Abstract
Several aldehyde dehydrogenase (ALDH) complexes have been purified from the membranes of acetic acid bacteria. The enzyme structures and the chemical nature of the prosthetic groups associated with these enzymes remain a matter of debate. We report here on the molecular and catalytic properties of the membrane-bound ALDH complex of the diazotrophic bacterium Gluconacetobacter diazotrophicus. The purified ALDH complex is a heterodimer comprising two subunits of 79.7 and 50 kDa, respectively. Reversed-phase high-pressure liquid chromatography (HPLC) and electron paramagnetic resonance spectroscopy led us to demonstrate, for the first time, the unequivocal presence of a pyrroloquinoline quinone prosthetic group associated with an ALDH complex from acetic acid bacteria. In addition, heme b was detected by UV-visible light (UV-Vis) spectroscopy and confirmed by reversed-phase HPLC. The smaller subunit bears three cytochromes c. Aliphatic aldehydes, but not formaldehyde, were suitable substrates. Using ferricyanide as an electron acceptor, the enzyme showed an optimum pH of 3.5 that shifted to pH 7.0 when phenazine methosulfate plus 2,6-dichlorophenolindophenol were the electron acceptors. Acetaldehyde did not reduce measurable levels of the cytochrome b and c centers; however, the dithionite-reduced hemes were conveniently oxidized by ubiquinone-1; this finding suggests that cytochrome b and the cytochromes c constitute an intramolecular redox sequence that delivers electrons to the membrane ubiquinone.
Gluconacetobacter diazotrophicus is a strict aerobe and a N2-fixing endophyte originally isolated from sugarcane (3, 8, 10, 19). This bacterium belongs to the recently introduced genus Gluconacetobacter of acetic acid bacteria (30). The oxidation of ethanol to acetic acid is the most distinctive feature of these bacteria. The process, located in the periplasmic space, is catalyzed by two membrane-bound enzyme complexes, alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH). These enzymes are usually recognized as quino-hemeproteins that contain pyrroloquinoline quinone (PQQ) and cytochromes c as prosthetic groups. This view of the ADH is fully supported by experimental data (1, 11, 12, 18, 21, 22, 27). On the other hand, the presence of PQQ in the ALDH of Acetobacter sp. strain BPR2001 (26) and Gluconacetobacter europaeus (29) has been questioned. Moreover, a purified preparation of ALDH that contains PQQ, but not cytochrome c, has been reported in Acetobacter polyoxogenes sp. nov. (9). A further complication arose from a report in Ga. europaeus (29) that suggested that a cytochrome b, a [2Fe:2S] cluster, and a molybdopterin center were additional prosthetic groups of ALDH. It has been reported that the ALDH enzymes from Acetobacter aceti (4) and Ga. europaeus (29) contain three subunits, whereas only two subunits were detected in Gluconobacter suboxydans (2), Acetobacter rances (15), and Acetobacter polyoxogenes (9).
We describe here the purification and molecular characterization of the two-subunit ALDH complex of Ga. diazotrophicus. Its prosthetic groups were analyzed by distinct alternative techniques that confirmed the presence of PQQ, one heme B, and three hemes C. In addition, the catalytic properties of the purified enzyme were examined.
MATERIALS AND METHODS
Chemicals and reagents.
QAE-Toyopearl was obtained from Supelco/Sigma-Aldrich Corp. (St. Louis, MO). Natriumlauryl-sulfate, acrylamide, bis-acrylamide, and Precision Plus protein standard were obtained from Bio-Rad Laboratories, Inc. (Hercules, CA). All other chemicals were obtained from Sigma-Aldrich Corp.
Strain, growth conditions, preparation of membranes, and culture methods.
Ga. diazotrophicus PAL5 (ATCC 49037) was grown at 30°C in LGIP medium supplemented with 0.75% ethanol, under the conditions described by Reis et al. (25), in a 60-liter working-volume Bioflow 5000 fermentor (New Brunswick Scientific), stirred at 120 rpm, and aerated at 60 liters of air min−1. The fermentor was seeded with 3 liters of an active culture. Cells were harvested in the early stationary phase (48 h). The procedures used for the disruption of cells and membrane preparation were described earlier (13). Protein in membranes and purified fractions were determined according to the method of Markwell et al. (20).
Purification of ALDH.
Cell membranes (∼10 mg of protein/ml) were suspended in a 0.01 M potassium phosphate buffer (pH 6.0; buffer KPB). Triton X-100 was added to the membrane suspension to a final concentration of 0.5% (vol/vol) and incubated at 4°C for 2 h under gentle stirring and thereafter centrifuged at 144,000 × g for 60 min. The supernatant containing ALDH activity was applied to a QAE-Toyopearl column (3 by 18 cm) previously equilibrated with KPB containing 0.1% Triton X-100. Nonretained protein was washed out with three bed volumes of the same buffer. ALDH was eluted with a 0 to 0.25 M NaCl linear gradient in the equilibration buffer. Fractions containing ferricyanide reductase activity with acetaldehyde, but not with ethanol, were collected and dialyzed overnight against 20 volumes of KPB containing 0.1% Triton X-100. Dialyzed ALDH fraction was applied to a HA-Ultrogel column (3 by 15 cm) previously equilibrated with KPB containing 0.1% Triton X-100. Nonretained protein was washed as described above. ALDH was eluted using a 0 to 0.25 M potassium phosphate (pH 6.0) linear gradient containing 0.1% Triton X-100. The active fractions were collected and concentrated 10-fold by ultrafiltration in an Amicon YM30 MW (Amicon Corp., Danvers, MA). The concentrated fractions were applied to a Sephacryl-S200 column (64 by 5 cm), previously equilibrated with five bed volumes of KPB containing 0.1% Triton X-100. The active fractions were pooled and concentrated as described above and stored at 4°C for further analyses.
The purified ALDH was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 16-by-14-cm slab gels with 10% polyacrylamide by the method of Goodhew et al. (14). For native PAGE, the same system was used, except that the polyacrylamide concentration was of 7.5%, and SDS was replaced by 0.1% Triton X-100. Native gels were stained with 0.05% Coomassie brilliant blue R-250 or alternatively, for enzyme activity (zymogram), in a medium containing 2.0 mM phenazine methosulfate (PMS), 0.34 mM nitroblue tetrazolium, and 100 mM acetaldehyde (1). The heme-catalyzed peroxidase stain assay described by Thomas et al. (28) was used to detect cytochrome c bands in SDS gels. For the sequence analysis, the protein bands were carefully excised from Coomassie blue-stained SDS gels and peptide sequenced as previously reported (17) The isoelectric point (pI) of the purified ALDH was determined by using the Phast system and gels with a pH range of 3.4 to 9.0 (Amersham Biosciences/GE Healthcare, United Kingdom).
Enzymatic activity assays.
The oxidoreductase activity of ALDH was measured spectrophotometrically in a Shimadzu UV-2401 PC UV-visible light (UV-Vis) spectrophotometer (Shimadzu, Kyoto, Japan) at pH 3.5 using potassium ferricyanide as an electron acceptor as described by Matsushita et al. (22) or at pH 7.0 using PMS plus 2,6-dichlorophenolindophenol (DCPIP) as electron acceptors, as described previously (8). One unit of the enzyme activity is the amount of enzyme that catalyzes the oxidation of 1.0 μmol of substrate per min under the indicated conditions.
Electron paramagnetic resonance (EPR) spectroscopy.
ALDH (6.0 mg of protein) in 500 μl of 10 mM phosphate buffer (pH 6.0) was transferred into a 3-mm (inner diameter) quartz tube and frozen in liquid nitrogen. CW-EPR spectra were recorded on a Bruker Elexsys E500 spectrometer at x-band (9.39 GHz) and 100-kHz modulation. The temperature at 5 or 10 K was maintained by an Oxford liquid helium continuous flow cryostat. The g-values were determined by measuring the magnetic field and the microwave frequency.
Determination of PQQ by reversed-phase HPLC.
The PQQ in the purified enzyme (3.0 mg of protein) was extracted with 90% methanol as described by Matsushita et al. (22) and determined by reverse-phase high-pressure liquid chromatography (HPLC). A chromatography system equipped with a photodiode array detector Waters model 996 (Milford, MA) and Millennium 2000 software (Microsoft Corporation) was used. Separation and identification of PQQ were performed in a Waters C18 Spherisorb S5 OD52 analytical column (4.6 by 150 mm) as described earlier (13). Eluted PQQ was detected at 275 nm, and its concentration was calculated from the area of the corresponding peak using commercial PQQ as a standard.
Determination of hemes.
UV-Vis difference spectra were recorded at room temperature in an OLIS-SLM DW 2000 spectrophotometer (Olis, Inc., Bogart, GA) using 1-cm light path cuvettes. Samples were reduced with sodium dithionite, whereas the references were oxidized with few grains of ammonium persulfate. The cytochromes content of the ALDH were determined at A550-540 using an extinction coefficient of 19.1 mM−1 cm−1 for cytochrome c and at A556-575 using an extinction coefficient of 21 mM−1 cm−1 for cytochrome b (7).
Extractable hemes from purified ALDH were determined by reversed-phase HPLC. Separation and identification of hemes were performed in a Waters Delta-Pak HPLC18 300 column (2 by 150 mm). Hemes were extracted from the purified ALDH (1.0 mg of protein) with acid-acetone and dissolved in a 0.5% trifluoroacetic acid-acetonitrile solution as described by Puustinen and Wikström (24). The samples were applied to the Delta Pak column, and the hemes were eluted by an acetonitrile gradient as described previously (13). Protoheme IX (Sigma), hemes B and O extracted from Escherichia coli, and hemes B and A extracted from mitochondrial particles from bovine heart were used as standards.
RESULTS
Purification of the membrane-bound ALDH.
ADH and ALDH complexes were extracted from membranes (10 mg of protein/ml) with 0.5% Triton X-100 in buffer KPB. During the first chromatography step in a QAE-Toyopearl column, the ADH and ALDH complexes were separated (not shown). A peak eluting at 50 mM NaCl contained all of the ADH activity (ethanol as substrate); however, this peak also showed activity on acetaldehyde. In contrast, the peak that eluted at 100 mM NaCl was very active for acetaldehyde and showed no activity on ethanol. Both peaks showed cytochrome absorption at 405 nm. Thus, fractions of the second cytochrome peak, specific for acetaldehyde, were pooled and subjected to further purification steps as described in Material and Methods. After starting with 3.0 g of membrane protein, at the end of the purification process 4.0 mg of homogenous ALDH was obtained (Table 1). During purification, stabilizing agents for ALDH, such as benzaldehyde and sucrose (2, 4, 15, 29), were unnecessary, and the resulting purified preparation could be stored for several weeks at 4.0°C, or for longer times at −70°C, without appreciable loss of the activity.
TABLE 1.
Purification summary of the membrane-bound ALDH
| Purification step | Total protein (mg) | Sp act (U/mg)a | Total activity (U) | Yield (%) |
|---|---|---|---|---|
| Membrane fraction | 3,000 | 5 | 15,000 | 100 |
| Solubilized fraction | 600 | 18 | 10,800 | 72 |
| QAE-Toyopearl | 250 | 32 | 8,000 | 53 |
| Ha-Ultrogel | 15 | 393 | 5,895 | 39 |
| Sephacryl S200 | 4 | 947 | 3,788 | 25 |
The activity was assayed under the standard conditions described in Materials and Methods.
Molecular properties of ALDH.
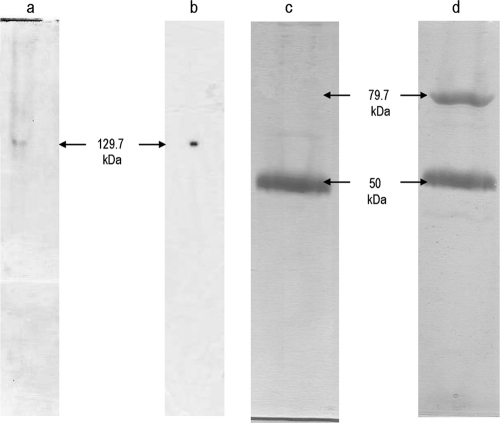
The homogeneity of the purified ALDH was analyzed by nondenaturing PAGE (Fig. 1). A single band with a molecular mass of 129 kDa was revealed by the Coomassie blue staining (Fig. 1a) or by the zymographic procedure revealing the specific dehydrogenase activity on acetaldehyde (Fig. 1b). Under denaturing conditions in SDS-PAGE, the enzyme showed the presence of two bands having an Mr of 79.7 and a mass of 50 kDa (Fig. 1d), as determined by mass spectroscopy (not shown). The heme-catalyzed peroxidase assay showed reaction of only the 50-kDa subunit (Fig. 1c). Since our SDS-PAGE gel was first developed for heme C and then for protein, the Coomassie stain gave unequal blue tones that reflected on the apparent weak staining of the 50-kDa hemeprotein (Fig. 1d).
FIG. 1.
PAGE of the purified aldehyde dehydrogenase complex from Ga. diazotrophicus. (a) Approximately 50 μg of purified protein was loaded on native gel (10%) and stained by Coomassie brilliant blue. (b) Aldehyde dehydrogenase activity was stained by zymography. Alternatively, the purified enzyme (50 μg of protein) was subjected to SDS-PAGE and first stained by the peroxidase assay with tetramethylbenzidine (c), followed by the Coomassie blue stain (d). The molecular mass of each band is indicated on the side in kilodaltons. The electrophoresis procedures and stain techniques are described in Materials and Methods.
The amino acid sequence of the two resolved polypeptides was determined, and protein identification was carried out by using Mascot Software, the BLAST facility of the National Center for Biotechnology Information ((http//ncbi.nlm.nih.gov/Blast), and The Institute for Genomic Research (http//www.tigr.org). Sequence analysis showed a perfect match with ALDH from Ga. europaeus (GenBank accession no. CAA69955.1) and A. polyoxogenes (SwissProt accession no. P17201.1), 75 to 100% identity with ALDH from A. pasteurianus (GenBank accession no. BAB97172.1), and 64 to 100% with ALDH from Go. suboxydans (GenBank accession no. BAD90575.1).
Cytochrome analysis.
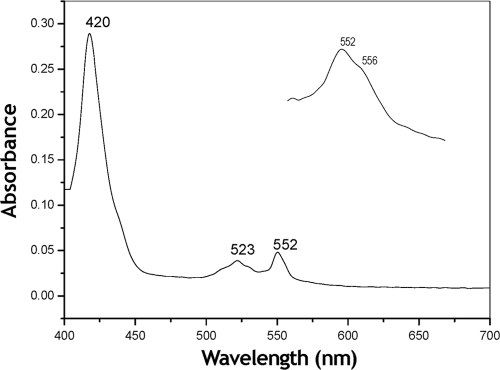
Visible light spectroscopy of the purified enzyme reduced by dithionite showed absorption maxima at 552, 523, and 420 nm that could be assigned to the characteristic α, β, and γ absorption bands, respectively, of the reduced c-type cytochromes (Fig. 2). In addition, the presence of a b-type cytochrome was strongly suggested by the existence of shoulders at 526 and 556 nm (inset in Fig. 2). The concentration of cytochrome c and of the putative cytochrome b in ALDH as calculated from the α-absorption bands were 23.3 and 7.23 nmol mg of protein−1, respectively, suggesting that the ALDH complex contains three cytochromes c and one cytochrome b. Spectral analysis of the purified ALDH reduced by its substrate acetaldehyde was also attempted; however, even after long-term incubation, the enzyme showed poor reduction levels of both cytochromes b and c. Thus, the appropriate difference spectrum of the acetaldehyde-reduced ALDH could not be obtained. A similar result was reported for the purified membrane ALDH of A. rances (15).
FIG. 2.
Visible-light absorption spectrum of the purified aldehyde dehydrogenase complex from Ga. diazotrophicus. The dithionite-reduced minus the persulfate-oxidized difference spectrum is depicted and was taken at an enzyme concentration of 0.3 mg ml−1 in 10 mM potassium phosphate buffer (pH 6.0) containing 0.1% Triton X-100. The inset shows the amplified α band of the spectrum.
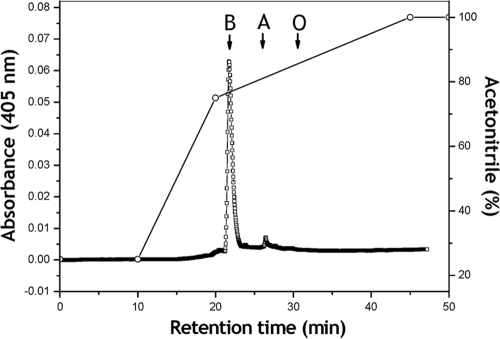
The presence of cytochrome b was confirmed by HPLC analysis of hemes extracted from the purified enzyme. A single major peak that eluted with the same retention time (i.e., 21.73 min) of the protoheme IX standard was obtained (Fig. 3). An approximate concentration of 6.5 nmol of cytochrome b per mg of protein was calculated from the area under the peak, which is close to the concentration calculated from the absorption spectrum of the dithionite reduced enzyme.
FIG. 3.
Reversed-phase HPLC chromatogram of heme extracted from the purified aldehyde dehydrogenase complex (0.4 mg of protein) from Ga. diazotrophicus. The system was calibrated with the following standards: hemes B and O extracted from Escherichia coli and hemes B and A extracted from submitochondrial particles from bovine heart and protohaem IX obtained from Sigma Chemical Co. The retention times for the standards were as follows: heme B, 21.73 min; heme A, 26.4 min; and heme O, 30.4 min.
EPR spectroscopy.
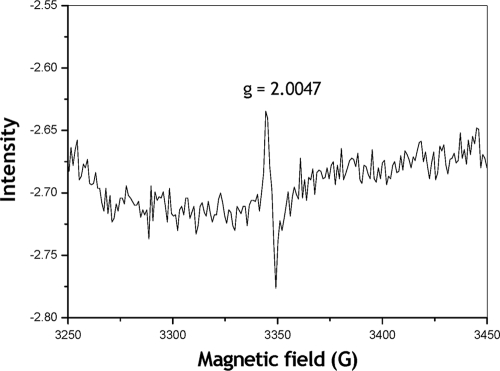
Earlier, this technique had been successfully applied to investigate the PQQ prosthetic group in bacterial ADHs complexes (16, 6). To our knowledge, ALDH complexes of acetic acid bacteria have yet to be analyzed by EPR spectroscopy. The X-Band CW-EPR spectrum of the purified ALDH in its reduced form showed a sharp signal centered at giso = 2.0047 (Fig. 4). Its g-value and lines width were typical for an organic radical, and it was assigned to the PQQ-semiquinone. Similar spectra were described for the PQQ moiety in purified ADH complexes from Pseudomonas aeruginosa (16) and Comamonas testosteroni (6).
FIG. 4.
EPR spectral analysis of the purified aldehyde dehydrogenase complex. The X-Band CW-EPR spectrum of the dithionite-reduced enzyme (6 mg of protein in 0.5 ml) was determined. The experimental conditions were as follows: microwave frequency, 9.39 GHz; temperature, 10 K; microwave power, 2.0 mW; and modulation amplitude, 0.0005 T.
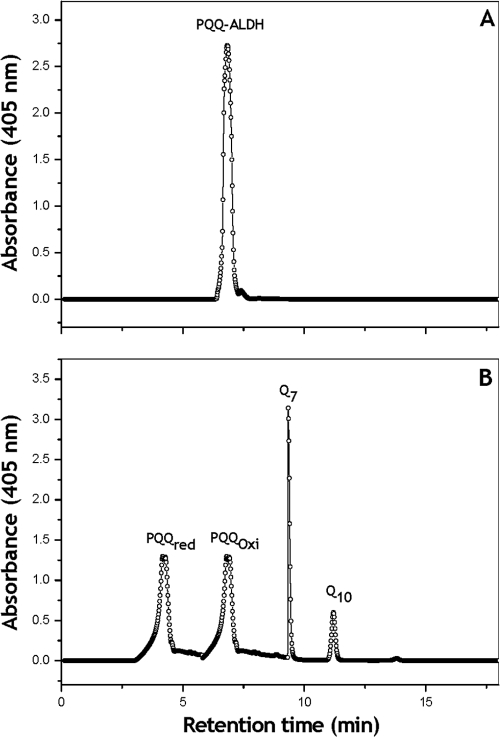
The presence of PQQ prosthetic group associated with the ALDH of Ga. diazotrophicus was further confirmed by HPLC analysis of methanol-extracted materials obtained from the purified enzyme (Fig. 5). A single major peak eluting with the same retention time (i.e., 4.28 min) of the authentic PQQ was obtained. The concentration of PQQ calculated from the area of the corresponding peak at 275 nm using commercial PQQ as standard was 7.0 nmol mg of protein−1, indicating that the purified ALDH complex contains one molecule of PQQ.
FIG. 5.
Reversed-phase HPLC of the PQQ. (A) Reversed-phase HPLC chromatogram of the methanol extracted quinones associated with the purified acetaldehyde dehydrogenase complex (3.0 mg of protein) of Ga. diazotrophicus. (B) The system was calibrated with commercial standards: PQQH2 (commercial form) and commercial PQQH2 oxidized with ammonium persulfate, Q7, and Q10 (retention times of 4.28, 6.8, 9.28, and 11.19 min, respectively).
Catalytic properties of ALDH.
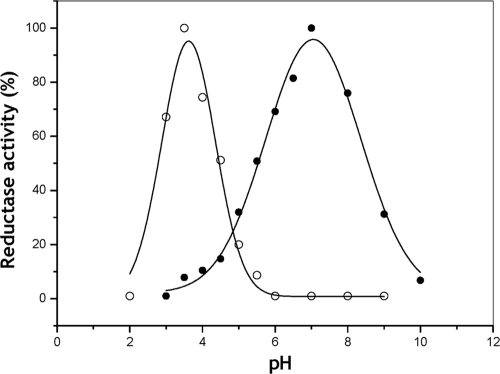
The activity of purified ALDH complex of Ga. diazotrophicus as a function of pH showed a sharp and symmetric activity peak with a maximum at pH 3.5 with potassium ferricyanide as electron acceptor (Fig. 6). The optimum pH shifted to a sharp 7.0 when PMS plus DCPIP were used as electron acceptors. ALDH possesses multiple redox centers, which are potentially reactive sites from which electrons can be withdrawn by different redox dye electron acceptors. Therefore, it is it likely that the response to pH obtained with the two different electron acceptor systems reflects the properties of particular redox centers within the enzyme. On the other hand, in native membranes of Ga. diazotrophicus, the aldehyde-ferricyanide reductase activity exhibited an optimum pH of 3.5 (not shown). Thus, the optimal pH determined for the ferricyanide reductase activity both in membranes and in the purified ALDH is similar to the pH range at which acetic acid bacteria usually produce vinegar.
FIG. 6.
Effect of pH on ferricyanide reductase (○) and DCPIP reductase (•) activities of the purified ALDH complex of Ga. diazotrophicus. The conditions used for the activity assays are described in Materials and Methods.
The relative oxidative activities of the purified enzyme toward several alcohols, aldehydes, and other organic compounds were examined. The enzyme showed rather narrow substrate specificity. Aliphatic aldehydes such as acetaldehyde (100%, relative activity), propionaldehyde (72.7%), and glutaraldehyde (26.4%) were good substrates. Formaldehyde and benzaldehyde were not oxidized at all. Other compounds, such as aliphatic alcohols and sugar aldehydes, were not substrates. Using acetaldehyde as a substrate and ferricyanide as an electron acceptor, the kinetic parameters at pH 3.5 and 30°C were as follows: Km = 3.3 × 10−3 M, kcat = 2.22 × 105 s−1, and kcat/Km = 6.72 × 106 M−1 s−1. The Km value was similar to those of the membrane-bound ALDH from A. aceti (i.e., 2.9 mM [4]), Go. suboxydans (i.e., 3.3 mM [2]), and A. rances (i.e., 1.0 mM [15]).
The pI of the purified ALDH complex was determined by the isoelectric focusing technique, and it was found to be 5.2 (data not shown), which is similar to that reported for the membrane ALDH purified from A. rances (15). The ferricyanide reductase activity of our purified ALDH was strongly inhibited by 1.0 mM HgCl2 or CuSO4 (not shown), whereas CaCl2, MgCl2, or EDTA at the same concentration did not affect enzyme activity (not shown).
Role of the ALDH cytochromes in redox catalysis.
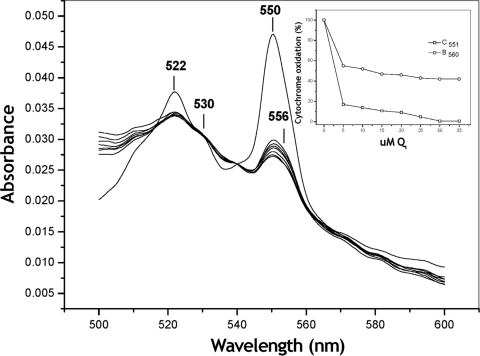
Since the reduction of the ALDH cytochromes by the physiological substrate aldehyde could not be observed, even after long-term incubation, an experiment was designed to evaluate the participation of ALDH cytochromes in the intramolecular electron transport to ubiquinone 1 (UQ1), which was used as an analog of the physiological UQ10 acceptor. The purified ALDH (50 μg of protein) was reduced in a stepwise fashion by dithionite, and the full reduced spectrum was recorded (Fig. 7). At this point, the oxidation of the ALDH complex was induced by successive additions of UQ1 (the concentrations are noted in the Fig. 7 inset). The spectra recorded after each UQ1 addition showed that both cytochromes c (ΔA550-540) and cytochrome b (ΔA556-570) were partially, but significantly, oxidized by UQ1. At the final titration point (35 μM UQ1), 20 and 32% of cytochromes c and b, respectively, remained reduced. These results strongly suggest that cytochrome b and the cytochromes c constitute an intramolecular redox sequence that delivers electrons to the endogenous UQ10 in the membrane.
FIG. 7.
Oxidation of cytochromes c and b associated with the aldehyde dehydrogenase complex by the electron acceptor UQ1. Purified ALDH (46 μg of protein) was stepwise reduced by limited amounts of dithionite, and the full reduced spectrum was traced. At this point, the oxidation of the ALDH complex was induced by successive additions of UQ1 (concentrations are noted in the inset). The oxidation of cytochromes c (ΔA550-540) and b (ΔA556-570) was measured and plotted against the UQ1 concentration reached after each addition (inset).
DISCUSSION
Oligomeric structure of ALDH complexes.
Several ALDH complexes have been purified from acetic acid bacteria (see Table 2 and the references therein); however, the results concerning the oligomeric structure and the chemical nature of the prosthetic groups associated with such enzymes are strikingly different, which suggests that among acetic acid bacteria, membrane-bound ALDH complexes are a heterogeneous group of enzymes. Table 2 shows that the molecular masses reported for the distinct ALDHs range between 90 and 157 kDa. Most frequently, the purified enzyme is a heteroligomer composed of either two (i.e., A. polyoxogenes, Go. suboxydans, A. rances, and Ga. diazotrophicus) or three subunits (i.e., A. aceti and Ga. europaeus). It is noteworthy that two distinct ALDH enzymes have been purified from membranes of A. aceti; one is a three-subunit ALDH, whereas the other is constituted by a single 125-kDa subunit. Although a full characterization is lacking, the enzymes seem to be quite distinct from each other in molecular structure, prosthetic groups, and catalytic properties (see Table 2 and references therein).
TABLE 2.
Properties of ALDH complexes purified from membranes of acetic acid bacteria
| Property | Findingsa for: |
||||||
|---|---|---|---|---|---|---|---|
| A. polyoxogenes | G. suboxydans | A. rances |
A. acetid |
Ga. europaeus | Ga. diazotrophicus | ||
| 1 | 2 | ||||||
| General features | |||||||
| Molecular mass (kDa) | 90 | 140 | 145 | 157 | 125 | ND | 129.7 |
| Subunit size (kDa) | 75, 19 | 86, 55 | 78, 66 | 75, 45, 14 | 125 | 79, 46, 17 | 79.7, 50 |
| Prosthetic groups | |||||||
| PQQ | YesA | YesA | YesA | YesA | ND | ND | 1BC |
| Heme C | NDD | 1DE | NDD | YesD | NDD | 3DF | 3DEF |
| Heme B | NDD | NDD | NDD | NDD | NDD | YesD | 1DG |
| [2Fe:2S] | ND | ND | ND | ND | ND | YesD | NDD |
| Molybdopterin | ND | ND | ND | ND | ND | YesH | ND |
| Catalytic properties | |||||||
| Aldehyde specificity | C2-C6 | C2-C6 | C2-C6 | C2-C6 | Broad | ND | C2-C6 |
| Optimal pHb | 7* | 4† | 5.1† | 5† | 8† | ND | 3.5†; 7* |
| Km (mM)c | 12 | 3.3 | 0.91 | 2.9 | 0.15 | 2.1 | 3.3 |
The source or reference(s) for the indicated strains were as follows: A. polyoxogenes(9), G. suboxydans (2, 5), A. rances (15), A. aceti (4, 5, 23), Ga. europaeus (29), and Ga. diazotrophicus (this study). ND, not determined or not detected. Determination methods are indicated by superscript capital letters as follows: A, fluorescence spectroscopy; B, HPLC; C, EPR; D, visible-light spectroscopy; E, redox titration; F, zymography; G, HPLC; H, deduced from amino acid sequence.
Electron acceptors: *, PMS plus DCPIP; †, ferricyanide.
Substrate: acetaldehyde.
Two ALDH complexes (1 and 2) have been purified from A. aceti.
It is also noteworthy that although our ALDH was purified as a two-subunit complex, sequence analysis of internal peptides of both subunits allowed us to localize the putative operon coding for the ALDH complex in the genome database of Ga. diazotrophicus (GenBank accession nos. AM889285.1, CAP57222.1, CAP57223.1, and CAP57224.1). The aldA-C putative operon of Ga. diazotrophicus is very similar to that present in Ga. europaeus (29); therefore, the ALDH complex of Ga. diazotrophicus to may contain a third 17-kDa subunit coded by the aldC gene (78% identity with aldG of Ga. europaeus; GenBank no. CAA69954.1) localized between the adhA and adhB genes. Thus, it seems that the third minor subunit in Ga. diazotrophicus is loosely bound to the membrane ALDH complex and probably removed upon detergent solubilization of the membrane. In any case, the dimeric structure constituted by the 79.5- and 50-kDa subunits seems to be the minimal structure required for catalysis in the ALDH complex of Ga. diazotrophicus; however, the lack of the 17-kDa subunit in the purified ALDH complex could explain the low reduction levels of cytochromes evoked by acetaldehyde, as discussed above.
Prosthetic groups of ALDH complexes.
The presence of PQQ in ALDHs has been questioned in two cases. (i) Takemura et al. (26), using chemical mutagenesis, obtained a PQQ-negative mutant of Acetobacter sp. strain BPR2001. That mutant was devoid of glucose dehydrogenase and ADH activities but contained close to normal membrane-bound ALDH activity; thus, these researchers concluded that PQQ is not a prosthetic group of ALDH complexes of acetic acid bacteria. However, the purification of this PQQ-independent ALDH was not attempted. (ii) Thurner et al. (29), based on the results obtained by Takemura et al. (26), extended that conclusion to the purified ALDH complex of Ga. europaeus; unfortunately, these authors did not explore the presence of PQQ in purified ALDH. In addition, the presence of PQQ in the one-subunit ALDH purified from A. aceti was not examined (23). In the present study, the unequivocal presence of PQQ in the purified ALDH complex was confirmed by three independent methods (i.e., fluorescence spectroscopy, EPR, and HPLC). A quantitative calibration with authentic PQQ indicated that 1 mol of PQQ is associated with the ALDH complex of Ga. diazotrophicus.
Hemes.
The presence of heme C in purified ALDH complexes is generally accepted. A single heme C was found to be associated with the ALDH of Go. suboxydans (2, 6). In the case of Ga. europaeus, the heme stain with tetramethylbenzidine was positive for both the 79- and the 46-kDa subunits (29). However, three CXXCH sequence motifs for heme C binding site were identified in the deduced amino acid sequence of the 46-kDa subunit, and none were identified in the sequence of the 79-kDa subunit (29). The ALDH of A. polyoxogenes (9) was apparently purified as a defective complex containing one 75-kDa subunit and one 19-kDa subunit and was therefore lacking the medium-size cytochrome c subunit (15). In addition, the one-subunit enzyme (125 kDa) purified from A. aceti was claimed to lack the typical absorption of a cytochrome c component (23). In the present study, the tetramethylbenzidine stain demonstrated the presence of heme C associated with the 50-kDa subunit only. The existence of the three CXXCH binding motifs for cytochrome c in the deduced amino acid sequence analysis supports the presence of three heme C groups in this subunit.
The presence of heme B associated with an ALDH complex was first suggested by Thurner et al. (29) on the basis of the difference spectrum of the purified ALDH of Ga. europaeus. In accordance with the findings of Thurner et al. (29), we detected heme B in the difference spectra of the ALDH purified from Ga. diazotrophicus and confirmed its chemical nature by HPLC analysis of acid-acetone extracted from the purified enzyme. Quantification of cytochromes associated with the purified enzyme, by visible light spectroscopy and HPLC, showed a 3:1 ratio for cytochromes c, and b, respectively.
[2Fe:2S] and molybdopterin.
The presence of [2Fe:2S] clusters in the ALDH complex of Ga. europaeus was hypothesized by Thurner et al. (29). These researchers observed a peak at 467 nm in the difference spectrum of the purified enzyme and found that the derived amino acid sequence of the 17-kDa protein contained eight conserved cysteine residues that apparently were involved in the binding of two putative [2Fe:2S] clusters. In this regard it is important to note that the spectra of distinct ALDH complexes previously purified (4, 9, 15), including our purified ALDH, do not contain such a spectral signal at 467 nm. We have to bear in mind that our purified ALDH is a heterodimer lacking the third-minor subunit; therefore, spectroscopic signals suggesting the presence of [2Fe:S] clusters are not expected. However, the deduced sequence analysis of the aldC gene coding for the putative 17-SIII subunit of Ga. diazotrophicus is identical to the corresponding gene in Ga europaeus (29). In any case, more experiments should be carried out to demonstrate the presence of [2Fe:2S] clusters in purified ALDHs.
In addition, Thurner et al. (29), on the basis of the deduced amino acid sequence of the ALDH complex of Ga. europaeus, proposed the presence of a molybdopterin cofactor to the 79-kDa subunit. Unfortunately, this proposal was not further validated by chemical analysis of the purified enzyme. Comparison of the consensus sequences of the PQQ and Mo-pterin binding domains with the amino acid deduced sequence of the ALDH from Ga. diazotrophicus showed equal probability that the prosthetic group of the ALDH from Ga. diazotrophicus is Mo-pterin or PQQ (i.e., 58 and 62%, respectively). In any case, the presence of molybdopterin in ALDHs of acetic acid bacteria must be confirmed by chemical techniques.
In conclusion, the ALDH complex of Ga. diazotrophicus was purified as a two-subunit quinoheme protein. The two prosthetic groups most likely associated with the major (79.7 kDa) subunit are one PQQ and one heme B. The minor subunit (50 kDa) is a hemeprotein containing three hemes C. Genome database analysis of Ga. diazotrophicus suggested that the membrane-bound complex contains a third 17-kDa putative subunit; however, its nature and function must be further confirmed. The enzyme is specific for aliphatic aldehydes (C2-C6) and is able to use ferricyanide or PMS plus DCPIP as electron acceptors having optimal pH values of 3.5 and 7.0, respectively. The UQ10 analogue, UQ1, was also able to accept electrons from the dithionite-reduced enzyme and that resulted in the partial but significant oxidation of the cytochromes b and c centers of the enzyme.
Acknowledgments
This study was supported in part by CONACYT grants 50672 and 41128 and by PAPIIT-UNAM grants IN204605 and IN212805.
We express our deep appreciation to Armando Gómez Puyou and Salvador Uribe for their generous help and criticism during preparation of the manuscript. We are grateful to Alejandra Abigail González-Valdez, Juan Manuel Méndez, Aurey Galván, and Juan Manuel Ortiz for technical assistance; Juan Manuel Barbosa Castillo and Ivett Rosas Arciniega for assistance with computer techniques; and María del Rocío Romualdo Martínez for secretarial assistance.
Footnotes
Published ahead of print on 27 August 2010.
REFERENCES
- 1.Adachi, O., E. Miyagawa, E. Shinagawa, K. Matsushita, and M. Ameyama. 1978. Purification and characterization of particulate alcohol dehydrogenase from Acetobacter aceti. Agric. Biol. Chem. 42:2331-2340. [Google Scholar]
- 2.Adachi, O., K. Tayama, E. Shinagawa, K. Matsushita, and M. Ameyama. 1980. Purification and characterization of membrane-bound aldehyde dehydrogenase from Gluconobacter suboxydans. Agric. Biol. Chem. 44:503-515. [Google Scholar]
- 3.Alvarez, B., and G. Martínez-Drets. 1995. Metabolic characterization of Acetobacter diazotrophicus. Can. J. Microbiol. 41:918-924. [Google Scholar]
- 4.Ameyama, M., K. Osada, E. Shinagawa, K. Matsushita, and O. Adachi. 1981. Purification and characterization of aldehyde dehydrogenase of Acetobacter aceti. Agric. Biol. Chem. 45:1889-1890. [Google Scholar]
- 5.Ameyama, M., and O. Adachi. 1982. Aldehyde dehydrogenase from acetic acid bacteria membrane-bound. Methods Enzymol. 89:491-497. [Google Scholar]
- 6.De Jong, G. A., H. A. Geerlof, J. Stoorvogel, J. A. Jongejan, S. De Vires, and A. Duine. 1995. Quinohaemoprotein ethanol dehydrogenase from Comamonas testosteroni. Purification, characterization, and reconstitution of the apoenzyme with pyrroloquinoline quinone analogues. Eur. J. Biochem. 230:899-905. [DOI] [PubMed] [Google Scholar]
- 7.Escamilla, J. E., R. Ramirez, I. P. Del Arenal, G. Zarzosa, and V. Linares. 1987. Expression of cytochrome oxidase in Bacillus cereus: effects of oxygen tension and carbon source. J. Gen. Microbiol. 133:3549-3555. [Google Scholar]
- 8.Flores-Encarnación, M., M. Contreras-Zentella, L. Soto-Urzúa, G. R. Aguilar, B. E. Baca, and J. E. Escamilla. 1999. The respiratory system and diazotrophic activity of Acetobacter diazotrophicus PAL5. J. Bacteriol. 181:6987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukaya, M., K. Tayama, H. Okumura, Y. Kawamura, and T. Beppu. 1989. Purification and characterization of membrane-bound aldehyde dehydrogenase from Acetobacter polyoxogenes. Appl. Microbiol. Biotechnol. 32:176-180. [Google Scholar]
- 10.Gillis, M., K. Kersters, B. Hoste, D. Janssens, R. M. Kroppenstedt, M. P. Stephan, K. R. S. Teixeira, J. Döbereiner, and J. De Ley. 1989. Acetobacter diazotrophicus sp. nov., a nitrogen-fixing acetic bacterium associated with sugarcane. Int. J. Syst. Bacteriol. 39:361-364. [Google Scholar]
- 11.Gomez-Manzo, S., M. Contreras-Zentella, A. Gonzalez-Valdez, M. E. Sosa-Torres, R. Arreguin-Espinoza, and E. Escamilla-Marvan. 2008. The PQQ-alcohol dehydrogenase of Gluconacetobacter diazotrophicus. Int. J. Food Microbiol. 12:71-78. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Manzo, S., A. Solano-Peralta, J. P. Saucedo-Vazquez, J. E. Escamilla-Marvan, P. M. H. Kroneck, and M. E. Sosa-Torres. 2010. The membrane-bound quinohemoprotein alcohol dehydrogenase from Gluconacetobacter diazotrophicus PAL5 carries a [2Fe-2S] cluster. Biochemistry 49:2409-2415. [DOI] [PubMed] [Google Scholar]
- 13.González, B., S. Martínez, J. L. Chávez, S. Lee, N. A. Castro, M. A. Domínguez, S. Gómez, M. L. Contreras, C. Kennedy, and J. E. Escamilla. 2006. Respiratory system of Gluconacetobacter diazotrophicus PAL5 Evidence for a cyanide-sensitive cytochrome bb and cyanide-resistant cytochrome ba quinol oxidases. Biochim. Biophys. Acta 1757:1614-1622. [DOI] [PubMed] [Google Scholar]
- 14.Goodhew, C. F., K. R. Brown, and G. W. Pettigrew. 1986. Heme staining in gels, as useful tool in the study of bacterial c-type cytochromes. Biochim. Biophys. Acta 852:288-294. [Google Scholar]
- 15.Hommel, R., and H. P. Kleber. 1990. Properties of the quinoprotein aldehyde dehydrogenase from Acetobacter rances. J. Gen. Microbiol. 136:1705-1711. [Google Scholar]
- 16.Kay, C., W. M. B. Mennenga, H. Görish, and R. Bitl. 2004. Characterization of the P cofactor radical in quinoprotein ethanol dehydrogenase of Pseudomonas aeruginosa by electron paramagnetic resonance spectroscopy. FEBS Lett. 564:69-72. [DOI] [PubMed] [Google Scholar]
- 17.Kinter, M., and N. E. Sherman. 2000. Mass spectrometric analysis of tryptic digests, p. 147-165. In D. M. Desiderio and N. M. M. Nibbering (ed.), Protein sequencing and identification using tandem mass spectrometry. John Wiley-Interscience, Inc., New York, NY.
- 18.Kondo, K., T. Beppu, and S. Horinouchi. 1995. Cloning, sequencing, and the characterization of the gene encoding the smallest subunit of the three component membrane bound alcohol dehydrogenase from Acetobacter pasteurianus. J. Bacteriol. 177:5048-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S., M. Flores-Encarnación, M. Contreras-Zentella, L. García-Flores, J. E. Escamilla, and C. Kennedy. 2004. Indole-3-acetic acid biosynthesis is deficient in Gluconacetobacter diazotrophicus strains with mutations in cytochrome c biogenesis genes. J. Bacteriol. 186:5384-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markwell, M. A. K., S. M. Haas, N. E. Tolbert, and L. L. Bieber. 1981. A modification of the Lowry procedure to simply protein determination in membranes and lipoprotein samples. Methods Enzymol. 72:269-303. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita, K., E. Takaki, M. Shinogawa, M. Ameyama, and O. Adachi. 1992. Ethanol oxidase respiratory chain of acetic acid bacteria. Reactivity with ubiquinone of pyrroloquinoline quinone-dependent alcohol dehydrogenase purified from Acetobacter aceti and Gluconobacter suboxydans. Biosci. Biotechnol. Biochem. 56:304-310. [Google Scholar]
- 22.Matsushita, K., T. Yakushi, Y. Takaki, H. Toyama, and O. Adachi. 1995. Generation mechanism and purification of an inactive form convertible in vivo to the active form of quinoprotein alcohol dehydrogenase in Gluconobacter suboxydans. J. Bacteriol. 177:6552-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muraoka, H., Y. Matabe, N. Ogasawara, and H. Takahashi. 1981. Purification and properties of coenzyme-independent aldehyde dehydrogenase from the membrane fraction of Acetobacter aceti. J. Ferment. Technol. 59:247-255. [Google Scholar]
- 24.Puustinen, A., and M. Wikström. 1991. The heme groups of cytochrome o from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis, V. M., F. L. Olivares, and J. Döbereiner. 1994. Improved methodology for isolation of Acetobacter diazotrophicus and confirmation of its endophytic habitat. World J. Microbiol. Biotechnol. 10:401-405. [DOI] [PubMed] [Google Scholar]
- 26.Takemura, H., T. Tsuchida, F. Yoshinga, K. Matsushita, and O. Adachi. 1994. Prosthetic group of aldehyde dehydrogenase in acetic acid bacteria is not pyrroloquinoline quinine. Biosci. Biotechnol. Biochem. 58:2082-2083. [Google Scholar]
- 27.Tayama, K., M. Fukaya, H. Okumura, Y. Kawamura, and T. Beppu. 1989. Purification and characterization of membrane-bound alcohol dehydrogenase from Acetobacter polyoxogenes. Appl. Microbiol. Biotechnol. 32:181-185. [Google Scholar]
- 28.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 29.Thurner, C., C. Vela, L. Meyer, L. Meile, and M. Teuber. 1997. Biochemical and genetic characterization of the acetaldehyde dehydrogenase complex from Acetobacter europaeus. Arch. Microbiol. 168:81-91. [DOI] [PubMed] [Google Scholar]
- 30.Yamada, Y., K. Hoshino, and T. Ishikawa. 1997. The phylogeny of acetic acid bacteria based on the partial sequences of 16S rRNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci. Biotechnol. Biochem. 61:1244-1251. [DOI] [PubMed] [Google Scholar]