Abstract
The alternative stress response sigma factor σH has a role in regulation of the osmotic stress response and in morphological differentiation in Streptomyces coelicolor A3(2). Its gene, sigH, is located in an operon with the gene that encodes its anti-sigma factor UshX (PrsH). However, no gene with similarity to an anti-anti-sigma factor which may have a role in σH activation by a “partner-switching” mechanism is located in the operon. By using a combination of several approaches, including pull-down and bacterial two-hybrid assays and visualization of the complex by native polyacrylamide electrophoresis, we demonstrated a direct interaction between UshX and the pleiotropic sporulation-specific anti-anti-sigma factor BldG. Osmotic induction of transcription of the sigHp2 promoter that is specifically recognized by RNA polymerase containing σH was absent in an S. coelicolor bldG mutant, indicating a role of BldG in σH activation by a partner-switching-like mechanism.
Streptomycetes are Gram-positive soil-dwelling bacteria that undergo a complex program of morphological differentiation involving the aerial hyphae erected on mycelial colonies which finally undergo septation to form chains of unigenomic spores. This process is accompanied by so-called physiological differentiation, characterized by the extensive production of biologically active secondary metabolites, including the majority of known antibiotics (10). In their natural habitat, streptomycetes are challenged with diverse nutritional and environmental stresses and regulation of the differentiation process is coupled with stress-related signals (51). The response to the stresses is mediated in many bacteria by alternative sigma factors of RNA polymerase, which after association with the catalytic core of RNA polymerase direct expression of genes encoding a large repertoire of stress proteins essential for overcoming these unfavorable conditions (18, 45). Alternative sigma factors are regulated at the transcriptional, translational, and posttranslational level. One common mechanism of regulation of their activity is the reversible interaction with their specific negative regulators, anti-sigma factors (17). The activity of anti-sigma factors can be regulated by a cascade of other proteins, as exemplified by the regulation of stress responses in Bacillus subtilis, where the binding of RsbW anti-sigma factors to σB is regulated by its antagonists, RsbV anti-anti-sigma factors, by a so-called “partner-switching” mechanism involving the interplay of various protein kinases and phosphatases (40, 45). A similar mechanism involving anti-sigma factor SpoIIAB and anti-anti-sigma factor SpoIIAA is used for regulation of B. subtilis sporulation-specific sigma factor σF (2, 16).
The large genome (8.67 Mbp) of the well-studied model organism Streptomyces coelicolor A3(2) has more than 12% regulatory genes, among them as many as 65 genes encoding sigma factors, including nine close homologues of the B. subtilis general stress response sigma factor σB (5, 15). Two of them, σF (SCO4035) and σN (SCO4034), have been shown to be implicated in the control of morphological differentiation (11, 44). σH (SCO5243) has been suggested to have a dual role in S. coelicolor, in regulation of the osmotic stress response and in morphological differentiation (24, 31, 46, 49). σB (SCO0600) is believed to be a master regulator governing osmotic and oxidative stress responses as well as morphological differentiation and to affect two other σB homologues, σL (SCO7278) and σM (SCO7314), which were shown to affect sporulation (35). In contrast, σK (SCO6520) has been recently shown to have an inhibitory role in differentiation (36), and no role could be assigned to σG (SCO7341) (28). DNA microarray analysis has revealed that transcription of six genes encoding these sigma factors (sigB, -H, -I, -K, -L, and -M) was induced by osmotic stress (22). These results underlined the connection between stress responses and differentiation in S. coelicolor. However, compared to the well-defined signal cascade system for activation of this type of sigma factor in B. subtilis (2, 16, 40, 45), in streptomycetes this regulation seems to be more complex, as phylogenetic analysis has revealed as many as 45 homologues of B. subtilis RsbW anti-sigma factors and 15 homologues of RsbV anti-anti-sigma factors (41). Four anti-sigma factors have been experimentally verified to interact with and inhibit activity of their partner sigma factors in S. coelicolor. They include UshX (PrsH, SCO5244) for σH (46, 47, 50), RsbA (SCO0599) for σB (34), RsmA (SCO7313) for σM (14), and RsfA (SCO4677) for σF (26) RsfA has also been found to interact with two homologues for anti-anti-sigma factors, SCO0781 and SCO0869 (26). The homologue of anti-anti-sigma factors, RsbV (SCO7325), has been shown to interact with RsbA and to be involved in activation of σB (34). One of the other homologues of B. subtilis anti-anti-sigma factor RsbV encoded by the bldG gene (SCO3549) has been shown to have a pleiotropic function in controlling both antibiotic production and differentiation in S. coelicolor A3(2) (7, 9). Downstream of bldG is the gene apgA (SCO3548) that encodes a homologue of B. subtilis anti-sigma factor RsbW, and these two genes are cotranscribed (7). BldG has been shown to interact with ApgA as a part of a regulatory system to control both antibiotic production and differentiation in S. coelicolor, suggesting that they form a partner-switching pair; however, no target sigma factor has been identified for this system (43).
We and others have previously identified and characterized the sigH gene encoding σH in S. coelicolor A3(2) that is inferred to have a dual role in regulation of the osmotic stress response and morphological differentiation (24, 31, 46, 49). This dual role of σH is underlined by functional characterization of two genes that belong to its regulon: the sporulation-specific ssgB gene essential for septation of aerial hyphae (23, 29, 48) and the glutamate synthase gene gltB having a role in the osmotic stress response in bacteria (30). The sigH gene is part of an operon comprising ushY, ushX, and sigH, and its expression is directed by four promoters, expressed differentially during development and in response to several stress conditions (31). The operon is autoregulated through the sigHp2 promoter, and UshX has been shown to be a specific anti-sigma factor for σH (46, 47, 50). In addition to the complex transcriptional regulation that involves the differentiation-specific bldD gene encoding a negative regulator inhibiting one of the sigH promoters (24), σH has been shown to undergo posttranslational processing during differentiation and under osmotic stress conditions (50). The first gene in the sigH operon, ushY, encodes a protein with no similarity to anti-anti-sigma factors (31). Its deletion does not affect differentiation and expression of the σH-dependent sigHp2 promoter, and its product did not interact with anti-sigma factor UshX and σH (J. Kormanec, unpublished results). Therefore, some other UshX antagonists, or σH-specific anti-anti-sigma factors, must be present in S. coelicolor. Here, we identify and characterize such an anti-anti-sigma factor candidate with a role in σH activation. By using several approaches, we show that BldG plays this role and interacts with the UshX anti-sigma factor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Conditions for Escherichia coli growth and transformation were as described previously (3). The bacteria were grown in Luria-Bertani (LB) medium. Where required, the media were supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, 34 μg/ml chloramphenicol, and 100 μg/ml streptomycin. Growth of strains was monitored by measurement of absorbance at 600 nm (OD600). Growth and manipulation of S. coelicolor A3(2) were carried out as described previously (25). For preparation of cell extracts from surface culture, 108 CFU of S. coelicolor M145 were spread on sterile cellophane membranes placed on minimal medium (MM) (25) in the presence of 0.5% (wt/vol) mannitol as a carbon source and grown at 30°C. For RNA isolation from liquid-grown cultures, 109 CFU of the particular S. coelicolor strain were inoculated in 50 ml of the liquid medium NMP (25) containing mannitol (0.5% wt/vol) as a carbon source, grown at 30°C to end of the exponential phase (20 h), and subjected to the following stress conditions: 0.5 M NaCl for 30 and 60 min or 1 M sucrose for 30 and 60 min.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotypes and relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| S. coelicolor M145 | Wild-type, prototrophic; SCP1− SCP2− Pgl+ | 25 |
| S. coelicolor ΔbldG 1DB | bldG null mutant in strain M145 | 6 |
| E. coli DH5α | F−supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1; host strain for plasmid cloning and propagation | Invitrogen |
| E. coli BL21(DE3)pLysS | F−ompT hsdS (rB− mB−) dcm+gal met λ (DE3) pLysS (Cmr); host strain for overexpression from pET plasmids | Novagen |
| E. coli BTH101 | F−cya99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1; adenylate cyclase-deficient host strain for BACTH system | 21 |
| Plasmids | ||
| pET28a | Kmr; expression plasmid with T7 lac promoter | Novagen |
| pET-bldG | Kmr; pET28a containing bldG gene under the T7 lac control | This study |
| pET-ApgA | Kmr; pET28a containing apgA gene under the T7 lac control | This study |
| pET-ushX1 | Kmr; pET28a containing ushX gene under the T7 lac control | 47 |
| pKT25 | Kmr; pSU40 derivative containing B. pertussis T25 fragment of adenylate cyclase for C-terminal fusions | 20 |
| pUT18C | Apr; pUC19 derivative containing B. pertusis T18 fragment of adenylate cyclase for C-terminal fusions | 20 |
| pKT25-zip | Kmr; pKT25 containing leucine zipper domain of the yeast GNC4 activator fused to the T25 fragment | 20 |
| pUT18C-zip | Apr; pUT18C containing leucine zipper domain of the yeast GNC4 activator fused to the T18 fragment | 20 |
| pKT25-ushX | Kmr; pKT25 containing the S. coelicolor ushX gene fused to the T25 fragment | This study |
| pKT25-ApgA | Kmr; pKT25 containing the S. coelicolor apgA gene fused to the T25 fragment | This study |
| pUT18C-bldG | Apr; pUT18C containing the S. coelicolor bldG gene fused to the T18 fragment | This study |
| pUT18C-sigH | Apr; pUT18C containing the S. coelicolor sigH gene fused to the T18 fragment | This study |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance.
Recombinant DNA techniques.
DNA manipulations in E. coli were done as described previously (3). Nucleotide sequencing was performed with the ABI PRISM dye terminator cycle sequencing ready reaction kit (Applied Biosystems) and analyzed on the Applied Biosystems model 373 DNA sequencer. Sequencing with DNA G+A and T+C sequence ladders for S1 nuclease mapping was performed by a chemical method (37).
Preparation of S. coelicolor cell extracts and pull-down assays.
Solid medium-grown mycelium of S. coelicolor M145 (from 10 plates grown for 3 days) was scraped from the cellophane, and all successive steps were carried out at 4°C. The mycelium was disrupted by grinding with purified acid-washed sea sand in the presence of liquid nitrogen in a mortar for about 5 min. Subsequently, the mixture was suspended in buffer T (20 mM Tris-Cl [pH 7.9], 0.01 mM EDTA, 10 mM MgCl2, 1 mM dithiothreitol [DTT], 20% [vol/vol] glycerol), and cell debris was removed by centrifugation for 30 min at 30,000 × g. The cell extract (16 mg of proteins) was loaded on 0.15 ml of a His-Tag Bind resin (Novagen, Madison, WI) column saturated with bound His-tagged UshX prepared as described previously (47). The column was washed successively with 1 ml of buffer T with 0.1, 0.5, and 1 M concentrations of NaCl, followed by elution with 0.4 ml elution buffer (20 mM Tris-Cl [pH 7.9], 0.5 M NaCl, 1 M imidazole). The sample was dialyzed overnight against 400 ml storage buffer (12.5 mM Tris-HCl [pH 7.9], 60 mM KCl, 1 mM EDTA, 1 mM DTT, 50% [vol/vol] glycerol) and stored at −20°C. Protein samples were separated by 17.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (33). Protein concentrations were determined according to the Bradford method (8) with bovine serum albumin (BSA) as a standard.
Protein identification by tandem mass spectrometry.
The protein sample (25 μl) was mixed 1:1 with the loading buffer (20 mM Tris-Cl [pH 7.9], 2 mM EDTA, 160 mM DTT, 2% [wt/vol] SDS, 0.1 mg/ml bromophenol blue, 20% [vol/vol] glycerol), 7 μl of freshly prepared solution of 133 mg/ml iodoacetamide was added, and the mixture was incubated for 20 min in the dark at room temperature. The sample was separated by 17.5% sodium SDS-PAGE. The selected protein band was excised from the gel and subjected to in-gel digestion with trypsin as described previously (32). The extracted peptide mixture was separated with a nanoAcquity UPLC system (Waters, Milford, MA) as described previously (19). The column was connected to PicoTip emitters (New Objective) mounted into the nanospray of a Q-Tof Premier mass spectrometer (Waters, United Kingdom). Data acquisition was performed in a data-dependent manner for the time of the separation, with up to three tandem mass spectrometry events collected at the same time. Data were processed by a ProteinLynx Global Server v. 2.2 (Waters, United Kingdom) and searched against the Uniprot_Sprot protein database (Swiss Institute of Bioinformatics) with the following criteria: fixed carbamidomethylation of Cys, variable Met oxidation, allowing tryptic fragments with one missed out, peptide mass tolerance of 100 ppm, and fragment mass tolerance of 0.1 Da. The results were validated by identification of three or more consecutive fragment ions from the same series.
Bacterial two-hybrid system to investigate protein-protein interactions.
The BACTH bacterial two-hybrid system that is based on the interaction-mediated reconstruction of adenylate cyclase (cya) activity in a cya-deficient E. coli strain (20) was used to assess the potential interactions between proteins. Two-hybrid expression plasmids were generated by PCR amplification of the whole genes by using chromosomal DNA from S. coelicolor M145 as a template and selected primers (Table 2) for a specific gene which introduced an XbaI site next to the translation initiation codon and an Acc65I site downstream of the stop codon, followed by digestion of the amplified fragments with XbaI and Acc65I and their cloning in pUT18C or pKT25 digested by the same restriction enzymes. To ensure higher fidelity of DNA synthesis during PCR, Pfu DNA polymerase (Stratagene) was used. The bldG gene was amplified using the primers BLDGDHDir and BLDGDHRev, and a 340-bp DNA fragment was cloned in pUT18C, resulting in pUT18C-bldG. The sigH gene was amplified using the primers SIGH2DHDir and SIGHDHRev and cloned as an 820-bp fragment in pUT18C, resulting in pUT18C-sigH. The ushX gene was amplified using the primers USHXDHDir and USHXDHRev and cloned as a 430-bp fragment in pKT25, resulting in pKT25-ushX. The apgA gene was amplified using the primers ORF3DHDir and ORF3DHRev and cloned as a 440-bp fragment into pKT25, resulting in pKT25-ApgA. The nucleotide sequences of all the genes were checked by sequencing. The interaction of fusion proteins encoded in pUT18C-zip and pKT25-zip (20) was used as a positive control. The combinations of the resulting pUT18C and pKT25 constructs were cotransformed into the cya-deficient strain E. coli BTH101 and screened on LB medium supplemented with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Ampicillin and kanamycin were added to select the plasmids, and streptomycin was added to select the strain. The colonies were screened after 2 days at 30°C. Quantitative measurements of β-galactosidase activities in the sets of three independent transformants for each plasmid combination were performed according to Miller (39). The statistical significance of data sets was analyzed by a two-tailed Student t test with a P of <0.05 considered significant.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| BLDGDHDir | GGGGTCTAGACGTGGACCTGTCCCTGTCGACCCG |
| BLDGDHRev | GGGGGTACCCGTCAGTCGGTGGCCGCCACCGC |
| SIGH2DHDir | GGGGTCTAGAGATGAGCGAGCACGAGCGACACGC |
| SIGHDHRev | GGGGGTACCGGTTACTCCTCGACGAGCAGCTTC |
| USHXDHDir | GGGGTCTAGAGGTGTCCCAGATCGCAGGCGAGC |
| USJXDHRev | GGGGGTACCCCTCACGTCGGCCCGGGTCCCGCG |
| ORF3DHDir | GGGGTCTAGACATGGCCACCGTCGAACTCCGTTTCAG |
| ORF3DHRev | GGGGGTACCGGTCAGATCGGTGGCAGCACCGCC |
| SCO3549Fw | CCCCCATATGGACCTGTCCCTGTCGACCCG |
| SCO3549Rv | CCCCAAGCTTCGTCAGTCGGTGGCCGCCACCGC |
| SCO3548Fw | CCCCCATATGGCCACCGTCGAACTCCGTTTCAGC |
| SCO3548Rv | CCCCAAGCTTGTCAGATCGGTGGCAGCACCGCC |
Overexpression of genes in E. coli T7 RNA polymerase system and protein purification.
Overexpression of S. coelicolor ushX in an E. coli pET28a system and native purification of His-tagged UshX on His-Tag Bind resin (Novagen) were done as described previously (47). Likewise, overexpression of the S. coelicolor bldG and SCO3548 genes was done in the E. coli pET28a system. The bldG gene was amplified from chromosomal DNA from S. coelicolor M145 by PCR using the primers SCO3549Fw and SCO3549Rv (Table 2), which introduced an NdeI site next to the translation initiation codon and a HindIII site downstream of the stop codon. To ensure higher fidelity of DNA synthesis during PCR, Pfu DNA polymerase (Stratagene) was used. After PCR amplification, the 340-bp bldG-containing PCR fragment was digested with NdeI and HindIII and cloned in pET28a cut with the same enzymes, resulting in pET-bldG. The apgA gene was similarly amplified using the primers SCO3548Fw and SCO3548Rv2 (Table 2) and cloned as a 430-bp NdeI-HindIII fragment into pET28a, resulting in pET-ApgA. The nucleotide sequences of both genes were checked by sequencing. The host strain for pET series expression plasmids, E. coli BL21(DE3)/pLysS, containing either plasmid was grown in LB medium containing chloramphenicol and kanamycin at 28°C to an OD600 of 0.5. Expression was induced with 1 mM IPTG for 3 h. Cells were harvested by centrifugation (15,000 × g, 10 min), and the purification of His-tagged proteins on His-Bind resin (Novagen, Madison, WI) was carried out under native conditions as described by the manufacturer. The eluted proteins were dialyzed overnight at 4°C against 100 volumes of the storage buffer and cleared by centrifugation at 30,000 × g for 10 min, and the supernatant was stored in aliquots at −20°C.
Native PAGE of proteins.
His-tagged proteins (at 100, 200, or 400 pmol of each protein), after purification by Ni2+ column chromatography, were incubated in the 15-μl reaction mixture containing binding buffer (40 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 20% [vol/vol] glycerol) at 30°C for 15 min. The mixture was immediately loaded on a nondenaturing polyacrylamide gel (6% [wt/vol] acrylamide, 0.16% [wt/vol] bisacrylamide in 25 mM Tris, 192 mM glycine, and 20% [vol/vol] glycerol) after a 1-h prerun at 5 mA and 4°C. The same buffer was used for electrophoresis. Electrophoresis was done 6 h at 4°C and 3 mA. The gel was stained by Coomassie brilliant blue R-250 as described previously (3).
RNA isolation and S1 nuclease mapping.
Total RNA was prepared from the cultures of S. coelicolor as previously described (27). The integrity of RNA was indicated by sharp rRNA bands after electrophoresis in agarose containing 2.2 M formaldehyde and staining with ethidium bromide (3). High-resolution S1 nuclease mapping was performed according to Kormanec (27). Samples (40 μg) of RNA (estimated spectrophotometrically) were hybridized to approximately 0.02 pmol DNA probe (1,300-bp BsiWI-PstI fragment uniquely labeled on the 5′ end at the BsiWI site), as described previously (46). The DNA fragment was labeled at its 5′ ends with [γ-32P]ATP (ICN; 4,500 Ci mmol−1) and T4 polynucleotide kinase as described previously (3). The protected DNA fragments were analyzed on DNA sequencing gels together with G+A and T+C sequencing ladders derived from the end-labeled fragment (37).
RESULTS
The anti-anti-sigma factor BldG interacts with the anti-sigma factor UshX.
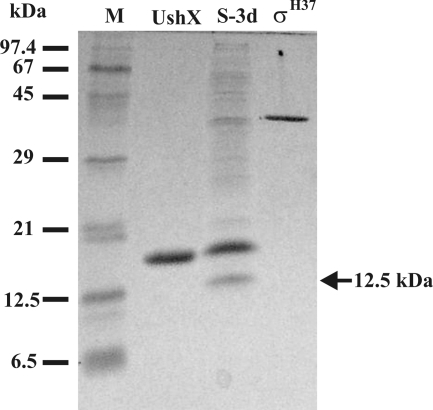
The sigH gene encoding a stress response alternative sigma factor in S. coelicolor is preceded by the ushX (prsH) gene encoding a σH-specific anti-sigma factor; however, no homologue of a gene encoding a proposed anti-anti-sigma factor has been found in the operon (31, 47, 50). Assuming a partner-switching-like mechanism of σH activation, an anti-anti-sigma factor interacting with UshX should be present in S. coelicolor. In order to identify an anti-anti-sigma factor interacting with UshX, we performed a pull-down assay on the metal chelate affinity column with the immobilized His-tagged UshX. As the level of σH dramatically increases during the development of S. coelicolor on solid media (50), we used cell extracts at different developmental stages prepared from the surface-grown strain on MM. In addition to a band of 37 kDa likely corresponding to σH, there was another dominant 12.5-kDa protein likely bound to UshX, the level of which increased during development. The 12.5-kDa protein band was missing in the negative-affinity control experiment in the absence of His-tagged UshX (data not shown). Based on this result, we selected the cell extract from S. coelicolor grown for 3 days, which corresponds to a time point when the aerial mycelium undergoes septation, as the best candidate for isolation and analysis of the 12.5-kDa proteins (Fig. 1). The protein was excised from the gel, and a tryptic digest of the protein was analyzed by electrospray tandem mass spectrometry. The analysis revealed a single match (Q9WVX8; RSBV_STRCO [UniProtKB]) with the proposed anti-anti-sigma factor SCO3549 encoded by the developmental bldG gene (7).
FIG. 1.
Pull-down assay of extract from S. coelicolor M145 grown for 3 days on solid MM. Samples were analyzed by 17.5% SDS-PAGE. Lane S-3d, eluted sample of proteins from S. coelicolor M145 extracts bound on the UshX column as described in Materials and Methods; lanes UshX and σH37, purified His-tagged proteins after Ni2+ affinity chromatography; lane M, molecular mass markers. The position of the 12.5-kDa protein band used for the tandem mass spectrometry is indicated by an arrow.
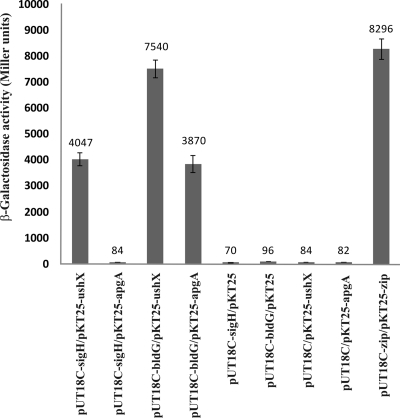
As a first strategy to confirm the interaction between BldG and UshX, we used a bacterial two-hybrid system (BACTH) based on functional complementation between Bordetella pertussis adenylate cyclase fragments, T18 and T25, expressed separately from two compatible plasmids (20). Full-length bldG and ushX genes were fused as C-terminal fusions in plasmids pUT18C and pKT25, respectively. In addition, the sigH gene encoding σH that has been previously shown to interact with UshX (47) and the anti-sigma factor gene apgA (SCO3548) previously verified to interact with BldG (43) were similarly cloned into plasmids pUT18C and pKT25, respectively. The resulting constructs (including appropriate negative and positive controls) were cotransformed into E. coli BTH101 and screened on LB medium with IPTG and X-Gal. Only transformants carrying the pairs sigH-ushX, bldG-ushX, and bldG-apgA showed a LacZ+ phenotype. The negative controls did not produce blue colonies. Moreover, transformants carrying a sigH and an apgA fusion also produced white colonies (data not shown). The results of quantitative β-galactosidase assays of the different transformants, shown in Fig. 2, confirmed that BldG and UshX physically interact with each other. Interestingly, based on the activity of β-galactosidase, this interaction seems to be stronger than the previously verified interaction between SigH and UshX (47) and even between BldG and ApgA (43). The interaction was comparable to that for the positive BACTH control leucine zipper interaction (20). In addition, the sigma factor σH and the anti-sigma factor ApgA likely do not interact, since the levels of β-galactosidase for the pair sigH-abpA were similar to those for the several sets of negative controls with the empty plasmids (Fig. 2).
FIG. 2.
Analysis of protein interactions using the BACTH system. Combinations of the plasmids indicated were transformed into E. coli BTH101, and β-galactosidase activity was estimated in triplicates as described in Materials and Methods. The error bars indicate the standard deviations from the mean, which is shown above each bar.
BldG interacts differentially with UshX and ApgA anti-sigma factors.
To corroborate the results from the two-hybrid system, the interaction between UshX and BldG was investigated by the analysis of their complex by native PAGE as previously performed for the interaction between S. coelicolor σH and UshX, where a complex in a 1:1 molar ratio was found (47). UshX, BldG, and ApgA were overproduced as N-terminal His-tagged proteins in an E. coli T7 RNA polymerase expression system. After isolation by native Ni2+ affinity chromatography, UshX was incubated at various molar ratios with BldG and samples were analyzed by native PAGE. In addition, BldG was similarly incubated with ApgA. The results of the native PAGE (Fig. 3) showed that BldG interacts with both ApgA and UshX. In both cases, a complex was visible as a new band, and at a particular ratio, freely interacting proteins disappeared. Interestingly, in contrast to ApgA, which formed a complex with BldG at an ∼1:1 molar ratio, UshX and BldG have been found to form a complex at an ∼1:2 molar ratio. The 1:1 molar ratio of the ApgA-BldG complex is consistent with the proposed heterotetrameric complex consisting of an ApgA dimer interacting with a BldG dimer (43). Assuming that BldG forms a dimer, it is likely that it may interact with one molecule of UshX. However, further study would be needed to explain more clearly this unusual binding.
FIG. 3.
Interaction of BldG with UshX and ApgA as monitored by native PAGE. Different molar ratios of the proteins indicated were incubated and analyzed by native PAGE as described in Materials and Methods. The numbers 1, 2, and 4 in the ratios correspond to 100, 200, and 400 pmol of the indicated protein used, respectively.
Induction of σH-dependent sigHp2 promoter is dependent upon BldG.
The S. coelicolor sigH operon is autoregulated by means of the sigHp2 promoter that is osmotically induced and specifically recognized by RNA polymerase containing σH (46, 47). To investigate whether BldG affects the activity of the promoter, S1 nuclease mapping was performed using RNA isolated from the wild-type S. coelicolor M145 and its isogenic bldG mutant strain, S. coelicolor ΔbldG 1DB (6) grown to exponential phase and induced by two highly osmotic conditions for two different time points. The sigHp2 promoter was induced by both osmotic conditions as previously described (31, 46), with the late kinetic where higher induction was at the later time points (60 min). However, no induction in activity was found in the bldG mutant, while the activities of the other three sigH promoters (sigHp1, sigHp3, and sigHp4) were not decreased under these conditions in the mutant strain, these acting as internal controls (Fig. 4). Interestingly, the activities of the other sigH promoters were even higher in the bldG mutant strain than in the wild-type S. coelicolor M145 strain. As the other sigH promoters are partially growth phase dependent (31), the differences between their activities in the two strains may be due to their partially different growth stages at the time point used for RNA isolation. However, we cannot rule out the presence of some other unknown mechanisms. These results indicated that bldG is essential for σH activation and that the binding of BldG to UshX likely influences the activation of σH during osmotic stress conditions.
FIG. 4.
High-resolution S1 nuclease mapping of the sigH operon in the wild-type strain S. coelicolor M145 (WT) and its isogenic bldG mutant, S. coelicolor ΔbldG 1DB. The 5′-end-labeled DNA fragment was hybridized with 40 μg RNA and treated with 120 U of S1 nuclease as described in Materials and Methods. RNA was isolated from both strains grown for 20 h in NMP liquid medium to the end of exponential phase (before stress) and subjected to the indicated stress conditions. Lane C, E. coli tRNA as a control; lanes A and T, G+A and T+C sequencing ladders (37). RNA-protected fragments corresponding to the sigHp1, sigHp2, sigHp3, and sigHp4 promoters (31) are indicated. All S1 nuclease mapping experiments were performed two times with independent sets of RNA with similar results.
DISCUSSION
The results of the present study, supported by three independent experiments, indicate that the S. coelicolor σH-specific anti-sigma factor UshX (PrsH) interacts with the previously characterized anti-anti-sigma factor BldG (7). In addition, the osmotic induction of σH activity measured through the activation of the σH-dependent sigHp2 promoter (31, 46, 47) is abolished in an S. coelicolor bldG mutant, indicating a role of BldG in σH activation by a partner-switching-like mechanism. The bldG gene belongs to the category of pleiotropic bld genes that affect formation of aerial hyphae and production of antibiotics in S. coelicolor (7, 9). Its Bld phenotype, with bald colonies lacking an aerial mycelium, suggests that the BldG-associated anti-sigma factor may interact with a sigma factor that is essential for development. This might be true for σH, which has a dual role in S. coelicolor, in regulation of the osmotic stress response and in morphological differentiation, affecting septation of aerial hyphae (24, 31, 46, 49), and its level dramatically increases during differentiation (50; J. Kormanec, unpublished results). However, in contrast to our earlier phenotypic analysis (46), a sigH deletion mutant was later reported to have no obvious phenotype, although different parental S. coelicolor A3(2) strains were used (35, 49).
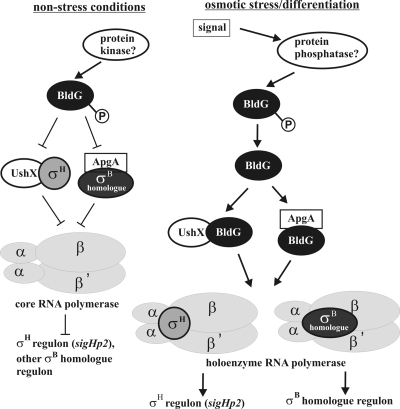
BldG has also been found to interact with another anti-sigma factor homologue, ApgA (SCO3548) (43), as verified also by our results. Based on these results, BldG has several anti-sigma partners to regulate, and in addition to σH, likely also other σB homologues, as σH does not interact with the other BldG-specific anti-sigma factor, ApgA (Fig. 3). Considering these results and published data about S. coelicolor σH, we suggest the following hypothesis of σH activation by a partner-switching-like mechanism (Fig. 5). At unstressed conditions, the basal level of σH is sequestered by its anti-sigma factor, UshX. After a signal related to osmotic stress, BldG is activated, likely by dephosphorylation with an unknown phosphatase, and its dephosphorylated form interacts with UshX anti-sigma factor to relieve σH, which in turn can associate with core RNA polymerase to form the holoenzyme recognizing the σH regulon, including the σH-dependent sigHp2 promoter that autoregulates the sigH operon. Additionally, dephosphorylated BldG interacts with the ApgA anti-sigma factor to similarly relieve another σB homologue for its activation. Actually, BldG has been found to be phosphorylated by wild-type S. coelicolor crude extracts. The level of phosphorylated BldG was highest during late vegetative growth and during aerial mycelium formation, subsequently falling during sporulation of aerial hyphae (6). This may be consistent with our model, in which σH activation likely occurs after osmotic stress and during the late stages of differentiation, where it accumulates and also undergoes posttranslational processing (50; J. Kormanec, unpublished results). Likewise, two genes of the σH regulon (ssgB and sigJ) have been shown to be activated at the later stages of development (29, 38).
FIG. 5.
Model of activation of σH activity by BldG in S. coelicolor. Under nonstressed conditions, σH is sequestered by the anti-sigma factor UshX and the σH regulon is not induced. After a signal related to an osmotic stress, BldG is activated by dephosphorylation with an unknown phosphatase and binds UshX to relieve σH, which in turn can associate with core RNA polymerase to form a holoenzyme transcribing genes belonging to the σH regulon, including the σH-dependent sigHp2 promoter that autoregulates the sigH operon. Additionally, dephosphorylated BldG interacts with ApgA anti-sigma factor to similarly relieve another σB homologue for its activation.
Reversible phosphorylation of BldG has been found to be critical for proper development of S. coelicolor (6). In the B. subtilis σB and σF systems, phosphorylation is carried out by the anti-sigma factors RsbW and SpoIIAB, respectively (1, 12, 13, 42).
However, like ApgA (43), UshX does not contain essential amino acid residues of the necessary HATPase_c kinase domain (Pfam accession no. PF02518) and so it is unlikely to phosphorylate BldG. We have consistently failed to observe BldG phosphorylation by UshX and ApgA (J. Kormanec, unpublished results). A similar situation has also been described for Mycobacterium tuberculosis σF, where the UshX homologue UsfX also lacks the conserved kinase domain and is unable to phosphorylate the σF-specific anti-anti-sigma factor RsfB (4). Therefore, another S. coelicolor RsbW homologue containing an active kinase domain might be the partner in this cascade for σH activation. There are several such candidates containing the necessary HATPase_c kinase domain in the S. coelicolor genome (41). Identification of a BldG-specific protein kinase and also the other BldG-specific sigma factor target would be necessary to characterize the regulatory circuits in S. coelicolor σH activation and to partially decipher the rather complex regulatory picture involving nine S. coelicolor σB homologues. These experiments are in progress.
Acknowledgments
We thank Ludovit Skultety for his assistance with the mass spectrometric analysis, Daniel Ladant for the bacterial two-hybrid system strain and plasmids, and Brenda Leskiw for providing the S. coelicolor bldG mutant strain. We thank Paul Dyson for critical reading of the manuscript.
This work was supported by VEGA grant 2/0104/09 from the Slovak Academy of Sciences and by the Slovak Research and Development Agency under contract APVV-0017-07. This publication is also the result of the TRANSMED project implementation, supported by the Research & Development Operational Programme funded by the ERDF.
Footnotes
Published ahead of print on 3 September 2010.
REFERENCES
- 1.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 2.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell-type specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. Wiley, New York, NY.
- 4.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bignell, D. R. D., L. H. Lau, C. R. Colvin, and B. K. Leskiw. 2003. The putative anti-anti-sigma factor BldG is post-translationally modified by phosphorylation in Streptomyces coelicolor. FEMS Microbiol. Lett. 225:93-99. [DOI] [PubMed] [Google Scholar]
- 7.Bignell, D. R. D., J. L. Warawa, J. L. Strap, K. F. Chater, and B. K. Leskiw. 2000. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146:2161-2173. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chater, K. F. 2000. Developmental decisions during sporulation in the aerial mycelium in Streptomyces, p. 33-48. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, DC.
- 11.Dalton, K. A., A. Thibessard, J. I. Hunter, and G. H. Kelemen. 2007. A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64:719-737. [DOI] [PubMed] [Google Scholar]
- 12.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Erringston, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 8:2653-2663. [DOI] [PubMed] [Google Scholar]
- 13.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaskell, A. A., J. C. Crack, G. H. Kelemen, M. I. Hutchings, and N. E. Le Brun. 2007. RsmA is and anti-sigma factor that modulates its activity through a [2Fe-2S] cluster cofactor. J. Biol. Chem. 282:31812-31820. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, M. Y., J. B. Bae, J. H. Park, and J. H. Roe. 2003. Isolation and characterization of Streptomyces coelicolor RNA polymerase, its sigma, and anti-sigma factors. Methods Enzymol. 370:73-82. [DOI] [PubMed] [Google Scholar]
- 16.Hecker, M., J. Pane-Farre, and U. Volker. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61:215-236. [DOI] [PubMed] [Google Scholar]
- 17.Helmann, J. B. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 18.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernychova, L., R. Toman, F. Ciampor, M. Hubalek, J. Vackova, A. Macela, and L. Skultety. 2008. Detection and identification of Copxiella burnetii based on the mass spectrometric analysis of the extracted proteins. Anal. Chem. 80:7097-7104. [DOI] [PubMed] [Google Scholar]
- 20.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. [PubMed] [Google Scholar]
- 22.Karoonuthaisiri, N., D. Weaver, J. Huang, S. N. Cohen, and C. M. Kao. 2005. Regional organization of gene expression in Streptomyces coelicolor. Gene 353:53-66. [DOI] [PubMed] [Google Scholar]
- 23.Keijser, B. J. F., E. E. E. Noens, B. Kraal, H. K. Koerten, and G. P. van Wezel. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225:59-67. [DOI] [PubMed] [Google Scholar]
- 24.Kelemen, G. H., P. H. Viollier, J. L. Tenor, L. Marri, M. J. Buttner, and C. J. Thompson. 2001. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40:804-814. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J., Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 26.Kim, E. S., J. Y. Song, D. W. Kim, K. F. Chater, and K. J. Lee. 2008. A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. J. Bacteriol. 190:7559-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kormanec, J. 2001. Analyzing the developmental expression of sigma factors with S1-nuclease mapping. Methods Mol. Biol. 160:481-494. [DOI] [PubMed] [Google Scholar]
- 28.Kormanec, J., D. Homerova, I. Barak, and B. Sevcikova. 1999. A new gene, sigG, encoding a putative alternative sigma factor of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 172:153-158. [DOI] [PubMed] [Google Scholar]
- 29.Kormanec, J., and B. Sevcikova. 2002. The stress-response sigma factor σH controls the expression of ssgB, a homologue of the sporulation-specific cell division gene ssgA in Streptomyces coelicolor A3(2). Mol. Genet. Genom. 267:536-543. [DOI] [PubMed] [Google Scholar]
- 30.Kormanec, J., and B. Sevcikova. 2002. Stress-response sigma factor σH directs expression of the gltB gene encoding glutamate synthase in Streptomyces coelicolor A3(2). Biochim. Biophys. Acta 1577:149-154. [DOI] [PubMed] [Google Scholar]
- 31.Kormanec, J., B. Sevcikova, N. Halgasova, R. Knirschova, and B. Rezuchova. 2000. Identification and transcriptional characterization of the gene encoding the stress-response sigma factor in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189:31-38. [DOI] [PubMed] [Google Scholar]
- 32.Labudova, M., J. Tomaskova, L. Skultety, J. Pastorek, and S. Pastorekova. 2009. The nucleoprotein of lymphocytic choriomeningitis virus facilitates spread of persistent infection through stabilization of the keratin network. J. Virol. 83:7842-7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lee, E. J., Y. H. Cho, H. S. Kim, B. E. Ahn, and J. H. Roe. 2004. Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186:8490-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, E. J., N. Karoonuthaisiri, H. S. Kim, J. H. Park, C. J. Cha, C. M. Kao, and J. H. Roe. 2005. A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 57:1252-1264. [DOI] [PubMed] [Google Scholar]
- 36.Mao, X.-M., Z. Zhou, X.-P. Hou, W.-J., Guan, and Y.-Q. Li. 2009. Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J. Bacteriol. 191:6473-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labelled DNA with base specific chemical cleavages. Methods Enzymol. 65:449-560. [DOI] [PubMed] [Google Scholar]
- 38.Mazurakova, V., B. Sevcikova, B., Rezuchova, and J. Kormanec. 2006. Cascade of sigma factors in streptomycetes: identification of a new extracytoplasmic function sigma factor σJ that is under the control of the stress-response sigma factor σH in Streptomyces coelicolor A3(2). Arch. Microbiol. 186:435-446. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcriptional factor of B. subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 41.Mittenhuber, G. 2002. A phylogenetic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. 4:427-452. [PubMed] [Google Scholar]
- 42.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 177:2912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parashar, A., K. R. Colvin, D. R. D. Bignell, and B. K. Leskiw. 2009. BldG and CO3548 interact antagonistically to control key developmental processes in Streptomyces coelicolor. J. Bacteriol. 191:2541-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potuckova, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 17:37-48. [DOI] [PubMed] [Google Scholar]
- 45.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Soneneshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 46.Sevcikova, B., O. Benada, O. Kofronova, and J. Kormanec. 2001. Stress-response sigma factor SigH is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 177:98-106. [DOI] [PubMed] [Google Scholar]
- 47.Sevcikova, B., and J. Kormanec. 2002. Activity of the Streptomyces coelicolor stress-response sigma factor SigH is regulated by an anti-sigma factor. FEMS Microbiol. Lett. 209:229-235. [DOI] [PubMed] [Google Scholar]
- 48.Sevcikova, B., and J. Kormanec. 2003. The ssgB gene, encoding a number of the regulon of stress-response sigma factor σH, is essential for aerial mycelium septation in Streptomyces coelicolor A3(2). Arch. Microbiol. 180:380-384. [DOI] [PubMed] [Google Scholar]
- 49.Viollier, P. H., G. H. Kelemen, G. E. Dale, K. T. Nguyen, M. J. Buttner, and C. J. Thompson. 2003. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47:699-714. [DOI] [PubMed] [Google Scholar]
- 50.Viollier, P. H., A. Weihofen, M. Folcher, and C. J. Thompson. 2003. Post-transcriptional regulation of the Streptomyces coelicolor stress responsive sigma factor, SigH, involves translational control, proteolytic processing, and an anti-sigma factor homologue. J. Mol. Biol. 325:637-649. [DOI] [PubMed] [Google Scholar]
- 51.Vohradsky, J., X. M. Li, G. Dale, M. Folcher, L. Nguyen, P. H. Viollier, and C. J. Thompson. 2000. Developmental control of stress stimulons in Streptomyces coelicolor revealed by statistical analysis of global gene expression patterns. J. Bacteriol. 182:4979-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]