Abstract
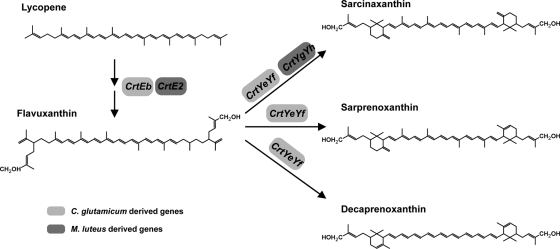
We report the cloning and characterization of the biosynthetic gene cluster (crtE, crtB, crtI, crtE2, crtYg, crtYh, and crtX) of the γ-cyclic C50 carotenoid sarcinaxanthin in Micrococcus luteus NCTC2665. Expression of the complete and partial gene cluster in Escherichia coli hosts revealed that sarcinaxanthin biosynthesis from the precursor molecule farnesyl pyrophosphate (FPP) proceeds via C40 lycopene, C45 nonaflavuxanthin, C50 flavuxanthin, and C50 sarcinaxanthin. Glucosylation of sarcinaxanthin was accomplished by the crtX gene product. This is the first report describing the biosynthetic pathway of a γ-cyclic C50 carotenoid. Expression of the corresponding genes from the marine M. luteus isolate Otnes7 in a lycopene-producing E. coli host resulted in the production of up to 2.5 mg/g cell dry weight sarcinaxanthin in shake flasks. In an attempt to experimentally understand the specific difference between the biosynthetic pathways of sarcinaxanthin and the structurally related ɛ-cyclic decaprenoxanthin, we constructed a hybrid gene cluster with the γ-cyclic C50 carotenoid cyclase genes crtYg and crtYh from M. luteus replaced with the analogous ɛ-cyclic C50 carotenoid cyclase genes crtYe and crtYf from the natural decaprenoxanthin producer Corynebacterium glutamicum. Surprisingly, expression of this hybrid gene cluster in an E. coli host resulted in accumulation of not only decaprenoxanthin, but also sarcinaxanthin and the asymmetric ɛ- and γ-cyclic C50 carotenoid sarprenoxanthin, described for the first time in this work. Together, these data contributed to new insight into the diverse and multiple functions of bacterial C50 carotenoid cyclases as key catalysts for the synthesis of structurally different carotenoids.
Carotenoids are natural pigments synthesized by bacteria, fungi, algae, and plants, and more than 750 different carotenoids have been isolated from natural sources (17). They possess important biological functions as protectants against light and oxygen excess in photosynthetic processes (32, 38), and they have been proposed to reduce the risk of certain cancers, cardiovascular disease, and Alzheimer disease due to their antioxidative properties (20, 46). The global market for carotenoids used as food colorants and nutritional supplements was estimated at approximately $935 million in 2005 (11). More than 95% of all natural carotenoids are based on a symmetric C40 phytoene backbone, and only a small number of C30 and even fewer C50 carotenoids have been discovered (42).
C50 carotenoids have multiple conjugated double bonds, and they contain at least one hydroxyl group; both these features contribute to strong antioxidative properties (17, 30, 32, 38). In nature, C50 carotenoids are synthesized by bacteria of the order Actinomycetales, and to date, only two different C50 carotenoid biosynthetic pathways have been described in the literature. The biosynthetic pathways of the ɛ-cyclic C50 carotenoid decaprenoxanthin [2,2′-bis-(4-hydroxy-3-methybut-2-enyl)-ɛ,ɛ-carotene] and the β-cyclic C50 carotenoid C.p.450 [2,2′-bis-(4-hydroxy-3-methybut-2-enyl)-β,β-carotene] have been elucidated in Corynebacterium glutamicum (22, 23) and in Dietzia sp. CQ4 (41), respectively. For both pathways, the common precursor, C40 lycopene, is synthesized from C15 farnesyl pyrophosphate (FPP) via the methylerythritol 4-phosphate (MEP) pathway, which is present in most eubacteria (33). Effective lycopene production has been achieved in genetically engineered noncarotenogenic hosts, such as Escherichia coli and Saccharomyces cerevisiae (9). Accordingly, the potential of using such biotechnologically relevant hosts for heterologous production of any lycopene-derived carotenoids has generated high interest.
The biosynthesis of cyclic C50 carotenoids from lycopene is catalyzed by lycopene elongase and carotenoid cyclases. Even though most carotenoids in plants and microorganisms exhibit cyclic structures, cyclization reactions were predominantly known for C40 pathways (45) catalyzed by monomeric enzymes that have been isolated from plants and bacteria (5, 16, 27, 29, 31, 36). In C. glutamicum, the genes crtYe, crtYf, and crtEb were identified as being involved in the conversion of lycopene to the ɛ-cyclic C50 carotenoid decaprenoxanthin (22, 44). Sequential elongation of lycopene into the acyclic C50 carotenoid flavuxanthin was catalyzed by the crtEb gene product lycopene elongase. Subsequent cyclization to decaprenoxanthin was catalyzed by a heterodimeric C50 carotenoid, ɛ-cyclase, encoded by crtYe and crtYf (22). C. glutamicum can synthesize both mono- and diglucosylated decaprenoxanthin; however, the genetic and enzymatic bases for glucosylation of decaprenoxanthin are unknown. Analogous to decaprenoxanthin, biosynthesis of the β-cyclic C50 carotenoid C.p.450 in Dietzia sp. CQ4 from lycopene involves lycopene elongase and C50 carotenoid β-cyclase activities (41).
While most cyclic carotenoids exhibit β-rings, ɛ-ring-containing pigments are common in higher plants (7), and carotenoids substituted only with γ-rings are rarely observed in plants and algae (14). To date, no biosynthetic pathway for γ-cyclic C50 carotenoids has been reported in the literature.
Micrococcus luteus NCTC2665 (the “Fleming strain”) is a Gram-positive bacterium belonging to the family Micrococcaceae within the order Actinomycetales. The carotenoids, including the γ-cyclic C50 sarcinaxanthin [(2R,6R,2′R,6′R)-(2,2′-bis(4-hydroxy-3-methyl-2-butenyl)-γ,γ-carotene)], synthesized by this bacterium have been identified and structurally elucidated (26). We recently isolated and characterized several wild-type M. luteus strains from the sea surface microlayer of the middle part of the Norwegian coast (39). Here, we report one additional such marine M. luteus isolate, designated Otnes7, forming color-intensive colonies indicating high sarcinaxanthin production levels. Both Otnes7 and NCTC2665 were used as M. luteus model strains, and the sarcinaxanthin biosynthetic gene clusters were cloned from both strains. The complete sarcinaxanthin biosynthetic pathway from lycopene was elucidated, including glucosylation, and we also explored the potential of using Otnes7-derived genes to achieve effective heterologous production of sarcinaxanthin in E. coli. The results add important new knowledge of the biosynthesis of C50 carotenoids, and in particular, they highlight the diverse functions of C50 carotenoid cyclases leading to synthesis of structurally different carotenoids.
MATERIALS AND METHODS
Bacteria, plasmids, standard DNA manipulations, and growth media.
The bacterial strains and plasmids used in this work are listed in Table 1. Bacteria were cultivated in Luria-Bertani (LB) broth (35), and recombinant E. coli cultures were supplemented with ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml) as appropriate. M. luteus and C. glutamicum strains were grown at 30°C and 225-rpm agitation for 24 h, and E. coli strains for cloning purposes were grown at 37°C and 225-rpm agitation overnight. For heterologous production of carotenoids, overnight cultures (100 ml) of recombinant E. coli cells grown at 30°C with 180-rpm agitation in shake flasks (500 ml) were diluted 1% in prewarmed medium with 0.5 mM of the Pm promoter inducer m-toluic acid (37) added unless otherwise indicated. In order to elucidate maximal sarcinaxanthin production yields, samples were initially taken after 16 h, 24 h, and 48 h and analyzed quantitatively as described below. The highest carotenoid abundance was typically observed after 48 h of cultivation, but it was only marginally higher than after 24 h of cultivation (data not shown). This is in agreement with analogous reports for heterologous production of zeaxanthin in E. coli (34), and therefore, production analyses were routinely performed by analyzing samples collected after 48 h. Standard DNA manipulations were performed according to the method of Sambrook et al. (35), and isolation of total DNA from M. luteus strains was performed as described previously (43).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | General cloning host | Gibco-BRL |
| E. coli XL1-Blue | Used as host for heterologous carotenoid production | Stratagene |
| M. luteus NCTC2665 | Wild-type strain | NCTC |
| M. luteus Otnes7 | Marine wild-type isolate | This work |
| C. glutamicum MJ-233C-MV10 | Tn31831 mutant of C. glutamicum MJ-233C; contains a wild-type crt gene cluster | 44 |
| Plasmids | ||
| pGEM-T | Ampr; standard cloning vector | Promega |
| pJBphOx | Ampr; pJB658 derivative with inducible Pm-xylS promoter/regulator system | 37 |
| pAC-LYC | Cmr; lycopene-producing plasmid containing crtEIB from P. ananatis; p15A ori | 8 |
| pCRT-EBIE2YgYh-2665 | pJBphOx with scFv-phOx gene replaced with the crtEBIE2YgYh region from strain Otnes7 | This work |
| pCRT-EBI-2665 | pJBphOx with scFv-phOx gene replaced with crtEBI from strain NCTC 2665 | This work |
| pCRT-E2YgYh-O7 | pJBphOx with scFv-phOx gene replaced with crtE2YgYh from strain Otnes7 | This work |
| pCRT-E2YgYh-2665 | pJBphOx with scFv-phOx gene replaced with crtE2YgYh from strain NCTC 2665 | This work |
| pCRT-E2Yg-O7 | pJBphOx with scFv-phOx gene replaced with crtE2Yg from strain Otnes7 | This work |
| pCRT-E2Yg-2665 | pJBphOx with scFv-phOx gene replaced with crtE2Yg from strain NCTC2665 | This work |
| pCRT-E2-O7 | pJBphOx with scFv-phOx gene replaced with crtE2 from strain Otnes7 | This work |
| pCRT-E2-2665 | pJBphOx with scFv-phOx gene replaced with crtE2 from strain NCTC2665 | This work |
| pCRT-YgYh-O7 | pJBphOx with scFv-phOx gene replaced with crtYgYh from strain Otnes7 | This work |
| pCRT-YgYh-2665 | pJBphOx with scFv-phOx gene replaced with crtYgYh from strain NCTC2665 | This work |
| pCRT-E2YgYhX-O7 | pJBphOx with scFv-phOx gene replaced with crtE2YgYhX from strain Otnes7 | This work |
| pCRT-E2-O7-YeYf-MJ | pJBphOx with scFv-phOx gene replaced with crtE2 from strain Otnes7 and YeYf from C. glutamicum | This work |
| pCRT-YeYfEb-MJ | pJBphOx with scFv-phOx gene replaced with crtYeYfEb from C. glutamicum | This work |
| pCRT-E2Yg-2665-Yf-MJ | pJBphOx with scFv-phOx gene substituted with crtE2Yg from strain Otnes7 and crtYf from C. glutamicum | This work |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
Vector construction. (i) pCRT-EBIE2YgYh-2665 and pCRT-EBI-2665.
The complete crtEBIE2YgYh gene cluster of M. luteus NCTC2665 was PCR amplified from genomic DNA by using the primer pair crtE-F (5′-TTTTTCATATGGGTGAAGCGAGGACGGG-3′) and crtYh-R (5′-TTTTTGCGGCCGCTCAGCGATCGTCCGGGTGGGG-3′). The crtEBI region of M. luteus NCTC2665 was PCR amplified from genomic DNA by using the primer pair crtE-F (see above) and crtI-R (5′-TTTTTGCGGCCGCTCATGTGCCGCTCCCCCCGG). The resulting PCR products (5,283 bp and 3,693 bp, respectively) were end digested with NdeI and NotI (the recognition sites are indicated in boldface in the primer sequences) and ligated into the corresponding sites of pJBphOx (37), yielding plasmids pCRT-EBIE2YgYh-2665 and pCRT-EBI-2665, respectively.
(ii) pCRT-E2YgYh-2665 and pCRT-E2YgYh-O7.
The crtE2YgYh regions of M. luteus strains NCTC2665 and Otnes7 were PCR amplified from genomic DNA using primers crtE2-F (5′-TTTTTCATATGATCCGCACCCTCTTCTG-3′) and crtYh-R (see above). The PCR products obtained (1,615 bp and 1,618 bp, respectively) were blunt-end ligated into the pGEM-T vector system (Promega, Madison, WI). The resulting plasmids were digested with NdeI and NotI (the recognition site is indicated in boldface in the primer), and the inserts were ligated into the corresponding sites of pJBphOx, yielding plasmids pCRT-E2YgYh-2665 and pCRT-E2YgYh-O7, respectively.
(iii) pCRT-E2YgYhX-O7.
The crtE2YgYhX region of M. luteus strain Otnes7 was PCR amplified from genomic DNA using primers crtE2-F (see above) and crtYX-R (5′-TTTTTCCTAGGAGATGGCCGCGAACATCCTG). In the resulting PCR product, crtYh and crtX were separated by or1008, encoding a putative protein with no assigned function. The PCR product was end digested with NdeI and BlnI (the recognition site is indicated in boldface in the primer), and the 3,085-bp fragment was ligated into the corresponding sites of pJBphOx, resulting in plasmid pCRT-E2YgYhX-O7.
(iv) pCRT-E2Yg-O7 and pCRT-E2Yg-2665.
The crtE2Yg coding regions of M. luteus strains NCTC2665 and Otnes7 were PCR amplified from chromosomal DNA using primers crtE2-F (see above) and crtYg-R (5′-TTTTTGCGGCCGCTCACCGGCTCCCCCGGTCGGTC-3′). The PCR products obtained were end digested with NdeI and NotI (the recognition site is indicated in boldface in the primer sequence), and the resulting 1,247-bp fragments were ligated into the corresponding sites of pJBphOx, resulting in plasmids pCRT-E2Yg-2665 and pCRT-E2Yg-O7, respectively.
(v) pCRT-E2-O7 and pCRT-E2-2665.
The crtE2 genes of M. luteus strains NCTC2665 and Otnes7 were PCR amplified from chromosomal DNA using primers crtE2-F (see above) and crtE2-R (5′-TTTTTGCGGCCGCTCATGCCGCCGCCCCCCGGG-3′). The resulting PCR products were end digested with NdeI and NotI (the recognition site is indicated in boldface in the primer sequence), and the 890-bp fragments were ligated into the corresponding sites of pJBphOx, resulting in plasmids pCRT-E2-2665 and pCRT-E2-O7, respectively.
(vi) pCRT-YgYh-O7 and pCRT-YgYh-2665.
The crtYgYh regions of M. luteus strains NCTC2665 and Otnes7 were PCR amplified from genomic DNA by using primers crtYg-F (5′-TTTTTCATATGATCTACCTGCTGGCCCT-3′) and crtYh-R (see above). The resulting 734-bp PCR products were end digested with NdeI (the restriction site is indicated in boldface in the primer sequence) and NotI and ligated into the corresponding sites of pJBphOx, resulting in plasmids pCRT-YgYh-2665 and pCRT-YgYh-O7, respectively.
(vii) pCRT-E2-O7-YeYf-MJ.
According to the gene sequences of crtE2 in M. luteus Otnes7 and crtYeYf in C. glutamicum MJ233-MV10, four primers, crtE2-F (5′-TGACCAACGACCGGTAGCGGAG-3′) and crtE2-i-R (5′-CCCATCCACTAAACTTAAACATCATGCCGCCGCCCCCCGG-3′), and crtYe-i-F (5′-TGTTTAAGTTTAGTGGATGGGTTGATCCCTATCATCGATATTTCAC-3′) and crtYf-R (5′-TTTTGCGGCCGCTTTTCCATCATGACTACGGCTTTTC), were used. Primers crtE2-i-R and crtYe-i-F contained homologous extensions of 21 bp (italics) at the 5′ ends as linker sequences in order to allow crossover PCR. The primer pair crtE2-F and crtE2-i-R was used to amplify a 1,227-bp fragment containing the crtE2 gene from genomic M. luteus DNA, and the primer pair crtYe-i-F and crtYf-R was used to amplify an 885-bp crtYeYf-containing fragment from genomic DNA of C. glutamicum MJ-233C-MV10. The resulting PCR fragments were used as templates for PCR with the primer pair crtE2-F and crtYe-R to amplify a hybrid DNA fragment (2,090 bp) containing crtE2 from M. luteus and crtYeYf from C. glutamicum connected by the 21-bp linker sequence. The resulting hybrid fragment was end digested with AgeI and NotI (the restriction site is indicated in boldface in the primer sequence), and the 2,070-bp fragment obtained was ligated into the corresponding sites of pJB658phOx, resulting in plasmid pCRT-E2-O7-YeYf-MJ.
(viii) pCRT-YeYfEb-MJ.
The crtYeYfEb genes from C. glutamicum strain MJ-233C-MV10 (Table 1) were PCR amplified from genomic DNA using primers crtYe-F1 (5′-TGGCTATCTCTAGAAAGGCCTACCCCTTAGGCTTTATGCAACAGAAACAATAATAATGGAGTCATGAACATATGATCCCTATCATCGATATTTCAC-3′) and crtYf-R (5′-TTTTGCGGCCGCCTGATCGGATAAAAGCAGAGTTATATC-3′). The resulting PCR product was digested with XbaI and NotI (the restriction site is indicated in boldface in the primer sequence), and the 1,789-bp fragment was ligated into the corresponding sites of pJBphOx, resulting in plasmid pCRT-YeYfEb-MJ.
All the constructed vectors were verified by DNA sequencing and transformed by electroporation (10) into the production host strains E. coli XL1-Blue and the lycopene-producing E. coli XL1-Blue(pAC-LYC) (8).
Extraction of carotenoids from bacterial-cell cultures.
To extract carotenoids from M. luteus strains, cells were harvested, washed with deionized H2O, and treated with lysozyme (20 mg/ml) and lipase (Fluka Chemicals, Germany) according to the method of Kaiser et al. (18), and the pigments were extracted with a mixture (7:3) of methanol and acetone. For recombinant E. coli strains, 50-ml aliquots of the cell cultures were centrifuged at 10,000 × g for 3 min, and the pellets were washed with deionized H2O; the cells were then frozen and thawed to facilitate extraction. Finally the pigments were extracted with 4 ml methanol-acetone (7:3) at 55°C for 15 min with thorough vortexing every 5 min. When necessary, up to three extraction cycles were performed to remove all visible colors from the cell pellet. When selective extraction for xanthophylls was desired, pure methanol was used. Butyhydroxytoluene (BHT) (0.05%) was added to the organic solvent to contribute to the stabilization of carotenoids (18). Samples for preparative high-performance liquid chromatography (HPLC) were in addition partitioned into 50% diethyl ether in petroleum ether. The collected upper phase was evaporated to dryness and dissolved in methanol.
Quantitative and qualitative LC-MS analyses of carotenoids in cell extracts.
Liquid chromatography-mass spectrometry (LC-MS) analyses of carotenoid-containing extracts were performed on an Agilent Ion Trap SL mass spectrometer equipped in front with an Agilent 1100 series HPLC system, including a diode array detector (DAD) for UV/visible (Vis) spectrum recording. Quantification of carotenoids was performed using the extracted wavelength chromatogram at peak λmax, 450 ± 16 nm for sarcinaxanthin and carotenoids with corresponding UV/Vis profiles and 470 ± 16 nm for lycopene and corresponding carotenoids, while MS detection was used to confirm the identities of known peaks for quantification and to determine the molecular masses of unknown carotenoids in the various cell extracts. Trans-beta-apo-8′-carotenal (Sigma) and lycopene (Fluka) were used as standards. They were dissolved in chloroform according to their solubilities and diluted in methanol. The correct concentrations of the prepared standard solutions were calculated from absorbance measurements of the solutions and by using the specific extinction coefficients E1%1cm, i.e., the absorption of 1% solution in a 1-cm cuvette at the maximum absorption wavelength, of 3,450 for lycopene and 2,590 for apocarotenal (15, 18). The standards were filtered through a syringe 0.2-μm polypropylene filter (Pall Gelman) and stored in amber glass vessels at −80°C under an N2 atmosphere if not used immediately.
Two HPLC protocols were used, a fast, high-throughput method for quantification of known carotenoids and a slow method with higher resolution for qualitative detection of all carotenoids in an extract permitting determination of the UV/Vis spectra and molecular masses of unknown carotenoids. A 2.1- by 30-mm Zorbax RR SB RP C18 column was used for the fast, high-throughput method. The carotenoids were eluted isocratically in methanol for 5 min. The column flow was kept at 0.4 ml/min, and 10 μl extract was injected for each run. The slow run method was run isocratically for 25 min with a mobile phase composition of MeOH/acetonitrile (7:3) with a 2.1- by 150-mm Zorbax SB RP C18 column using a flow rate of 250 μl/min. Ten or 20 μl extract was injected depending on the concentrations of carotenoids in the various extracts. The mass spectrometer was operated in positive scan mode using chemical ionization. The settings of the atmospheric pressure chemical ionization (APCI) source were 325°C dry temperature, 350°C vaporizer temperature, 50 lb/in2 nebulizer pressure, and 5.0 liter/min dry gas.
Purification of carotenoids.
For purification of carotenoids, preparative HPLC was performed on an Agilent preparative HPLC 1100 series system equipped with two preparative HPLC pumps, a preparative autosampler, and a preparative fraction collector. The mobile phases were methanol in channel 1 and acetonitrile in channel 2. Samples (2 ml) were injected at a flow rate of 20 ml/min into a Zorbax RP C18 21- by 250-mm preparative LC column. Online MS analysis was performed by splitting the flow 1:200 after the column using an Agilent LC flow splitter, and a makeup flow of 1 ml methanol/min was used to carry the analytes to the mass spectrometer with less than a 15-s delay. The diode array detector was used to trigger fraction collection.
Carotenoid structure determination by NMR.
When appropriate, nuclear magnetic resonance (NMR) was used for carotenoid structure determination. All NMR spectra were recorded on a Bruker Avance 600-MHz instrument fitted with an inverse triple resonance cryoprobe (TCI) using CDCl3 as a solvent with trimethylsilyl (TMS) as an internal reference. 1H and 13C signals were unambiguously assigned with the aid of in-phase correlation spectroscopy (ip-SCOPY), heteronuclear single-quantum coherence (HSQC), heteronuclear multiple-bond correlation (HMBC), nuclear Overhauser effect spectroscopy (NOESY), and HSQC-total-correlation spectroscopy (TOCSY) experiments.
RESULTS
Analysis of carotenoids produced by M. luteus NCTC2665 and a new marine M. luteus isolate designated Otnes7.
As a basis for the current studies, we characterized the major carotenoids synthesized by the M. luteus wild-type strain NCTC2665. In addition, we report here one new selected marine isolate, designated Otnes7, classified as an M. luteus strain by 16S rRNA sequence analysis (performed by NCIMB Ltd., Scotland) and forming colonies on LB agar plates with higher color intensity than other wild-type M. luteus strains (data not shown). We chose to include Otnes7 as an alternative model strain in this study, in particular to investigate whether its genes might also be favorable for efficient heterologous sarcinaxanthin production in E. coli strains (see below).
Cell extracts from shake flask cultures of strain NCTC2665 were analyzed by LC-MS, and one major peak (peak 3) (Fig. 1A) was identical to that of sarcinaxanthin purified and structurally identified by NMR earlier from wild-type M. luteus strains (39). In addition, two minor peaks, peak 1 and peak 2, were identified, with the same absorption spectra as sarcinaxanthin (Fig. 1A). The retention time of peak 2 was equal to that of sarcinaxanthin monoglucoside identified by NMR earlier (39), while peak 1 was more polar and therefore was predicted to represent sarcinaxanthin diglucoside (Table 2). An analogous analysis of strain Otnes7 revealed that it produced the same type of carotenoids; however, the total carotenoid level was higher (190 μg/g cell dry weight [CDW]) than that of NCTC2665 cells (145 μg/g CDW) under the conditions tested, which was in agreement with the different colony color intensities of the two strains.
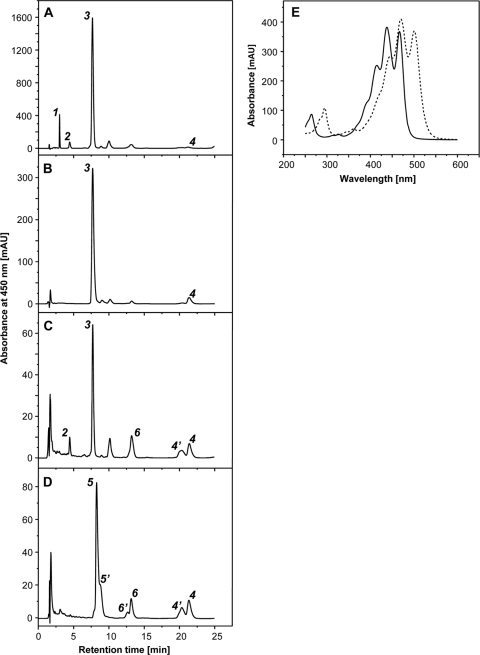
FIG. 1.
(A to D) HPLC elution profiles of carotenoids extracted from M. luteus strains Otnes7 (A), E. coli(pAC-LYC)(pCRT-E2YgYh-O7) (B), E. coli(pAC-LYC)(pCRT-E2YgYhX-O7) (C), and E. coli(pAC-LYC)(pCRT-E2-O7) (D). Peak 1, sarcinaxanthin diglucoside; peak 2, sarcinaxanthin monoglucoside; peak 3, sarcinaxanthin; peak 4, lycopene; peak 5, flavuxanthin; peak 6, nonaflavuxanthin; Peaks 4′, 5′, and 6′ are the cis isomers of 4, 5, and 6, respectively. (E) Absorption spectra of carotenoids from peaks 1, 2, and 3 (solid line) and peaks 4, 5, and 6 (dashed line). AU, arbitrary units.
TABLE 2.
Characteristics of carotenoids extracted from M. luteus strain Otnes7 and from recombinant E. coli strainsa
| Carotenoid (trivial name) | λmax (nm) in the HPLC eluent | Molecular mass (Da) | Retention time (min) |
|---|---|---|---|
| Sarcinaxanthin diglucoside | 414 438 467 | 1,028 | 3.0 |
| Sarcinaxanthin monoglucoside | 414 438 467 | 886 | 4.5 |
| Sarcinaxanthin | 414 438 467 | 704 | 7.7 |
| Flavuxanthin | 445 470 501 | 704 | 8.2 |
| Nonaflavuxanthin | 445 470 501 | 620 | 13.2 |
| Lycopene | 445 470 501 | 536 | 21.3 |
| Decaprenoxanthin | 414 438 467 | 704 | 10.1 |
| Sarprenoxanthin | 414 438 467 | 704 | 8.9 |
The carotenoids were dissolved in methanol and separated by HPLC using the Zorbax C18 150- by 30-mm column (see Materials and Methods). All these extracted carotenoids have a characteristic three-peak absorption profile (Fig. 1E), and all λmax values are given.
Genetic characterization and heterologous expression of the M. luteus sarcinaxanthin biosynthetic gene cluster.
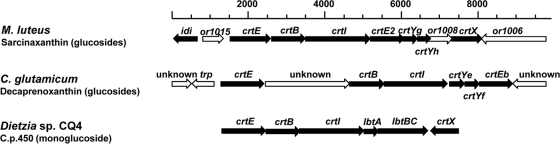
The genome sequence of M. luteus strain NCTC2665 (accession number NC_012803) has been deposited in the databases, and just before the submission of this paper, an accompanying publication appeared in the scientific literature (47). In silico screening of the sequence data resulted in the identification of a putative carotenoid biosynthesis gene cluster consisting of a total of nine open reading frames, or1007 to or1015. The genetic organization of the carotenoid (crt) genes in M. luteus displayed certain similarities to the previously published biosynthetic gene clusters for the C50 carotenoids C.p.450 and decaprenoxanthin in Dietzia sp. (41) and C. glutamicum (22), respectively (Fig. 2). The deduced M. luteus gene products displayed between 27% and 55% primary sequence identity to enzymes of the decaprenoxanthin and C.p.450 biosynthetic pathways. Based on these sequence analyses, the M. luteus genes crtE (encoding geranyl geranyl pyrophosphate [GGPP] synthase), crtB (encoding phytoene synthase), crtI (encoding phytoene desaturase), crtE2 (encoding lycopene elongase), crtYg (encoding the C50 cyclase subunit), crtYh (encoding the C50 cyclase subunit), and crtX (encoding glycosyl transferase) were assigned (Table 3). In addition, or1008 and or1015 encoded putative proteins with no assigned functions. In an attempt to identify relevant transcription initiation elements, the “neural network promoter prediction” method (http://www.fruitfly.org/seq_tools/promoter.html) was applied to the entire crt gene cluster, including or1015 and the upstream region (Fig. 2). By far the highest score was observed for a nucleotide sequence located 112 to 62 bp upstream of the crtE start codon, suggesting that the gene cluster is transcribed as a polycistronic operon from this promoter.
FIG. 2.
Chromosomal organization of the M. luteus sarcinaxanthin biosynthetic gene cluster presented in this study. The analogous C. glutamicum and Dietzia sp. biosynthetic gene clusters for the C50 carotenoids decaprenoxanthin and C.p.450, respectively, are included for comparison. Genes indicated by white arrows are suggested not to be involved in carotenoid biosynthesis.
TABLE 3.
M. luteus sarcinaxanthin biosynthetic genes and primary sequence comparison with respective homologues from biosynthesis of decaprenoxanthin and C.p.450 in C. glutamicum and Dietzia sp. CQ4, respectively
| ORFa | Gene name | Predicted gene product |
C. glutamicum |
Dietzia sp. CQ4 |
||
|---|---|---|---|---|---|---|
| Homologue | Primary sequence identity (%) | Homologue | Primary sequence identity (%) | |||
| or1007 | crtX | Glycosyl transferase (CrtX) | None | CrtX | 43 | |
| or1008 | Unknown | |||||
| or1009 | crtYh | C50 γ-cyclase subunit (CrtYh) | CrtYf | 31 | LbtBCb | 38 |
| or1010 | crtYg | C50 γ-cyclase subunit (CrtYg) | CrtYe | 32 | LbtA | 36 |
| or1011 | crtE2 | Lycopene elongase (CrtE2) | CrtEb | 50 | LbtBCc | 52 |
| or1012 | crtI | Phytoene desaturase (CrtI) | CrtI | 43 | CrtI | 53 |
| or1013 | crtB | Phytoene synthase (CrtB) | CrtB | 41 | CrtB | 48 |
| or1014 | crtE | GGPP synthase (CrtE) | CrtE | 31 | CrtE | 33 |
ORF, open reading frame.
The N-terminal region of LbtBC (amino acids 1 to 134) is homologous to those of M. luteus CrtYh and C. glutamicum CrtYf, respectively.
The C-terminal region of LbtBC (amino acids 135 to 432) is homologous to those of M. luteus CrtE2 and C. glutamicum CrtEb, respectively.
To experimentally verify that the identified M. luteus gene cluster encoded an active sarcinaxanthin biosynthetic pathway, the entire crtEBIE2YgYh region from NCTC2665 was cloned in frame and under the transcriptional control of the positively regulated Pm-xylS promoter/regulator system in plasmid pJBphOx (37). This expression vector has many favorable properties useful for regulated expression of genes and pathways at relevant levels in Gram-negative bacteria (1). The resulting plasmid, pCRT-EBIE2YgYh-2665 (Table 1), was transformed into the noncarotenogenic E. coli host strain XL1-Blue, and the recombinant strain was analyzed for carotenoid production under Pm-induced conditions (0.5 mM m-toluic acid) over 48 h (see Materials and Methods). LC-MS analysis of cell extracts revealed a small peak at a retention time, absorption spectrum, and molecular mass identical to those of sarcinaxanthin identified in the M. luteus strains (see above). The recombinant E. coli strain produced small amounts of sarcinaxanthin (10 to 15 μg/g CDW). No sarcinaxanthin was detected in plasmid-free cells, thus confirming that the identified gene cluster encodes a sarcinaxanthin biosynthetic pathway from farnesyl pyrophosphate. The biological role of the crtX gene was experimentally confirmed (see below).
Sarcinaxanthin production levels in E. coli can be increased 150-fold (2.5 mg/g CDW) by expressing Otnes7-derived crtE2YgYh genes in a lycopene-producing host.
To investigate the reason for the poor sarcinaxanthin production levels obtained in E. coli, we established a recombinant strain, E. coli(pCRT-EBI-2665), expressing the crtE, crtB, and crtI genes from NCTC2665, encoding enzymes assumed to catalyze the conversion of FPP into lycopene (Fig. 2 and Table 3). Analysis of this recombinant strain under Pm-induced conditions (0.5 mM toluic acid) confirmed that it produced lycopene as the sole carotenoid. However, the lycopene production yield was low (8 to 12 μg/g CDW) and in the same range as the sarcinaxanthin production yield obtained when the complete crtEBIE2YgYh gene cluster was expressed (see above). These data suggested that lycopene synthesis might be a bottleneck for efficient sarcinaxanthin production in this E. coli host. Therefore, E. coli XL1-Blue was transformed with plasmid pAC-LYC (6, 8) harboring the Pantoea ananatis crtEIB genes encoding three enzymes for biosynthesis of lycopene from isoprenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). LC-MS analysis confirmed that the resulting strain, E. coli(pAC-LYC), accumulated lycopene (1.8 mg/g CDW) as the sole carotenoid, and therefore, further carotenoid production experiments were performed using XL1-Blue(pAC-LYC) as a host.
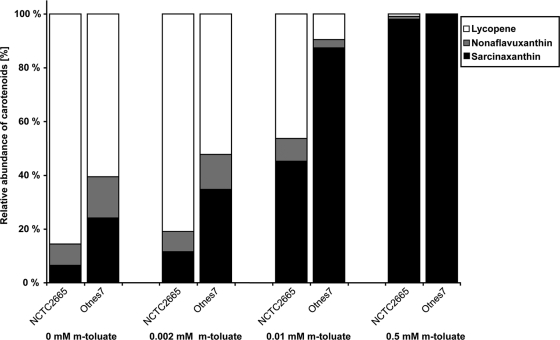
We then established E. coli(pAC-LYC)(pCRT-E2YgYh-2665) expressing the crtE2, crtYg, and crtYh genes from NCTC2665 (Table 1). LC-MS analysis of cell extracts revealed a total maximum carotenoid accumulation of 2.3 mg/g CDW, and about 98% of the total carotenoid produced was identified as sarcinaxanthin (Fig. 3, 0.5 mM inducer). These data demonstrated that the M. luteus NCTC2665 crtE2YgYh gene products could effectively convert lycopene into sarcinaxanthin. This result also confirmed that M. luteus genes were not efficient for lycopene production under the conditions tested. The reason for this is unknown, and it was not further investigated in the present study.
FIG. 3.
Relative carotenoid abundances in extracts from E. coli(pAC-LYC)(pCRT-E2YgYh-O7) and E. coli(pAC-LYC)(pCRT-E2YgYh-2665) overexpressing crtE2, crtYg, and crtYh genes from M. luteus strains Otnes7 and NCTC2665 (Table 1) cultivated in the presence of various Pm inducer concentrations (0, 0.002, 0.01, and 0.5 mM m-toluic acid). The fractions of sarcinaxanthin, lycopene, and intermediates are indicated. Samples (three replicates) were analyzed after 48 h of cultivation to ensure maximum sarcinaxanthin production levels (see Materials and Methods).
It was of interest to test if Otnes7 genes could contribute to more efficient sarcinaxanthin production levels in this host. Therefore, the alternative strain E. coli(pAC-LYC)(pCRT-E2YgYh-O7) expressing the crtE2, crtYg, and crtYh genes from Otnes7 was established. The total carotenoid production level (2.5 mg/g CDW) of the resulting recombinant strain was slightly higher than that of the analogous strain E. coli(pAC-LYC)(pCRT-E2YgYh-2665) (2.3 mg/g CDW). Interestingly, we noticed that all (100%) of the total carotenoid produced by E. coli(pAC-LYC)(pCRT-E2YgYh-O7) was sarcinaxanthin and no lycopene was present in these cells (Fig. 3). This demonstrated very efficient lycopene conversion by using Otnes7-derived genes, indicating that lycopene synthesis is a bottleneck for further improved sarcinaxanthin production under the conditions tested. Therefore, to further compare the efficiency of using Otnes7- versus NCTC2665-derived biosynthetic genes, production analyses were performed with different Pm inducer concentrations (Fig. 3). The results demonstrated that the strain E. coli(pAC-LYC)(pCRT-E2YgYh-O7) produced sarcinaxanthin at higher levels than E. coli(pAC-LYC)(pCRT-E2YgYh-2665) under all induction conditions tested, thus confirming that Otnes7 genes may be preferable for efficient sarcinaxanthin production in the E. coli host. DNA sequence analysis of the cloned Otnes7 crtE2YgYh fragment revealed a total of 24 nucleotide substitutions compared to the corresponding NTCT2665 DNA sequence, corresponding to four amino acid substitutions in CrtE2, five in CrtYg, and two substitutions plus one insertion in CrtYh. Whether these sequence variations positively affect the expression levels or the catalytic properties of the respective proteins remains unknown, and it was not further investigated here.
The M. luteus crtE2 gene product catalyzes in vivo conversion of lycopene to C45 nonaflavuxanthin and C50 flavuxanthin.
To elucidate the individual biosynthetic steps in the conversion of lycopene to sarcinaxanthin, we established strain E. coli(pAC-LYC)(pCRT-E2-2665) expressing the crtE2 genes from NCTC2665 (Table 1) and analyzed it for carotenoid production. Two different carotenoids were accumulated in the cells in addition to lycopene (Fig. 1D); all three compounds shared identical UV/Vis profiles. No sarcinaxanthin was detected. The minor carotenoid had a molecular mass of 620 Da, indicating a C45 xanthophyll compared with reported values (620 to 624 Da) (3), and the major carotenoid had a molecular mass of 704 Da, indicating a C50 xanthophyll (704 to 738 Da) (3). The major carotenoid was purified by preparative HPLC and analyzed by NMR (see Table S1 in the supplemental material). Inspection of 1H, 13C, and HSQC spectra revealed chemical shifts in agreement with reported data for the acyclic C50 carotenoid flavuxanthin (24). The minor carotenoid was identified as nonaflavuxanthin on the basis of the UV/Vis profile and the mass (Table 2). These results verified that the M. luteus crtE2 gene encodes a lycopene elongase catalyzing the sequential elongation of the C40 carotenoid lycopene via the C45 carotenoid nonaflavuxanthin to the C50 carotenoid flavuxanthin. A similar analysis using the analogous strain E. coli(pAC-LYC)(pCRT-E2-Otnes7) gave the same conclusion (data not shown). We noticed that the relative conversion of lycopene was substantially higher in the latter strain (79% versus 23%), which was in agreement with the generally effective sarcinaxanthin production obtained when Otnes7 genes were expressed (Fig. 3).
The M. luteus crtYg and crtYh genes together encode an active C50 carotenoid cyclase catalyzing cyclization of C50 flavuxanthin to C50 sarcinaxanthin in vivo.
To investigate if crtYg encoded any cyclase activity, we constructed and analyzed the recombinant strains E. coli(pAC-LYC)(pCRT-E2Yg-O7) and E. coli(pAC-LYC)(pCRT-E2Yg-2665) expressing the crtE and crtYg genes from NCTC2665 and Otnes7, respectively (Table 1). The carotenoids produced by both strains were flavuxanthin, nonaflavuxanthin, and lycopene, and their relative abundances were similar to those in strains E. coli(pAC-LYC)(pCRT-E2-O7) and E. coli(pAC-LYC)(pCRT-E2-2665), respectively. No sarcinaxanthin was detected in either of the strains. These data thus implied that the CrtYg and CrtYh polypeptides must function together as an active carotenoid cyclase catalyzing cyclization of flavuxanthin to sarcinaxanthin in vivo. To analyze the specificity of this carotenoid cyclase, we established the recombinant strains E. coli(pAC-LYC)(pCRT-YgYh-O7) and E. coli(pAC-LYC)(pCRT-YgYh-2665) expressing the crtYg and crtYh genes from NCTC2665 and Otnes7, respectively (Table 1). The aim was to analyze whether the CrtYgYh cyclases could catalyze cyclization of lycopene in vivo. HPLC analysis showed that both strains accumulated lycopene as the only carotenoid (data not shown), confirming that the crtYgYh gene products cannot use lycopene as a substrate. These data confirmed that the CrtYg and CrtYh polypeptides together constitute an active γ-type C50 carotenoid cyclase catalyzing cyclization of flavuxanthin to sarcinaxanthin in vivo.
The M. luteus crtX gene encodes an active glycosyl transferase that can be used to produce monoglucosylated sarcinaxanthin in E. coli.
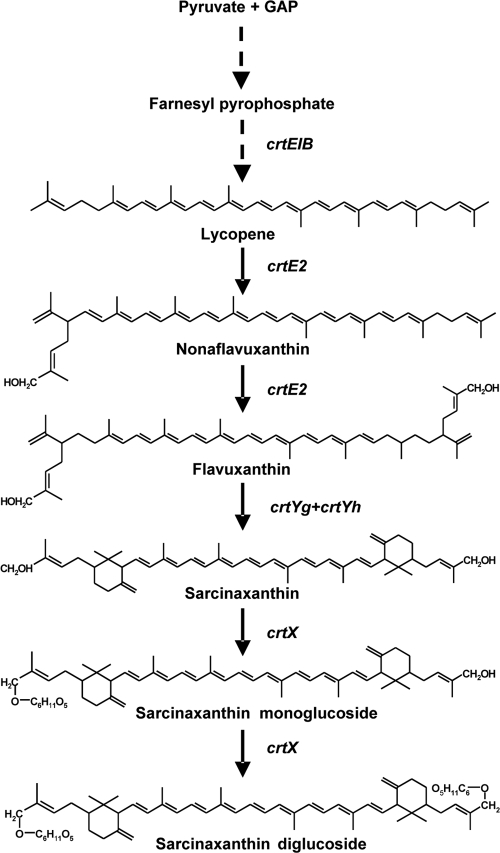
Downstream of crtYh is or1008, encoding a putative protein with no significant homology to any known proteins, followed by or1007, which encodes a polypeptide sharing 46% primary sequence identity with the putative glucosyl transferase protein CrtX from Dietzia sp. CQ4 (41). To our knowledge, no analogous gene has been found in the C. glutamicum genome sequence, and yet this bacterium can synthesize glucosylated decaprenoxanthin (24). The or1007 gene is depicted here as crtX (Table 3), and to unravel its biological function, we constructed and analyzed the recombinant strain E. coli(pAC-LYC)(pCRT-E2YgYhX-O7), which expresses the crtE2, crtYg, crtYh, and crtX genes from Otnes7 (Table 1). The resulting HPLC profile (Fig. 1C) revealed sarcinaxanthin as the major carotenoid (peak 3), but an additional, more polar carotenoid was eluted earlier (peak 2) and had a retention time and absorption spectrum identical to those of sarcinaxanthin monoglucoside from M. luteus Otnes 7 (Fig. 1C and E). Another minor peak was observed with the same retention time as sarcinaxanthin diglucoside produced by M. luteus strains (39); however, the amount detected was too small for a confident analysis of the mass and absorption spectrum. About 10% of the total sarcinaxanthin produced was glucosylated both in M. luteus wild-type strains and when heterologously produced in E. coli under the conditions tested. These results confirmed that crtX encodes an active glycosyl transferase that is necessary for the glucosylation of sarcinaxanthin. Based on all the accumulated data, we could deduce the complete biosynthetic pathway of sarcinaxanthin and its glucosides from FPP and via lycopene in M. luteus, as presented in Fig. 4.
FIG. 4.
Elucidated biosynthetic pathway for the individual steps in the formation of sarcinaxanthin and its glucosides from lycopene. CrtEBI, GGPP synthase, phytoene synthase, and phytoene desaturase; CrtE2, lycopene elongase; CrtYg plus CrtYf, C50 carotenoid γ-cyclase; CrtX, C50 carotenoid glycosyl transferase.
Expression of a hybrid operon containing the decaprenoxanthin cyclase genes crtYe and crtYf leads to production of three different C50 carotenoids in E. coli.
The biosynthetic pathway for decaprenoxanthin in C. glutamicum is reported to involve cyclization of flavuxanthin by an ɛ-cyclic C50 cyclase encoded by crtYe and crtYf. At this point, the biochemical functions of the C50 cyclase proteins in M. luteus and C. glutamicum seemed to represent the major difference between the biosynthetic pathways of sarcinaxanthin and decaprenoxanthin. To experimentally verify this, we established and analyzed E. coli(pAC-LYC)(pCRT-E2-O7-YeYf-MJ) expressing a hybrid operon containing crtE2 from Otnes7 and crtYe and crtYf from C. glutamicum MJ233-MV10. We expected this recombinant strain to produce decaprenoxanthin. Surprisingly, LC-MS analysis revealed that three different cyclic C50 carotenoids were accumulated (Fig. 5), exhibiting the same UV/Vis absorbance spectrum and mass (Fig. 1E). The retention time for peak 1 was identical to that of sarcinaxanthin. Peaks 1, 2, and 3 represented 35%, 46%, and 19%, respectively, of the total carotenoid content of the recombinant cells. No lycopene was detected.
FIG. 5.
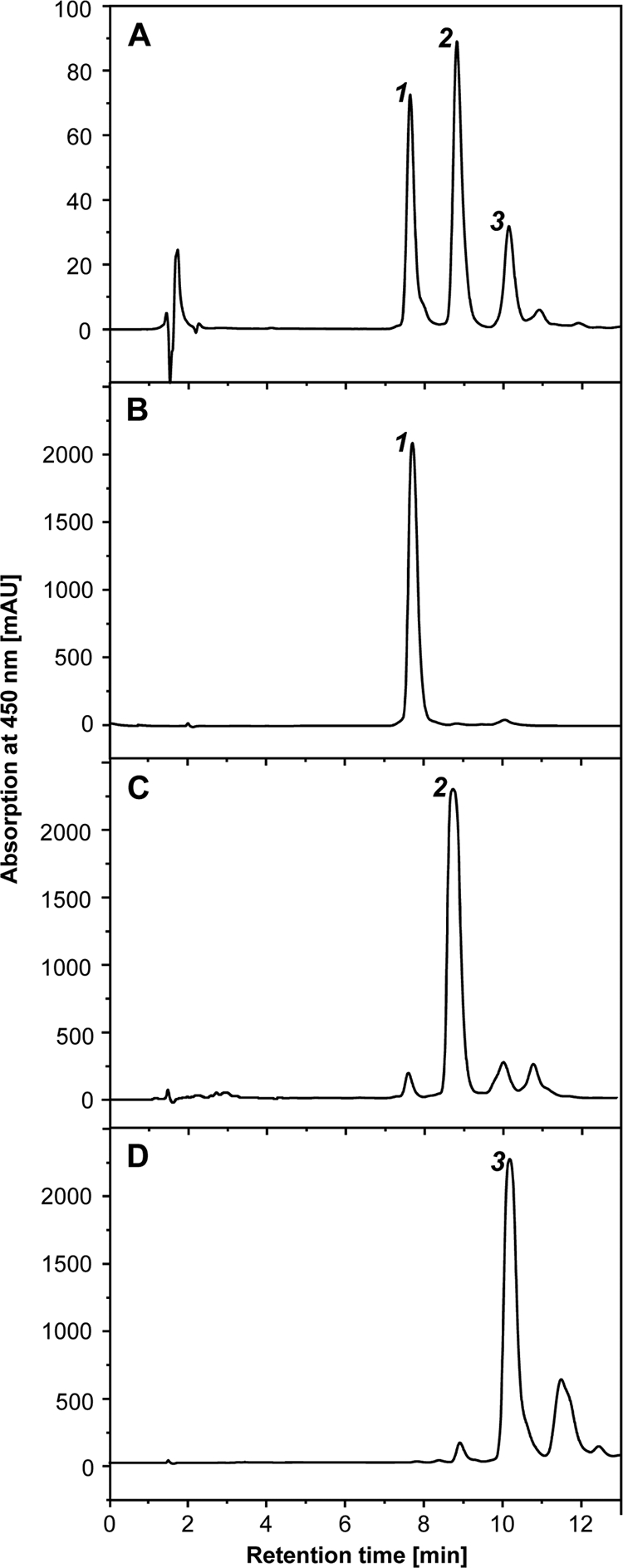
HPLC elution profiles of the carotenoids extracted from E. coli(pAC-LYC)(pCRT-E2 ml-YeYfcg) (A); purified peak 1, sarcinaxanthin (B); peak 2, sarprenoxanthin (C); and peak 3, decaprenoxanthin (D).
All three fractions were then purified by preparative HPLC and analyzed by NMR. The NMR data confirmed the identity of peak 1 as sarcinaxanthin (25), while peak 3 was identified as decaprenoxanthin (12, 13) (see Table S1 in the supplemental material). The NMR data for the purified peak 2 (Table 4), representing the major product, indicated a molecule that could not be clearly distinguished from a 1:1 mixture of sarcinaxanthin and decaprenoxanthin, so its existence had to be confirmed by chromatographic methods. The purified fractions were reanalyzed by LC using the slow run method (see Materials and Methods) to control its purity, and the corresponding chromatogram clearly shows that fraction 2 is a distinct carotenoid different from fractions 1 and 3 (Fig. 5B and C). Peak 2 was identified as a new bicyclic asymmetric C50 carotenoid with the systematic name 2,2′-bis(4-hydroxy-3-methyl-2-butenyl)-γ,ɛ-carotene. The compound exhibits one ɛ-cyclic and one γ-cyclic structure and thus appears to be a structural combination of sarcinaxanthin and decaprenoxanthin. This is in total agreement with the polarities of the three carotenoids (Fig. 5), and the compound was named sarprenoxanthin (Fig. 5 and 6). To our knowledge, this carotenoid has not been previously reported in the literature.
TABLE 4.
1H and 13C NMR assignments of sarprenoxanthina
| Position | 1H | Multiplicityb | 13C |
|---|---|---|---|
| C-1 | 36.0 | ||
| CH-2 | 1.36 | m | 44.2 |
| CH2-3 | 1.66 | m | 28.7 |
| 2.07 | m | ||
| CH-4 | 5.45 | m | 121.4 |
| C-5 | 134.4 | ||
| CH-6 | 2.44 | d (10.4) | 56.6 |
| CH-7 | 5.33 | dd (15.4, 10.4) | 130.3 |
| CH-8 | 6.15 | d (15.4) | 138.1 |
| C-9 | 135.3 | ||
| CH-10 | 6.14 | m | 130.4 |
| CH-11 | 6.62 | m | 124.9 |
| CH-12 | 6.36 | d (14.9) | 137.4 |
| C-13 | 136.4 | ||
| CH-14 | 6.25 | m | 132.4 |
| CH-15 | 6.63 | m | 130.0 |
| Me-16 | 0.93 | s; trans to C-7 | 26.8 |
| Me-17 | 0.74 | s; cis to C-7 | 16.3 |
| Me-18 | 1.53 | s | 23.1 |
| Me-19 | 1.93 | s | 13.16 |
| Me-20 | 1.97 | s | 12.8 |
| CH2-1″ | 1.81 | m | 28.0 |
| 2.27 | m | ||
| CH-2″ | 5.41 | m | 126.1 |
| C-3″ | 135.2 | ||
| CH2-4″ | 4.01 | s | 69.2 |
| Me-C-3″ | 1.67 | s | 13.8 |
| C-1′ | 39.3 | ||
| CH-2′ | 1.28 | m | 48.5 |
| CH2-3′ | 1.19 | m | 28.9 |
| 1.73 | m | ||
| CH2-4′ | 2.04 | m | 36.3 |
| 2.35 | ddd (13.4, 4.2, 2.6) | ||
| C-5′ | 150.4 | ||
| CH-6′ | 2.48 | d (9.9) | 58.5 |
| CH-7′ | 5.83 | dd (15.5, 9.9) | 128.4 |
| CH-8′ | 6.12 | d (15.5) | 137.6 |
| C-9′ | 135.4 | ||
| CH-10′ | 6.12 | m | 130.7 |
| CH-11′ | 6.62 | m | 124.9 |
| CH-12′ | 6.34 | d (14.9) | 137.4 |
| C-13′ | 136.4 | ||
| CH-14′ | 6.25 | m | 132.4 |
| CH-15′ | 6.63 | m | 130.0 |
| Me-16′ | 0.95 | s; trans to C-7′ | 27.7 |
| Me-17′ | 0.73 | s; cis to C-7′ | 15.3 |
| CH2-18′ | 4.53 | s; cis to C-6′ | 108.1 |
| 4.76 | s; trans to C-6′ | ||
| Me-19′ | 1.98 | s | 13.21 |
| Me-20′ | 1.97 | s | 12.8 |
| CH2-1″″ | 1.73 | m | 28.4 |
| 2.26 | m | ||
| CH-2″″ | 5.43 | m | 126.2 |
| C-3″″ | 135.3 | ||
| CH2-4″″ | 4.03 | s | 69.1 |
| Me-C-3″″ | 1.66 | s | 13.8 |
Recorded in CDCl3. Chemical shift values are expressed as δ values (ppm) from TMS.
s, singlet; d, doublet; dd, doublet of doublets; ddd, doublet of doublet of doublets; m, multiplet. J coupling values (Hz) are shown in parentheses.
FIG. 6.
Diverse biochemical functions of the M. luteus and the C. glutamicum C50 carotenoid cyclases. Biosynthesis of both sarcinaxanthin and decaprenoxanthin involves conversion of lycopene to flavuxanthin catalyzed by the lycopene elongases CrtEb and CrtE2 in M. luteus and C. glutamicum, respectively. The M. luteus CrtYgYh polypeptides constitute a γ-cyclase specifically converting flavuxanthin into sarcinaxanthin. In contrast, the C. glutamicum CrtYgYh polypeptides constitute both γ-cyclase and ɛ-cyclase activities and can convert flavuxanthin into three different C50 carotenoids; decaprenoxanthin, sarcinaxanthin, and sarprenoxanthin.
To rule out the possibility that these results were due to any unforeseen functions of the hybrid operon as such, we established the analogous strain E. coli(pAC-LYC)(pCRT-YeYfEb-MJ) expressing the decaprenoxanthin genes crtYe, crtYf, and crtEb from C. glutamicum (Fig. 2). The resulting chromatographic profile and absorbance spectrum were the same as in the hybrid construct, with relative abundance, mass, UV/Vis profile, and retention time identical to those of the carotenoids (data not shown). Moreover, DNA sequencing confirmed that the expressed crt genes were wild-type C. glutamicum sequences. Together, these results revealed that the crtYe and crtYf genes encoded a multifunctional C50 carotenoid cyclase that could catalyze the synthesis of three different bicyclic carotenoids. To our knowledge, this is the first report that C. glutamicum genes can be used to synthesize sarcinaxanthin and sarprenoxanthin.
DISCUSSION
In this paper, we report the cloning, characterization, and functional expression of the sarcinaxanthin biosynthetic gene cluster from M. luteus NCTC2665, consisting of six genes (crtE, crtB, crtI, crtE2, crtYg, and crtYh) encoding enzymes involved in the conversion of FPP into sarcinaxanthin. By expressing single genes and combinations of genes under the control of the positively regulated Pm-xylS promoter/regulator system in E. coli hosts, we discovered that sarcinaxanthin biosynthesis from the precursor FPP proceeds via lycopene, nonaflavuxanthin, and flavuxanthin to sarcinaxanthin (Fig. 4). M. luteus strains synthesized both mono- and diglucosylated derivatives of sarcinaxanthin, and we demonstrated that the crtX gene located proximal to the sarcinaxanthin genes encoded glycosyl transferase activity for the glucosylation of sarcinaxanthin (Fig. 2 and Table 3). No analogous glycosyl transferase-encoding gene has been identified in the natural decaprenoxanthin producer C. glutamicum. The reported presence of decaprenoxanthin glucosides in lycopene-producing E. coli cells expressing the C. glutamicum crtEb, crtYe, and crtYf genes led to the concomitant suggestion that host-encoded enzymes might be responsible for the glucosylation of decaprenoxanthin in C. glutamicum and E. coli (22). Based on their proximal localization and similar orientations, together with promoter prediction analyses, we propose that the sarcinaxanthin biosynthetic genes, including crtX, are cotranscribed from a common promoter located upstream of crtE in M. luteus (Fig. 2). The translational stop codons of crtB, crtI, crtE2, and crtYg overlap the translational start codons of their respective downstream genes, which may allow translational coupling to ensure equimolar expression and/or proper folding of the products. Secondary-structure analysis of the deduced gene products revealed six transmembrane helices for the M. luteus lycopene elongase CrtE2, and both CrtYg and CrtYh exhibited three transmembrane helices, indicating that they are transmembrane proteins (data not shown). This is in accordance with previous findings for carotenoid genes in other bacteria (4).
A new marine M. luteus isolate designated Otnes7 was included, together with the well-known M. luteus model strain NCTC2665. By expressing Otnes7 genes in a lycopene-producing E. coli host, we achieved complete conversion of the lycopene into production of 2.5 mg/g CDW sarcinaxanthin in shake flask cultures. Our data presented here indicate that Otnes7 genes may be favorable compared to M. luteus NCTC2665 genes when high-level heterologous production of sarcinaxanthin is desirable. Whether these differences are due to variations in the biochemical properties of the enzymes or, alternatively, different expression levels of the corresponding genes remains unknown. Our production experiments were done in small-scale shake flasks and without any optimization of growth conditions. We believe that conducting controlled high-cell-density cultivations of the recombinant E. coli strains under controlled conditions should further increase sarcinaxanthin production yields. However, this was not within the scope of the present study. In any case, our results implied that engineering E. coli host strains for higher lycopene production is presumably the immediate bottleneck for achieving increased sarcinaxanthin production.
The M. luteus CrtYg and CrtYh polypeptides were shown here to catalyze the in vivo γ-cyclization of the linear C50 carotenoid flavuxanthin to bicyclic sarcinaxanthin, and this cyclization reaction is independent of the prior stepwise lycopene elongation. In C. glutamicum, the C50 ɛ-cyclization reaction was reported by Krubasik et al. (22) to also be catalyzed by a heterodimeric protein encoded by crtYe and crtYf. However, in a separate report by these authors (24), different carotenoid intermediates were identified in C. glutamicum strains, and thus, an aberrant decaprenoxanthin pathway with a variant reaction sequence was proposed. Here, lycopene is transformed via monocyclic C45 nonaprene to bicyclic C50 sarcinene by two separate elongation/cyclization steps before decaprenoxanthin is formed by two sequential hydroxylation reactions. To our knowledge, the genetic basis for this alternative pathway is unknown. It should be noted that sarcinene has also been detected in M. luteus cell extracts (28), but whether this is relevant to sarcinaxanthin biosynthesis remains unclear. No sarcinene or nonaprene intermediates were detected in the present study. In Dietzia sp. CQ4, the conversion of the linear C50 carotenoid intermediate C.p.496 to the bicyclic C.p.450 has been reported to be catalyzed by a β-cyclase encoded by lbtA and the lbtB region in the lbtBC genes. The lbtB-encoded β-cyclase subunit appeared to be fused with a lycopene elongase (encoded by the lbtC region in the lbtBC genes) (41). The authors proposed a two-step sequential reaction consisting of the stepwise elongation of lycopene by the addition of two C5 units and subsequent C50 β-cyclization, analogous to the reaction cascade in M. luteus proposed here. The M. luteus CrtYgYh enzyme represents the first specific C50 carotenoid γ-cyclase that has been reported in the scientific literature.
Three different types of carotenoid cyclases are currently known, and based on sequence homology, the C50 carotenoid cyclases from Dietzia sp. and C. glutamicum have been proposed to be members of the CrtYcd-type cyclases (21). In contrast to other crtYcd-type cyclases, the C50 carotenoid cyclases are not involved in lycopene cyclization (41). Taking this together with our data presented here, we suggest that the C50 cyclases constitute a novel subtype of carotenoid cyclases conferring β-, ɛ-, and γ-cyclization of C50 carotenoids.
Carotenoid cyclases usually produce only one kind of ring structure (2), but lycopene cyclase from the cyanobacterium Prochlorococcus marinus has both β- and ɛ-cyclase activities (40). Surprisingly, construction and expression of a hybrid operon carrying the M. luteus crtE2 gene and the C50 carotenoid ɛ-cyclase genes crtYeYf from C. glutamicum resulted in accumulation of three different C50 carotenoids in E. coli: ɛ-cyclic decaprenoxanthin, γ-cyclic sarcinaxanthin, and a new molecule containing both ɛ- and γ-rings, here designated sarprenoxanthin (Fig. 6). This result proved that the C. glutamicum cyclase encoded by crtYeYf can function both as an ɛ- and a γ-cyclase when expressed heterologously in E. coli, and thus, it has not one but multiple catalytic functions under the conditions tested. It was plausible to assume that the formation of sarprenoxanthin might be an artifact of overexpression; however, the same carotenoid products were detected in extracts of cells grown under noninduced conditions (data not shown). These results are contradictory to previous reports (22, 24), as neither sarcinaxanthin nor sarprenoxanthin has been identified in cell extracts of C. glutamicum strains or in cell extracts of recombinant E. coli strains expressing decaprenoxanthin biosynthetic genes.
In summary, we have unraveled the biosynthetic pathway of the γ-bicyclic C50 carotenoid sarcinaxanthin in M. luteus, including the function of the crtX gene product as sarcinaxanthin glycosyl transferase. In particular, our studies have generated new and important insight into the diverse catalytic functions of natural carotenoid cyclases as a novel class of enzymes directing biosynthesis of structurally different C50 carotenoids. Recently, the C40 carotenoid biosynthetic pathway of Brevibacterium linens was redesigned and extended by recruitment of heterologous genes leading to production of unexpected carotenoids in E. coli hosts (19). As we see it, C50 cyclases should represent interesting targets for future synthetic biology approaches aiming at generating structurally diverse carotenoids with interesting properties that might not be present in nature.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Research Council of Norway and Promar.
Plasmid pAC-LYC was kindly provided by Francis X. Cunningham, Jr., Department of Cell Biology and Molecular Genetics, University of Maryland.
Footnotes
Published ahead of print on 27 August 2010.
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Brautaset, T., R. Lale, and S. Valla. 2009. Positively regulated bacterial expression systems. Microb. Biotechnol. 2:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton, G. 1998. Overview of carotenoid biosynthesis, p. 13-147. In G. Britton, S. Liaan-Jensen, and H. Pfander (ed.), Carotenoids, vol. 3: biosynthesis and metabolism. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 3.Britton, G., S. Liaaen-Jensen, and H. Pfander (ed.). 2004. Handbook. Birkhäuser Verlag, Basel, Switzerland.
- 4.Cunningham, F. X., and E. Gantt. 1998. Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol Plant Mol. Biol. 49:557-583. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham, F. X., Jr., D. Chamovitz, N. Misawa, E. Gantt, and J. Hirschberg. 1993. Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of beta-carotene. FEBS Lett. 328:130-138. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, F. X., Jr., and E. Gantt. 2007. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth. Res. 92:245-259. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, F. X., Jr., B. Pogson, Z. Sun, K. A. McDonald, D. DellaPenna, and E. Gantt. 1996. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8:1613-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, F. X., Jr., Z. Sun, D. Chamovitz, J. Hirschberg, and E. Gantt. 1994. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell 6:1107-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A., S.-H. Yoon, S.-H. Lee, J.-Y. Kim, D.-K. Oh, and S.-W. Kim. 2007. An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl. Microbiol. Biotechnol. 77:505-512. [DOI] [PubMed] [Google Scholar]
- 10.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, P. D., and P. M. Bramley. 2004. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43:228-265. [DOI] [PubMed] [Google Scholar]
- 12.Fukuoka, S., Y. Ajiki, T. Ohga, Y. Kawanami, and K. Izumori. 2004. Production of dihydroxy C50-carotenoid by Aureobacterium sp. FERM P-18698. Biosci. Biotechnol. Biochem. 68:2646-2648. [DOI] [PubMed] [Google Scholar]
- 13.Gerspacher, M. and H. Pfander. 1989. C45- and C50-carotenoids: synthesis of an optically active cyclic C20-building block and of decaprenoxanthin (=(2R,6R,2′R,6′R)-2′,2′-Bis(4-hydroxy-3-methylbut-2-enyl)-epsilon,epsilon- carotene). Helv. Chim. Acta 72:151-157. [Google Scholar]
- 14.Goodwin, T. 1980. The Biochemistry of the carotenoids, 2nd ed., vol. 1. Chapman and Hall, London, United Kingdom.
- 15.Harker, M., and P. M. Bramley. 1999. Expression of prokaryotic 1-deoxy-D-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett. 448:115-119. [DOI] [PubMed] [Google Scholar]
- 16.Hundle, B., M. Alberti, V. Nievelstein, P. Beyer, H. Kleinig, G. A. Armstrong, D. H. Burke, and J. E. Hearst. 1994. Functional assignment of Erwinia herbicola Eho10 carotenoid genes expressed in Escherichia coli. Mol. Gen. Genet. 245:406-416. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, H., C. L. Braun, and H. Ernst. 2008. The chemistry of novel xanthophyll carotenoids. Am. J. Cardiol. 101:50D-57D. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser, P., P. Surmann, G. Vallentin, and H. Fuhrmann. 2007. A small-scale method for quantitation of carotenoids in bacteria and yeasts. J. Microbiol. Methods 70:142-149. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. H., Y. H. Park, C. Schmidt-Dannert, and P. C. Lee. 2010. Redesign, reconstruction, and directed extension of the Brevibacterium linens C40 carotenoid pathway in Escherichia coli. Appl. Environ. Microbiol. 76:5199-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsh, V. A., S. T. Mayne, U. Peters, N. Chatterjee, M. F. Leitzmann, L. B. Dixon, D. A. Urban, E. D. Crawford, and R. B. Hayes. 2006. A prospective study of lycopene and tomato product intake and risk of prostate cancer. Cancer Epidemiol. Biomarkers Prev. 15:92-98. [DOI] [PubMed] [Google Scholar]
- 21.Klassen, J. L. 2010. Phylogenetic and evolutionary patterns in microbial carotenoid biosynthtesis and revealed comparative genomics. PloS One 5:e11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krubasik, P., M. Kobayashi, and G. Sandmann. 2001. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur. J. Biochem. 268:3702-3708. [DOI] [PubMed] [Google Scholar]
- 23.Krubasik, P., and G. Sandmann. 2000. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 263:423-432. [DOI] [PubMed] [Google Scholar]
- 24.Krubasik, P., S. Takaichi, T. Maoka, M. Kobayashi, K. Masamoto, and G. Sandmann. 2001. Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch. Microbiol. 176:217-223. [DOI] [PubMed] [Google Scholar]
- 25.Lanz, M., B. Bartels, and H. Pfander. 1997. Total synthesis of (all-E,2R,6R,2′R,6′R)- and (all-E,2R,6S,2′R,6′S)-2,2′-Bis(4-hydroxy-3-methylbut-2-enyl)-gamma,gamma-carotene (Sarcinaxanthin). Helv. Chim. Acta 80:804-817. [Google Scholar]
- 26.Liaaen-Jensen, S., O. B. Weeks, R. H. Strang, and D. Thirkell. 1967. Identity of the C-50-carotenoid dehydrogenans-P439 and sarcinaxanthin. Nature 214:379-380. [DOI] [PubMed] [Google Scholar]
- 27.López-Nieto, M. J., J. Costa, E. Peiro, E. Mendez, M. Rodriguez-Saiz, J. L. de la Fuente, W. Cabri, and J. L. Barredo. 2004. Biotechnological lycopene production by mated fermentation of Blakeslea trispora. Appl. Microbiol. Biotechnol. 66:153-159. [DOI] [PubMed] [Google Scholar]
- 28.Mathews, M. M., and W. R. Sistrom. 1960. The function of the carotenoid pigments of Sarcina lutea. Arch. Microbiol. 35:139-146. [DOI] [PubMed] [Google Scholar]
- 29.Matsumura, H., H. Takeyama, E. Kusakabe, J. G. Burgess, and T. Matsunaga. 1997. Cloning, sequencing and expressing the carotenoid biosynthesis genes, lycopene cyclase and phytoene desaturase, from the aerobic photosynthetic bacterium Erythrobacter longus sp. strain Och101 in Escherichia coli. Gene 189:169-174. [DOI] [PubMed] [Google Scholar]
- 30.Miller, N. J., J. Sampson, L. P. Candeias, P. M. Bramley, and C. A. Rice-Evans. 1996. Antioxidant activities of carotenes and xanthophylls. FEBS Lett. 384:240-242. [DOI] [PubMed] [Google Scholar]
- 31.Misawa, N., M. Nakagawa, K. Kobayashi, S. Yamano, Y. Izawa, K. Nakamura, and K. Harashima. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172:6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naguib, Y. M. 2000. Antioxidant activities of astaxanthin and related carotenoids. J. Agric. Food Chem. 48:1150-1154. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Concepcion, M., and A. Boronat. 2002. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130:1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruther, A., N. Misawa, P. Boger, and G. Sandmann. 1997. Production of zeaxanthin in Escherichia coli transformed with different carotenogenic plasmids. Appl. Microbiol. Biotechnol. 48:162-167. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Schumann, G., H. Nurnberger, G. Sandmann, and H. Krugel. 1996. Activation and analysis of cryptic crt genes for carotenoid biosynthesis from Streptomyces griseus. Mol. Gen. Genet. 252:658-666. [DOI] [PubMed] [Google Scholar]
- 37.Sletta, H., A. Nedal, T. E. Aune, H. Hellebust, S. Hakvag, R. Aune, T. E. Ellingsen, S. Valla, and T. Brautaset. 2004. Broad-host-range plasmid pJB658 can be used for industrial-level production of a secreted host-toxic single-chain antibody fragment in Escherichia coli. Appl. Environ. Microbiol. 70:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sommerburg, O. G., W. G. Siems, J. S. Hurst, J. W. Lewis, D. S. Kliger, and F. J. van Kuijk. 1999. Lutein and zeaxanthin are associated with photoreceptors in the human retina. Curr. Eye Res. 19:491-495. [DOI] [PubMed] [Google Scholar]
- 39.Stafsnes, M. H., K. D. Josefsen, G. Kildahl-Andersen, S. Valla, T. E. Ellingsen, and P. Bruheim. 2010. Isolation and characterization of marine pigmented bacteria from Norwegian coastal waters and screening for carotenoids with UVA-blue light absorbing properties. J. Microbiol. 48:16-23. [DOI] [PubMed] [Google Scholar]
- 40.Stickforth, P., S. Steiger, W. R. Hess, and G. Sandmann. 2003. A novel type of lycopene epsilon-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch. Microbiol. 179:409-415. [DOI] [PubMed] [Google Scholar]
- 41.Tao, L., H. Yao, and Q. Cheng. 2007. Genes from a Dietzia sp. for synthesis of C40 and C50 beta-cyclic carotenoids. Gene 386:90-97. [DOI] [PubMed] [Google Scholar]
- 42.Tobias, A. V., and F. H. Arnold. 2006. Biosynthesis of novel carotenoid families based on unnatural carbon backbones: a model for diversification of natural product pathways. Biochim. Biophys. Acta 1761:235-246. [DOI] [PubMed] [Google Scholar]
- 43.Tripathi, G., and S. K. Rawal. 1998. Simple and efficient protocol for isolation of high molecular weight DNA from Streptomyces aureofaciens. Biotechnol. Lett. 12:629-631. [Google Scholar]
- 44.Vertès, A. A., Y. Asai, M. Inui, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1994. Transposon mutagenesis of coryneform bacteria. Mol. Gen. Genet. 245:397-405. [DOI] [PubMed] [Google Scholar]
- 45.Wang, C., M. K. Oh, and J. C. Liao. 2000. Directed evolution of metabolically engineered Escherichia coli for carotenoid production. Biotechnol. Prog. 16:922-926. [DOI] [PubMed] [Google Scholar]
- 46.Wang, W., L. Shinto, W. E. Connor, and J. F. Quinn. 2008. Nutritional biomarkers in Alzheimer's disease: the association between carotenoids, n-3 fatty acids, and dementia severity. J. Alzheimers Dis. 13:31-38. [DOI] [PubMed] [Google Scholar]
- 47.Young, M., V. Artsatbanov, H. R. Beller, G. Chandra, K. F. Chater, L. G. Dover, E. B. Goh, T. Kahan, A. S. Kaprelyants, N. Kyrpides, A. Lapidus, S. R. Lowry, A. Lykidis, J. Mahillon, V. Markowitz, K. Mavromatis, G. V. Mukamolova, A. Oren, J. S. Rokem, M. C. Smith, D. I. Young, and C. L. Greenblatt. 2010. Genome sequence of the Fleming strain of Micrococcus luteus, a simple free-living actinobacterium. J. Bacteriol. 192:841-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.