Abstract
When they are available, Sinorhizobium meliloti utilizes C4-dicarboxylic acids as preferred carbon sources for growth while suppressing the utilization of some secondary carbon sources such as α- and β-galactosides. The phenomenon of using succinate as the sole carbon source in the presence of secondary carbon sources is termed succinate-mediated catabolite repression (SMCR). Genetic screening identified the gene sma0113 as needed for strong SMCR when S. meliloti was grown in succinate plus lactose, maltose, or raffinose. sma0113 and the gene immediately downstream, sma0114, encode the proteins Sma0113, an HWE histidine kinase with five PAS domains, and Sma0114, a CheY-like response regulator lacking a DNA-binding domain. sma0113 in-frame deletion mutants show a relief of catabolite repression compared to the wild type. sma0114 in-frame deletion mutants overproduce polyhydroxybutyrate (PHB), and this overproduction requires sma0113. Sma0113 may use its five PAS domains for redox level or energy state monitoring and use that information to regulate catabolite repression and related responses.
Sinorhizobium, Rhizobium, Bradyrhizobium, and Azorhizobium (rhizobia) are important nitrogen-fixing prokaryotes. These grow in the soil as free-living organisms but can also live as nitrogen-fixing symbionts inside roots of plants belonging to the family Leguminosae (8, 11, 21, 33, 41, 54). Rhizobia are able to utilize a wide range of compounds as carbon sources, such as sugars, amino acids, and tricarboxylic acid (TCA) cycle intermediates. Studies have shown that the C4-dicarboxylic TCA cycle intermediates succinate, fumarate, and malate support high rates of growth in laboratory medium and are used by rhizobia in preference to carbon sources including glucose, fructose, galactose, lactose, and myo-inositol (23, 26, 44, 63).
The phenomenon of Sinorhizobium meliloti utilizing succinate and similar C4-dicarboxylic acids in preference to other compounds is called succinate-mediated catabolite repression (SMCR) (9). One of the first reports of catabolite repression in S. meliloti (then Rhizobium meliloti) showed that S. meliloti exhibited diauxic growth in a medium containing 0.2% succinate and 0.2% lactose as carbon sources (63). This study also showed that the production of β-galactosidase was repressed when succinate and lactose were present together and that it increased to higher levels after succinate had been exhausted from the medium.
Succinate and other C4-dicarboxylic acids are sensed and transported by the Dct (dicarboxylate transport) system which is encoded by dctA, dctB, and dctD (48, 66, 69-71). DctB is activated by the presence of C4-dicarboxylic acids and autophosphorylates. Activated DctB phosphorylates DctD, which then binds upstream of dctA along with σ54-RNA polymerase to initiate transcription (70). dctA encodes the permease required for transport of succinate and other C4-dicarboxylic acids. When succinate is in abundance, S. meliloti will preferentially import this carbon source for metabolism and inhibit other sugar transport systems, like those needed for α- and β-galactoside transport, using inducer exclusion or expulsion (9, 16).
Model organisms such as Escherichia coli and Bacillus subtilis use the phosphotransferase system (PTS) to phosphorylate and transport glucose into the cell and establish catabolite repression. However, S. meliloti does not have a complete PTS; rather, it contains a select set of PTS proteins. These include (i) PtsP (EINtr), which is phosphorylated by phosphoenolpyruvate; (ii) Hpr, which is phosphorylated on His22 by EINtr-P; (iii) two EIIA-like proteins, ManX and EIIANtr, each of which can be phosphorylated by Hpr-His22P; and (iv) HprK, a regulatory protein kinase which can phosphorylate Hpr on Ser53. S. meliloti lacks a typical enzyme I, as well as the PTS transport-related proteins EIIB and EIIC (2). Thus, the S. meliloti PTS proteins do not function as part of a classical PTS pathway of phosphorylation and transport. However, they are involved in regulating SMCR (2). Presumably, other proteins that carry information about the metabolic status of the cell must input information into the PTS, and the PTS also likely interacts with downstream proteins. The system of signal transduction that regulates SMCR in S. meliloti is interesting because the favored carbon sources enter S. meliloti not through the PTS, as they do in model organisms like E. coli and B. subtilis, but rather through DctA. Because of these differences from standard models, it is important to better understand the regulation of catabolite repression in S. meliloti. Such information will shed light on the control of global gene expression not only in this organism but also in other alphaproteobacteria, most of which contain only a partial PTS, and in other bacteria that preferentially catabolize TCA cycle intermediates.
To this end, we aimed to find mutations that affect core components needed for SMCR. We hypothesized that such mutations should affect the succinate-mediated repression of at least several secondary carbon sources. Three disaccharides which are transported independently and cleaved to monomers by different enzymes were chosen as secondary carbon sources: an α-galactoside (raffinose), a β-galactoside (lactose), and an α-glucoside (maltose). To find genes involved in the succinate-regulated catabolism of these sugars, we utilized random transposon mutagenesis of S. meliloti. One mutant strain exhibited a relaxation of SMCR when grown in succinate plus raffinose, lactose, or maltose. This strain had an insertion in sma0113, which encodes the histidine kinase of a two-component regulatory system. This paper describes the isolation and initial characterization of sma0113 mutants and characterization of sma0114, the neighboring gene, which encodes a response regulator.
MATERIALS AND METHODS
Growth media.
S. meliloti strains were grown in tryptone-yeast (TY) broth, and E. coli strains were grown in lysogeny broth (LB) (6, 49). SOC medium was used during the initial incubation of bacterial cells after transformation by electroporation (2, 49). M9 minimal medium was supplemented with cobalt chloride at 5 ng/ml (2, 49). Antibiotics were used as follows: ampicillin, 50 μg/ml (Ap50); gentamicin, 30 μg/ml for E. coli and 15 μg/ml (Gm15) for S. meliloti; neomycin, 200 μg/ml (Nm200) or 100 μg/ml (Nm100); spectinomycin, 100 μg/ml (Sp100); streptomycin, 500 μg/ml (Sm500) or 250 μg/ml (Sm250); kanamycin, 25 μg/ml (Km25); tetracycline, 10 μg/ml (Tc10). S. meliloti strains were grown on either M9 minimal medium plus 0.4% glycerol or TY agar plates at 30°C, and E. coli strains were grown on LB agar plates incubated at 37°C. The same temperatures were used for liquid cultures.
Transposon mutagenesis.
S. meliloti wild-type strain Rm1021 was mutagenized by mating with E. coli strain MM294a/pRK609 (Table 1). Plasmid pRK609 is a self-transmissible plasmid used to deliver TnphoA. The two strains were mixed together on a TY plate without antibiotics and incubated for 24 h at 30°C. With a toothpick, swaths of bacteria were picked from the plate, suspended in 100 μl of M9 minimal medium to give moderately turbid suspensions, and spread on M9 plates containing 0.2% succinate plus 0.02% lactose, 60 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), Sm500, and Nm200. Using this method, approximately 30,000 colonies were screened and 98 strains, which were dark blue, were retained for the study.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference(s) or source |
|---|---|---|
| S. meliloti strains | ||
| Rm1021 | Wild type (Smr) | 37 |
| RB111 | Rm1021::Δhpr (Smr) (in-frame deletion) | 2 |
| CAP43 | Rm1021 smc02324::pCAP114 (Smr Nmr) | 1 |
| CAP95 | RB111 smc02324::pCAP114 (Smr Nmr) | 3 |
| CAP149 | PG30 smc02324::pCAP114 (Smr Nmr) | This work |
| DG3155 | Rm1021 phbA::Tn5-233 (Smr Spr) | This work |
| DG3159 | PG22 smc02324::pCAP77 | This work |
| PG22 | Rm1021::Δsma0113 (Smr) (in-frame deletion) | This work |
| PG24 | Rm1021::Δsma0114 (Smr) (in-frame deletion) | This work |
| PG26 | Rm1021::Δsma0113Δsma0114 (Smr) (in-frame deletion) | This work |
| PG30 | PG22 Δsma0113 Δhpr (Smr) (in-frame deletion) | This work |
| RB68 | Rm1021 sma0113::TnphoA (Smr Nmr) | This work |
| PG76 | PG24 smc02324::pPG138 (Smr Nmr) | This work |
| PG79 | PG24 smc02324::pPG139 (Smr Nmr) | This work |
| PG92 | PG22 smc02324::pPG155 (Smr Nmr) | This work |
| PG94 | PG22 smc02324::pPG156 (Smr Nmr) | This work |
| PG98 | PG22 smc02324::pCAP87 (Smr Nmr) | This work |
| PG99 | PG24 smc02324::pCAP87 (Smr Nmr) | This work |
| PG100 | PG26 smc02324::pCAP87 (Smr Nmr) | This work |
| PG104 | PG26 smc02324::pPG158 (Smr Nmr) | This work |
| PG105 | Rm1021 smc02324::pCAP87 (Smr Nmr) | This work |
| E. coli strains | ||
| MM294a | Host for plasmid pRK609 | 4, 19 |
| XL1Blue | Used to harbor and propagate plasmids (Tcr) | Stratagene |
| Plasmids | ||
| pJQ200SK | Suicide plasmid containing Bacillus subtilis sacB (Gmr) | 46 |
| pGEM-T Easy | T/A cloning vector for cloning PCR products (Apr) | Promega |
| pCAP77 | Suicide vector pMB438 containing smc02324 with trp terminator (Kmr Nmr) | 1 |
| pCAP87 | Suicide vector pMB438 containing smc02324 with trp terminator and trp promoter (Kmr Nmr) | 1 |
| pCAP114 | pCAP77 encoding Hpr-S53A (Nmr) | 1 |
| pMB438 | Suicide vector (Kmr Nmr) | 5 |
| pMB393 | Broad-host-range plasmid (Spr) | 5 |
| pRB88 | Suicide vector pJQ200SK containing central 228-bp deletion region of hpr (Gmr) | 2 |
| pRK609 | Used to deliver TnphoA (Cmr Kmr Nmr) | 19, 32 |
| pPG44 | pGEM-T Easy with sma0113 177-bp left flanking region (Apr) | This work |
| pPG45 | pGEM-T Easy with sma0113 135-bp right flanking region (Apr) | This work |
| pPG46 | Suicide vector pJQ200SK containing 177-bp/135-bp flanking regions of sma0113 (Gmr) | This work |
| pPG47 | pGEM-T Easy with sma0114 207-bp left flanking region (Apr) | This work |
| pPG52 | pGEM-T Easy with sma0114 175-bp right flanking region (Apr) | This work |
| pPG53 | Suicide vector pJQ200SK containing 207-bp/175-bp flanking regions of sma0114 (Gmr) | This work |
| pPG67 | Broad-host-range plasmid with trp terminator (Tcr) | This work |
| pPG138 | pCAP87 encoding Sma0114 (Nmr) | This work |
| pPG139 | pCAP87 encoding Sma0114D57A (Nmr) | This work |
| pPG155 | pCAP87 encoding Sma0113 (Nmr) | This work |
| pPG156 | pCAP87 encoding Sma0113H670K (Nmr) | This work |
| pPG158 | pCAP87 encoding Sma0113 and Sma0114 (Nmr) | This work |
Inverse PCR to identify the TnphoA insertion site.
Identification of the TnphoA insertion site followed the standard inverse PCR protocol with the following specific primers and restriction enzymes (42). Genomic DNA was isolated from strain RB68 and digested with XhoI and SalI. The digested DNA was ligated into circles, and the regions flanking the TnphoA insertion were amplified by inverse PCR with primers 111 (5′-GGCAAGCTGGGCTCTATTCAG-3′) and 116 (5′-GAACGTTACCATGTTAGGAGGTC-3′). The PCR fragments were gel purified and subcloned into pGEM-T Easy (Promega) and sequenced by standard methods.
Analysis of diauxic growth.
S. meliloti starter cultures were grown in M9 minimal medium with 0.4% glycerol and streptomycin (Sm250) at 30°C. Cells were washed and resuspended in M9 minimal medium without a carbon source. In order to ensure that all of the strains were in exactly the same medium, a master mix was created that contained M9 minimal medium, 0.05% succinate plus the secondary carbon source at 0.1%, and appropriate antibiotics. All of the strains in an experiment were resuspended in master mix to an initial optical density at 595 nm (OD595) of 0.005. Cell density was measured in 100-μl samples in a 96-well plate with a Bio-Rad 550 plate reader at 595 nm (the path length was 0.32 cm).
Automated growth curves were done in a Bio-Tek Synergy HT-1 Multi-Detection Microplate reader (Bio-Tek Instruments, Winooski, VT). When grown in a 24-well culture plate (Becton Dickinson), cells were resuspended in a total volume of 450 μl per well. Cells in 48-well culture plates were resuspended in a total volume of 225 μl per well (the path length was 0.30 cm). OD595 measurements were taken every 15 min using the KC4 v.3.4 software (Bio-Tek), which was run automatically using Automate software (Unisyn Software, Los Angeles, CA).
Construction of S. meliloti strains.
S. meliloti strains PG22, PG24, and PG26 contain in-frame deletions of sma0113, sma0114, and both sma0113 and sma0114, respectively. The general protocols used for constructing each were similar. To construct the sma0113 deletion, two plasmids were constructed that contained an upstream flanking region of sma0113 and a downstream flanking region. The upstream flanking region of the gene was amplified by PCR using primer 265 (5′-GGTCTAGAGCGGGATGAGGAGAAGTG-3′), which contains a native XbaI site (underlined), and primer 266 (5′-GCATGCCTCCGCGTTGTAGCCCACGAG-3′), which contains an added SphI site. The resulting 177-bp amplification product was inserted into T/A cloning vector pGEM-T Easy to create plasmid pPG44. The downstream flanking region of the gene was amplified by PCR using primers 267 (5′-GCATGCGTCATGGTCCCAGCGCGGACC-3′), which contains an added SphI site, and primer 268 (5′-CAGGCTGCAGCGCGAGGTCTCGAC-3′), which contains a native PstI site. The resulting 135-bp amplification product was inserted into pGEM-T Easy to create plasmid pPG45. pPG44 was digested with XbaI and SphI to remove the upstream flanking region. pPG45 was digested with SphI and PstI to remove the downstream flanking region. sacB suicide plasmid pJQ200SK was digested with XbaI and PstI (46). A triple-fragment ligation was performed. The left and right flanks were ligated to each other at the SphI sites, the upstream end of this fragment was ligated to the XbaI site of pJQ200SK, and the downstream end was ligated to the PstI site of pJQ200SK. The resulting plasmid, pPG46, contained the sma0113 gene with an in-frame deletion of bases 180 to 2427.
pPG46 was transformed into strain Rm1021 by electroporation (2.2 kV, 1-mm-gap cuvettes). These cells were plated on TY Gm15 Sm500 plates to select for pPG46 integration into sma0113. A gentamicin/streptomycin-resistant colony was then grown at 30°C overnight in TY with only Sm500 to allow the plasmid to undergo a second recombination event, leaving behind the sma0113 gene with the in-frame deletion. This overnight culture was plated onto a TY plate supplemented with 5% sucrose and Sm500. Cells that lost the plasmid and its sacB gene should grow on the sucrose plates and yield gentamicin-sensitive colonies because sucrose is toxic in the presence of sacB (46). Individual sucrose-resistant colonies were tested on a TY Gm15 Sm500 plate. Colonies that were gentamicin sensitive were tested further by PCR screening. A colony with the sma0113 in-frame deletion was called strain PG22. The in-frame deletion of sma0113 was confirmed by sequencing the appropriate PCR product amplified from the chromosome.
The sma0114 in-frame deletion was constructed in the same manner as the sma0113 in-frame deletion. The upstream flank of sma0114 was amplified by PCR using primer 271 (5′-GTCATGGTCCCAGCGCGGACC-3′) and primer 269 (5′-GCATGCGGATTCGTCTTCGACCACG-3′) containing an added SphI site. The resulting 207-bp amplification product was inserted into pGEM-T Easy to create plasmid pPG47. The downstream flank of sma0114 was amplified by PCR using primer 290 (5′-GCATGCCCGAACATCCCCTTGCTGACGAAG-3′) containing an added SphI site and primer 268. The resulting 175-bp amplification product was inserted into pGEM-T Easy to create plasmid pPG52. pPG47 was digested with SphI and SalI (SalI site was part of the pGEM-T Easy multicloning site). pPG52 was digested with SphI and PstI. Plasmid pJQ200SK was digested with SalI and PstI. A triple-fragment ligation was performed, and the two flanking regions were annealed at the SphI sites and the SalI and PstI sites ligated the flanking regions to pJQ200SK. The resulting vector, pPG53, contained sma0114 with an in-frame deletion of bases 136 to 283. Integration of pPG53 and replacement of the native copy of sma0114 were done as described above for sma0113. A colony with the sma0114 in-frame deletion was called strain PG24. The in-frame deletion of sma0113 was confirmed by sequencing the appropriate PCR product amplified from the chromosome.
Strain PG26, the sma0113 sma0114 double-deletion mutant, was created using sma0113 in-frame deletion strain PG22 as the host strain and the sma0114 deletion plasmid pPG53 to replace sma0114. The integration of pPG53 and replacement of the native copy of sma0114 were done as described for sma0113.
Genetic complementation of sma0113 and sma0114 mutations.
The in-frame deletion mutations of sma0113 and sma0114 were complemented by inserting wild-type copies of the genes into the rhamnose-binding protein gene (smc02324) using pCAP87. pCAP87 is a suicide plasmid containing a constitutive trp promoter upstream of the multiple cloning site and an internal fragment of smc02324 which targets the plasmid to an operon that encodes a rhamnose transport system (1, 35, 45). Wild-type copies of sma0113 and sma0114 were cloned into plasmid pCAP87. All pCAP87-based plasmids were electroporated into competent S. meliloti strains. These cells were plated onto TY Nm200 Sm500 plates to screen for plasmid integration. Individual colonies were restreaked onto M9 Nm200 Sm500 plates supplemented with 0.2% rhamnose. The colonies that had diminished growth on rhamnose were screened by PCR to confirm that plasmids recombined into the smc02324 gene. This method complemented the mutations of strains PG22 (Δsma0113), PG24 (Δsma0114), and PG26 (Δsma0113 Δsma0114), creating strains PG92, PG76, and PG104, respectively (1).
Plasmid pCAP87, without any gene insertion, was electroporated into strains Rm1021, PG22, PG24, and PG26, creating strains PG105, PG98, PG99, and PG100, respectively, which were used as negative controls. All of the genes used for complementation were verified by sequencing. Their positions and orientations in the smc02324 gene were verified by PCR using primer 408 (5′-GCGGCCGCGCGGCATCAAGGTCATCTCC-3′), which binds to the integrated vector (1), and primer 268 (5′-CAGGCTGCAGCGCGAGGTCTCGAC-3′), which binds downstream of sma0114.
Construction of site-directed mutant strains.
Site-directed mutants were constructed using the overlap extension method with Advantage Taq polymerase (Clontech) (22, 49). The sma0114-D57A product was ligated into pCAP87 to create plasmid pPG139. Plasmid pPG139 was electroporated into strain PG24, which recombined into smc02324 to create PG79. The sma0113-H670K product was ligated into pCAP87 to create plasmid pPG156. Plasmid pPG156 was electroporated into strain PG22, which recombined into smc02324 to create strain PG94. All of the genes altered by site-directed mutagenesis were verified by sequencing.
Strain construction for epistasis experiments.
An in-frame deletion of the hpr gene was introduced into strain PG22 (Δsma0113) using the suicide plasmid pRB88 to give strain PG30 (Δsma0113 Δhpr). Plasmid pCAP114 was introduced into the smc02324 gene of strain PG30 to give strain CAP149, a Δsma0113 Δhpr double-mutant strain that contained a single copy of hpr-S53A expressed from its native promoter. A control strain containing only the vector pCAP77 in smc02324 was constructed by integrating pCAP77 into the smc02324 gene of PG22. This resulted in strain DG3159.
RT-PCR experiments.
Colonies of strains used in reverse transcription (RT)-PCR were picked off freshly streaked (48 h) M9 plus glycerol (0.4%) plus Sm250 plus Nm100 plates and grown overnight with shaking at 30°C in 150-by-18-mm tubes containing 3 ml of M9 plus succinate plus Sm250 to mid-exponential phase (OD595, ∼ 0.1). Cultures were diluted from 1:10 to 1:20 in 3 ml of the same medium and allowed to grow under the same conditions until they reached an OD595 of 0.1 to 0.2. Cell density was measured in 100-μl samples in a 96-well plate with a Bio-Rad 550 plate reader at 595 nm. A 0.75-ml culture volume was added to 0.75 ml of Qiagen RNAlater. Cells were pelleted, and total RNA was extracted using a Promega SV RNA isolation kit, skipping the recommended DNase treatment step. Instead, an Ambion TurboDNA-free kit was used to remove contaminating DNA from the RNA samples following purification. RT was done with 50 ng of total RNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase from NEB and a pool of primers (0.25 μg of each in a 25-μl reaction mixture) to make cDNA copies of sma0114, phbA (smc03879), phaZ (smc02770), and smc00128 transcripts (see the supplemental material for primer sequences). Controls were generated the same way but without M-MLV reverse transcriptase in order to assess the levels of contaminating DNA. All of the experiments reported in this paper were free of contaminating DNA. One microliter of each RT reaction mixture or of the corresponding no-M-MLV control was subjected to 32 cycles of PCR using primers designed to amplify cDNA arising from sma0114, phbA, phaZ, and smc00128 transcripts (see the supplemental material for primer sequences). The cycling parameters were 95°C for 20 s, 60°C for 20 s, and 72°C for 45 s. Using this protocol, the amount of PCR product was proportional to the amount of input cDNA. Levels of sma0114, phbA, and phaZ RT-PCR products were normalized to the level of the constitutively expressed smc00128 product (30, 40).
Analysis of PHB levels.
A 1.5-ml volume of each culture used for RT-PCR was centrifuged, and the pellets were suspended in a solution of 10% ethanol plus 0.5 μg/ml Nile Red, which was diluted 1:2,000 from a 1-mg/ml stock made in dimethyl sulfoxide. After a 30-min incubation, cells were washed twice with distilled water and resuspended in distilled water. A 100-μl volume of each suspension was transferred to a 96-well microtiter plate, and the OD595 and Nile Red fluorescence were determined in a Bio-Tek Synergy HT-1 Multi-Detection Microplate reader. For Nile Red fluorescence determinations, excitation was at 570 nm and emission was measured at 620 nm. The fluorescence was normalized by dividing it by the OD595 of the sample. This method is based on previously published assays (47, 55). A transposon insertion in the 86th codon of phbA (smc08379) was isolated and transduced into strain Rm1021 to give strain DG3155. This strain was used as a negative control when measuring polyhydroxybutyrate (PHB) levels.
β-Galactosidase assays.
S. meliloti cells were grown in M9 medium supplemented with 0.4% glycerol and streptomycin. After cells had reached stationary phase, they were washed with M9 medium without a carbon source and resuspended in 50 ml M9 medium with streptomycin supplemented with 0.2% succinate, 0.2% lactose, or a combination of 0.2% succinate plus 0.2% lactose in a 250-ml flask. The cells were incubated at 30°C, and growth was monitored by determining the OD595s of 100-μl samples. Samples were removed, the OD595 was recorded at each time point for later use in plotting data, and the pellets were frozen. Pellets were later resuspended in Z buffer, and β-galactosidase assays were conducted as previously described (38). OD415 readings from the first 5 to 30 min of a kinetic assay were used to calculate the slope of the linear portion of the curve by a least-squares fit method. This rate was divided by the OD595 of the cell pellet suspension to give β-galactosidase units per unit of mass. β-Galactosidase activity was plotted as a function of culture OD595 determined by growth curve measurements.
Nodulation assay.
Alfalfa (Medicago sativa cv. AS13R) seeds were sterilized and sprouted as described previously (15). Seedlings were then placed on basic nodulation medium (BNM) agar slants in 20-mm glass tubes and inoculated with 500 μl of a suspension of S. meliloti (13). Suspensions were made by washing 1 ml of TY-grown culture with 1 ml of BNM and resuspending the pellet in 10 ml of BNM. Plant growth tubes were loosely capped and incubated in a growth chamber with an 8/16-h dark/light cycle at 26°C. Thirty days after inoculation, nodules were counted and plant shoots were cut above the cotyledons, dried, and weighed.
RESULTS
Screening to find genes involved in SMCR.
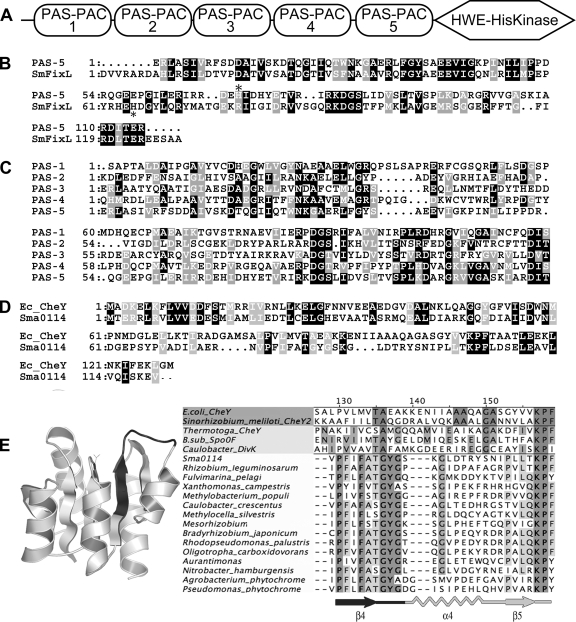
In order to identify genes involved in SMCR, we mutagenized strain Rm1021 with TnphoA. When mutagenized bacteria were plated on M9 succinate plus lactose plus 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), most of the colonies were light blue because succinate inhibited the expression of endogenous β-galactosidase. We screened approximately 30,000 colonies and isolated 98 dark blue strains which expressed β-galactosidase in the presence of succinate plus lactose. The strains underwent further screening to eliminate those that were β-galactosidase constitutive, as well as those unable to grow on succinate. All of the mutants were able to grow on succinate, but one mutant, strain RB97, was dark blue on M9 minimal agar with 0.2% succinate plus X-Gal, indicating that it constitutively expressed β-galactosidase. Fifty-one of the remaining strains were further analyzed by growth in three different liquid M9 minimal media containing succinate plus lactose, maltose, or raffinose to check for relief of succinate-imposed diauxie; the remaining 46 strains remained untested. One mutant, strain RB68, showed a shortened diauxic lag in all three media. The insertion site of the transposon in RB68, identified by inverse PCR, was in sma0113, a putative histidine kinase on the pSymA megaplasmid. sma0113 encodes an 853-amino-acid protein that contains five tandem PAS-PAC domains and a recently described HWE kinase domain (28) (Fig. 1). PAS-PAC domains often contain cofactors such as heme, flavin adenine dinucleotide, or flavin mononucleotide. They are frequently present in signaling proteins that respond to light, redox potential, oxygen, energy status, or electron transport chain activity (7, 39, 51, 60, 61). In some cases, they are involved in responses to TCA cycle intermediates such as fumarate and succinate (24, 25, 29, 39). In Sma0113, the PAS1 and PAS4 domains are the most similar to each other (35.5% identical), while the PAS1 and PAS3 domains are the least similar (19.8% identical). PAS5 is 35.8% identical to the O2-sensing PAS domain in the S. meliloti FixL protein, though PAS5 lacks the histidine residue that covalently binds heme in FixL (Fig. 1B and C) (60). sma0114 is downstream of sma0113, and together these genes form an apparent operon. sma0114 encodes a CheY-like protein and thus lacks the DNA-binding domain seen in many response regulators (Fig. 1D) (34, 50, 57, 59, 65).
FIG. 1.
Overview of the Sma0113 and Sma0114 proteins. (A) Diagram of the Sma0113 showing the five PAS domains and the HWE kinase domain. (B) ClustalX plot showing similarity between the PAS-5 domain of Sma0113 and the PAS domain of S. meliloti FixL. The asterisk indicates the histidine in FixL that is covalently bound to heme. (C) ClustalX plot showing similarity among the five PAS domains of Sma0113. (D) ClustalX plot showing similarity between Sma0114 and E. coli CheY. (E, left) A model of the Sma0114 protein based on coordinates of the MicA protein of Streptococcus pneumoniae (Protein Data Bank entry 1nxo). The PFxFATGY motif is shaded dark gray. (E, right) The β4-α4-β5 regions of Sma0114 and other proteins are compared. Shown is an alignment of the primary sequences of the β4-α4-β5 regions of five single-domain response regulators (RRs) whose structures have been solved, followed by Sma0114 and 14 similar RR domains. The 14 were selected by picking the top Sma0114 BLAST matches that were from differing genera. The proteins used were Sma0114 and proteins from Rhizobium leguminosarum (ZP_02295076.1), Fulvimarina pelagi (ZP_01438889.1), Xanthomonas campestris (NP_638306.1), Methylobacterium populi (YP_001923188.1), Caulobacter crescentus (NP_421809.1), Methylocella silvestris (ZP_02956328.1), Mesorhizobium loti (YP_673186.1), Bradyrhizobium japonicum (NP_771385.1), Rhodopseudomonas palustris (YP_781621.1), Oligotropha carboxidovorans (EDT30613.1), Aurantimonas sp. (ZP_01226142.1), Nitrobacter hamburgensis (YP_571619.1), Agrobacterium tumefaciens (Q8UDG1), and Pseudomonas putida (Q8VRN6).
Deletion of sma0113 causes a relaxation of SMCR.
Strains PG22 (Δsma0113), PG24 (Δsma0114), and PG26 (Δsma0113 Δsma0114), containing unmarked in-frame deletions, were constructed using sacB counterselection.
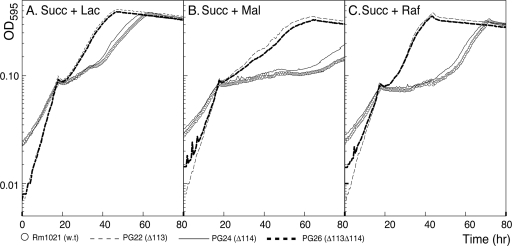
Strains PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) exhibited the same weak SMCR phenotype as RB68 (sma0113::TnphoA) in M9 medium containing succinate plus lactose, maltose, or raffinose (Fig. 2). In M9 succinate plus lactose, strains PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) had a shorter lag period than strain Rm1021 (wild type) once succinate was exhausted and before growth on lactose began (Fig. 2A). Strains PG22 and PG26 grown in M9 succinate plus maltose also showed a shortening of the diauxic lag compared to Rm1021 (Fig. 2B). The weakened SMCR was most apparent when these strains were grown in succinate plus raffinose, an α-galactoside. In that case, the lag period of these mutants was almost four times shorter than in strain Rm1021 (wild type) (Fig. 2C). Thus, PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) had a generalized relaxation of SMCR.
FIG. 2.
Deletion of sma0113 results in relief of SMCR in succinate plus lactose, maltose, or raffinose. Strains Rm1021 (wild type), PG22 (Δsma0113), PG24 (Δsma0114), and PG26 (Δsma0113 Δsma0114) were grown in M9 medium containing succinate plus one of three secondary carbon sources. Strains PG22 and PG26 showed a relaxation of SMCR, as indicated by the shortened diauxic lag. The curves were time shifted to align them at the point of succinate exhaustion in order to more readily compare diauxic lag lengths.
Strain PG24 (Δsma0114) did not show altered diauxic growth in M9 succinate plus lactose, maltose, or raffinose (Fig. 2). The growth phenotype, including the length of the diauxic lag, under these conditions was the same as that seen in strain Rm1021 (wild type). These results are discussed later.
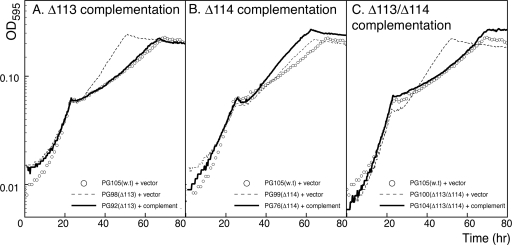
Complementation of the Δsma0113 mutation restores the wild-type SMCR phenotype during growth on mixed carbon sources.
Complementation of the deletions in strains PG22, PG24, and PG26 was carried out by using a trp promoter to constitutively express wild-type genes as described in Material and Methods and presented in Table 1. Each complementing strain was grown in M9 minimal medium with succinate plus lactose as described previously. A complementing sma0113 gene was able to restore the wild-type SMCR phenotype to the Δsma0113 mutant (stain PG92 in Fig. 3 A). The Δsma0113 Δsma0114 double-mutant strain was complemented by the addition of the sma0113 and sma0114 genes together (strain PG104 in Fig. 3C). In addition, normal growth was restored in M9 succinate plus maltose or raffinose to the Δsma0113 and Δsma0113 Δsma0114 mutant strains by complementation (data not shown). The sma0114 deletion strain did not have a discernible SMCR phenotype. When the sma0114 gene was overexpressed in the Δsma0114 mutant (strain PG76), the phenotype remained the same as in the deletion mutant (Fig. 3B). RT-PCR experiments confirmed that the sma0114 genes were indeed overexpressed (see Fig. 5B and the supplemental material).
FIG. 3.
Complementation of mutant strains. Mutations were complemented with wild-type genes inserted into the rhamnose-binding protein gene smc02324. Cultures were grown in M9 minimal medium with 0.05% succinate plus 0.1% lactose. (A) Complementation of the Δsma0113 mutation. (B) Complementation of the Δsma0114 mutation. (C) Complementation of the Δsma0113 Δsma0114 double mutation. The curves were time shifted to align them at the point of succinate exhaustion in order to more readily compare diauxic lag lengths.
Nonphosphorylatable Sma0113 mutants exhibit relaxed SMCR during diauxic growth on succinate plus secondary carbon sources.
The two site-directed mutant strains PG94 (sma0113-H670K) and PG79 (sma0114-D57A) were constructed to make each protein unphosphorylatable by altering the residues that are phosphorylated in HWE two-component kinases (28) and response regulators (18, 58). Strain PG94 (sma0113-H670K) had the same relaxed SMCR phenotype as strain PG22 (Δsma0113), indicating that phosphorylation is needed for this protein to signal in the bacteria (see Fig. S2A in the supplemental material). Strain PG79 (sma0114-D57A) did not show an SMCR phenotype (see Fig. S2B in the supplemental material).
SMCR is relaxed in strains with Δsma0113 mutations, as shown by β-galactosidase expression during growth in M9 succinate plus lactose.
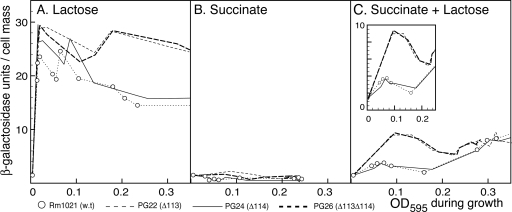
Plate assays and diauxic growth curves showed a relaxation of SMCR in strains PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) grown in M9 succinate plus lactose. We wanted to test biochemically whether the genes for lactose utilization are upregulated by lactose when succinate is present. This was done by measuring endogenous β-galactosidase during growth in M9 succinate plus lactose. Cells were pregrown in M9 minimal medium plus glycerol and then transferred to M9 medium with succinate only, lactose only, or succinate plus lactose. When transferred to M9 medium with succinate as the sole carbon source, strains PG22 (Δsma0113), PG24 (Δsma0114), and PG26 (Δsma0113 Δsma0114) expressed a basal level of β-galactosidase, <2 U (Fig. 4 B). In M9 plus 0.2% lactose, β-galactosidase was induced rapidly in all four strains and was then maintained at high levels throughout exponential growth. However, the β-galactosidase levels in strains PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) stayed high as they went into stationary phase, while the β-galactosidase levels in strains Rm1021 and PG24 (Δsma0114) dropped (Fig. 4A). Interesting differences between the strains were seen when they were grown in M9 medium containing succinate plus lactose. β-Galactosidase was induced early in both PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) even though succinate was present (Fig. 4C). There was only a small corresponding increase in β-galactosidase in strains PG24 (Δsma0114) and Rm1021 (wild type). During the second phase of diauxie, when lactose is consumed, β-galactosidase levels increased at equal rates in all four strains (Fig. 4C). These results indicated that the sma0113 mutation in strains PG22 (Δsma0113) and PG26 (Δsma0113 Δsma0114) resulted in a relaxation of SMCR, which allowed lactose to induce lac genes even when succinate was present.
FIG. 4.
Deletion of sma0113 results in relief of SMCR, as measured by β-galactosidase production. In the representative experiment plotted here, the indicated strains were grown to stationary phase in M9 plus 0.4% glycerol and then diluted in M9 minimal medium containing lactose, succinate, or succinate plus lactose. (A) Endogenous β-galactosidase activity during growth in M9 minimal medium plus 0.2% lactose plotted against the OD595 of cultures as they grew. (B) Endogenous β-galactosidase activity during growth in M9 minimal medium plus 0.2% succinate plotted against the OD595 of cultures as they grew. (C). Endogenous β-galactosidase activity during growth in M9 minimal medium with 0.2% succinate plus 0.2% lactose plotted against the OD595 of cultures as they grew. The main graphs are scaled the same. The inset in panel C shows rescaled data from the first half of the succinate-plus-lactose experiment. Data points are explicitly shown for strain Rm1021. For the other curves, lines are drawn directly from data point to data point but the points have been left out for the sake of clarity.
Epistatic analysis of sma0113 and hpr-S53A.
Previous work has shown that the PTS protein Hpr is important for regulation of SMCR in S. meliloti. In order to explore the regulatory relationship between Sma0113 and Hpr, a Δsma0113 hpr-S53A double mutant (strain CAP149) was constructed. The SMCR phenotypes produced by the two mutations, when present singly, are quite different. The Δsma0113 mutation results in weakened SMCR, and the hpr-S53A allele results in very strong SMCR (3). On M9 medium plates containing succinate plus lactose and X-Gal, control strain CAP43 (wild type) showed a normal SMCR phenotype, strain DG3159 (Δsma0113) showed weak SMCR, strain CAP95 (hpr-S53A) showed very strong SMCR, and strain CAP149 (Δsma0113 hpr-S53A) demonstrated strong SMCR (data not shown). These results showed that Hpr was epistatic to Sma0113, indicating that Hpr acts downstream of Sma0113 in regulating SMCR.
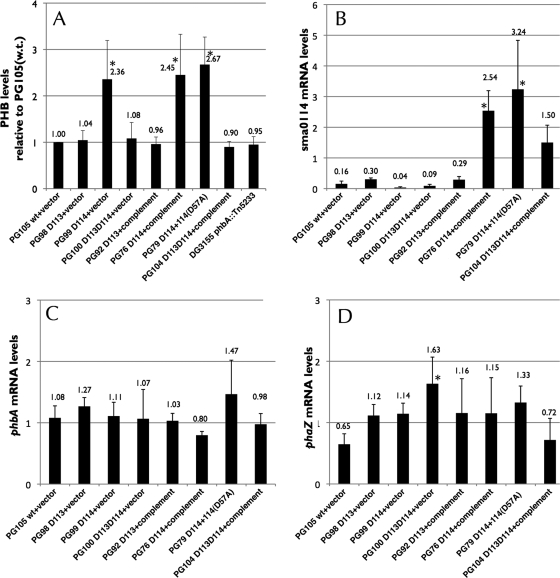
PHB levels are regulated by Sma0114.
During the course of our work on control of SMCR by the PTS in S. meliloti, we discovered that levels of the carbon storage compound PHB were often affected by mutations in hpr, manX, and hprK, which encode components of the PTS (unpublished results). Accumulation of PHB occurs when S. meliloti has extra carbon relative to other major nutrients like nitrogen and/or it has excess reducing capacity and needs to regenerate NAD(P)H (10, 14). Thus, PHB is a storage compound for both carbon and reducing power. Given that Sma0113 regulates SMCR, as does the PTS, we compared levels of PHB in wild-type S. meliloti with strains with altered sma0113 and sma0114 during growth in M9 succinate medium (Fig. 5 A). Strain PG105 (wild type) showed little or no PHB accumulation during growth in M9 succinate, and neither did strain PG98 (Δsma0113). However, strain PG99 (Δsma0114) showed relatively high levels of PHB during growth. Interestingly, strain PG100 (Δsma0113Δsma0114), which lacks both Sma0113 and Sma0114, did not show increased levels of PHB. Thus, the PHB phenotype caused by deleting sma0114 requires functional Sma0113. Heterologous overexpression of either sma0114 (in strain PG76) or sma0114-D57A (in strain PG79) did not fix the overproduction of PHB associated with the Δsma0114 mutation.
FIG. 5.
PHB and mRNA levels in select isogenic strains of S. meliloti. (A) PHB levels during growth in M9 succinate. Levels have been normalized to that found in strain PG105 (wild type). The mean and standard deviation from four independent experiments are shown. Asterisks indicate levels that are significantly different from those found in strain PG105 at a P value of <0.05. Note that the phbA mutant strain and other strains have ∼1 U of background Nile Red staining due to cell membrane and other hydrophobic compounds. (B) Levels of sma0114 mRNA during growth in M9 succinate as determined by RT-PCR. The mean and standard deviation from three independent experiments are shown. In each experiment, mRNA levels were measured two or three times. Asterisks indicate levels that are significantly different from those found in strain PG105 at a P value of <0.05. (C) Levels of phbA mRNA during growth in M9 succinate as determined by RT-PCR. The mean and standard deviation from three independent experiments are shown. In each experiment, mRNA levels were measured two or three times. No strain was found to express phbA at levels significantly different than that found in strain PG105. (D) Levels of phaZ mRNA during growth in M9 succinate as determined by RT-PCR. The mean and standard deviation from three independent experiments are shown. In each experiment, mRNA levels were measured two or three times. D113 and D114 in the genotypes represent in-frame deletions of sma0113 and sma0114, respectively. Asterisks indicate levels that are significantly different from those found in strain PG105 at a P value of <0.05. For a typical RT-PCR experiment, see Fig. S1 in the supplemental material.
Expression of the phbA and phaZ genes, which encode proteins involved in the synthesis and depolymerization of PHB, was roughly equivalent in the strains under study (Fig. 5C and D). This indicated that the Δsma0114 mutation likely acted to increase PHB levels through nontranscriptional mechanisms.
Mutations in sma0113 or sma0114 do not affect symbiosis with M. sativa.
Strains Rm1021 (wild type), PG22 (Δsma0113), and PG24 (Δsma0114) were tested to see if symbiosis was altered when they were grown in association with M. sativa. All of the strains produced functional nodules, and the mutations did not significantly affect nodulation numbers or shoot weight (see Fig. S3 in the supplemental material).
DISCUSSION
The genes and proteins needed for SMCR in S. meliloti are important regulators of carbon choice, carbon metabolism, and other physiological traits (2, 9, 16). In this study, we identified and then characterized a two-component regulatory system in S. meliloti composed of Sma0113 and Sma0114. Of these, Sma0113 is clearly involved in control of SMCR. sma0113, located on the pSymA megaplasmid, encodes a sensor histidine kinase that is part of a two-component regulatory system. Sma0113 contains five tandem PAS domains and an HWE histidine kinase domain (28). PAS domains can have various roles, which include sensing light, oxygen, redox potential, carboxylic acids, electron transport, or proton motive force (60). The genome of S. meliloti is predicted to contain a total of 23 proteins with at least one PAS domain: 5 are on pSymA, 6 are on pSymB, and 12 are on the chromosome (52). Of the 23 proteins, 7 have multiple PAS domains and 3 (Sma0113, Sma1548, and Smb20868) have five PAS domains, which is the most found in S. meliloti. Sma0113 also possesses an HWE histidine kinase domain (28). There are eight proteins with this domain in S. meliloti; half of these (Sma0113, Smc01507, Smb20515, and ExsG) also have PAS domains (64).
Strains lacking Sma0113 were shown to have shortened diauxic lags when grown in minimal medium with succinate plus lactose, maltose, or raffinose, suggesting that the Sma0113 protein is needed to provide strong catabolite repression in response to at least several different secondary carbon sources. Studies with a site-directed mutant form of Sma0113 in which the conserved histidine residue at position 670 was substituted with lysine indicated that this conserved phosphorylation site was necessary for protein function or signal transduction. A strain carrying this mutation showed the same relaxed SMCR phenotype as a strain lacking Sma0113 altogether.
In wild-type S. meliloti, β-galactosidase levels were low when the bacteria were grown on succinate alone and were high when they were grown solely on lactose. When grown in medium with succinate and lactose together, wild-type S. meliloti repressed the expression of β-galactosidase while succinate was present; once succinate was exhausted, β-galactosidase expression was upregulated for growth on lactose (Fig. 4). Strains lacking Sma0113 induced β-galactosidase early during diauxic growth when succinate was still present (Fig. 4). Moreover, the sma0113 mutant strains failed to downregulate lacZ upon entering stationary phase, suggesting that Sma0113 is also required for proper regulation of lacZ in response to signals other than succinate.
Downstream of sma0113 is sma0114, which encodes a single-domain, CheY-like, response regulator. The lack of a DNA-binding domain, which is present in many response regulators, suggests that Sma0114 signals through protein-protein interactions (Fig. 1D) (52). Sma0114 contains a conserved aspartate residue, Asp57, which is phosphorylated in CheY-like proteins upon activation, as well as Asp13, which is involved in metal binding and is required for the phosphorylation of CheY-like proteins (34, 50). An S. meliloti strain lacking sma0114 had phenotypically normal SMCR. In addition, introduction of a site-directed mutation that rendered the critical aspartate 57 residue unphosphorylatable did not affect any of the SMCR phenotypes tested. These results were puzzling and suggested that Sma0114 may not be the cognate response regulator of Sma0113. However, there are reasons to think that it may interact with Sma0113. First, the sma0114 gene is immediately downstream of sma0113 and the two likely form an operon. Second, the PHB phenotype seen in the sma0114 deletion requires Sma0113. Lastly, Sma0114 and similar response regulators have a common motif (PFxFA[T/S]GY) in the region containing the β4 strand and the following loop (Fig. 1E). In many response regulators, this region is important for activation, allosteric control, and binding of protein partners (12, 36, 56). Response regulators with this motif are found almost exclusively in the alphaproteobacteria, and they are commonly associated with PAS domain-containing HWE kinases (P. P. Garcia, unpublished results). Redundancy, or cross talk, may explain the apparent lack of phenotype seen when sma0114 is mutated in S. meliloti. In the case of redundancy, another response regulator may exist that normally interacts with Sma0113 and carries out signaling to the same downstream targets as Sma0114. In the case of cross talk, another response regulator may be phosphorylated by Sma0113, which then carries out signaling to downstream proteins when Sma0114 is absent. Both redundancy and cross talk have been reported in the literature (17, 53, 68). Alternatively, Sma0114 may not be the cognate response regulator of Sma0113 but rather act as a regulator of Sma0113 and influence its phosphorylation of other response regulators. In another alphaproteobacterium, Caulobacter crescentus, phosphorylation of multiple response regulators by a single histidine kinase and inhibition of kinases by response regulators are well established (31, 43). There are 40 histidine kinases and 58 response regulators encoded in the S. meliloti genome (20, 62). Thus, the possibility of response regulators being redundant or phosphorylated by more than one histidine kinases is not unreasonable (27, 31).
The deletion of sma0114 caused upregulation of PHB synthesis. Interestingly, the PHB upregulation in the Δsma0114 mutant strain required functional Sma0113 because a strain lacking both sma0114 and sma0113 had normal levels of PHB. RT-PCR experiments indicated that the overproduction of PHB was not caused by altered transcription of phb genes because neither phbA or phaZ levels were affected by the sma0114 deletion. This suggests that the increase in PHB in the sma0114 deletion may have been caused by altered activities of enzymes involved in its synthesis or degradation or by changes in metabolite levels in the cell.
There are several possibilities for how Sma0113 might affect SMCR. Clearly, one or more of the five PAS domains must be involved in sensing signals that are then transduced via the HWE kinase domain to downstream targets that directly or indirectly impact SMCR. PAS domains may affect kinase activity through altering the oligomerization state of the sensor kinase (39), or they may, in some cases, interact directly with the catalytic subdomain of the kinase domain (67). The nature of the signals sensed by the PAS domains is unclear. One possibility is that succinate or another TCA cycle C4-dicarboxylic acid is directly sensed by Sma0113. In E. coli, a PAS domain in the histidine kinase DcuS binds fumarate and other C4-dicarboxylic acids and transduces information to downstream targets that are needed to transport and metabolize fumarate and related carbon compounds (24, 25, 29). Another possibility is that Sma0113 senses carbon or energy availability by monitoring the levels of reducing equivalents or by monitoring electron transport. PAS domain-containing proteins, and other proteins, have been shown to act in this capacity. For example, energy taxis, which is used for movement toward favored carbon or energy sources, is an example where two-component systems transmit information about whether cells are well supplied with readily usable carbon (61). Sma0113 may operate on a similar principle in S. meliloti. When reducing and carbon equivalents are plentiful or flow through the electron transport chain is sufficient, S. meliloti cells may perceive that they are sated and repress the utilization of secondary carbon sources.
Supplementary Material
Acknowledgments
This work was supported by Department of Energy contracts DE-FG02-01ER15175 and DE-FG02-06ER15805 to D.J.G., by a University of Connecticut Research Foundation grant to D.J.G., by a Heinz Herrmann Graduate Fellowship in Cell Biology from the University of Connecticut's Graduate School to R.M.B., and by a Department of Education GAANN fellowship to P.P.G.
Footnotes
Published ahead of print on 3 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arango Pinedo, C., and D. J. Gage. 2009. Plasmids that insert into the rhamnose utilization locus, rha: a versatile tool for genetic studies in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 17:201-210. [DOI] [PubMed] [Google Scholar]
- 2.Arango Pinedo, C., R. M. Bringhurst, and D. J. Gage. 2008. Sinorhizobium meliloti mutants lacking phosphotransferase system enzyme HPr or EIIA are altered in diverse processes, including carbon metabolism, cobalt requirements, and succinoglycan production. J. Bacteriol. 190:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arango Pinedo, C., and D. J. Gage. 2009. HPrK regulates succinate-mediated catabolite repression in the Gram negative symbiont Sinorhizobium meliloti. J. Bacteriol. 191:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backman, K., and H. W. Boyer. 1983. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene 26:197-203. [DOI] [PubMed] [Google Scholar]
- 5.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. Biotechniques 29:240-245. [DOI] [PubMed] [Google Scholar]
- 6.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 9.Bringhurst, R. M., and D. J. Gage. 2002. Control of inducer accumulation plays a key role in succinate-mediated catabolite repression in Sinorhizobium meliloti. J. Bacteriol. 184:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cevallos, M. A., S. Encarnación, A. Leija, Y. Mora, and J. Mora. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-beta-hydroxybutyrate. J. Bacteriol. 178:1646-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dénarié, J., F. Debelle, and J. C. Prome. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65:503-535. [DOI] [PubMed] [Google Scholar]
- 12.Dyer, C. M., and F. W. Dahlquist. 2006. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J. Bacteriol. 188:7354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhardt, D. W., E. M. Atkinson, and S. R. Long. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256:998-1000. [DOI] [PubMed] [Google Scholar]
- 14.Encarnación, S., M. Dunn, K. Willms, and J. Mora. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gage, D., T. Bobo, and S. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gage, D. J., and S. R. Long. 1998. α-Galactoside uptake in Rhizobium meliloti: isolation and characterization of agpA, a gene encoding a periplasmic binding protein required for melibiose and raffinose utilization. J. Bacteriol. 180:5739-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara, D., T. Yamashino, and T. Mizuno. 2004. Genome-wide comparison of the His-to-Asp phosphorelay signaling components of three symbiotic genera of rhizobia. DNA Res. 11:57-65. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch, A. M. 1992. Developmental biology of legume nodulation. New Phytologist 122:211-237. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 23.Hornez, J., M. Timinouni, C. Defives, and J. Derieux. 1994. Unaffected nodulation and nitrogen fixation in carbohydrate pleiotropic mutants of Rhizobium meliloti. Curr. Microbiol. 28:225-229. [Google Scholar]
- 24.Janausch, I. G., I. Garcia-Moreno, D. Lehnen, Y. Zeuner, and G. Unden. 2004. Phosphorylation and DNA binding of the regulator DcuR of the fumarate-responsive two-component system DcuSR of Escherichia coli. Microbiology 150:877-883. [DOI] [PubMed] [Google Scholar]
- 25.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 26.Jelesko, J. G., and J. A. Leigh. 1994. Genetic characterization of a Rhizobium meliloti lactose utilization locus. Mol. Microbiol. 11:165-173. [DOI] [PubMed] [Google Scholar]
- 27.Jenal, U., and M. Y. Galperin. 2009. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr. Op. Microbiol. 12:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J. Bacteriol. 186:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kneuper, H., I. G. Janausch, V. Vijayan, M. Zweckstetter, V. Bock, C. Griesinger, and G. Unden. 2005. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 280:20596-20603. [DOI] [PubMed] [Google Scholar]
- 30.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Gen. Genet. 272:1-17. [DOI] [PubMed] [Google Scholar]
- 31.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 32.Long, S., S. McCune, and G. C. Walker. 1988. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 170:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long, S. R. 1996. Rhizobium symbiosis: nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukat, G. S., B. H. Lee, J. M. Mottonen, A. M. Stock, and J. B. Stock. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 266:8348-8354. [PubMed] [Google Scholar]
- 35.Mauchline, T. H., J. E. Fowler, A. K. East, A. L. Sartor, R. Zaheer, A. H. Hosie, P. S. Poole, and T. M. Finan. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U. S. A. 103:17933-17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McEvoy, M. M., A. Bren, M. Eisenbach, and F. W. Dahlquist. 1999. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J. Mol. Biol. 289:1423-1433. [DOI] [PubMed] [Google Scholar]
- 37.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Möglich, A., R. A. Ayers, and K. Moffat. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris, J., and J. E. Gonzalez. 2009. The novel genes emmABC are associated with exopolysaccharide production, motility, stress adaptation, and symbiosis in Sinorhizobium meliloti. J. Bacteriol. 191:5890-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mylona, P., K. Pawlowski, and T. Bisseling. 1995. Symbiotic nitrogen fixation. Plant Cell 7:869-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul, R., T. Jaeger, S. Abel, I. Wiederkehr, M. Folcher, E. G. Biondi, M. T. Laub, and U. Jenal. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole, P. S., A. Blyth, C. J. Reid, and K. Walters. 1994. myo-inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 140:2787-2795. [Google Scholar]
- 45.Poysti, N. J., and I. J. Oresnik. 2007. Characterization of Sinorhizobium meliloti triose phosphate isomerase genes. J. Bacteriol. 189:3445-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 47.Ratcliff, W. C., S. V. Kadam, and R. F. Denison. 2008. Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol. Ecol. 65:391-399. [DOI] [PubMed] [Google Scholar]
- 48.Ronson, C. W., P. M. Astwood, B. T. Nixon, and F. M. Ausubel. 1987. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 15:7921-7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Sanders, D. A., B. L. Gillece-Castro, A. M. Stock, A. L. Burlingame, and D. E. Koshland, Jr. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 264:21770-21778. [PubMed] [Google Scholar]
- 51.Sarand, I., S. Osterberg, S. Holmqvist, P. Holmfeldt, E. Skarfstad, R. E. Parales, and V. Shingler. 2008. Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ. Microbiol. 10:1320-1334. [DOI] [PubMed] [Google Scholar]
- 52.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva, J. C., A. Haldimann, M. K. Prahalad, C. T. Walsh, and B. L. Wanner. 1998. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc. Natl. Acad. Sci. U. S. A. 95:11951-11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaink, H. P. 1995. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu. Rev. Phytopathol. 33:345-368. [DOI] [PubMed] [Google Scholar]
- 55.Spiekermann, P., B. H. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbuchel. 1999. A sensitive, viable-colony staining method using Nile Red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 56.Stock, A. M., and J. Guhaniyogi. 2006. A new perspective on response regulator activation. J. Bacteriol. 188:7328-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stock, A. M., J. M. Mottonen, J. B. Stock, and C. E. Schutt. 1989. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature 337:745-749. [DOI] [PubMed] [Google Scholar]
- 58.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 59.Stock, A. M., and A. H. West. 2003. Response regulator proteins and their interactions with histidine protein kinases, p. 237-270. In M. Inouye and R. Dutta (ed.), Histidine kinases in signal transduction. Academic Press, Inc., New York, NY.
- 60.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 62.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 63.Ucker, D. S., and E. R. Signer. 1978. Catabolite-repression-like phenomenon in Rhizobium meliloti. J. Bacteriol. 136:1197-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ulrich, L. E., and I. B. Zhulin. 2007. MiST: a microbial signal transduction database. Nucleic Acids Res. 35:D386-D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Y. P., K. Birkenhead, B. Boesten, S. Manian, and F. O'Gara. 1989. Genetic analysis and regulation of the Rhizobium meliloti genes controlling C4-dicarboxylic acid transport. Gene 85:135-144. [DOI] [PubMed] [Google Scholar]
- 67.Yamada, S., H. Sugimoto, M. Kobayashi, A. Ohno, H. Nakamura, and Y. Shiro. 2009. Structure of PAS-linked histidine kinase and the response regulator complex. Structure 17:1333-1344. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448-1456. [DOI] [PubMed] [Google Scholar]
- 69.Yarosh, O. K., T. C. Charles, and T. M. Finan. 1989. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol. Microbiol. 3:813-823. [DOI] [PubMed] [Google Scholar]
- 70.Yurgel, S. N., and M. L. Kahn. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28:489-501. [DOI] [PubMed] [Google Scholar]
- 71.Yurgel, S. N., and M. L. Kahn. 2005. Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J. Bacteriol. 187:1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.