Abstract
Streptomyces ambofaciens synthesizes the macrolide antibiotic spiramycin. The biosynthetic gene cluster for spiramycin has been characterized for S. ambofaciens. In addition to the regulatory gene srmR (srm22), previously identified (M. Geistlich et al., Mol. Microbiol. 6:2019-2029, 1992), three putative regulatory genes had been identified by sequence analysis. Gene expression analysis and gene inactivation experiments showed that only one of these three genes, srm40, plays a major role in the regulation of spiramycin biosynthesis. The disruption of srm22 or srm40 eliminated spiramycin production while their overexpression increased spiramycin production. Expression analysis was performed by reverse transcription-PCR (RT-PCR) for all the genes of the cluster in the wild-type strain and in the srm22 (srmR) and srm40 deletion mutants. The results from the expression analysis, together with the ones from the complementation experiments, indicated that Srm22 is required for srm40 expression, Srm40 being a pathway-specific activator that controls most, if not all, of the spiramycin biosynthetic genes.
Streptomyces species are Gram-positive, soil-inhabiting, filamentous bacteria that undergo a complex process of morphological differentiation and produce a great variety of secondary metabolites, including antibiotics with important applications in human medicine and in agriculture. These secondary metabolites are synthesized by complex pathways that utilize primary metabolites as building blocks. The genes required for the biosynthesis of a particular compound are generally clustered, and their expression is coregulated. Secondary metabolite biosynthesis is often activated in a growth phase-dependent manner and is genetically controlled at several levels (5). Pleiotropic regulatory genes can control the onset of production of several secondary metabolites produced by one Streptomyces strain. In some cases, these pleiotropic regulators also have an influence on morphological differentiation (12, 38, 39). For most of the secondary metabolite biosynthetic pathways, there is a specific level of control exerted by pathway-specific regulatory proteins. These regulatory proteins are encoded by genes generally located within the biosynthetic gene cluster. Many of these regulatory proteins belong to the SARP (Streptomyces antibiotic regulatory protein) family of DNA-binding proteins (46). But regulatory proteins belonging to other families, such as the LAL family (large ATP-binding regulators of the LuxR family) (18) and families comprising two-component histidine kinase and response regulator pairs (1), LysR-like regulators (17), TetR regulators (9), and γ-butyrolactone receptors (44), could also play a role as pathway-specific regulators. In addition, proteins that do not belong to large recognized families have been found to regulate the expression of biosynthetic genes (20). The expression of biosynthetic gene clusters could be regulated by a single pathway-specific regulator, for instance, PikD, which belongs to a LAL family and is a positive regulator for the pikromycin biosynthetic gene cluster in Streptomyces venezuelae (47). But a complex regulatory cascade involving several regulatory proteins encoded by genes in the cluster could also control antibiotic biosynthesis, as for tylosin biosynthesis in Streptomyces fradiae (15) or alpomycin biosynthesis in Streptomyces ambofaciens (2, 9).
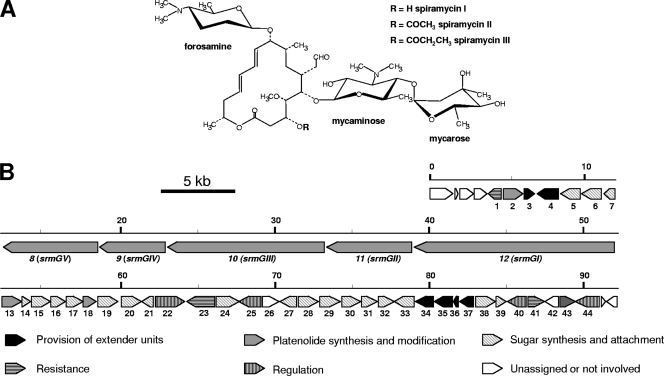
The macrolide antibiotic spiramycin is produced by S. ambofaciens (35). The spiramycin molecule consists of a polyketide lactone ring (platenolide) synthesized by a type I polyketide synthase (PKS), to which three deoxyhexoses (mycaminose, forosamine, and mycarose, in that order) are attached successively (Fig. 1 A). The entire spiramycin biosynthetic gene cluster has previously been cloned and sequenced (20, 25, 28) (Fig. 1B). The transcriptional activator SrmR (Srm22), encoded by a gene within the spiramycin biosynthetic gene cluster of S. ambofaciens, has been shown to be required for the transcription of at least two of the genes involved in spiramycin biosynthesis, srmGI (srm12) and srmX (srm21) (20). Three other putative regulatory genes located within the spiramycin biosynthetic gene cluster have been identified by sequence analysis (25). Neither Srm22 (SrmR) nor any of the putative regulators belong to an identified family of proteins involved in the pathway-specific regulation of antibiotic production. In this study, we demonstrated that two of these genes are not involved in the regulation of spiramycin biosynthesis but that the transcription of the biosynthetic genes is controlled by Srm22 (SrmR) and the newly identified Srm40 regulator.
FIG. 1.
Structure of spiramycins and genetic organization of the spiramycin biosynthetic gene cluster. (A) Structure of spiramycins. (B) Genetic organization of the spiramycin biosynthetic gene cluster. The proposed functions of the gene products in spiramycin biosynthesis are indicated by various filling patterns.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
All strains and plasmids used in this study are described in Table 1. Standard media and culture conditions were used (27, 40). The following antibiotics were incorporated in the medium when required for selection: ampicillin (Amp), thiostrepton (Tsr), apramycin (Apr), hygromycin B (Hyg), puromycin (Pur), and kanamycin (Kan). For spiramycin production, S. ambofaciens strains were grown in MP5 liquid medium as previously described (34). The detection and quantification of spiramycin were performed by bioassay and high-performance liquid chromatography (HPLC) as previously described (21). Spiramycins I, II, and III were used as standards for quantification by HPLC. The nomenclature used for the genes of the spiramycin biosynthetic gene cluster is different from the one published earlier (25) but is the one used by Nguyen et al. (32).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 23877 | Wild-type S. ambofaciens strain | ATCC |
| OSC2 | Derivative of S. ambofaciens ATCC 23877 devoid of pSAM2 | 36 |
| SPM249 | srm22::Ωhyg in S. ambofaciens ATCC 23877 | This work |
| SPM508 | Δsrm25::att3-Ωaac in S. ambofaciens OSC2 | This work |
| SPM107 | Δsrm40::att3-aac in S. ambofaciens OSC2 | This work |
| DH5α | E. coli general cloning host strain | Promega |
| S17.1 | E. coli strain for conjugation from E. coli to S. ambofaciens | 41 |
| DY330 | E. coli strain used for PCR-targeted mutagenesis | 48 |
| KS272/pKOBEG | E. coli strain used for PCR-targeted mutagenesis | 13 |
| Plasmids | ||
| pGEMT-Easy | Ampr; E. coli vector for cloning PCR products | Promega |
| pCR2.1 | Ampr; E. coli vector for cloning PCR products | Invitrogen |
| pMBL18 | Ampr; E. coli cloning vector | 31 |
| pOJ260 | Aprr; replicative vector in E. coli; nonreplicative in Streptomyces; used for gene disruption in S. ambofaciens | 6 |
| pUWL201 | Ampr Tsrr; E. coli-Streptomyces shuttle vector for gene expression in S. ambofaciens under the control of ermE*p | 19 |
| pOSV211 | Ampr Aprr; att3-Ωaac cassette (36) cloned into the vector pGP704Not (13, 30), the template for PCR amplification of att3-aac | This work |
| pOSV234 | Ampr Aprr; att3-aac cassette (34) cloned into the vector pGP704Not (13, 30), the template for PCR amplification of att3-aac | 32 |
| pOSV238 | Hygr; E. coli vector derived from pBK-CMV (Stratagene) by replacing the neo gene with the hyg gene | 25 |
| pOS49.32 | Ampr Kanr; internal fragment of srm22 cloned into pRC2.1 | This work |
| pOS49.43 | Ampr Kanr; derived from pOS49.32; Ωhyg into srm22 | This work |
| pOS49.50 | Aprr Hygr; insert from pOS49.43 cloned into pOJ260 | This work |
| pSPM5 | Ampr; cosmid (vector pWED1) from the S. ambofaciens gene library containing part of the spiramycin cluster | 25 |
| pSPM36 | Ampr Purr; cosmid (vector pWED2) from the S. ambofaciens gene library containing part of the spiramycin cluster | 25 |
| pSPM75 | Ampr Tsrr; coding sequence of srm40 cloned into pUWL201 | This work |
| pSPM107 | Ampr Aprr Purr; srm40 inactivation (Δsrm40::att3-aac) by PCR targeting into pSPM36 | This work |
| pSPM502 | Ampr; 15.1-kb BglII-AseI/Klenow fragment from pSPM5 cloned into pMBL18 digested by BamHI/Klenow | This work |
| pSPM504 | Hygr; 15.1-kb insert from pSPM502 cloned into pOSV238 | This work |
| pSPM508 | Aprr Hygr; srm25 inactivation (Δsrm25::att3-Ωaac) by PCR targeting into pSPM504 | This work |
| pSPM520 | Ampr; fragment of 2 kb containing the short form of the srm22 coding sequence cloned into pGEM-T Easy | This work |
| pSPM521 | Ampr; fragment of 2.2 kb containing the long form of the srm22 coding sequence cloned into pGEM-T Easy | This work |
| pSPM522 | Ampr; fragment of 2.6 kb containing the long form of the srm22 coding sequence and its promoter cloned into pGEM-T Easy | This work |
| pSPM523 | Ampr Tsrr; insert from pSPM520 (HindIII-BamHI fragment) cloned into pUWL201 | This work |
| pSPM524 | Ampr Tsrr; insert from pSPM521 (HindIII-BamHI fragment) cloned into pUWL201 | This work |
| pSPM525 | Ampr Tsrr; insert from pSPM522 (HindIII-BamHI fragment) cloned into pUWL201 | This work |
| pSPM527 | Ampr; pSPM521 with a 4-bp insertion at the XhoI restriction site | This work |
| pSPM528 | Ampr Tsrr; insert from pSPM527 (HindIII-BamHI fragment) cloned into pUWL201 | This work |
DNA manipulation and bacterial transformation.
DNA extraction and manipulation and transformation of Escherichia coli and Streptomyces were performed according to standard protocols (27, 40).
Isolation of total RNA from Streptomyces mycelium.
Mycelium was separated from fermentation broth and was washed with diethyl pyrocarbonate (DEPC)-treated water by vacuum filtration through glass fiber filters. The mycelium was rapidly collected, and about 0.4 g of mycelium was added to the mixture of 0.4 g glass beads from Sigma (106 μm and finer; acid washed), 400 μl RNase-free water, 500 μl acid phenol, and 400 μl macaloid. The mixture was shaken twice in a Fast Prep machine from Bio 101 (Savant) (force, 6.5; 30 s) and was then centrifuged. The supernatant obtained was extracted by acid phenol and then precipitated overnight with NaCl (0.2 M final concentration) and 1 volume of isopropanol. Nucleic acid preparations were treated with DNase I (DNA-free kit; Ambion), according to the manufacturer's instructions.
Gene expression analysis by RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed with a Qiagen OneStep RT-PCR kit using 1 μg of total RNAs as a template. The conditions were as follows: for cDNA synthesis, 50°C for 30 min, followed by 95°C for 15 min, and for amplification, 5 cycles at 97°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by 20 cycles at 97°C for 1 min, 58°C for 1 min, and 72°C for 1 min. Primers (20- to 22-mers; average melting temperature [Tm], 65°C to 70°C) (see the supplemental material) were designed to generate PCR products of approximately 400 bp, except for srm9 (srmGIV), srm36, and hrdB, where specific primers amplified fragments of 317 bp, 151 bp, and 177 bp, respectively. The gene hrdB, encoding the major sigma factor, was used as a control, as it is expressed at a constant level (11). All RT-PCR products were purified with s QIAquick PCR purification kit (Qiagen). With each pair of primers, negative-control experiments were carried out in the absence of reverse transcriptase (with DNA polymerase alone) to confirm that the amplified products were derived from RNA templates and not from chromosomal DNA, which might contaminate RNA preparations.
Targeted disruption of srm22, srm25, and srm40.
The copy of srm22 (srmR) was inactivated by gene replacement with a copy of the gene disrupted by the Ωhyg hygromycin resistance cassette. A DNA fragment internal to the srm22 coding sequence was amplified by PCR using the SRMR1 and SRMR2 primers (see the supplemental material). The 1.5-kb PCR product was inserted into the vector pCR2.1, leading to plasmid pOS49.32. pOS49.32 was then digested by BstEII and treated by the Klenow enzyme. The cassette Ωhyg (7) was obtained as a BamHI fragment and then blunt ended by treatment with the Klenow enzyme. The hygromycin resistance cassette Ωhyg was introduced in the digested plasmid pOS49.32, leading to plasmid pOS49.43, in which the genes srm22 and hyg are in the same orientation. Plasmid pOS49.43 was finally digested by Asp718I and XbaI and the insert fragment, blunt ended by Klenow treatment, and cloned into the plasmid pOJ260, previously digested by EcoRV, yielding pOS49.50. S. ambofaciens protoplasts were then transformed by alkali-denatured pOS49.50 DNA according to the method of Oh and Chater (33). Hygr clones were selected; among them, Aprs clones were screened. The Aprs clones probably resulted from a double recombination event leading to the replacement of srm22 by srm22::Ωhyg. This was checked by Southern analysis (data not shown). One such clone (srm22::Ωhyg) was retained for further analysis and named SPM249.
The disruptions of srm25 and srm40 were obtained by PCR targeting (13, 22, 48). For the inactivation of srm25, a 15.1-kb fragment from pSPM5 containing srm25 and flanking genes was first cloned into pMBL18, yielding pSPM502, and then the insert of pSPM502 was cloned into pOSV238, yielding pSPM504. The excisable cassette att3-Ωaac (36) was amplified using pOSV211 as a template and the primer set EDR3/EDR4 (see the supplemental material). Electrocompetent KS272/pKOBEG cells containing the plasmid pSPM504 were transformed with the purified PCR product. Aprr clones in which most of the srm25 coding sequence had been replaced by the att3-Ωaac cassette through λ-RED recombination were obtained. The resulting plasmid, pSPM508, was introduced into S. ambofaciens OSC2 via protoplast transformation, and apramycin selection was applied. Aprr transformants were screened for sensitivity to hygromycin, indicating a double-crossover allelic exchange in Streptomyces. This was confirmed by PCR and Southern blot analysis (data not shown). One clone with the expected structure for srm25 inactivation (Δsrm25::att3-Ωaac) was retained and named SPM508.
For the inactivation of srm40, the excisable cassette att3-aac was amplified using pOSV234 as a template and the primer set KF32/KF33 (see the supplemental material). The resulting PCR product was used to transform the E. coli strain DY330 (48) containing the cosmid pSPM36, which carries the target gene. Aprr clones in which most of the srm40 coding sequence had been replaced by the att3-aac cassette through λ-RED recombination were obtained. The resulting recombinant cosmid, pSPM107, was introduced into S. ambofaciens OSC2 via protoplast transformation, and apramycin selection was applied. Aprr transformants were screened for sensitivity to puromycin, indicating a double-crossover allelic exchange. This was confirmed by PCR amplification and Southern blot analysis (data not shown). One Δsrm40::att3-aac S. ambofaciens mutant strain was retained and named SPM107.
Construction of plasmids for expression of srm22 and srm40.
For the expression of srm22, three plasmids were constructed, containing the different forms of srm22. These forms were obtained by PCR using chromosomal DNA from S. ambofaciens OSC2 as a template and oligonucleotides, containing either BamHI or HindIII restriction sites, as primers (see the supplemental material). Oligonucleotides EDR39 and EDR42 were used to amplify the short form of srm22, starting with the ATG codon proposed by Geistlich et al. (20). The resulting 2-kb PCR product was cloned into the plasmid pGEMT-Easy, leading to the plasmid pSPM520. Oligonucleotides EDR40 and EDR42 were used to amplify the long form of the srm22 gene, starting from the most upstream ATG codon. The resulting 2.2-kb PCR product was inserted into the plasmid pGEMT-Easy, yielding the plasmid pSPM521. Oligonucleotides EDR41 and EDR42 were used to amplify the long form of srm22 together with its own promoter region. The resulting 2.6-kb PCR product was introduced into the plasmid pGEMT-Easy, yielding the plasmid pSPM522. The three plasmids pSPM520, pSPM521, and pSPM522 were digested by BamHI and HindIII, and their inserts containing the various forms of the srm22 gene were cloned into pUWL201 digested by BamHI and HindIII, leading to the plasmids pSPM523, pSPM524, and pSPM525, respectively. In all cases, the various forms of srm22 are under the control of the ermE*p promoter.
A plasmid containing a frameshift mutation in the region between the start codon proposed by Geistlich et al. (20) and the most upstream start codon was constructed. This plasmid, pSPM527, was obtained by digestion of plasmid pSPM521 with XhoI, treatment with the Klenow enzyme, and self-ligation. The insert BamHI-HindIII of pSPM527 was then inserted into the plasmid pUWL201 cut with the same enzymes, yielding the plasmid pSPM528.
For the expression of srm40, the srm40 coding sequence was amplified with the primers KF30 and KF31 (see the supplemental material). The resulting 1.5-kb PCR product was cloned into pUWL201 digested by BamHI and HindIII, yielding pSPM75, in which srm40 is expressed under the control of the ermE*p promoter.
RESULTS
Putative regulatory genes present in the spiramycin biosynthetic gene cluster.
The gene cluster directing spiramycin biosynthesis is presented in Fig. 1. The gene srmR (srm22) has previously been shown to encode an activator required for transcription from the srmGI (srm12) and srmX (srm21) promoters (20). Besides srm22, the sequence analysis of the spiramycin cluster revealed the presence of three other putative regulatory genes: srm25, srm40, and srm44.
The deduced product of srm25 (465 amino acid residues) showed high sequence similarity to members of the HflX subfamily of GTPases. Proteins from this family act as molecular switches, modulating diverse cellular processes in response to conformational changes induced by GTP hydrolysis. Their exact role remains sometimes elusive, but members of the HflX family have been shown to have pleiotropic action (8). In particular, Srm25 resembles the product of tylV from the tylosin biosynthetic gene cluster (63% identity and 76% similarity). The gene tylV also encodes a putative GTPase and might play a regulatory role in tylosin biosynthesis, as the inactivation of this gene was associated with reduced tylosin production (16, 42).
The deduced product of srm40 (387 amino acid residues) showed high sequence similarity to the product of acyB2 (69% identity and 77% similarity) from Streptomyces thermotolerans, the producer of carbomycin (3), and to the product of tylR (43% identity and 57% similarity) from S. fradiae (4). AcyB2 is required for carbomycin production and is supposed to play a regulatory role. TylR has been identified as a pathway-specific activator of tylosin production, directly controlling the expression of most of the biosynthetic genes (4, 42).
The deduced product of srm44 (503 amino acid residues) belongs to the GntR family of transcriptional regulators and more precisely to the MocR subfamily of transcriptional regulators containing a DNA-binding helix-turn-helix domain and an aminotransferase domain (COG1167) (37). Recent results (H. C. Nguyen, E. Darbon, S. Lautru, and J.-L. Pernodet, unpublished data) have shown that the genes srm42 and srm43 were not involved in spiramycin biosynthesis. Therefore, the fact that srm44 belongs to the spiramycin gene cluster is questionable.
Time course of the expression of spiramycin biosynthetic genes and putative regulatory genes.
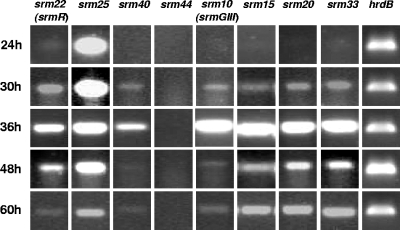
Under the culture conditions used in liquid medium, spiramycin is detectable after 40 to 48 h of cultivation and the spiramycin concentration continues to increase at least until 72 h. To correlate this observation with the expression of biosynthetic and putative regulatory genes, the expression of the regulatory gene srm22 (srmR), of the three putative regulatory genes (srm25, srm40, and srm44), and of four biosynthetic genes (srm10 [srmGIII], srm15, srm20, and srm33) was studied at different cultivation time points by RT-PCR for the strain S. ambofaciens OSC2. The gene srm10 encodes one of the proteins of the PKS involved in the biosynthesis of the platenolide (10, 28). The products of srm15, srm20, and srm33 were proposed to be involved in the biosynthesis of mycaminose, forosamine, and mycarose, respectively (25). Total RNAs were prepared from OSC2 after growth for 24 h, 30 h, 36 h, 48 h, and 60 h, i.e., before and after the onset of spiramycin production, and used as a template for gene expression analysis by RT-PCR. The results of these experiments are presented in Fig. 2.
FIG. 2.
Gene expression analysis of some genes from the spiramycin biosynthetic gene cluster. Gene expression was analyzed by RT-PCR in OSC2 at 24 h, 30 h, 36 h, 48 h, and 60 h. The transcript of hrdB was used as a control.
For the biosynthetic genes, the transcripts were not detected at 24 h, they were detected at a low level at 30 h, they seemed to be very abundant at 36 h, and then they decreased but were still detected after 48 h and 60 h of cultivation. This is consistent with the time course of spiramycin production. For the regulatory gene srm22, the transcripts were present in very small amounts at 24 h. Transcription seemed to increase, to reach a maximum at 36 h, and then to decrease. This is in agreement with the idea that Srm22 is an activator required for the expression of the biosynthetic genes (C. M. Farnet, A. Staffa, and X. Yang, U.S. patent application US 2003/0113874 A1). No RT-PCR products were detected for the putative regulatory gene srm44, suggesting that it does not play a major role in the regulation of spiramycin biosynthesis. The putative regulatory gene srm25 was always abundantly transcribed, even if its transcription seemed to decrease with time. For the third putative regulatory gene, srm40, the transcripts were not detected at 24 h, but they were detected at 30 h, relatively abundant at 36 h, and then detected in smaller amounts at 48 and 60 h. Its transcription pattern was quite similar to those of the biosynthetic genes.
These experiments showed that four biosynthetic genes involved in the synthesis of the different components of the spiramycin molecule, the polyketide macrolactone and the three sugars, had similar transcription patterns and were all abundantly transcribed at 36 h, i.e., a few hours before the detection of spiramycin in the culture medium. The expression profiles of srm22 and srm40 are compatible with a regulatory role for these genes. From the expression profile of srm25, it is difficult to draw evidence concerning its putative regulatory role. The fact that no transcript could be detected for srm44 is barely compatible with a regulatory role for this gene in spiramycin production. As other experiments showed that srm42 and srm43 were not involved in spiramycin biosynthesis (Nguyen et al., unpublished), we therefore considered that srm44 was probably not part of the spiramycin gene cluster and its role was not further studied.
Test of the involvement of the putative regulatory genes in the regulation of spiramycin biosynthesis.
In order to probe their involvement in the regulation of spiramycin biosynthesis, the putative regulatory genes srm25 and srm40 were inactivated by PCR targeting, followed by gene replacement. The resulting mutant strains were called SPM508 (Δsrm25::att3-Ωaac) and SPM107 (Δsrm40::att3-aac). Inactivation of srm22 was also performed by replacing in the S. ambofaciens chromosome the wild-type gene with a copy of the gene interrupted by the cassette Ωhyg. The resulting strain was called SPM249 (srm22::Ωhyg). These mutant strains were cultivated under the condition of spiramycin production. The culture supernatants were analyzed for spiramycin production, and the results are presented in Table 2.
TABLE 2.
Spiramycin production by various strains
| Strain | Characteristic(s) | Spiramycin concn (μM/g [DCW] ± SD)a |
|---|---|---|
| OSC2 | Wild type | 75.2 ± 2.5 |
| SPM249 | srm22::Ωhyg | ND |
| SPM107 | srm40::att3-aac | ND |
| SPM508 | srm25::att3-Ωaac | 60.6 ± 1.4 |
| SPM249 (pUWL201) | srm22::Ωhyg harboring the empty cloning vector | ND |
| SPM249 (pSPM523) | srm22::Ωhyg; overexpression of the short form of srm22 | ND |
| SPM249 (pSPM524) | srm22::Ωhyg; overexpression of the long form of srm22 | 47.3 ± 1.3 |
| SPM249 (pSPM528) | srm22::Ωhyg; overexpression of the long form of srm22 with a frameshift mutation | ND |
| SPM249 (pSPM525) | srm22::Ωhyg; overexpression of the long form of srm22; presence of the srm22 promoter region | 132.5 ± 5.9 |
| SPM107 (pUWL201) | srm40::att3-aac harboring the empty cloning vector | ND |
| SPM107 (pSPM75) | srm40::att3-aac; overexpression of srm40 | 135.2 ± 6.9 |
| OSC2 (pUWL201) | Wild-type strain harboring the empty cloning vector | 120.8 ± 6.8 |
| OSC2 (pSPM525) | Wild-type strain; overexpression of the long form of srm22; presence of the srm22 promoter region | 168.1 ± 4.3 |
| OSC2 (pSPM75) | Wild-type strain; overexpression of srm40 | 288.4 ± 8.6 |
| SPM249 (pSPM75) | srm22::Ωhyg; overexpression of srm40 | 273.5 ± 9.3 |
| SPM107 (pSPM525) | srm40::att3-aac; overexpression of the long form of srm22; presence of the srm22 promoter region | ND |
DCW, dry cell weight; ND, not detectable.
As expected, the strain SPM249 (srm22::Ωhyg) was unable to produce spiramycin, as observed by Geistlich et al. for the srmR (srm22) mutant strain that they studied (20). The mutant strain SPM107 (Δsrm40::att3-aac) was also unable to produce spiramycin. This indicated that srm40 is also a major regulatory gene, essential for spiramycin biosynthesis. For the mutant strain SPM508 (Δsrm25::att3-Ωaac), spiramycin production was still observed. The level of spiramycin produced by SPM508 was slightly lower than that produced by the wild-type strain, and the difference in spiramycin production is statistically significant (Student test; P = 0.0009); this might indicate a minor role for srm25 in regulation but could also be due to variability in the level of spiramycin production between different clones. The roles of the genes srm22 and srm40 in regulation were further studied.
Overexpression of srm22 and srm40 in the S. ambofaciens mutant and wild-type strains.
To confirm that the inactivation of srm22 and srm40 was the sole reason for the loss of spiramycin production in SPM249 and SPM107, the genes srm22 and srm40 were introduced and expressed into the corresponding mutant strains. The vector used was pUWL201, a multicopy plasmid in which the genes can be expressed under the control of the strong constitutive promoter ermE*p. For srm22, Geistlich et al. (20) had mapped the transcription start point, and they proposed for the srm22 coding sequence an initiation codon located 442 bp downstream from the transcription start point. A close examination of the sequence revealed that it was possible to extend the coding frame by 151 bp upstream of this initiation codon and to start from another ATG codon. To identify the codon used for translation initiation, several constructions were made in pUWL201. They contained the short form of srm22 (beginning 39 bp upstream of the ATG codon predicted by Geistlich et al. [20]) (pSPM523), the long form of srm22 (beginning 38 bp upstream of the most upstream ATG codon) (pSPM524), or the long form of srm22 into which a +1 frameshift mutation was introduced by adding 4 base pairs at a position located between the most upstream start codon and the start codon proposed by Geistlich et al. (20) (pSPM528). The plasmids pSPM523, pSPM524, and pSPM528 were introduced into the srm22 disruption mutant SPM249. pSPM523, carrying the short form of srm22, did not restore spiramycin production; pSPM524, carrying the long form of srm22, restored production, but its derivative with the frameshift mutation, pSPM528, did not restore the production (Table 2). This indicated that the most upstream initiation codon is most probably the one used for srm22 expression.
The level of spiramycin production obtained when pSPM524 was introduced into SPM249 (the srm22 disruption mutant) was not as good as the one obtained with the wild-type strain. Another construction was made, in which the long form of srm22, together with its promoter region (beginning 144 bp upstream of the transcription start point), was cloned into pUWL201. When this plasmid, pSPM525, was introduced into SPM249, it restored spiramycin production and increased it by 1.8-fold compared to the level for the wild-type strain (Table 2).
For the overexpression of srm40, a single construction, pSPM75, was made, in which a DNA fragment, containing the complete srm40 coding region and beginning 92 bp upstream of the initiation codon, was cloned into pUWL201 under the control of ermE*p. When the plasmid pSPM75, expressing Srm40, was introduced into the srm40 deletion mutant (strain SPM107), it restored and increased 1.8-fold the production of spiramycin (Table 2).
The plasmids pSPM525 and pSPM75 were also introduced into the wild-type strain OSC2, where they increased spiramycin production by factors of 2.2 and 3.8, respectively (Table 2). These results indicate that Srm22 (SrmR) and Srm40 are required for spiramycin biosynthesis and that they act as activators.
Gene transcription analysis of the strains OSC2, SPM249, and SPM107.
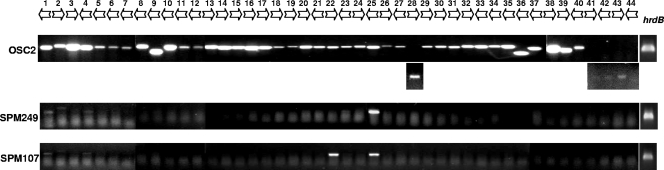
The transcription of 44 genes from the region of the spiramycin biosynthetic cluster was analyzed by RT-PCR in S. ambofaciens OSC2. Total RNA was extracted after 36 h of cultivation, as the time course of biosynthetic gene expression suggested that these genes were abundantly transcribed at this time (Fig. 2). We normally used 25 cycles to detect transcripts. Whenever 25 cycles did not yield a product, analysis was repeated at 27 cycles to distinguish low-level transcription (positive at 27 cycles and negative at 25 cycles). The results of the expression analysis by RT-PCR are presented in Fig. 3.
FIG. 3.
Gene expression analysis of all the genes from the spiramycin biosynthetic gene cluster. Transcripts from three strains were analyzed by RT-PCR: OSC2, the srm22-disrupted strain (SPM249), and the srm40-disrupted strain (SPM107). Total RNAs were extracted from all strains after 36 h of cultivation. Twenty-five cycles of PCR were routinely employed; whenever this generated no product, analysis was repeated at 27 cycles to detect low-level transcripts. The transcript of hrdB was used as a control.
In the wild-type strain OSC2, all genes studied were transcribed at 36 h, with the exception of srm44, which was silent, as previously observed at all time points (Fig. 2). For detection of the transcripts of srm28, srm41, srm42, and srm43, 27 PCR cycles were required to obtain a detectable signal.
To identify the genes whose transcription is controlled by Srm22 or Srm40, the same type of analysis was performed with the mutant strains SPM249 and SPM107 with inactivation of srm22 and srm40, respectively. In the srm22 mutant, SPM249, all genes tested were switched off, except srm25, for which a band was clearly visible, and srm1, srm2, and srm4, for which very faint bands were observed. In the srm40 mutant, SPM107, srm25 was expressed, and srm22 was also expressed, but no amplification was observed for most of the genes, and very faint bands were observed for a few of them (srm1, srm2, and srm4).
Taken together, these results indicated that most of the genes of the spiramycin cluster are not transcribed when Srm22 or Srm40 is absent. The transcription of the few genes which did not appear totally dependent on Srm22 or Srm40 was nevertheless decreased in the absence of these activators. These results also suggest that srm22 is not dependent on Srm40 for its transcription but that srm40 requires Srm22 to be transcribed. This is in agreement with the time course of their expression, srm22 being transcribed before srm40 (Fig. 2).
Hierarchy of the regulators.
The results of the transcription analysis suggested that Srm22 might activate the transcription of srm40, whose product, in its turn, might activate the transcription of most, if not all, of the genes involved in spiramycin biosynthesis. To confirm the role and the hierarchy of Srm22 and Srm40 in the regulation of spiramycin production, the srm40 and srm22 genes were overexpressed in the srm22- and srm40-disrupted mutants, respectively. For this purpose, pSPM525 (overexpressing Srm22) and pSPM75 (overexpressing Srm40) were introduced into the SPM107 (deletion of srm40) and SPM249 (disruption of srm22) mutant strains, respectively. The results of spiramycin production are presented in Table 2. Spiramycin was produced when srm40 was overexpressed in the srm22 mutant. However, no spiramycin was detected when srm22 was overexpressed in the srm40 mutant. This showed that srm22 requires the presence of srm40 to exert its activator role on spiramycin biosynthesis.
Taken together, the results of the expression analysis involving the phenotypes of the srm22 and srm40 mutants and the possibilities of restoring spiramycin production in these mutants led us to propose that Srm22 positively controls the transcription of the srm40 gene. Srm40 positively controls the transcription of most, if not all, of the genes implicated in spiramycin biosynthesis. This regulatory cascade is schematically presented in Fig. 4.
FIG. 4.
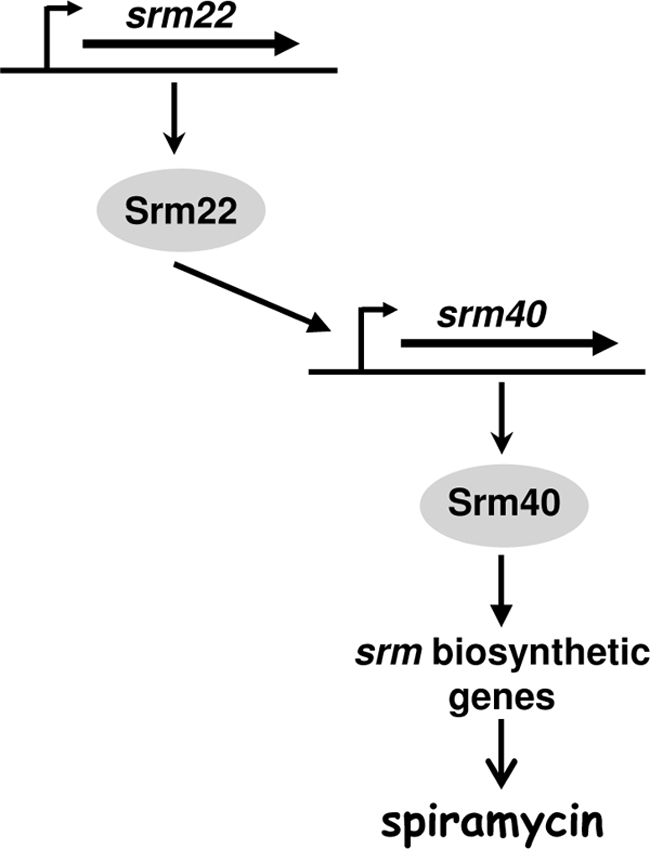
Proposed model for the regulation of spiramycin biosynthesis in S. ambofaciens.
DISCUSSION
In this work, we have shown that two genes of the spiramycin biosynthetic gene cluster, srm22 (srmR) and srm40, encode regulatory proteins activating the expression of spiramycin biosynthetic genes. These two activators act at the transcription level.
Concerning Srm22 (SrmR), our results indicate that it is longer than was predicted by Geistlich et al. (20) and is composed of 650 amino acids. The work of Geistlich et al. (20) demonstrated a regulatory role for srm22 (srmR). At that time, SrmR (Srm22) did not present any significant similarity to proteins in databases and these authors proposed that this protein could be the prototype of a new class of regulatory proteins. Now, numerous homologues of Srm22 are present in databases, and most of them are found in actinobacteria. The N-terminal part of Srm22 presents some similarity with the GAF domain. This domain is present in phytochromes and cGMP-specific phosphodiesterases, and proteins with a GAF domain are frequently involved in signal transduction. The C-terminal part of Srm22 presents similarity with protein from the COG2508 family. These proteins are thought to be involved in the regulation of secondary metabolism. The protein most similar to SrmR is CdaR (39% identity and 54% similarity) from Streptomyces coelicolor. The cdaR gene product is known to positively regulate genes for the biosynthesis of the calcium-dependent antibiotic (CDA), and its activity might be modulated by phosphorylation (26). In addition, CdaR expression was shown to be repressed by AbsA2 and activated by ppGpp (24, 29). This might provide indications for the study of the regulation of Srm22 expression.
The regulator Srm40 is a protein of 387 amino acids in which no known conserved domain was detected. Srm40 is highly similar to AcyB2 from S. thermotolerans and to TylR from S. fradiae. These two proteins act as activators of the production of the macrolide antibiotics carbomycin in S. thermotolerans and tylosin in S. fradiae, respectively.
The gene srm44 is most probably located outside the cluster. Other experiments showed that the inactivation of srm42 or srm43 had no effect on spiramycin biosynthesis (Nguyen et al., unpublished), suggesting that they were not part of the cluster. No transcription of srm44 was observed at different times during growth of S. ambofaciens. Therefore, this gene is probably not playing a major role in the regulation of spiramycin biosynthesis, although we cannot exclude that it might play a role under growth conditions that have not been explored in this work. The case of srm25 is intriguing, as this gene is located among biosynthetic genes, but its expression is not regulated as one of the biosynthetic genes: it was found to be transcribed at all time points studied, and its expression is independent from Srm22 and Srm40. A homologue of srm25, tylV, is present in the tylosin cluster in S. fradiae. The transcription of tylV is activated by TylS, a regulator of tylosin biosynthesis (42), but the role of TylV is not clear and TylV is not part of the regulatory cascade controlling tylosin biosynthesis (15). The inactivation of tylV decreases tylosin production in S. fradiae (16). Similarly, a slight decrease of spiramycin production was observed for the strain SPM508, in which srm25 is deleted. However, this could be due to variability in the level of spiramycin production between different clones, and this is not sufficient for a major role in the regulation of spiramycin biosynthesis to be attributed to Srm25.
During growth in the spiramycin production medium, the transcript of srm22 is the first to be detected, and then the transcripts of srm40 and of the biosynthetic genes are detected. All regulatory and biosynthetic genes are transcribed at 36 h of cultivation, before spiramycin could be detected in the production medium. Then, the transcription of the regulatory and biosynthetic genes decreases, but transcripts are nevertheless detected at 60 h. The timing of their transcription and the results of complementation experiments showed that the two activators Srm22 and Srm40 act in cascade, Srm22 activating the transcription of srm40 and Srm40 activating the transcription of the spiramycin biosynthetic genes. However, we could not rule out the possibility that one or more genes in the spiramycin cluster also directly require Srm22 (in addition to Srm40) for their expression.
The involvement of two pathway-specific activators in the regulatory cascade controlling spiramycin biosynthesis is reminiscent of the regulation of prodiginin biosynthesis in S. coelicolor A3(2). In this strain, RedZ activates the transcription of redD; RedD activates the expression of the red structural genes (23, 45). The involvement of several activators might offer multiple opportunities for the input of diverse regulatory signals. For example, it should be noted that the rare codon UUA is present in the srm22 transcript as in the redZ transcript, providing an opportunity for translational regulation (12).
Homologues of these two genes, srm22 and srm40, were found associated in other actinobacterial species. In Streptomyces eurythermus, in the gene cluster directing the biosynthesis of the macrolide antibiotic angolamycin (GenBank accession no. EU232693), the products of the convergent orf5 and orf6 genes are similar to Srm22 (38% identity and 51% similarity) and Srm40 (45% identity and 60% similarity), respectively. In Streptomyces mycarofaciens, in the gene cluster directing the biosynthesis of the macrolide antibiotic midecamycin (N. Midoh, S. Hoshiko, and T. Murakami, U.S. patent application 2006/0121577 A1) (GenBank accession no. BD420675), the products of the convergent orf27 and orf28 genes are similar to Srm40 (66% identity and 74% similarity) and Srm22 (74% identity and 82% similarity), respectively. In Micromonospora carbonacea, in the cluster directing the biosynthesis of the macrolide antibiotic rosaramicin (Farnet et al., U.S. patent application US 2003/0113874 A1) (GenBank accession no. AX697977), the products of the convergent orf14 and orf15 genes are similar to Srm22 (40% identity and 54% similarity) and Srm40 (46% identity and 59% similarity), respectively. The presence of genes encoding these two regulators in various macrolide biosynthetic gene clusters suggests that the cascade of regulation in which they participate might be conserved in all these actinobacterial species. In addition, homologues of Srm22 and Srm40 are encoded by neighbor genes in the genomes of Streptomyces sp. strain C (genes SSNG_03326 and SSNG_03327), of Streptomyces sp. strain Mg1 (genes SSAG_07397 and SSAG_07398), and of Micromonospora sp. strain M42 (genes MCBG_00153 and MCBG_00159).
The spiramycin biosynthetic genes are related to those involved in the biosynthesis of erythromycin or tylosin, two macrolides for which the biosynthesis has been extensively studied. However, the regulatory mechanisms controlling the expression of these biosynthetic clusters are very different. No regulatory gene was found in the erythromycin biosynthetic gene cluster. Recently, however, an activator of erythromycin biosynthesis was characterized (14). It is encoded by the bldD gene, located 1.5 Mb away from the biosynthetic gene cluster. Comparison of BldD expression levels in the wild-type and the erythromycin-overproducing strains of S. erythraea suggests that during strain improvement, mutations were introduced in genes that regulate BldD expression (14).
In contrast to the erythromycin cluster, the tylosin biosynthetic gene cluster from S. fradiae contains multiple regulatory genes encoding γ-butyrolactone-binding protein homologues (TylP and TylQ), SARP regulators (TylS and TylT), and putative regulators which do not belong to large families of regulatory proteins (TylR and TylU). TylT is not essential for tylosin production, but detailed studies of the regulation of tylosin biosynthesis have shown that the products of the other five regulatory genes are involved in a complex regulatory cascade (for a review, see reference 15). At the bottom of this cascade is TylR, which directly activates the transcription of the tylosin biosynthetic genes. During the empirical strain improvement program for enhancing tylosin production by S. fradiae, only one of the five regulatory genes, tylQ was altered, having undergone a single point mutation that inactivated its product (43). As TylQ is a repressor of tylR expression, this mutation was highly beneficial for tylosin production. Interestingly, another single-nucleotide mutation affecting tylQ was also observed in an independent strain improvement program by genome shuffling (49).
The situation encountered for the regulation of spiramycin biosynthesis is much simpler than that encountered for tylosin biosynthesis. However, it should be noted that similar regulators, TylR and Srm40, presenting 43% sequence identity, are located at the bottoms of both regulatory cascades and are involved in the activation of the transcription of the biosynthetic genes. In S. ambofaciens, further studies are required for knowledge of how the expression of srm22 is regulated and how the two regulatory proteins activate the transcription of their target genes. It might also be of interest to know if the regulatory genes have been altered during the strain improvement program that led to the overproducing strain used for industrial spiramycin production.
Supplementary Material
Acknowledgments
F.K. was supported by a CNRS-BDI fellowship. H.C.N. received fellowships from the Vietnamese Ministry of Education and from the Université Paris-Sud. This work was supported in part by Sanofi-Aventis, by the European Union through the Integrated Project ActinoGEN (CT-2004-0005224), and by the Pôle de Recherche et d'Enseignement Supérieur UniverSud Paris.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adamidis, T., P. Riggle, and W. Champness. 1990. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigle, B., X. Pang, B. Decaris, and P. Leblond. 2005. Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J. Bacteriol. 187:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arisawa, A., N. Kawamura, H. Tsunekawa, K. Okamura, H. Tone, and R. Okamoto. 1993. Cloning and nucleotide sequences of two genes involved in the 4″-O-acylation of macrolide antibiotics from Streptomyces thermotolerans. Biosci. Biotechnol. Biochem. 57:2020-2025. [DOI] [PubMed] [Google Scholar]
- 4.Bate, N., A. R. Butler, A. R. Gandecha, and E. Cundliffe. 1999. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6:617-624. [DOI] [PubMed] [Google Scholar]
- 5.Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 6.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 7.Blondelet-Rouault, M. H., J. Weiser, A. Lebrihi, P. Branny, and J. L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190:315-317. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. D. 2005. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 83:738-746. [DOI] [PubMed] [Google Scholar]
- 9.Bunet, R., M. V. Mendes, N. Rouhier, X. Pang, L. Hotel, P. Leblond, and B. Aigle. 2008. Regulation of the synthesis of the angucyclinone antibiotic alpomycin in Streptomyces ambofaciens by the autoregulator receptor AlpZ and its specific ligand. J. Bacteriol. 190:3293-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgett, S., S. Kuhstoss, R. N. Rao, M. A. Richardson, and P. R. Rosteck, Jr. August 1999. Platenolide synthase gene. U.S. patent 5,945,320.
- 11.Buttner, M. J., K. F. Chater, and M. J. Bibb. 1990. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2). J. Bacteriol. 172:3367-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chater, K. F., and G. Chandra. 2008. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in streptomyces. J. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 13.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chng, C., A. M. Lum, J. A. Vroom, and C. M. Kao. 2008. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc. Natl. Acad. Sci. U. S. A. 105:11346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cundliffe, E. 2008. Control of tylosin biosynthesis in Streptomyces fradiae. J. Microbiol. Biotechnol. 18:1485-1491. [PubMed] [Google Scholar]
- 16.Cundliffe, E., N. Bate, A. Butler, S. Fish, A. Gandecha, and L. Merson-Davies. 2001. The tylosin-biosynthetic genes of Streptomyces fradiae. Antonie Van Leeuwenhoek 79:229-234. [DOI] [PubMed] [Google Scholar]
- 17.DeSanti, C. L., and W. R. Strohl. 2003. Characterization of the Streptomyces sp. strain C5 snp locus and development of snp-derived expression vectors. Appl. Environ. Microbiol. 69:1647-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Schrijver, A., and R. De Mot. 1999. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145(6):1287-1288. [DOI] [PubMed] [Google Scholar]
- 19.Doumith, M., P. Weingarten, U. F. Wehmeier, K. Salah-Bey, B. Benhamou, C. Capdevila, J. M. Michel, W. Piepersberg, and M. C. Raynal. 2000. Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. Mol. Gen. Genet. 264:477-485. [DOI] [PubMed] [Google Scholar]
- 20.Geistlich, M., R. Losick, J. R. Turner, and R. N. Rao. 1992. Characterization of a novel regulatory gene governing the expression of a polyketide synthase gene in Streptomyces ambofaciens. Mol. Microbiol. 6:2019-2029. [DOI] [PubMed] [Google Scholar]
- 21.Gourmelen, A., M. H. Blondelet-Rouault, and J. L. Pernodet. 1998. Characterization of a glycosyl transferase inactivating macrolides, encoded by gimA from Streptomyces ambofaciens. Antimicrob. Agents Chemother. 42:2612-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guthrie, E. P., C. S. Flaxman, J. White, D. A. Hodgson, M. J. Bibb, and K. F. Chater. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144:727-738. [DOI] [PubMed] [Google Scholar]
- 24.Hesketh, A., W. J. Chen, J. Ryding, S. Chang, and M. Bibb. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karray, F., E. Darbon, N. Oestreicher, H. Dominguez, K. Tuphile, J. Gagnat, M. H. Blondelet-Rouault, C. Gerbaud, and J. L. Pernodet. 2007. Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 153:4111-4122. [DOI] [PubMed] [Google Scholar]
- 26.Khanin, R., V. Vinciotti, V. Mersinias, C. P. Smith, and E. Wit. 2007. Statistical reconstruction of transcription factor activity using Michaelis-Menten kinetics. Biometrics 63:816-823. [DOI] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 28.Kuhstoss, S., M. Huber, J. R. Turner, J. W. Paschal, and R. N. Rao. 1996. Production of a novel polyketide through the construction of a hybrid polyketide synthase. Gene 183:231-236. [DOI] [PubMed] [Google Scholar]
- 29.McKenzie, N. L., and J. R. Nodwell. 2007. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 189:5284-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, H. C., F. Karray, S. Lautru, J. Gagnat, A. Lebrihi, T. D. Ho Huynh, and J. L. Pernodet. 2010. Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob. Agents Chemother. 54:2830-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, S. H., and K. F. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pernodet, J. L., M. T. Alegre, M. H. Blondelet-Rouault, and M. Guerineau. 1993. Resistance to spiramycin in Streptomyces ambofaciens, the producer organism, involves at least two different mechanisms. J. Gen. Microbiol. 139:1003-1011. [DOI] [PubMed] [Google Scholar]
- 35.Pinnert-Sindico, S. 1954. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. Ann. Inst. Pasteur (Paris) 87:702-707. [PubMed] [Google Scholar]
- 36.Raynal, A., F. Karray, K. Tuphile, E. Darbon-Rongere, and J. L. Pernodet. 2006. Excisable cassettes: new tools for functional analysis of Streptomyces genomes. Appl. Environ. Microbiol. 72:4839-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 38.Rigali, S., H. Nothaft, E. E. Noens, M. Schlicht, S. Colson, M. Muller, B. Joris, H. K. Koerten, D. A. Hopwood, F. Titgemeyer, and G. P. van Wezel. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237-1251. [DOI] [PubMed] [Google Scholar]
- 39.Rigali, S., F. Titgemeyer, S. Barends, S. Mulder, A. W. Thomae, D. A. Hopwood, and G. P. van Wezel. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 42.Stratigopoulos, G., N. Bate, and E. Cundliffe. 2004. Positive control of tylosin biosynthesis: pivotal role of TylR. Mol. Microbiol. 54:1326-1334. [DOI] [PubMed] [Google Scholar]
- 43.Stratigopoulos, G., and E. Cundliffe. 2002. Inactivation of a transcriptional repressor during empirical improvement of the tylosin producer, Streptomyces fradiae. J. Ind. Microbiol. Biotechnol. 28:219-224. [DOI] [PubMed] [Google Scholar]
- 44.Takano, E. 2006. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9:287-294. [DOI] [PubMed] [Google Scholar]
- 45.White, J., and M. Bibb. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, D. J., Y. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y. X., K. Perry, V. A. Vinci, K. Powell, W. P. Stemmer, and S. B. del Cardayre. 2002. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644-646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.