Abstract
Glycopeptidolipids (GPLs) are one of the major glycolipid components present on the surface of Mycobacterium avium complex (MAC) that belong to opportunistic pathogens distributed in the natural environment. The serovars of MAC, up to around 30 types, are defined by the variable oligosaccharide portions of the GPLs. Epidemiological studies show that serovar 4 is the most prevalent type, and the prognosis of pulmonary disease caused by serovar 4 is significantly worse than that caused by other serovars. However, little is known about the biosynthesis of serovar 4-specific GPL, particularly the formation of the oligosaccharide portion that determines the properties of serovar 4. To investigate the biosynthesis of serovar 4-specific GPL, we focused on one segment that included functionally unknown genes in the GPL biosynthetic gene cluster of a serovar 4 strain. In this segment, a putative hemolytic protein gene, hlpA, and its downstream gene were found to be responsible for the formation of the 4-O-methyl-rhamnose residue, which is unique to serovar 4-specific GPL. Moreover, functional characterization of the hlpA gene revealed that it encodes a rhamnosyltransferase that transfers a rhamnose residue via 1→4 linkage to a fucose residue of serovar 2-specific GPL, which is a key pathway leading to the synthesis of oligosaccharide of serovar 4-specific GPL. These findings may provide clues to understanding the biological role of serovar 4-specific GPL in MAC pathogenicity and may also provide new insights into glycosyltransferase, which generates structural and functional diversity of GPLs.
The genus Mycobacterium has a unique feature in the cell envelope that contains a multilayered structure consisting of peptidoglycan, mycolyl-arabinogalactan complex, and surface glycolipids (8, 12). It is known that these components play a role in protection from environmental stresses, such as antimicrobial agents and host immune responses (8, 12). Some of them are recognized as pathogenic factors related to mycobacterial diseases, such as tuberculosis and leprosy (8, 12). In case of nontuberculous mycobacteria that are widely distributed in the natural environment as opportunistic pathogens, glycopeptidolipids (GPLs) are abundantly present on the cell envelope as surface glycolipids (34). GPLs have a core structure in which a fatty acyl-tetrapeptide is glycosylated with 6-deoxy-talose (6-d-Tal) and O-methyl-rhamnose (O-Me-Rha) (2, 5, 13). This structure is common to all types of GPLs, and GPLs with this structure that have not undergone further glycosylation are termed non-serovar-specific GPLs (nsGPLs) (2, 5, 13). Structural diversity generated by further glycosylations, such as rhamnosylation, fucosylation, and glucosylation, is observed for the oligosaccharide portion linked to the 6-d-Tal residue of nsGPLs from Mycobacterium avium complex (MAC), a member of the nontuberculous mycobacteria consisting of two species, M. avium and M. intracellulare (2, 5, 34). Consequently, these nsGPLs with varied oligosaccharides lead to the formation of the serovar-specific GPLs (ssGPLs) that define around 30 types of MAC serovars (10).
The properties of MAC serovars are known to be notably different from each other and also to be closely associated with the pathogenicity of MAC (3, 6, 18, 30, 31, 32). Various epidemiological studies indicate that serovar 4 is the most prevalent type and is also one of the serovars frequently isolated from AIDS patients (1, 20, 33, 36). Additionally, pulmonary MAC disease caused by serovar 4 is shown to exhibit a poorer prognosis than that caused by other serovars (23). With respect to host immune responses to MAC infection, serovar 4-specific GPL is reported to have characteristic features that are in contrast to those of other ssGPLs (21, 30). Structurally, serovar 4-specific GPL contains a unique oligosaccharide in which the oligosaccharide of serovar 2-specific GPL is further glycosylated with 4-O-methyl-rhamnose (4-O-Me-Rha) residue through a 1→4 linkage (Table 1) (24). Therefore, it is thought that the presence of 4-O-Me-Rha and its linkage position are important in exhibiting the specificity of biological activities. The biosynthesis of the oligosaccharide portion in several ssGPLs is currently being clarified (15, 16, 17, 25, 26), while that of serovar 4-specific GPL is still unresolved. In this study, we have focused on the genomic region predicted to be associated with GPL biosynthesis in the serovar 4 strain and explored the key genes responsible for the formation of 4-O-Me-Rha that might determine the specific properties of MAC serovar 4.
TABLE 1.
Oligosaccharide structures of serovar 2- and 4-specific GPLs
MATERIALS AND METHODS
Bacterial strains, culture conditions, and DNA manipulation.
Table 2 indicates the bacterial strains and vectors used in this study. MAC strains were grown in Middlebrook 7H9 broth (Difco) with 0.05% Tween 80 supplemented with 10% Middlebrook ADC enrichment (BBL). For GPL production, Mycobacterium smegmatis strains were cultured in 2× YT broth (16 g tryptone, 10 g yeast extract, and 5 g NaCl per liter) with 0.2% Tween 80. DNA manipulation of M. smegmatis strains was conducted as previously described (27). PCR amplification was done by two-step PCR using TaKaRa LA Taq with GC buffer, with the following program: denaturation at 98°C for 20 s and annealing-extension at 68°C for an appropriate time depending on the length of the targeted region. Escherichia coli strain DH5α was used for the routine manipulation and propagation of plasmid DNA. Antibiotics were added as required: kanamycin, 50 μg/ml for E. coli and 25 μg/ml for M. smegmatis; hygromycin B, 150 μg/ml for E. coli and 75 μg/ml for M. smegmatis. Oligonucleotide primers used in this study are listed in Table 3.
TABLE 2.
Bacterial strains and vectors used in this study
| Strain or vector | Characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli DH5α | Cloning host | TaKaRa |
| M. smegmatis mc2155 | Expression host | 29 |
| M. intracellulare ATCC 35767 | MAC serovar 4 strain | 35 |
| M. avium JATA51-01 | Source of the rtfA gene | 26 |
| Vectors | ||
| pYM301a | Site-specific integrating mycobacterial vector carrying an hsp60 promoter cassette | 25 |
| pMV261a | E. coli-Mycobacterium shuttle vector carrying an hsp60 promoter cassette with an AflII site | This study |
| pMVΔmtfF | Source of mdhtA, merA, and gtfD genes | 26 |
| pMV-rtfA-mdhtA-merA-gtfD | pMV261a carrying rtfA, mdhtA, merA, and gtfD genes | This study |
| pYM-hlpA | pYM301a carrying the hlpA gene | This study |
| pYM-hlpA-orf2 | pYM301a carrying the hlpA gene and ORF2 | This study |
| pYM-orf3-orf4-orf5 | pYM301a carrying ORF3, ORF4, and ORF5 | This study |
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequencea | Restriction site |
|---|---|---|
| RTFA-S | 5′-CGGGATCCCATGAAATTTGCTGTGGCAAG-3′ | BamHI |
| RTFA-A | 5′-AACTGCAGCTCAGCGACTTCGCTGCGCTTC-3′ | PstI |
| MDHTA-S2 | 5′-GCTCTAGACTGCAGAAAAACCAACTTCTACTGCCTGACCTG-3′ | PstI |
| GTFD-A2 | 5′-GGAATTCTTAAGTCTACGGTTCTGCGCTTCGTTCTTTG-3′ | AflII |
| HLPA-S | 5′-GGAATTCGTGACAACGACGCCACCAGT-3′ | EcoRI |
| HLPA-A | 5′-CCATCGATACTACGCTGCCGCGCTAGGCG-3′ | ClaI |
| ORF2-A | 5′-CCCAAGCTTCTCAGACTCTAACGTACAGTTC-3′ | HindIII |
| ORF3-S | 5′-CACCTGCAGAAATGACCGCCACAACCAGGGC-3′ | PstI |
| ORF5-A | 5′-GCAGAATTCCTACGGCGCCAATTCGATGAG-3′ | EcoRI |
| GTFB-S1 | 5′-GGAACTCCTGCACCTTGGGGCCGT-3′ | |
| MDHTA-A2 | 5′-GGTGCGGGTCAACGTAGAGGTG-3′ |
Underlining indicates the restriction site.
Construction of expression vectors.
For generation of the serovar 2-specific GPL (GPL-S2)-producing strain, the vector possessing rtfA, mdhtA, merA, and gtfD genes was constructed. The rtfA gene was amplified from genomic DNA of M. avium strain JATA51-01 using primers RTFA-S and RTFA-A. The mdhtA, merA, and gtfD genes were amplified as one operon from the previously constructed vector pMVΔmtfF using primers MDHTA-S2 and GTFD-A2 (26). After construction of pMV261a, in which an AflII site was introduced into pMV261, the above two PCR products were digested with each restriction enzyme and cloned into the BamHI-PstI and PstI-AflII sites of pMV261a to give pMV-rtfA-mdhtA-merA-gtfD (Table 2). The fragments for construction of expression vectors were amplified from genomic DNA of MAC serovar 4 strain (ATCC 35767) using the following primers: HLPA-S and HLPA-A for hlpA, HLPA-S and ORF2-A for hlpA-orf2, and ORF3-S and ORF5-A for orf3-orf4-orf5. These PCR products were digested with each restriction enzyme and cloned into the EcoRI-ClaI, EcoRI-HindIII, and PstI-EcoRI sites of pYM301a to give pYM-hlpA, pYM-hlpA-orf2, and pYM-orf3-orf4-orf5, respectively (Table 2).
Isolation and purification of GPLs.
To isolate whole-lipid extracts, harvested bacterial cells were mixed with CHCl3/CH3OH (2:1 [vol/vol[) for several hours at room temperature. The extracts in organic phase were separated by adding water and evaporated until dry. To remove the lipid components except for GPLs, the whole-lipid extracts were subjected to mild alkaline hydrolysis to prepare the crude GPLs as previously described (27, 28). For analytical thin-layer chromatography (TLC), crude GPLs on silica gel 60 plates (Merck) were developed with CHCl3/CH3OH/H2O (30:8:1 [vol/vol/vol]), followed by spraying with 10% H2SO4 and charring. Purified GPLs were prepared from crude GPLs by preparative TLC on the same plates and extracted from the bands corresponding to each GPL. To determine the linkage position of sugar moieties, perdeuteriomethylation was performed for purified GPLs as previously described (7, 11, 15).
GC-MS and MALDI-TOF MS analysis.
Purified and perdeuteriomethylated GPLs were hydrolyzed in 2 M trifluoroacetic acid (2 h, 120°C), and the released sugars were reduced with NaBD4 and then acetylated with pyridine/acetic anhydride (1:1 [vol/vol]) at room temperature overnight. The resulting alditol acetates were analyzed by gas chromatography-mass spectrometry (GC-MS) with a GCMS-QP2010 (Shimadzu) equipped with a SP-2380 column (Supelco) using helium gas. The temperature program was from 52 to 172°C with an increase in temperature of 40°C/min, 172 to 223°C at 3°C/min, and then 223 to 270°C at 40°C/min. To determine the total mass of the purified GPLs, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectra were acquired with an Ultraflex II (Bruker Daltonics). Samples were dissolved in chloroform-methanol (2:1 [vol/vol]) at a concentration of 1 mg/ml, 1 μl was applied directly to the sample plate, and then 1 μl of 10 mg/ml 2,5-dihydroxybenzoic acid in chloroform/methanol (1:1 [vol/vol]) was added as a matrix. The purified GPL was analyzed in the reflectron mode with an accelerating voltage operating in a positive mode of 20 kV (17).
Nucleotide sequence accession number.
The 6.8-kb genomic region amplified from the MAC serovar 4 strain (ATCC 35767) by using primers GTFB-S1 and MDHTA-A2 has been deposited in the DDBJ nucleotide sequence database under accession no. AB550236.
RESULTS
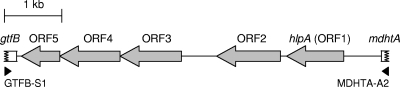
Previously, the A5 strain, one of the MAC serovar 4 strains, was reported to contain a genomic region similar to the GPL biosynthetic gene cluster identified in other serovars (22). However, to date, there are no studies clarifying the biosynthetic pathways involved in the formation of 4-O-Me-Rha, which is unique to serovar 4-specific GPL. To explore this glycosylation pathway, we focused on one segment interposed with the gtfB and mdhtA genes whose organization was shown to vary in strains of other serovars (14, 22). In this study, using another serovar 4 strain, ATCC 35767, whose genomic information is unknown, we designed various primers for PCR amplification of a focused segment based on the sequences from other serovar strains. After the testing of primer pairs, a 6.8-kb fragment was amplified with primers GTFB-S1 and MDHTA-A2 (Fig. 1). Nucleotide sequences of the amplified fragments were similar to that of the GPL biosynthetic gene cluster from the A5 strain (94% identity in nucleotide sequences) (GenBank accession no. AY130970.1). This segment contains five complete open reading frame (ORF) genes (Fig. 1): the ORF1 gene, similar to a putative hemolytic protein gene (hlpA) previously found in the GPL biosynthetic gene cluster of the serovar 2 strain (69% identity in amino acid sequences) (GenBank accession no. AF125999.1) (14); the ORF2 gene, an undefined gene showing low similarity to some O-methyltransferases; and the ORF3, ORF4, and ORF5 genes, with amino acid sequences almost identical to those of three proteins, including GtfTB, which were previously identified as biosynthetic enzymes for serovar 8-specific GPL (GenBank accession no. AB437139.1) (25).
FIG. 1.
Organization of the 6.8-kb genomic segment isolated from MAC serovar 4 strain (ATCC 35767). Filled triangles indicate the primers used for PCR amplification.
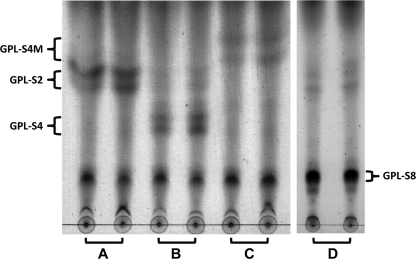
Prior to functional analysis of each ORF, it was necessary to prepare a strain producing the substrate for the enzymes participating in the biosynthesis of serovar 4-specific GPL. Since serovar 4-specific GPL has a structure in which the terminal Fuc residue of serovar 2-specific GPL is further glycosylated with 4-O-Me-Rha, we created a recombinant M. smegmatis strain (termed MS-S2) by introducing the plasmid vector pMV-rtfA-mdhtA-merA-gtfD possessing M. avium rtfA, mdhtA, merA, and gtfD genes, which were previously shown to convert nsGPLs to serovar 2-specific GPL with a terminal Fuc residue (termed GPL-S2) (26). For five ORFs, we first examined the function of the ORF1 (termed hlpA) and its downstream ORF2 gene by TLC analysis of recombinant strains, because these have not been functionally defined and it is difficult to predict the role of each gene. In comparison with the profile of the control strain (MS-S2/pYM301a) (Fig. 2, lane A), the new products (GPL-S4) were observed for the strain with the hlpA gene introduced (MS-S2/pYM-hlpA) (Fig. 2, lane B). Moreover, when the expression vector covering both hlpA and ORF2 was introduced into MS-S2 (MS-S2/pYM-hlpA-orf2), another new product (GPL-S4M) appeared (Fig. 2, lane C). These observations indicated that GPL-S2 was converted to structurally different compounds by the expression of hlpA and that the compounds generated by hlpA were further modified by ORF2. As for the ORF3, ORF4, and ORF5 genes, which show a high similarity to the biosynthetic genes for serovar 8-specific GPL, we further generated a strain having three ORFs (MS-S2/pYM-orf3-orf4-orf5) and examined the GPL production by TLC analysis (Fig. 2, lane D). The results indicated the appearance of known product GPL-S8, previously shown to have a sugar residue of serovar 8-specific GPL, with no GPL-S4 and GPL-S4M (25), demonstrating that the enzymes encoded by three ORFs might act on the serovar 1-specific GPL which was produced as a precursor of GPL-S2 and subsequently yielded GPL-S8.
FIG. 2.
TLC analysis of crude GPL extracts from recombinant M. smegmatis strains MS-S2/pYM301a (A), MS-S2/pYM-hlpA (B), MS-S2/pYM-hlpA-orf2 (C), and MS-S2/pYM-orf3-orf4-orf5 (D). GPL extracts were prepared from the total lipid fraction after a mild alkaline hydrolysis step. Each recombinant strain was tested by two samples derived from independent colonies. Samples were spotted and developed in CHCl3-CH3OH-H2O (30:8:1 [vol/vol/vol]).
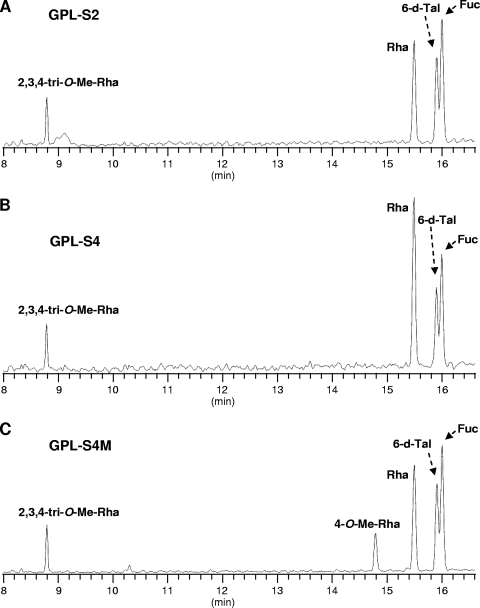
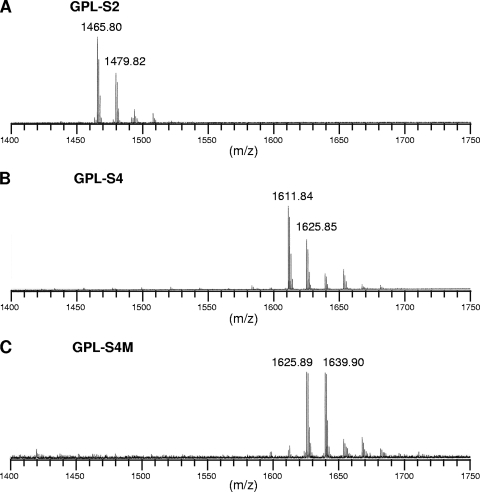
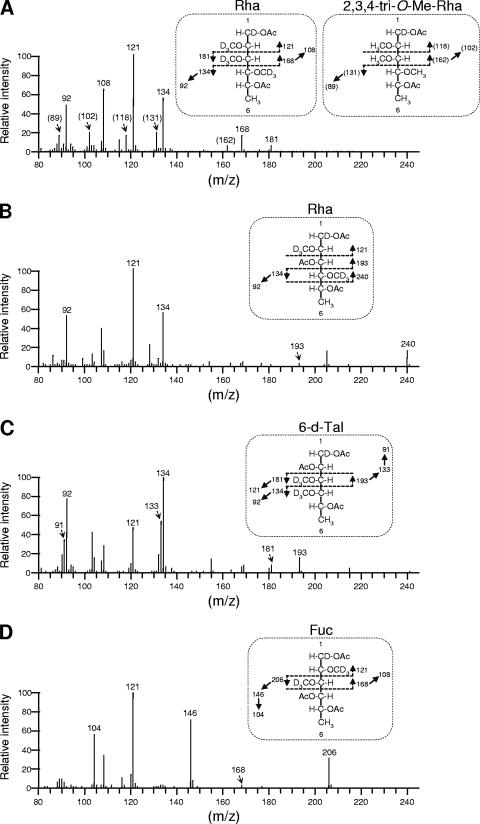
Because the compounds produced by hlpA and ORF2 were structurally unidentified, we performed a GC-MS analysis of the products GPL-S2, GPL-S4, and GPL-S4M, which were purified from recombinant strain MS-S2/pYM301a, MS-S2/pYM-hlpA, and MS-S2/pYM-hlpA-orf2, respectively. Although two spots were seen for each product, this might be due to a different methylation pattern for the fatty acid portion, which is often observed with GPL biosynthesis of M. smegmatis and does not affect oligosaccharide structure (19, 25). In GC-MS profiles of GPL-S2 and GPL-S4, the classes of the detected sugar residues, Fuc, 6-d-Tal, Rha, and 2,3,4-tri-O-Me-Rha, were found to be identical to each other (Fig. 3 A and B). However, it was observed that the intensity of the Rha residue in GPL-S4 was higher than that of the other sugars, while in GPL-S2, the intensity of the Rha residue was lower than that of Fuc, indicating that the proportion of Rha content in GPL-S4 was relatively large compared to that in GPL-S2. These results implied that the hlpA gene mediates the transfer of an additional Rha residue to GPL-S2. In contrast, the profiles of GPL-S4M showed the presence of 4-O-Me-Rha that is specifically observed for serovar 4-specific GPL (Fig. 3C), demonstrating that ORF2 encodes a rhamnosyl 4-O-methyltransferase and that both genes are responsible for the formation of the unique sugar residue of serovar 4-specific GPL. Furthermore, we confirmed the molecular masses of products GPL-S2, GPL-S4, and GPL-S4M by MALDI-TOF MS analysis (Fig. 4). Each product contained two main pseudomolecular ions ([M + Na]+) with 14 mass unit differences, indicating the presence or absence of the methyl group in the fatty acid portion as described above. Thus, the results revealed that the mass unit difference between GPL-S2 (m/z 1,465.80, 1,479.82) and GPL-S4 (m/z 1,611.84, 1,625.85) was 146 and that between GPL-S2 and GPL-S4M (m/z 1,625.89, 1,639.90) was 160, demonstrating that the Rha and 4-O-Me-Rha residues were further added to the GPL-S2 to yield GPL-S4 and GPL-S4M, respectively.
FIG. 3.
GC-MS of alditol acetate derivatives from GPL-S2 (A), GPL-S4 (B), and GPL-S4M (C), which were purified from GPL extracts of recombinant M. smegmatis strains MS-S2/pYM301a, MS-S2/pYM-hlpA, and MS-S2/pYM-hlpA-orf2, respectively.
FIG. 4.
MALDI-TOF MS of GPL-S2 (A), GPL-S4 (B), and GPL-S4M (C), which were purified from GPL extracts of recombinant M. smegmatis strains MS-S2/pYM301a, MS-S2/pYM-hlpA, and MS-S2/pYM-hlpA-orf2, respectively.
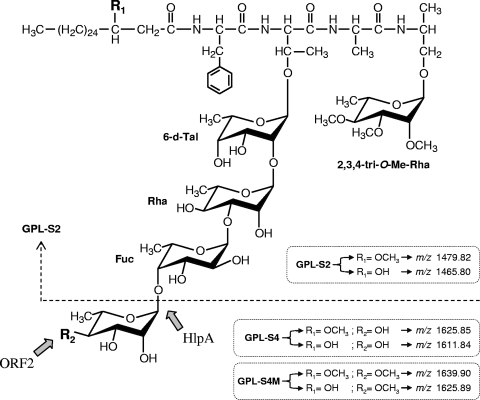
The results from TLC, GC-MS, and MALDI-TOF MS analyses strongly suggested that hlpA and ORF2 are involved in the formation of 4-O-Me Rha. However, it is not clear whether the hlpA gene product functions as a glycosyltransferase that transfers a Rha via 1→4 linkage to a Fuc residue, which is observed only for serovar 4-specific GPL. To elucidate the function of hlpA, we determined the linkage of sugar moieties of GPL-S4 produced by recombinant strain MS-S2/pYM-hlpA (Fig. 2, lane B). The purified GPL-S4 was subjected to perdeuteriomethylation followed by GC-MS and gave four peaks corresponding to Fuc, 6-d-Tal, Rha, and 2,3,4-tri-O-Me-Rha (data not shown). The spectra of Rha and 6-d-Tal demonstrated that the linkage position between these two sugar residues is commonly observed in the oligosaccharide of all ssGPLs, and position C-3 of Rha is linked to the next one, which is consistent with the data previously reported (Fig. 5 B and C) (25, 26). In addition, the detection of fragment ions at m/z 121, 168, and 206 in spectra of Fuc indicated that its positions C-2 and C-3 were deuteriomethylated (Fig. 5D), meaning that position C-1 of Fuc is linked to position C-3 of Rha and position C-4 of Fuc is linked to the next one. These observations were supported by the fact that GPL-S4 was structurally based on the oligosaccharide of serovar 2-specific GPL. The peak of 2,3,4-tri-O-Me-Rha was found to include mixed fragment ions (Fig. 5A). A group of fragment ions corresponding to the spectra of 2,3,4-tri-O-Me-Rha linked to alaninol of tetrapeptide was observed. The remaining fragment ions at m/z 121, 134, 168, and 181 indicate the presence of deuteriomethyl groups at positions C-2, C-3, and C-4 of the other Rha that is linked at the terminus of oligosaccharide in GPL-S4. These results indicate that position C-1 of terminal Rha is linked to position C-4 of Fuc. Accordingly, the oligosaccharide structures of GPL-S4 were determined to have Rha-(1→4)-Fuc-(1→3)-Rha-(1→2)-6-d-Tal at d-allo-Thr, demonstrating that hlpA encodes a rhamnosyltransferase that transfers a Rha residue via 1→4 linkage to a Fuc residue of serovar 2-specific GPL (Fig. 6).
FIG. 5.
GC-MS spectra and fragment ion assignments of 2,3,4-tri-O-Me-Rha (A), Rha (A and B), 6-d-Tal (C), and Fuc (D), which are derived from alditol acetates of sugars released from deuteriomethylated GPL-S4. Ac, acetate; D, deuterium.
FIG. 6.
Proposed structures and biosynthetic pathways for GPL-S2, GPL-S4, and GPL-S4M. Parentheses indicate structural differences between three compounds, which are deduced from MALDI-TOF MS analyses {pseudomolecular ions ([M + Na]+)}.
DISCUSSION
It is known that serovar 4 is the most prevalent type, and serovar 4-specific GPL, particularly its oligosaccharide portion, plays a role in exhibiting the specific properties that belong to pathogenic factors. However, to date, the biosynthesis of its oligosaccharide portion has not been clarified. In this study, structural determination of three recombinant products, GPL-S2, GPL-S4, and GPL-S4M, revealed that hlpA and its downstream gene (ORF2) in the GPL biosynthetic gene cluster are involved in the formation of 4-O-methyl Rha, which is unique to serovar 4-specific GPL (Fig. 6). Previously, it was reported that the GPL biosynthetic gene cluster of MAC serovar 2 strains contained one gene whose amino acid sequences are similar to that of hlpA with 69% identity (14). This has been regarded as the gene not associated with GPL biosynthesis, because its amino acid sequences are similar to those of hemolytic proteins distributed in some species of bacteria (4). Thus, as shown in Fig. 6, it was surprising that hlpA from serovar 4 was found to encode a rhamnosyltransferase that plays a critical role in the pathway leading from serovar 2-specific GPL to serovar 4-specific GPL. For mycobacterium species, a BLAST analysis of HlpA revealed that its homologues are seen only in MAC serovar 2 and not in other species, including Mycobacterium tuberculosis. When we tested the function of HlpA from serovar 2, it did not serve as a glycosyltransferase like HlpA from serovar 4 (data not shown). At present, the function of HlpA from serovar 2 is still unclear, because the biosynthesis of the oligosaccharide portion in serovar 2-specific GPL has been fully elucidated (14, 26). The oligosaccharide structure of serovar 2-specific GPL is basic for several ssGPLs, including serovar 4-specific GPL. In the biosynthetic gene cluster of serovar 2-specific GPL, several insertion sequence (IS) elements are observed, raising the possibility that the HlpA from serovar 2 is retained through genomic alterations that induce biosynthetic changes from other ssGPLs to serovar 2-specific GPL. Therefore, HlpA in the serovar 2 strain originally might function as a glycosyltransferase in the biosynthesis of oligosaccharides of other serovars.
Most HlpA homologues are putatively categorized as hemolytic proteins because they are similar to one protein from Prevotella intermedia, which is actually proved to have hemolytic activity (4). Since the amino acid sequences of HlpA show 38% identity and 54% similarity to the above protein of P. intermedia, we predicted that HlpA also possesses hemolytic activity as an additional function. However, none was detected when hlpA was expressed in M. smegmatis and E. coli by plate assay using a sheep blood agar plate (data not shown). A BLAST analysis of HlpA homologues showed that they also contained a partial motif of some glycosyltransferases and methyltransferases. Therefore, it is envisaged that the evolutionary ancestor of HlpA might have lost hemolytic activity in MAC or, conversely, have been altered to retain it in P. intermedia in the course of phylogenetic evolution between these bacterial species.
Serovar 4 strains, including ATCC 35767, have been recognized as strains producing the serovar 4-specific GPL but not the serovar 8-specific GPL (24, 35). However, as shown in Fig. 1, we found that the GPL biosynthetic gene cluster contains three known genes (the ORF3, ORF4, and ORF5 genes) previously identified as biosynthetic genes responsible for the formation of 4,6-O-(1-carboxyethylidene)-3-O-methyl glucose residue in the oligosaccharide of serovar 8-specific GPL (25). TLC analysis showed that overexpression of three ORFs potentially produces the serovar 8-specific GPL, including the 4,6-O-(1-carboxyethylidene)-3-O-methyl glucose residue (Fig. 2, lane D), demonstrating that in the serovar 4 strain, there is inefficient expression of three genes, which might be caused by genomic alterations affecting their transcription, resulting in the loss of serovar 8-specific GPL. Moreover, HlpA homologues are often found in several species of cyanobacteria but not in other bacterial groups and mycobacterium species, implying the occurrence of a certain kind of “horizontal gene transfer” between these environmental bacteria. Thus, MAC seemed to incorporate foreign genes or realign preexisting genes to modify the oligosaccharide structures of GPLs for their survival in a varied environment. In terms of sugar composition and linkage affecting the properties of ssGPLs, the functional aspects of the 4-O-methyl-Rha residue, which influence the interactions with the host cell, are still unclear. In addition, the sugar linkage Rha-(1→4)-Fuc is seen only in serovar 4-specific GPL and not in other ssGPLs, suggesting that it might generate unique properties that differ notably from those generated by other sugar linkages. Also, the rarity of this sugar linkage could be one of the factors that define the specificity of MAC serovar 4, which would be resolved by further studies, including the generation of an hlpA knockout mutant. For functional characterization of hlpA and ORF2, we have adopted the gene expression experiment using the M. smegmatis strain. Further enzymatic analyses, such as in vitro testing with recombinant proteins, would confirm our results. Taken together, these findings may contribute to understanding the mechanism for generation of structural and functional diversity of GPLs as well as their biological role in MAC.
Acknowledgments
This research was partially supported by a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Science and Technology of Japan and by a Research on Emerging and Re-Emerging Infectious Diseases grant from the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print on 3 September 2010.
REFERENCES
- 1.Askgaard, D. S., S. B. Giese, S. Thybo, A. Lerche, and J. Bennedsen. 1994. Serovars of Mycobacterium avium complex isolated from patients in Denmark. J. Clin. Microbiol. 32:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspinall, G. O., D. Chatterjee, and P. J. Brennan. 1995. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51:169-242. [DOI] [PubMed] [Google Scholar]
- 3.Barrow, W. W., T. L. Davis, E. L. Wright, V. Labrousse, M. Bachelet, and N. Rastogi. 1995. Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infect. Immun. 63:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beem, J. E., W. E. Nesbitt, and K. P. Leung. 1999. Cloning of Prevotella intermedia loci demonstrating multiple hemolytic domains. Oral Microbiol. Immunol. 14:143-152. [DOI] [PubMed] [Google Scholar]
- 5.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium: characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. [PubMed] [Google Scholar]
- 6.Birkness, K. A., W. E. Swords, P. H. Huang, E. H. White, C. S. Dezzutti, R. B. Lal, and F. D. Quinn. 1999. Observed differences in virulence-associated phenotypes between a human clinical isolate and a veterinary isolate of Mycobacterium avium. Infect. Immun. 67:4895-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorndal, H., C. G. Hellerqvist, B. Lindberg, and S. Svensson. 1970. Gas-liquid chromatography and mass spectrometry in methylation analysis of polysaccharides. Angew. Chem. Internat. Ed. Engl. 9:610-619. [Google Scholar]
- 8.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 9.Camphausen, R. T., R. L. Jones, and P. J. Brennan. 1986. Structure and relevance of the oligosaccharide hapten of Mycobacterium avium serotype 2. J. Bacteriol. 168:660-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 58:2018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 12.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 13.Daffe, M., M. A. Laneelle, and G. Puzo. 1983. Structural elucidation by field desorption and electron-impact mass spectrometry of the C-mycosides isolated from Mycobacterium smegmatis. Biochim. Biophys. Acta 751:439-443. [DOI] [PubMed] [Google Scholar]
- 14.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 149:2797-2807. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein, T. M., F. S. Silbaq, D. Chatterjee, N. J. Kelly, P. J. Brennan, and J. T. Belisle. 1998. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J. Bacteriol. 180:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara, N., N. Nakata, S. Maeda, T. Naka, M. Doe, I. Yano, and K. Kobayashi. 2007. Structural characterization of a specific glycopeptidolipid containing a novel N-acyl-deoxy sugar from Mycobacterium intracellulare serotype 7 and genetic analysis of its glycosylation pathway. J. Bacteriol. 189:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara, N., N. Nakata, T. Naka, I. Yano, M. Doe, D. Chatterjee, M. McNeil, P. J. Brennan, K. Kobayashi, M. Makino, S. Matsumoto, H. Ogura, and S. Maeda. 2008. Structural analysis and biosynthesis gene cluster of an antigenic glycopeptidolipid from Mycobacterium intracellulare. J. Bacteriol. 190:3613-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horgen, L., E. L. Barrow, W. W. Barrow, and N. Rastogi. 2000. Exposure of human peripheral blood mononuclear cells to total lipids and serovar-specific glycopeptidolipids from Mycobacterium avium serovars 4 and 8 results in inhibition of TH1-type responses. Microb. Pathog. 29:9-16. [DOI] [PubMed] [Google Scholar]
- 19.Jeevarajah, D., J. H. Patterson, M. J. McConville, and H. Billman-Jacobe. 2002. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148:3079-3087. [DOI] [PubMed] [Google Scholar]
- 20.Julander, I., S. Hoffner, B. Petrini, and L. Ostlund. 1996. Multiple serovars of Mycobacterium avium complex in patients with AIDS. APMIS 104:318-320. [PubMed] [Google Scholar]
- 21.Kano, H., T. Doi, Y. Fujita, H. Takimoto, I. Yano, and Y. Kumazawa. 2005. Serotype-specific modulation of human monocyte functions by glycopeptidolipid (GPL) isolated from Mycobacterium avium complex. Biol. Pharm. Bull. 28:335-339. [DOI] [PubMed] [Google Scholar]
- 22.Krzywinska, E., and J. S. Schorey. 2003. Characterization of genetic differences between Mycobacterium avium subsp. avium strains of diverse virulence with a focus on the glycopeptidolipid biosynthesis cluster. Vet. Microbiol. 91:249-264. [DOI] [PubMed] [Google Scholar]
- 23.Maekura, R., Y. Okuda, A. Hirotani, S. Kitada, T. Hiraga, K. Yoshimura, I. Yano, K. Kobayashi, and M. Ito. 2005. Clinical and prognostic importance of serotyping Mycobacterium avium-Mycobacterium intracellulare complex isolates in human immunodeficiency virus-negative patients. J. Clin. Microbiol. 43:3150-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNeil, M., A. Y. Tsang, and P. J. Brennan. 1987. Structure and antigenicity of the specific oligosaccharide hapten from the glycopeptidolipid antigen of Mycobacterium avium serotype 4, the dominant mycobacterium isolated from patients with acquired immune deficiency syndrome. J. Biol. Chem. 262:2630-2635. [PubMed] [Google Scholar]
- 25.Miyamoto, Y., T. Mukai, Y. Maeda, M. Kai, T. Naka, I. Yano, and M. Makino. 2008. The Mycobacterium avium complex gtfTB gene encodes a glucosyltransferase required for the biosynthesis of serovar 8-specific glycopeptidolipid. J. Bacteriol. 190:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto, Y., T. Mukai, Y. Maeda, N. Nakata, M. Kai, T. Naka, I. Yano, and M. Makino. 2007. Characterization of the fucosylation pathway in the biosynthesis of glycopeptidolipids from Mycobacterium avium complex. J. Bacteriol. 189:5515-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto, Y., T. Mukai, F. Takeshita, N. Nakata, Y. Maeda, M. Kai, and M. Makino. 2004. Aggregation of mycobacteria caused by disruption of fibronectin-attachment protein-encoding gene. FEMS Microbiol. Lett. 236:227-234. [DOI] [PubMed] [Google Scholar]
- 28.Patterson, J. H., M. J. McConville, R. E. Haites, R. L. Coppel, and H. Billman-Jacobe. 2000. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275:24900-24906. [DOI] [PubMed] [Google Scholar]
- 29.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 30.Sweet, L., and J. S. Schorey. 2006. Glycopeptidolipids from Mycobacterium avium promote macrophage activation in a TLR2- and MyD88-dependent manner. J. Leukoc. Biol. 80:415-423. [DOI] [PubMed] [Google Scholar]
- 31.Sweet, L., W. Zhang, H. Torres-Fewell, A. Serianni, W. Boggess, and J. Schorey. 2008. Mycobacterium avium glycopeptidolipids require specific acetylation and methylation patterns for signaling through toll-like receptor 2. J. Biol. Chem. 283:33221-33231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tassell, S. K., M. Pourshafie, E. L. Wright, M. G. Richmond, and W. W. Barrow. 1992. Modified lymphocyte response to mitogens induced by the lipopeptide fragment derived from Mycobacterium avium serovar-specific glycopeptidolipids. Infect. Immun. 60:706-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang, A. Y., J. C. Denner, P. J. Brennan, and J. K. McClatchy. 1992. Clinical and epidemiological importance of typing of Mycobacterium avium complex isolates. J. Clin. Microbiol. 30:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vergne, I., and M. Daffe. 1998. Interaction of mycobacterial glycolipids with host cells. Front. Biosci. 3:d865-876. [DOI] [PubMed] [Google Scholar]
- 35.Wayne, L. G., R. C. Good, A. Tsang, R. Butler, D. Dawson, D. Groothuis, W. Gross, J. Hawkins, J. Kilburn, M. Kubin, K. H. Schroder, V. A. Silcox, C. Smith, M. F. Thorel, C. Woodley, and M. A. Yakrus. 1993. Serovar determination and molecular taxonomic correlation in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 43:482-489. [DOI] [PubMed] [Google Scholar]
- 36.Yakrus, M. A., and R. C. Good. 1990. Geographic distribution, frequency, and specimen source of Mycobacterium avium complex serotypes isolated from patients with acquired immunodeficiency syndrome. J. Clin. Microbiol. 28:926-929. [DOI] [PMC free article] [PubMed] [Google Scholar]