, 1 μg/μl BSA, 13 μM dATP, 13 μM dTTP, 13 μM dGTP, [α-32P]dCTP in a 15-μl (final volume) mixture.
Along with the 21-nt short-patch repair intermediate, several fainter bands, mostly in the range from 22 to 40 nt, were detected (Fig. 7C, middle panel, and Fig. 8A), demonstrating that there was replication beyond the lesion site. Whether these bands were true long-patch repair intermediates or instead reflected the existence of certain polymerase activities in A. fulgidus extracts could not be determined from the results of this study.
DISCUSSION
For several archaea, including the hyperthermophilic organism A. fulgidus, there is evidence that the genome is organized into histone-containing nucleosome-like structures (9, 29, 66). Thus, these organisms may serve as prokaryotic model systems for DNA metabolism in eukaryotic cells, including human cells. Furthermore, substantially increased levels of hydrolytic depurination and deamination are expected in DNA in organisms living at elevated temperatures (16, 36, 38), although certain chromosomal proteins confined to (hyper)thermophiles may provide protection against such damage (66). Here, for the first time, we describe the entire BER pathway in a euryarchaeon, the hyperthermophilic organism A. fulgidus, including characterization of the wild-type enzyme initiating repair of uracil in DNA (Fig. 6); this organism has quite a different enzymology than the hyperthermophilic crenarchaeon P. aerophilum, the only archaeon investigated previously (50).
Previously, several lines of evidence supported the notion that Afung is the principal UDG enzyme of A. fulgidus. First, different agents cause similar levels of inhibition of rAfung and UDG activity in cell extract (31). Second, measurements of enzyme activity as a function of temperature have shown that rAfung covers the whole temperature range (20 to 100°C) (31) for UDG activity detected in A. fulgidus cell extracts. (I. Knævelsrud, S. Kazazic, N.-K. Birkeland, and S. Bjelland, unpublished results). Third, the other DNA glycosylase enzymes of A. fulgidus that have been characterized, AfalkA and Afogg (4, 8, 35), as well as the less-characterized homologue (ORF AF1692) of the E. coli Nth enzyme, do not exhibit uracil-releasing activity (31). Here we provide conclusive evidence that Afung is the principal and probably only UDG enzyme of A. fulgidus, both for removal of uracil incorporated opposite adenine during replication and for removal of deaminated cytosine, based on the finding that nearly 100% of the UDG activity present in cell extract was depleted by rabbit serum raised against rAfungH (Fig. 2). Since adenine deamination is a minor reaction compared to cytosine deamination (36), it is somewhat surprising that uracil repair in A. fulgidus seems to rely only on Afung, whereas hypoxanthine repair may rely on at least two proteins, AfalkA (40) and an endonuclease V homologue (39). Although improbable, the possibility that there are very different repair mechanisms for uracil cannot be completely ruled out. For example, no gene encoding a member of any UDG family has been detected in the genome of Methanothermobacter thermautotrophicus, in which a member of the exonuclease III/xth nuclease family (Mth212 protein) is the possible initiator of uracil repair. In addition to conventional exonuclease III-like activities, Mth212 exhibits a specific endonuclease activity that nicks dsDNA at the 5′ side of a 2′-deoxyuridine residue (18).
Previous calculations based on activity measurements showed that an A. fulgidus cell contains ∼1,000 Afung molecules, which is equivalent to ∼9.5 × 10−4 molecules per G·C base pair (31). This number is somewhat higher than that for E. coli, which contains ∼300 Ung molecules per cell (17, 37), resulting in ∼1.3 × 10−4 molecules per G·C base pair. Nevertheless, since the turnover numbers for rAfung, wtAfung, and Ung are ∼3, ∼100 (Table 1), and ∼800 uracil residues released per min, respectively, E. coli should have a significant overcapacity for uracil excision compared to A. fulgidus (31). Our reexamination of the cellular Afung content using SDS-PAGE and Western blot analysis (Fig. 3) showed that there were ∼500 molecules per cell or ∼4.8 × 10−4 molecules per G·C base pair, which largely confirmed the results based on the activity measurements (31). However, the decrease in Km and the increase in kcat/Km for uracil of wtAfung compared to rAfung (Table 1) indicate that the in vivo enzyme is more efficient than previously anticipated (31), suggesting that uracil recognition and binding and/or excision are facilitated by accessory factors.
It is tempting to speculate that the lower capacity for uracil excision exhibited by A. fulgidus evolved to confer resistance to a high steady-state level of uracil in DNA. In E. coli, the capacity for efficient uracil excision results in genome fragmentation with increased DNA uracil levels (34). However, it is important to precisely determine the steady-state concentration of uracil in A. fulgidus DNA, where the additional protection of the nucleoprotein structure provided by the Alba protein coat (66), which is restricted to hyperthermophiles and thermophiles, also may provide extra protection against deamination. Future investigations should also determine the contribution of nonmutagenic dUMP incorporation compared to cytosine deamination (3, 43) in the formation of uracil moieties in A. fulgidus DNA, as well as measure A. fulgidus dUTPase efficiency (32).
We have also suggested a three-dimensional structure of Afung (Fig. 5A and B) based on a previously described crystal structure for a family 4 UDG (24) and the Tmung structure by homology modeling, which indicates the active site amino acid residues involved in coordination of an extrahelical (flipped-out) DNA uracil (Fig. 5C) and hence explains the limited substrate specificity of rAfung (31).
In addition to A. fulgidus, several euryarchaea, as well as the only known parasitic hyperthermophile, Nanoarchaeum equitans (65), contain only one UDG (Table 2). Like A. fulgidus, N. equitans contains a family 4 ORF. At present, the information available indicates that family 1 and 2 UDGs are confined to mesophilic organisms (44), whereas family 4 to 6 UDGs and MIG enzymes are present in (hyper)thermophiles (Table 2). For instance, three UDGs belonging to different families have been characterized in the crenarchaeote P. aerophilum (Table 2), while three ORFs belonging to UDG families 4, 5, and 6 have been identified in Sulfolobus solfataricus (7, 49, 69), and one of the corresponding proteins has been characterized (11).
TABLE 2.
Characterized UDGs of organisms that thrive at high temperaturesa
| Organism | Protein | ORF | Family | Mr | No. of amino acids | UDG activityb | Reference(s) |
|---|---|---|---|---|---|---|---|
| Euryarchaeota | |||||||
| Archaeoglobus fulgidus | Afungc | AF2277 | 4 | 22,718 | 199 | + (ssDNA TC > G > A) | 31 |
| Methanobacterium thermoautotrophicum | MthMIG | pFV1-Orf10 | MIG | 25,386 | 221 | + (G > C) − (ssDNA) | 23 |
| Methanocaldococcus jannaschii | MjUDG | MJ1434 | 6 | 25,938 | 220 | + (TCGA) | 7 |
| Pyrococcus furiosus | PfuUDG | PF1385 | 4 | 22,335 | 196 | + (ssDNA GA) | 28 |
| Crenarchaeota | |||||||
| Pyrobaculum aerophilum | PaUDGac | PAE0651 | 4 | 21,563 | 196 | + (G > A) | 49, 51 |
| PaUDGb | PAE1327 | 5 | 24,914 | 226 | + (G > A > ssDNA) | 49 | |
| PaMIG | PAE3199 | MIG | 26,446 | 230 | + (G) − (ATC) | 70 | |
| Sulfolobus solfataricus | UDG1 | SSO2275 | 4 | 24,367 | 216 | + (G > ssDNA > A) | 11 |
| Bacteria | |||||||
| Thermotoga maritima | TmUDG | TM0511 | 4 | 21,501 | 192 | + (GA) | 47 |
| Thermus thermophilus | TtUDGa | TTHA0718 | 4 | 22,966 | 205 | + (C > G > A, ssDNA) | 59 |
| TtUDGb | TTHA1149 | 5 | 24,286 | 219 | + (G > C > A) − (ssDNA) | 59 |
Families 1, 2, and 3 have not been identified in hyperthermophiles.
The opposite bases are indicated in parentheses.
The protein interacts with PCNA (proliferating cell nuclear antigen; sliding clamp) homologues, as demonstrated for PaUDGa or suggested for Afung because of a putative PCNA-binding motif (69).
All UDGs are monofunctional DNA glycosylases (Fig. 6, step 1), and uracil removal is normally followed by AP endonuclease (Fig. 6, step 2b) and 5′-dRP lyase (Fig. 6, step 3b) activities that prime polymerization (Fig. 6, step 4) and ligation (Fig. 6, step 5), as recently indicated by experiments using BER components and cell extract of the crenarchaeon P. aerophilum (50). This is true for bacterial systems as well as most eukaryotic systems investigated, including mammalian cells (30), which have an inefficient 3′-phosphodiesterase (61) and an efficient 5′-dRP lyase activity, the latter of which is a function of DNA polymerase β (41, 58), which is not present in A. fulgidus (29). Somewhat surprisingly, our results demonstrate that following uracil excision by the Afung protein of A. fulgidus cell extract, the AP site is processed predominantly by a lyase (Fig. 6, step 2a) function (Fig. 7A and B). We found that this process may also occur in Thermoplasma acidophilum, another euryarchaeon investigated by our research group (M. N. Moen, I. Knævelsrud, G. T. Haugland, K. Grøsvik, N.-K. Birkeland, I. Leiros, A. Klungland, and S. Bjelland, unpublished results). The putative AP lyases of the A. fulgidus BER pathway are the Afogg protein with confirmed AP lyase activity (8) and the Afnth protein, which has been characterized structurally but has not been characterized biochemically yet (55) and which contains lysine and aspartate at positions equivalent to Lys120 and Asp138 of E. coli Nth, which are believed to be the catalytic residues in the β-elimination reaction. The AP endonuclease activity is probably a function of the A. fulgidus exonuclease III homologue Afxth (also called Af_Exo), which recently was characterized biochemically and structurally (52). The 3′-phosphodiesterase activity may also be a function of the Afxth protein (Fig. 6, step 3a). Based on the results of our experiments it is difficult to decide whether a 5′-dRP lyase function (Fig. 6, step 3b) is present in A. fulgidus. Indeed, the efficient activities of AP lyase and 3′-phosphodiesterase are sufficient to prime polymerization and ligation in short-patch BER. Long-patch repair could be involved in the processing of AP endonuclease-incised DNA, although our results do not definitely demonstrate that this type of repair occurs in A. fulgidus. Similar to our results, lyase activity was found to predominate over hydrolytic endonuclease activity for AP site incision in Schizosaccharomyces pombe (2). The same may be true for Saccharomyces cerevisiae (22), which has an efficient 3′-phosphodiesterase that has been demonstrated to be a function of the Apn2 protein (64).
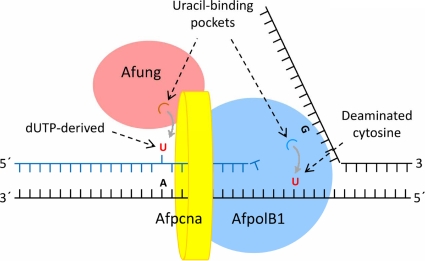
Although, as described above, A. fulgidus lacks a polymerase β homologue, the polymerase catalyzing the gap-filling reaction in short-patch BER (Fig. 6, step 4) should be an enzyme with similar functional characteristics. Interestingly, an ORF (AF0693m) probably coding for a family B polymerase (putative protein designated AfpolB2) that is like polymerase β without proliferating cell nuclear antigen (PCNA)-binding motif, as well as the ExoI, ExoII, and ExoIII motifs (relevant for the 3′→5′ exonuclease function), is present in the A. fulgidus genome (Table 3 ), and this protein appears to be similar to the recently characterized B2 repair polymerase of P. aerophilum (50). The PCNA-binding and proofreading abilities of AfpolB1 indicate that it has a role in DNA replication (6, 53, 54), which is also true for members of the family D polymerases (Table 3) (42, 56). Furthermore, since Afung contains a PCNA-binding motif (69), it probably removes uracil from DNA incorporated opposite adenine during replication, as suggested for mammalian UNG2 (27). Indeed, physical and functional interactions between UDG and PCNA from the euryarchaeon Pyrococcus furiosus were recently demonstrated (28). Like the results for other family B DNA polymerases of archaea, sequence alignments indicated the presence of an N-terminal uracil-binding pocket in AfpolB1 that is able to recognize uracil in the template strand and cause termination of DNA synthesis before the polymerase function inserts a noncognate adenine into DNA (Fig. 9 and Table 3) (6, 15). The stalled DNA polymerase(s) may then dissociate from Afpcna, followed by reassembly of template strands and initiation of BER, in which some sort of rotation of the replication clamp may help Afung being transferred to the lesion site. Both AfpolB1 and Afung may always be attached to Afpcna, serving as uracil sensors. Indeed, it is tempting to speculate that the uracil-binding pocket of archaeon family B polymerases evolved to avoid generation of single-strand breaks in front of an advancing replication fork by protecting uracil from UDG attack and offering time for reassembly of template strands before initiation of BER. Accordingly, an interesting recent study showed that UDG is profoundly inhibited by simultaneous binding of both PCNA and family B polymerase, indicating that when the PCNA-polymerase complex encounters uracil in DNA templates, BER is switched off to protect the complex from a repair pathway that is dangerous in the context of ssDNA formed during replication (14). The working model shown in Fig. 9 is supported by reports that there are larger complexes containing proteins required for both replication and short- and long-patch BER, including human UNG2, APE1, DNA polymerases β and δ, XRCC1, PCNA, and DNA ligase (1).
TABLE 3.
DNA polymerases of A. fulgidus
| Protein | ORF (accession no.) | Family | Mr | No. of amino acids | Exonuclease (3′→5′) | PCNA binding | U-binding pocket | Proposed function(s) |
|---|---|---|---|---|---|---|---|---|
| AfpolB1 | AF0497 (O29753) | B | 89,850 | 781 | +a | +a | +b | Replication and repair |
| AfpolB2 | AF0693m | B | 75,330 | 645 | − | − | − | Repair |
| AfpolD | AF1722 (O28552) | D | 129,368 | 1,143 | +b | +a | − | Replication |
| AF1790 (O28484) | 54,586 | 488 |
FIG. 9.
Model for Afung-initiated repair of misincorporated uracil and detection of deaminated cytosine in DNA during replication. Afung contains a PCNA-binding motif (69), indicating that there is PCNA-dependent postreplicative removal of uracil, where Afpcna (AF0335 [accession no. O29912]) is a homotrimer. Interactions of AfpolB1 with Afpcna have been demonstrated experimentally (42, 53, 54). Detection and accommodation of uracil by AfpolB1 probably interrupt chain elongation, which may promote reassembly of template strands and transfer of Afung to the lesion site initiating BER, as shown in Fig. 6. See text for further details.
The A. fulgidus genome contains two ORFs (29) that exhibit homology to human ligases I, III, and IV (26 to 29% identity at the amino acid level). The putative Aflig1 and Aflig2 proteins (556 and 313 amino acid residues, respectively) both seem to contain the expected active site lysine, as well as the six sequence motifs (motifs I, III, IIIa, and IV to VI) conserved in DNA ligases and RNA capping enzymes (12) and are thus reasonable candidates to carry out the last step (step 5) in the BER pathway (Fig. 6).
In conclusion, the present results suggest an enzymology for AP site excision in (hyper)thermophilic archaea that is different than that in most other systems, including mammalian cells; i.e., the AP site is incised by an efficient AP lyase and further processed by an efficient 3′-phosphodiesterase to form the fully processed gap required for single-nucleotide insertion and ligation (Fig. 6). The finding that repair product formation is stimulated by ATP and ADP similarly in vitro raises the question of whether ADP is more important in vivo because of its higher heat stability. Our goal is to assign certain gene products of A. fulgidus to the different replication and repair functions by performing in vitro reconstitution experiments using purified recombinant proteins and possibly by inhibiting certain reactions carried out by cell extract using antibodies to the corresponding proteins.
Acknowledgments
This work was supported by Research Council of Norway grant 148997/432.
We are indebted to M. Madsen and E. Skårland for technical assistance and to S. Boiteux, B. Kavli, and G. Slupphaug for providing materials.
Footnotes
Published ahead of print on 7 May 2010.
REFERENCES
- 1.Akbari, M., M. Otterlei, J. Peña-Diaz, P. A. Aas, B. Kavli, N. B. Liabakk, L. Hagen, K. Imai, A. Durandy, G. Slupphaug, and H. E. Krokan. 2004. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 32:5486-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alseth, I., H. Korvald, F. Osman, E. Seeberg, and M. Bjørås. 2004. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 32:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, S., T. Heine, R. Sneve, I. König, H. E. Krokan, B. Epe, and H. Nilsen. 2005. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis 26:547-555. [DOI] [PubMed] [Google Scholar]
- 4.Birkeland, N.-K., H. Ånensen, I. Knævelsrud, W. Kristoffersen, M. Bjørås, F. T. Robb, A. Klungland, and S. Bjelland. 2002. Methylpurine DNA glycosylase of the hyperthermophilic archaeon Archaeoglobus fulgidus. Biochemistry 41:12697-12705. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chalov, S. E., O. L. Voronina, O. N. Sergienko, and V. G. Lunin. 2003. Thermostable DNA-polymerase from the thermophilic archaeon microorganism Archaeoglobus fulgidus VC16 and its features. Biochemistry (Mosc.) 68:301-308. [DOI] [PubMed] [Google Scholar]
- 7.Chung, J. H., E. K. Im, H.-Y. Park, J. H. Kwon, S. Lee, J. Oh, K.-C. Hwang, J. H. Lee, and Y. Jang. 2003. A novel uracil-DNA glycosylase family related to the helix-hairpin-helix DNA glycosylase superfamily. Nucleic Acids Res. 31:2045-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, J. H., M.-J. Suh, Y. I. Park, J. A. Tainer, and Y. S. Han. 2001. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat. Res. 486:99-111. [DOI] [PubMed] [Google Scholar]
- 9.Čuboňová, L., K. Sandman, S. J. Hallam, E. F. DeLong, and J. N. Reeve. 2005. Histones in Crenarchaea. J. Bacteriol. 187:5482-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. http://www.pymol.org.
- 11.Dionne, I., and S. D. Bell. 2005. Characterization of an archaeal family 4 uracil DNA glycosylase and its interaction with PCNA and chromatin proteins. Biochem. J. 387:859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty, A. J., and S. W. Suh. 2000. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 28:4051-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emmerhoff, O. J., H.-P. Klenk, and N.-K. Birkeland. 1998. Characterization and sequence comparison of temperature-regulated chaperonins from the hyperthermophilic archaeon Archaeoglobus fulgidus. Gene 215:431-438. [DOI] [PubMed] [Google Scholar]
- 14.Emptage, K., R. O'Neill, A. Solovyova, and B. A. Connolly. 2008. Interplay between DNA polymerase and proliferating cell nuclear antigen switches off base excision repair of uracil and hypoxanthine during replication in archaea. J. Mol. Biol. 383:762-771. [DOI] [PubMed] [Google Scholar]
- 15.Fogg, M. J., L. H. Pearl, and B. A. Connolly. 2002. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 9:922-927. [DOI] [PubMed] [Google Scholar]
- 16.Frederico, L. A., T. A. Kunkel, and B. R. Shaw. 1990. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 29:2532-2537. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 18.Georg, J., L. Schomacher, J. P. J. Chong, A. I. Majerník, M. Raabe, H. Urlaub, S. Müller, E. Ciirdaeva, W. Kramer, and H.-J. Fritz. 2006. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 34:5325-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouet, P., E. Courcelle, D. I. Stuart, and F. Métoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 20.Grogan, D. W. 2004. Stability and repair of DNA in hyperthermophilic archaea. Curr. Issues Mol. Biol. 6:137-144. [PubMed] [Google Scholar]
- 21.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 22.Hanna, M., B. L. Chow, N. J. Morey, S. Jinks-Robertson, P. W. Doetsch, and W. Xiao. 2004. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair 3:51-59. [DOI] [PubMed] [Google Scholar]
- 23.Horst, J.-P., and H.-J. Fritz. 1996. Counteracting the mutagenic effect of hydrolytic deamination of DNA 5-methylcytosine residues at high temperature: DNA mismatch N-glycosylase Mig.Mth of the thermophilic archaeon Methanobacterium thermoautotrophicum THF. EMBO J. 15:5459-5469. [PMC free article] [PubMed] [Google Scholar]
- 24.Hoseki, J., A. Okamoto, R. Masui, T. Shibata, Y. Inoue, S. Yokoyama, and S. Kuramitsu. 2003. Crystal structure of a family 4 uracil-DNA glycosylase from Thermus thermophilus HB8. J. Mol. Biol. 333:515-526. [DOI] [PubMed] [Google Scholar]
- 25.Jeon, S.-J., and K. Ishikawa. 2003. A novel ADP-dependent DNA ligase from Aeropyrum pernix K1. FEBS Lett. 550:69-73. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 27.Kavli, B., O. Sundheim, M. Akbari, M. Otterlei, H. Nilsen, F. Skorpen, P. A. Aas, L. Hagen, H. E. Krokan, and G. Slupphaug. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277:39926-39936. [DOI] [PubMed] [Google Scholar]
- 28.Kiyonari, S., M. Uchimura, T. Shirai, and Y. Ishino. 2008. Physical and functional interactions between uracil-DNA glycosylase and proliferating cell nuclear antigen from the euryarchaeon Pyrococcus furiosus. J. Biol. Chem. 283:24185-24193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klenk, H.-P., R. A. Clayton, J.-F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 30.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16:3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knævelsrud, I., P. Ruoff, H. Ånensen, A. Klungland, S. Bjelland, and N.-K. Birkeland. 2001. Excision of uracil from DNA by the hyperthermophilic Afung protein is dependent on the opposite base and stimulated by heat-induced transition to a more open structure. Mutat. Res. 487:173-190. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W.H. Freeman, New York, NY.
- 33.Koulis, A., D. A. Cowan, L. H. Pearl, and R. Savva. 1996. Uracil-DNA glycosylase activities in hyperthermophilic micro-organisms. FEMS Microbiol. Lett. 143:267-271. [DOI] [PubMed] [Google Scholar]
- 34.Kouzminova, E. A., and A. Kuzminov. 2004. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol. Microbiol. 51:1279-1295. [DOI] [PubMed] [Google Scholar]
- 35.Leiros, I., M. P. Nabong, K. Grøsvik, J. Ringvoll, G. T. Haugland, L. Uldal, K. Reite, I. K. Olsbu, I. Knævelsrud, E. Moe, O. A. Andersen, N.-K. Birkeland, P. Ruoff, A. Klungland, and S. Bjelland. 2007. Structural basis for enzymatic excision of N1-methyladenine and N3-methylcytosine from DNA. EMBO J. 26:2206-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl, T., S. Ljungquist, W. Siegert, B. Nyberg, and B. Sperens. 1977. DNA N-glycosidases. Properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 252:3286-3294. [PubMed] [Google Scholar]
- 38.Lindahl, T., and B. Nyberg. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13:3405-3410. [DOI] [PubMed] [Google Scholar]
- 39.Liu, J., B. He, H. Qing, and Y. W. Kow. 2000. A deoxyinosine specific endonuclease from hyperthermophile, Archaeoglobus fulgidus: a homolog of Escherichia coli endonuclease V. Mutat. Res. 461:169-177. [DOI] [PubMed] [Google Scholar]
- 40.Mansfield, C., S. M. Kerins, and T. V. McCarthy. 2003. Characterisation of Archaeoglobus fulgidus AlkA hypoxanthine DNA glycosylase activity. FEBS Lett. 540:171-175. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto, Y., and K. Kim. 1995. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269:699-702. [DOI] [PubMed] [Google Scholar]
- 42.Motz, M., I. Kober, C. Girardot, E. Loeser, U. Bauer, M. Albers, G. Moeckel, E. Minch, H. Voss, C. Kilger, and M. Koegl. 2002. Elucidation of an archaeal replication protein network to generate enhanced PCR enzymes. J. Biol. Chem. 277:16179-16188. [DOI] [PubMed] [Google Scholar]
- 43.Nilsen, H., I. Rosewell, P. Robins, C. F. Skjelbred, S. Andersen, G. Slupphaug, G. Daly, H. E. Krokan, T. Lindahl, and D. E. Barnes. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5:1059-1065. [DOI] [PubMed] [Google Scholar]
- 44.Pearl, L. H. 2000. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 460:165-181. [DOI] [PubMed] [Google Scholar]
- 45.Piersen, C. E., A. K. McCullough, and R. S. Lloyd. 2000. AP lyases and dRPases: commonality of mechanism. Mutat. Res. 459:43-53. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sandigursky, M., and W. A. Franklin. 1999. Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr. Biol. 9:531-534. [DOI] [PubMed] [Google Scholar]
- 48.Sandigursky, M., and W. A. Franklin. 2000. Uracil-DNA glycosylase in the extreme thermophile Archaeoglobus fulgidus. J. Biol. Chem. 275:19146-19149. [DOI] [PubMed] [Google Scholar]
- 49.Sartori, A. A., S. Fitz-Gibbon, H. Yang, J. H. Miller, and J. Jiricny. 2002. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 21:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sartori, A. A., and J. Jiricny. 2003. Enzymology of base excision repair in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 278:24563-24576. [DOI] [PubMed] [Google Scholar]
- 51.Sartori, A. A., P. Schär, S. Fitz-Gibbon, J. H. Miller, and J. Jiricny. 2001. Biochemical characterization of uracil processing activities in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 276:29979-29986. [DOI] [PubMed] [Google Scholar]
- 52.Schmiedel, R., E. B. Kuettner, A. Keim, N. Sträter, and T. Greiner-Stöffele. 2009. Structure and function of the abasic site specificity pocket of an AP endonuclease from Archaeoglobus fulgidus. DNA Repair 8:219-231. [DOI] [PubMed] [Google Scholar]
- 53.Seybert, A., D. J. Scott, S. Scaife, M. R. Singleton, and D. B. Wigley. 2002. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res. 30:4329-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seybert, A., and D. B. Wigley. 2004. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. EMBO J. 23:1360-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shekhtman, A., L. McNaughton, R. P. Cunningham, and S. M. Baxter. 1999. Identification of the Archaeoglobus fulgidus endonuclease III DNA interaction surface using heteronuclear NMR methods. Structure 7:919-930. [DOI] [PubMed] [Google Scholar]
- 56.Shen, Y., X.-F. Tang, and I. Matsui. 2003. Subunit interaction and regulation of activity through terminal domains of the family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 278:21247-21257. [DOI] [PubMed] [Google Scholar]
- 57.Slupphaug, G., B. Kavli, and H. E. Krokan. 2003. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 531:231-251. [DOI] [PubMed] [Google Scholar]
- 58.Sobol, R. W., R. Prasad, A. Evenski, A. Baker, X.-P. Yang, J. K. Horton, and S. H. Wilson. 2000. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature 405:807-810. [DOI] [PubMed] [Google Scholar]
- 59.Starkuviene, V., and H.-J. Fritz. 2002. A novel type of uracil-DNA glycosylase mediating repair of hydrolytic DNA damage in the extremely thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 30:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stetter, K. O.1992. The genus Archaeoglobus, p. 707-711. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 61.Suh, D., D. M. Wilson III, and L. F. Povirk. 1997. 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 25:2495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, Y., M. S. Seo, J. H. Kim, Y. J. Kim, G. A. Kim, J. I. Lee, J.-H. Lee, and S.-T. Kwon. 2008. Novel DNA ligase with broad nucleotide cofactor specificity from the hyperthermophilic crenarchaeon Sulfophobococcus zilligii: influence of ancestral DNA ligase on cofactor utilization. Environ. Microbiol. 10:3212-3224. [DOI] [PubMed] [Google Scholar]
- 63.Sung, J.-S., and D. W. Mosbaugh. 2003. Escherichia coli uracil- and ethenocytosine-initiated base excision DNA repair: rate-limiting step and patch size distribution. Biochemistry 42:4613-4625. [DOI] [PubMed] [Google Scholar]
- 64.Unk, I., L. Haracska, S. Prakash, and L. Prakash. 2001. 3′-Phosphodiesterase and 3′→5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 21:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Söll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U. S. A. 100:12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White, M. F., and S. D. Bell. 2002. Holding it together: chromatin in the Archaea. Trends Genet. 18:621-626. [DOI] [PubMed] [Google Scholar]
- 67.Wibley, J. E. A., T. R. Waters, K. Haushalter, G. L. Verdine, and L. H. Pearl. 2003. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell 11:1647-1659. [DOI] [PubMed] [Google Scholar]
- 68.Wilbur, W. J., and D. J. Lipman. 1983. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. U. S. A. 80:726-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, H., J.-H. Chiang, S. Fitz-Gibbon, M. Lebel, A. A. Sartori, J. Jiricny, M. M. Slupska, and J. H. Miller. 2002. Direct interaction between uracil-DNA glycosylase and a proliferating cell nuclear antigen homolog in the crenarchaeon Pyrobaculum aerophilum. J. Biol. Chem. 277:22271-22278. [DOI] [PubMed] [Google Scholar]
- 70.Yang, H., S. Fitz-Gibbon, E. M. Marcotte, J. H. Tai, E. C. Hyman, and J. H. Miller. 2000. Characterization of a thermostable DNA glycosylase specific for U/G and T/G mismatches from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol. 182:1272-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao, A., F. C. Gray, and S. A. MacNeill. 2006. ATP- and NAD+-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol. Microbiol. 59:743-752. [DOI] [PubMed] [Google Scholar]