Abstract
Salmonella enterica, a common food-borne pathogen, differentially regulates the expression of multiple genes during the infection cycle. These genes encode systems related to motility, adhesion, invasion, and intestinal persistence. Key among them is a type three secretion system (T3SS) encoded within Salmonella pathogenicity island 1 (SPI1). In addition to the SPI1 T3SS, other systems, including flagella and type 1 fimbriae, have been implicated in Salmonella pathogenesis. In this study, we investigated the dynamic expression of the flagellar, SPI1, and type 1 fimbrial genes. We demonstrate that these genes are expressed in a temporal hierarchy, beginning with the flagellar genes, followed by the SPI1 genes, and ending with the type 1 fimbrial genes. This hierarchy could mirror the roles of these three systems during the infection cycle. As multiple studies have shown that extensive regulatory cross talk exists between these three systems, we also tested how removing different regulatory links between them affects gene expression dynamics. These results indicate that cross talk is critical for regulating gene expression during transitional phases in the gene expression hierarchy. In addition, we identified a novel regulatory link between flagellar and type 1 fimbrial gene expression dynamics, where we found that the flagellar regulator, FliZ, represses type 1 fimbrial gene expression through the posttranscriptional regulation of FimZ. The significance of these results is that they provide the first systematic study of the effect of regulatory cross talk on the expression dynamics of flagellar, SPI1, and type 1 fimbrial genes.
Salmonella enterica causes a large number of diseases ranging from self-limiting gastroenteritis to life-threatening systemic infection (21, 62). Previous studies have identified multiple factors involved in Salmonella pathogenesis, including those related to motility, adhesion, invasion, and intestinal persistence (12, 18, 39, 43, 45, 55, 66, 83, 86-88). Key among them is a type 3 secretion system (T3SS) encoded within a 40-kb region of the chromosome called Salmonella pathogenicity island 1 (SPI1) (46-48, 51, 63, 76). The SPI1 T3SS functions as a molecular hypodermic needle, enabling Salmonella to inject proteins into host cells (11-13). These injected proteins both commandeer the actin cytoskeleton to facilitate the invasion of host cells and induce inflammatory diarrhea (27, 29, 33, 52, 59, 92).
In addition to the SPI1 T3SS, other systems, including flagella and type 1 fimbriae, have been implicated in Salmonella pathogenesis (31, 36, 75). Briefly, flagella are long helical filaments attached to rotary motors embedded within the membrane that enable the bacterium to swim in liquids and swarm over surfaces (9). Flagella are thought to facilitate invasion by enabling Salmonella to swim to sites of invasion (39, 75). In addition to motility, flagellin activates the expression of proinflammatory cytokines (26, 60, 61, 77, 84). Type 1 fimbriae, on the other hand, are hairlike appendages that carry adhesins specific for mannosylated glycoproteins on eukaryotic cell surfaces (2, 25, 54, 78). They are thought to be involved in pathogenesis by facilitating binding to intestinal epithelial cells (4, 24, 35, 40, 78). As with flagella, type 1 fimbriae do not appear to play a direct role in intestinal invasion but rather are thought to contribute to intestinal colonization and persistent infections (1, 19).
Multiple studies have shown that extensive regulatory cross talk exists between these three systems (5, 10, 22, 38, 53, 58, 72, 79). While the molecular details have been studied extensively (Fig. 1), the role and significance of these cross talk interactions are still relatively unknown. As all three systems play unique and potentially mutually exclusive roles during the infection cycle, we hypothesized that regulatory cross talk controls their dynamic expression. In particular, regulatory cross talk controls the timing of induction and the duration of gene expression for these three systems. To test the hypothesis, we monitored SPI1, flagellar, and type 1 fimbrial gene expression dynamics in a number of mutants where different regulatory links between these three systems had been selectively removed. Based on these results, we demonstrate that there is a natural hierarchy in the expression dynamics of the three, beginning with flagella, followed by the SPI1 T3SS, and ending with type 1 fimbriae. Our results also indicate that the regulatory cross talk between the three systems serves to tune the timing of gene expression with regard to their temporal activation and deactivation.
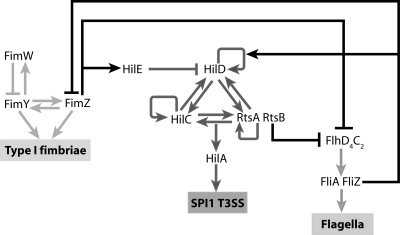
FIG. 1.
Coordinate regulation of the flagellar, SPI1, and type 1 fimbrial genes. The master regulator for flagellar gene expression is FlhD4C2 (9). The FlhD4C2 complex, in turn, activates two additional regulators, FliA and FliZ, encoded within the fliAZ operon. FliA is a flagellum-specific alternate sigma factor essential for the expression of the motor, filament, and chemotaxis genes. FliZ is a posttranslational activator of FlhD4C2 (71). FliZ also activates HilD (38, 41, 53, 57, 82) and represses FimZ posttranscriptionally (this study). In the SPI1 T3SS, HilD, HilC, and RtsA form three interlocking positive-feedback loops where all three activate each other's and their own expression (23). In addition, they can independently activate HilA expression. HilA is required for the expression of the genes encoding the SPI1 T3SS. SPI1 gene expression is negatively regulated by HilE, which binds to HilD and prevents it from activating the SPI1 promoters. RtsB, encoded within the same operon as RtsA, binds to the PflhDC promoter and represses motility (22). In type 1 fimbriae, FimW and FimZ form a coupled feedback loop where they can activate their own and each other's expression (72). They can also independently activate the expression of the PfimA promoter, which controls the expression of genes encoding type 1 fimbriae. FimY and FimW also participate in a negative-feedback loop, where FimY activates FimW expression and FimW represses FimY expression. FimZ also binds to the PflhDC promoter and represses the expression of the flagellar genes (10) and induces the expression of HilE to repress SPI1 gene expression (5, 72).
MATERIALS AND METHODS
General techniques and growth conditions.
All experiments were performed in Luria-Bertani (LB) broth at 37°C unless noted otherwise. Antibiotics were added at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol at 20 μg/ml, kanamycin at 40 μg/ml, and tetracycline at 15 μg/ml. All experiments involving growth of strains carrying the helper plasmid pKD46 or pCP20 were performed at 30°C as previously described (14). Loss of these temperature-sensitive plasmids was achieved by growth at 42°C. Removal of the antibiotic from the FRT-Cm/Kan-FRT insert was achieved by passing pCP20 through the isolated mutants (8). Enzymes were purchased from Fermentas or New England Biolabs and used according to the manufacturer's recommendations. Primers were purchased from IDT Inc.
Strain and plasmid construction.
Bacterial strains and plasmids are described in Tables 1 and 2, respectively. All S. enterica serovar Typhimurium strains are isogenic derivatives of strain 14028 (American Type Culture Collection). The S. enterica serovar Typhimurium generalized transducing phage P22 HT105/1 int-201 was used in all transductional crosses (15). Chromosomal mutations were introduced using standard λ Red recombination in cells carrying the helper plasmid pKD46 as described by Datsenko and Wanner (14). All mutations were checked using primers that target sequences outside the deleted region. Prior to removal of the antibiotic resistance marker, the constructs resulting from this procedure were moved into a clean wild-type background (14028) by P22 transduction.
TABLE 1.
List of strains used in this study
| Strain | Genotype or characteristica | Source or referenceb |
|---|---|---|
| 14028 | Wild-type S. enterica serovar Typhimurium | ATCCc |
| JS481 | Δ(invH-avrA)2916::Cm (called ΔSPI1) | 20 |
| CR201 | ΔfliZ::FRT | 71 |
| CR222 | ΔflhDC::FRT | 71 |
| CR312 | ΔfimZ::Cm | 72 |
| CR322 | ΔfimY::FRT | 72 |
| CR334 | ΔfimYZ::FRT | 72 |
| CR314 | ΔfimW::Cm | 72 |
| CR800 | ΔrtsB::Cm | |
| CR801 | ΔrtsB::FRT | |
| CR802 | ΔfimZ::Cm ΔfliZ::FRT | |
| CR803 | ΔfimZ::FRT ΔfliZ::FRT | |
| CR804 | ΔPfimZ::tetRA | |
| CR805 | ΔPfimZ::tetRA ΔfimY::FRT | |
| CR806 | ΔPfimZ::tetRA ΔfimYZ::FRT | |
| CR807 | ΔrtsB::Cm ΔfimZ::FRT | |
| CR808 | ΔrtsB::FRT ΔfimZ::FRT | |
| CR809 | ΔfimYZ::FRT ΔfimW::FRT |
All Salmonella strains are isogenic derivatives of S. enterica serovar Typhimurium strain 14028.
Strains are from this study unless specified otherwise.
ATCC, American Type Culture Collection.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristic | Reference or sourcea |
|---|---|---|
| pKD46 | bla PBADgam beto exo pSC101 oriTS | 14 |
| pKD3 | bla FRT cm FRT oriR6K | 14 |
| pCP20 | bla cat cI857 λPRflp pSC101 oriTS | 8 |
| pSS009 | kan luxCDABE ori p15a | 71 |
| pSS010 | kan PflgA-luxCDABE ori p15a | 71 |
| pSS077 | kan PhilA-luxCDABE ori p15a | |
| pSS222 | kan PfimA-luxCDABE ori p15a | |
| pPROTet.E | cm PLtetO-1ori ColE1 | Stratagene |
| pSS013 (pFliZ) | cm PLtetO-1fliZ ori ColE1 | 71 |
| pRtsB | cm PLtetO-1rtsB ori ColE1 | |
| pFimZ | cm PLtetO-1fimZ ori ColE1 | |
| pPROBE-Venus | kan venus ori p15a | 73 |
| PfimA-Venus | kan PfimA-venus ori p15a | 73 |
Plasmids were made in this study unless specified otherwise.
Standard “scarred” FLP recombination target (FRT) mutants were produced as previously described, using pKD3 as the PCR template (14). Strain CR800 (ΔrtsB::Cm) was made using primers SS217F (TTTT AGC GTT TTT ATC TTC CTC TCG TCA TCA ATA TGT TAA GTG TAG GCT GGA GCT GCT TC) and SS217R (AGT TGC CTT GCC TAC CAC TCT ACC AAC ATT TTA GGA AAA ACA TAT GAA TAT CCT CCT TAG). The antibiotic marker was removed by passing pCP20 through strain CR800, resulting in strain CR801 (ΔrtsB::FRT).
Strain CR803 (ΔfimZ::FRT ΔfliZ::FRT) was constructed by transducing the antibiotic resistance marker from CR312 to CR201 using P22 transduction (resulting in strain CR802) and then removing the resistance marker using pCP20. Strain CR804 (PfimZ::tetRA) was constructed by replacing the native PfimZ promoter with a tetRA element from transposon Tn10. The tetRA element was amplified using primers SS178F (CCA TTA AAT GTA AAT ATT TCA CAT AAA ATT AAT ATT TAC AAG AGT AGG GAA CTG CCA) and SS178R (CTG TTA TGC GTC CTT CGT TTT ATA ATA AGC GTC AGA CAC CCT AAG CAC TTG TCT CCT), with TH8094 as the template (44). The resulting strain, CR804 (PfimZ::tetRA), places FimZ under the control of a tetracycline-inducible promoter. Strains CR805 (ΔPfimZ::tetRA ΔfimY::FRT) and CR806 (ΔPfimZ::tetRA ΔfimYZ::FRT) were constructed by transducing the tetRA element from CR804 into strains CR322 and CR334, respectively.
Strain CR808 (ΔrtsB::FRT ΔfliZ::FRT) was made by first transducing the chloramphenicol resistance marker from strain CR800 (ΔrtsB::Cm) to CR201 (ΔfliZ::FRT) and then removing the antibiotic resistance marker from the resulting strain, CR807 (ΔrtsB::Cm ΔfliZ::FRT). Strain CR809 (ΔfimYZ::FRT ΔfimW::FRT) was made by first transducing the chloramphenicol resistance marker from strain CR314 (ΔfimW::Cm) into strain CR334 (ΔfimYZ::FRT), resulting in the ΔfimYZ::FRT ΔfimW::Cm strain, and then removing the resistance marker from the ΔfimYZ::FRT ΔfimW::Cm strain by using pCP20.
Transcriptional fusions of the promoters of interest were made in the following manner. The PhilA promoter was amplified using primers PhilAF (GGG GGA TCC ACT TGT CAT CGC TAT GAT GA) and PhilAR (GGG GAA TTC ACA GGA TTA AAA TGT GGC AT). The PfimA promoter was amplified using primers SS104F (TTT GGT ACC AAA TCT GTG AGG CCG GAT TG) and SS104R (GGG GAA TTC GTA GAG GTC ATT AAT TTA TG). The PCR fragments from both were digested with KpnI and EcoRI (underlined sequences) and cloned into the multiple-cloning site of plasmid pSS009. The resulting plasmids were called pSS077 and pSS222, respectively.
Expression vectors for RtsB and FimZ were constructed by PCR amplifying the gene of interest and cloning it in the multiple-cloning site of the vector, pPROTet.E. The resulting arrangement put the gene under a constitutively active PLtetO-1 promoter. The rtsB gene was amplified using primers SS198F (GGC GAA TTC TTA TAA GGA GGA AAA ACA TTT GAG ATA TCT GAC AAT GCA) and SS198R (TTT GGT ACC TTA CGT AAT ATC GAC TGA TA). The fimZ gene was amplified using primers SS106F (GGG GAA TTC TAA CAG TCT GAG GCA TAC AA) and SS106R (TTT GGT ACC TTA CAA TAA TTC GTG TGA TT). The resulting plasmids were called pRtsB and pFimZ, respectively. Prior to transformations in wild-type and mutant strains, all constructs were verified by sequencing.
Fluorescence and luminescence assays.
Endpoint fluorescent measurements and dynamic luminescence measurements were made using a Tecan Safire2 microplate reader. For fluorescence endpoint measurements for type 1 fimbrial gene expression (PfimA promoter activity), a 1-ml culture was grown at 37°C overnight and then subcultured 1:1,000 in fresh medium and grown under static conditions for 24 h at 37°C. One hundred microliters was then transferred to a 96-well microplate, and the relative fluorescence and optical density at 600 nm (OD600) were measured. The fluorescence readings, given as relative fluorescence units (RFU), were normalized with the OD600 to account for cell density. All endpoint experiments were done on three different days, with six repeats on each day. The average values and the standard deviations of the data are reported. Statistical analysis was performed using Student's t test, where the reported P values are based on the average fluorescence value for each separate day.
Dynamic luminescence experiments were performed as follows. Cells were grown overnight at 37°C in LB without salt and with vigorous shaking. The overnight culture was then subcultured 1:500 in fresh LB medium (with 1% salt). A 100-μl aliquot of the subcultured cells was then transferred to a 96-well plate and covered with a Breathe-Easy membrane to prevent evaporation. Luminescence and optical density readings were then taken every 20 min. All experiments were done in three independent repeats, with six samples in each experiment. The average values and the standard deviations of the data at each time point are reported.
RESULTS
Flagellar, SPI1, and type 1 fimbrial genes are expressed in a temporal hierarchy.
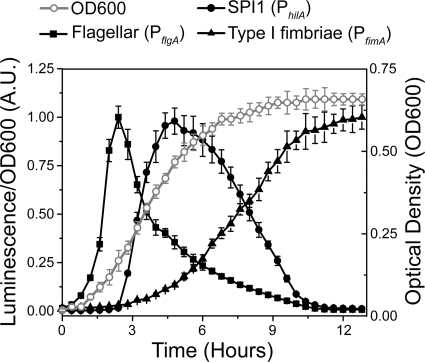
Our governing hypothesis is that regulatory cross talk controls the dynamic expression of flagellar, SPI1, and type 1 fimbrial genes. As a first step toward testing this hypothesis, we measured gene expression dynamics for the three systems in wild-type cells. In these experiments, we first grew the cells overnight in Luria-Bertani (LB) medium without salt and then subcultured them into fresh LB-1% NaCl medium in the absence of shaking, thus inducing a transition from SPI1-repressing to SPI1-inducing conditions. Growth under low-oxygen and high-salt conditions has previously been shown to induce SPI1 gene expression in vitro (3, 51). To measure gene expression dynamics, we employed plasmid-based transcriptional fusions of the PflgA, PhilA, and PfimA promoters to the luciferase operon luxCDABE from Photorhabdus luminescens (71, 89). The PflgA promoter controls the expression of the genes encoding the structural components of the flagellar P-ring protein (65) and thus provides a proxy measure for flagellar gene expression. The PhilA promoter controls the expression of HilA, the master regulator for the genes encoding the SPI1 T3SS (56), and similarly provides a proxy for the expression of SPI1 genes (20). The PfimA promoter controls the expression of the six genes encoding type 1 fimbriae (69, 70). The reason that we chose bacterial luciferase as opposed to other reporters for these experiments is that it is sensitive to dynamic changes in promoter activity (30, 71).
As shown in Fig. 2, we found that flagellar, SPI1, and type 1 fimbrial genes are expressed in a temporal hierarchy. Specifically, the cells first express flagellar genes, followed by SPI1 genes and, lastly, type 1 fimbrial genes. Hierarchical expression, however, is not entirely surprising as previous studies have shown that the growth phase plays an important role in the timing of activation of these three systems (7, 42, 67). In particular, the flagellar genes are maximally expressed during the early log phase, the SPI1 genes during the late exponential phase, and the type 1 fimbrial genes upon entry into stationary phase. The immediate question then is whether regulatory cross talk plays any role in dictating this transcriptional hierarchy.
FIG. 2.
Dynamic expression of the flagellar, SPI1, and type 1 fimbrial genes. Time course dynamics of the PflgA (flagellar), PhilA (SPI1), and PfimA (type 1 fimbrial) promoter activities in wild-type cells as determined using luciferase transcriptional reporters. For reference purposes, the optical density (OD600) was plotted to illustrate how each system is activated during different phases of growth. In these experiments, cells were first grown overnight at 37°C in LB without salt and then subcultured 1:500 in fresh LB-1% salt. Cells were then grown statically with luminescence, and optical density readings were taken every 20 min. Average promoter activities from three independent experiments on separate days are reported. For each experiment, six samples were tested. Error bars denote standard deviations. A.U., arbitrary units.
Regulatory cross talk tunes gene expression dynamics.
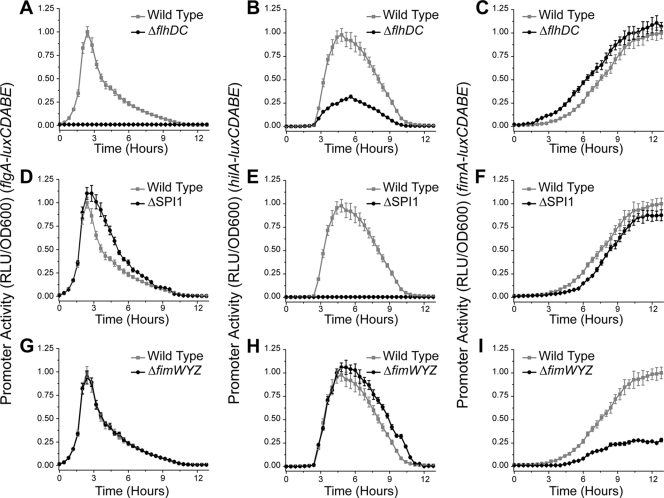
To determine whether the transcriptional hierarchy is due to cross talk, we measured gene expression dynamics in mutants where each system was selectively inactivated. First, we measured how inactivating flagellar gene expression using the ΔflhDC mutant affected SPI1 and type 1 fimbrial gene expression. FlhD and FlhC form the FlhD4C2 complex, the master regulator of flagellar gene expression in enteric bacteria (37, 85). Deleting flhD and flhC shuts off flagellar gene expression (Fig. 3 A). As shown in Fig. 3B, SPI1 gene expression is significantly attenuated in the ΔflhDC mutant. However, the dynamics of SPI1 gene expression are unchanged; the times when SPI1 gene expression is first turned on and then off are the same in the ΔflhDC mutant as in the wild type. The only change in gene expression is that the magnitude is reduced. With regard to type 1 fimbriae, we found that gene expression was induced prematurely by roughly an hour in the ΔflhDC mutant compared to that in the wild type (Fig. 3C).
FIG. 3.
Effect of transcriptional cross talk on flagellar, SPI1, and type 1 fimbrial gene expression dynamics. (A to C) The flagellar genes amplify SPI1 gene expression and delay type 1 fimbrial gene expression. PflgA (A), PhilA (B), and PfimA (C) promoter activities in the wild type and the ΔflhDC mutant are shown. (D to F) The SPI1 genes reduce the duration of flagellar gene expression and accelerate the induction of type 1 fimbrial gene expression. PflgA (D), PhilA (E), and PfimA (F) promoter activities in the wild type and the ΔSPI1 mutant are shown. (G to I) Type 1 fimbrial genes do not affect flagellar gene expression and reduce the duration of SPI1 gene expression. PflgA (G), PhilA (H), and PfimA (I) promoter activities in the wild type and the ΔfimYZW mutant are shown. Experiments were performed as described for Fig. 2. The mutants were also tested to see whether they affected growth. However, no change in optical density as a function of time was observed (data not shown).
Next, we tested the effect of inactivating SPI1 gene expression. To do this, we employed a mutant where the entire pathogenicity island was deleted (ΔSPI1) (20). Deleting SPI1 inactivates PhilA promoter activity (Fig. 3E). The reason is that hilD is required for the transcription of hilA (20, 74). When we measured gene expression in the ΔSPI1 mutant, we found that the flagellar genes were expressed for an hour longer than they were in the wild type (Fig. 3D). In the case of the type 1 fimbrial genes (Fig. 3F), we found that their induction was delayed by 30 min in the ΔSPI1 mutant compared to that in the wild type.
Last, we tested the effect of repressing type 1 fimbrial gene expression. Here, we employed the ΔfimWYZ mutant. This mutant lacks the three regulatory proteins known to directly regulate type 1 fimbrial gene expression (72, 80, 81, 90). While PfimA promoter activity is not completely repressed in this mutant (Fig. 3I), these three proteins are the ones that would most likely participate in any transcriptional cross talk. The other known genes in the type 1 fimbrial cluster encode the structural elements of type 1 fimbriae. When we measured gene expression dynamics in the ΔfimWYZ mutant, we observed no change in flagellar gene expression (Fig. 3G). However, in the case of the SPI1 genes (Fig. 3H), we found that they are expressed for an hour longer in the ΔfimWYZ mutant than in the wild type.
Collectively, these results demonstrate that the transcriptional hierarchy is not due to regulatory cross talk alone, as initially hypothesized, but rather is controlled predominantly by the growth phase and external regulatory factors. Regulatory cross talk, on the other hand, appears to tune gene expression dynamics. In particular, cross talk primarily regulates the transitions between the different phases of gene expression, with the notable exception being the requirement of flagellar gene expression for maximal SPI1 gene expression. Next, we tested the role of specific regulators in establishing this cross talk.
FliZ regulates the magnitude of SPI1 gene expression and the timing of type 1 fimbrial gene expression.
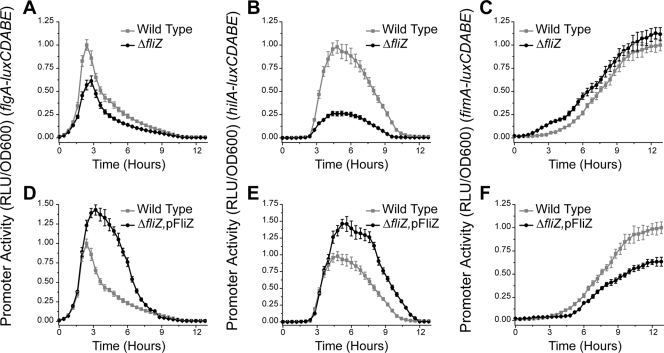
The flagellar regulator FliZ is a posttranslational activator of FlhD4C2 (71). In addition, FliZ is a positive regulator of SPI1 gene expression (38, 41, 53, 57, 82). While the specific mechanism is unknown, FliZ-dependent activation of SPI1 gene expression is known to occur through HilD (41, 53). To test the specific role of FliZ in regulatory cross talk, we measured flagellar, SPI1, and type 1 fimbrial gene expression dynamics in the wild type, a ΔfliZ mutant, and a ΔfliZ mutant constitutively expressing FliZ from a plasmid.
Comparing gene expression dynamics in these different strains, we found that FliZ is a positive regulator of flagellar and SPI1 gene expression and a negative regulator of type 1 fimbrial gene expression. In particular, we found that deleting FliZ decreases the relative magnitudes of flagellar and SPI1 gene expression but does not affect their dynamics (Fig. 4 A and B). Moreover, deleting FliZ accelerates the induction of type 1 fimbrial gene expression (Fig. 4C). We also found that overexpressing FliZ increases both the magnitudes and durations of flagellar and SPI1 gene expression (Fig. 4D and E), whereas it represses and delays type 1 fimbrial gene expression (Fig. 4F). Comparison of these results with those obtained using the ΔflhDC mutant suggests that the effect of flagellar gene expression on SPI1 and type 1 fimbrial gene expression is due to FliZ.
FIG. 4.
FliZ controls the magnitude of SPI1 gene expression and the dynamics of type 1 fimbrial gene expression. (A to C) Deleting FliZ represses flagellar and SPI1 gene expression and accelerates the induction of type 1 fimbrial genes. PflgA (A), PhilA (B), and PfimA (C) promoter activities in the wild type and the ΔfliZ mutant are shown. (D to F) Overexpressing FliZ increases the magnitude of flagellar and SPI1 gene expression and delays the induction of type 1 fimbrial genes. PflgA (D), PhilA (E), and PfimA (F) promoter activities in the wild type and the ΔfliZ mutant constitutively expressing FliZ from a PLtetO-1 promoter on a plasmid (pFliZ) are shown. Experiments were performed as described for Fig. 2.
Repression of type 1 fimbrial gene expression by FliZ is through FimZ.
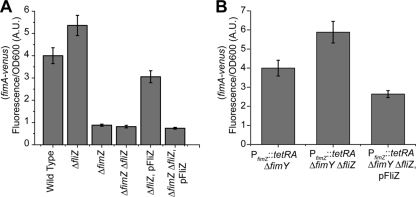
While FliZ has previously been shown to regulate SPI1 gene expression in a number of studies (38, 41, 53, 57, 82), its effect on type 1 fimbrial gene expression has not previously been reported to the best of our knowledge. Therefore, to further characterize this FliZ-dependent repression of type 1 fimbrial gene expression, we sought to determine the genetic target. To do this, we first compared PfimA promoter activities in the wild type, a ΔfimZ mutant, a ΔfliZ mutant, a ΔfimZ ΔfliZ mutant, a ΔfliZ mutant constitutively expressing FliZ from a plasmid, and a ΔfimZ ΔfliZ mutant also containing the FliZ plasmid. We performed endpoint measurements after 24 h of growth. In these experiments, we employed promoter fusions to the fluorescent protein Venus on a plasmid that were identical to the fusions used for the luciferase reporters (32). The reason for using a fluorescent reporter rather than bacterial luciferase is that it is much more stable and thus provides a better measure of integrated promoter activity, as is desired in an endpoint measurement (32).
As shown in Fig. 5 A, deleting FliZ increases the activity of the PfimA promoter (P < 0.01). Similarly when FliZ is expressed from a constitutive promoter on a plasmid in an otherwise ΔfliZ background, PfimA promoter activity is repressed (P < 0.01). However, in the ΔfimZ mutant, we did not observe any additional change in promoter activity when FliZ was also removed (P > 0.20). When FliZ was expressed from a plasmid in an otherwise ΔfimZ ΔfliZ background, we observed a minor decrease in PfimA promoter activity, though the effect was only marginally significant (P = 0.04). We also performed similar experiments with FimY and found that FliZ is still able to regulate the PfimA promoter in the absence of FimY (data not shown). These results suggest that FliZ does not directly repress PfimA promoter activity. We next tested whether FliZ represses type 1 fimbrial gene expression through FimZ. To test this hypothesis, we employed a strain where FimZ was constitutively expressed. Specifically, we replaced the PfimZ promoter with the tetracycline-inducible tetRA element from transposon Tn10 (PfimZ::tetRA). This arrangement decouples FimZ expression from its native regulation and causes it to be constitutively expressed from its native chromosomal locus in the presence of tetracycline. In addition, we also deleted FimY so that regulation was dependent entirely on FimZ (FimW is a negative regulator of type 1 fimbrial gene expression and operates through FimY [72]). When we tested the effect of FliZ in a PfimZ::tetRA ΔfimY genetic background, we found that deleting FliZ increases PfimA promoter activity (P < 0.01) whereas constitutively expressing FliZ represses it (P < 0.01) (Fig. 5B). We also performed similar experiments with FimY and found that FliZ has no effect (data not shown). These results indicate that FliZ regulates type 1 fimbrial gene expression through FimZ. One possibility is that FliZ posttranscriptionally regulates FimZ in a manner similar to FlhD4C2 and HilD. Alternatively, FliZ may prevent FimZ from activating the PfimA promoter.
FIG. 5.
FliZ regulates type 1 fimbrial gene expression though FimZ. (A) FliZ is unable to regulate PfimA promoter activity in the absence of FimZ. PfimA promoter activities in the wild type, the ΔfimZ mutant, the ΔfliZ mutant, the ΔfimZ ΔfliZ mutant, the ΔfliZ mutant expressing FliZ from the constitutive PLtetO-1 promoter on a plasmid (pFliZ), and the ΔfimZ ΔfliZ mutant harboring pFliZ are shown. (B) FliZ regulates FimZ posttranscriptionally. PfimA promoter activities in the PfimZ::tetRA ΔfimY mutant, the PfimZ::tetRA ΔfimY ΔfliZ mutant, and the PfimZ::tetRA ΔfimY ΔfliZ mutant expressing FliZ from the constitutive PLtetO-1 promoter on a plasmid are shown. In the genetic background PfimZ::tetRA, FimZ is under the control of a tetracycline-inducible promoter. Overnight cultures were subcultured 1:1,000 in fresh LB and then grown statically at 37°C for 24 h. FimZ expression was induced by addition of 15 μg/ml tetracycline. Fluorescence and optical density (OD600) values were then measured for each sample. Average promoter activities from three independent experiments on separate days are reported. For each experiment, six samples were tested. Error bars denote standard deviations.
RtsB directly regulates the dynamics of flagellar gene expression and indirectly regulates the dynamics of SPI1 and type 1 fimbrial gene expression through FliZ.
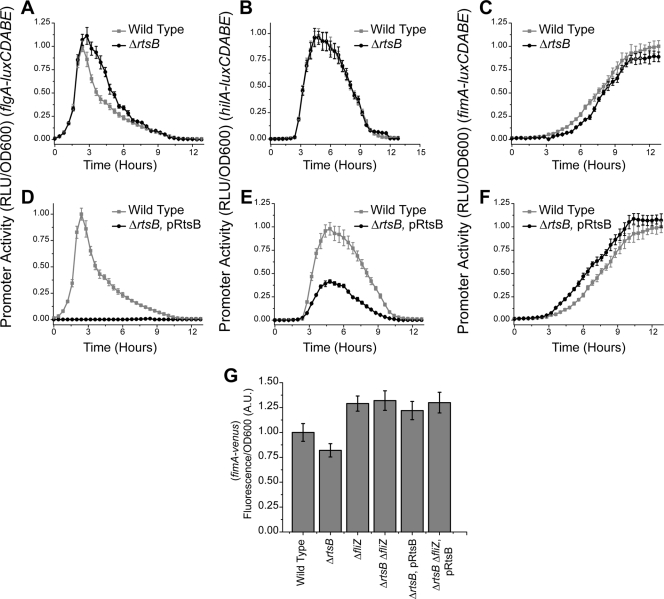
RtsB has previously been shown to bind to the PflhDC promoter and repress flagellar gene expression (22). While RtsB does not reside within SPI1, it is located in the same operon as RtsA, an AraC-like regulator of SPI1 gene expression. This two-gene operon requires HilD for expression, and its expression is correlated with the other SPI1 genes (20, 22). To understand how RtsB contributes to transcriptional cross talk, we measured the PflgA, PhilA, and PfimA promoter activities in the wild type, an ΔrtsB mutant, and an ΔrtsB mutant where RtsB was constitutively expressed from a plasmid. Comparing gene expression dynamics in these different mutants, we found that RtsB represses the dynamics of both flagellar and SPI1 gene expression and accelerates type 1 fimbrial gene expression. In the ΔrtsB mutant, we found that flagellar genes are expressed for roughly an hour longer (Fig. 6 A) and that the induction of type 1 fimbrial gene expression is weakly delayed (Fig. 6C). No change in SPI1 gene expression, however, was observed (Fig. 6B). When RtsB is constitutively expressed, we found that it completely inhibits flagellar gene expression (Fig. 6D). It also represses SPI1 gene expression (Fig. 6E) and accelerates the induction of type 1 fimbrial genes by roughly 1 h (Fig. 6F).
FIG. 6.
RtsB controls the dynamics of flagellar and type 1 fimbrial gene expression. (A to C) Deleting RtsB increases the duration of flagellar gene expression and slows the induction of type 1 fimbrial genes. PflgA (A), PhilA (B), and PfimA (C) promoter activities in the wild type and the ΔrtsB mutant are shown. (D to F) Overexpressing RtsB inactivates flagellar gene expression and accelerates the induction of type 1 fimbrial genes. PflgA (D), PhilA (E), and PfimA (F) promoter activities in the wild type and the ΔrtsB mutant constitutively expressing RtsB from the constitutive PLtetO-1 promoter on a plasmid (pRtsB) are shown. Dynamic luminescence experiments were performed as described for Fig. 2. (G) RtsB regulates type 1 fimbrial gene expression through FliZ. PfimA promoter activities in the wild type, the ΔrtsB mutant, the ΔfliZ mutant, the ΔfliZ ΔrtsB mutant, and the ΔrtsB and ΔfliZ ΔrtsB mutants constitutively expressing RtsB are shown. Endpoint fluorescence experiments were performed as described for Fig. 5.
As we observed similar dynamic responses in the ΔrtsB and ΔSPI1 mutants, we conclude that the effect of SPI1 genes on flagellar and type 1 fimbrial gene expression is most likely due to RtsB. To explain our overexpression experiments, we note the following. While RtsB has been shown to directly repress flagellar gene expression, it is not believed to directly regulate SPI1 gene expression (22). Most likely, its effect on SPI1 gene expression is through FliZ. In particular, when RtsB is constitutively expressed, SPI1 gene expression dynamics mirror those in the ΔflhDC mutant. Similarly, RtsB is likely also affecting type 1 fimbrial gene expression through FliZ. To prove this, we compared PfimA promoter activities in the wild type, the ΔrtsB mutant, the ΔfliZ mutant, the ΔfliZ ΔrtsB mutant, and the ΔrtsB and ΔfliZ ΔrtsB mutants constitutively expressing RtsB, using endpoint measurements after 24 h of growth. Consistent with our hypothesis, we found that RtsB has no effect on type 1 fimbrial gene expression in the absence of FliZ (Fig. 6G). Specifically, we found no change in PfimA promoter activity when RtsB was deleted (P > 0.8) or expressed on a plasmid (P > 0.95). However, we qualify this analysis by noting that RtsB itself has only a marginal statistical effect on PfimA promoter activity as determined using endpoint measurements (P = 0.03).
FimZ regulates the dynamics of SPI1 gene expression and is the link between the type 1 fimbrial genes and the flagellar and SPI1 genes.
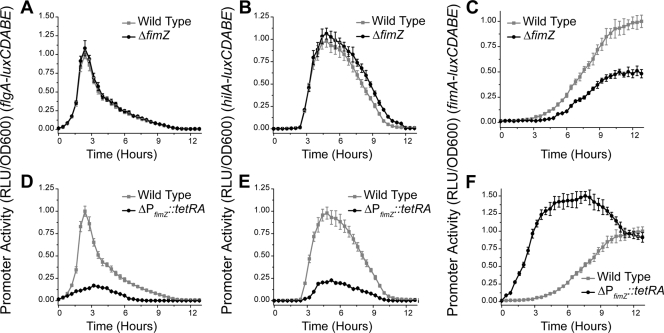
FimZ is a positive regulator of type 1 fimbrial gene expression (90, 91). FimZ has also been shown to bind the PflhDC promoter and repress flagellar gene expression (10). In addition, it represses SPI1 gene expression by increasing the expression of HilE, a negative regulator of SPI1 gene expression (72). To determine the contribution of FimZ to regulatory cross talk, we compared gene expression dynamics in the wild type, the ΔfimZ mutant, and a strain where FimZ is constitutively expressed (ΔPfimZ::tetRA).
In the ΔfimZ mutant, we found that flagellar gene expression is unchanged (Fig. 7 A), whereas the SPI1 genes were expressed for an additional 30 min relative to their expression in the wild type (Fig. 7B). We also found that type 1 fimbrial gene expression was reduced roughly 2-fold (Fig. 7C). When FimZ was constitutively expressed, however, we found that both the flagellar and SPI1 genes were repressed (Fig. 7D and E). Also, as expected, type 1 fimbrial gene expression was significantly accelerated when FimZ was constitutively expressed.
FIG. 7.
FimZ controls the dynamics of SPI1 gene expression. (A to C) Deleting FimZ increases the duration of SPI1 gene expression. PflgA (A), PhilA (B), and PfimA (C) promoter activities in the wild type and the ΔfimZ mutant are shown. (D to F) Overexpressing FimZ represses both flagellar and SPI1 gene expression. PflgA (D), PhilA (E), and PfimA (F) promoter activities in the wild type and the PfimZ::tetRA mutant, where FimZ is under the control of a tetracycline-inducible promoter, are shown. FimZ expression was induced by addition of 15 μg/ml tetracycline. Experiments were performed as described for Fig. 2.
Comparison of these results with those obtained using the ΔfimWYZ mutant suggests that FimZ is the link between type 1 fimbrial genes and flagellar and SPI1 gene expression as similar dynamic behaviors were observed in the two mutants. Interestingly, we found that deleting FimZ had no significant effect on flagellar gene expression. This is probably due to the fact that, under physiological conditions, the type 1 fimbrial genes are expressed long after the flagellar genes no longer are. Only when FimZ is constitutively expressed is repression seen. Similarly, the repression of the SPI1 gene is likely due to a combination of increased HilE expression and reduced flagellar gene expression.
DISCUSSION
Salmonella enterica needs to coordinate the expression of a diverse number of cellular systems during the infection cycle (28, 31, 34, 36, 50, 63, 75). In this study, we investigated the dynamic regulation of three of these systems, namely, flagella, the SPI1 T3SS, and type 1 fimbriae. We were able to demonstrate that these three systems are expressed sequentially during in vitro growth. This hierarchy in gene expression could mirror the roles of these three systems during the infection cycle. According to this simplified model, Salmonella first needs to swim to the sites of invasion in the distal small intestine. Logically then, the flagellar genes are expressed first. Upon reaching its target sites for invasion, Salmonella stops synthesizing flagella as movement is no longer required and starts to synthesize the SPI1 T3SSs necessary for invasion. Once the bacterium enters the stationary phase, it stops synthesizing the SPI1 T3SSs and begins to express the type 1 fimbrial genes involved in intestinal colonization and persistence (1, 19, 49). In vivo, we imagine that this last step is necessary only in those bacteria that were unable to breach the intestinal epithelium.
As multiple studies have shown that extensive regulatory interactions exist between these three systems (5, 10, 22, 38, 72), we hypothesized that regulatory cross talk may govern the hierarchy. However, we found that transcriptional hierarchy is controlled predominately by external factors, external in the sense that the hierarchy is not due to known interactions among these three systems. Cross talk, rather, appears to tune the hierarchy. In particular, cross talk between these three systems is critical for regulating gene expression during transition phases in the hierarchy. The one exception is the effect of FliZ on SPI1 gene expression dynamics.
Among the regulators underlying this cross talk, we found that FliZ is the most significant as it regulates both SPI1 and type 1 fimbrial gene expression, where the latter is a novel finding of this study. Moreover, FliZ's effect on SPI1 gene expression is profound, reducing the expression roughly 3-fold. While FimZ also regulates flagellar and SPI1 gene expression, the effects are minor and are really seen only when the regulator is constitutively expressed. Interestingly, unlike RtsB and FimZ, FliZ does not appear to directly regulate transcription in these three systems. Rather, FliZ appears to function through another transcription factor, be it FlhD4C2 (71), HilD (41), or FimZ. While the underlying mechanism of action of FliZ is still unclear, our data imply that it is posttranslational in all three systems.
One outstanding question concerns the physiological role of this regulatory cross talk between the flagellar, SPI1, and type 1 fimbrial genes. In particular, cross talk has a relatively minor role in regulating the hierarchical expression of these three systems, at least under the conditions investigated in this study. As a comparison, the most well-characterized example of regulatory cross talk involves the hierarchical expression of carbohydrate transport and metabolic genes during growth on mixed substrates. In that case, transcriptional cross talk is used to enforce a strict hierarchy in carbohydrate utilization (17, 64). While we also cannot discount that other factors associated with the flagellar, SPI1, and type 1 fimbrial systems are involved in regulating the transcriptional hierarchy, we expect that the hierarchy is not due to cross talk but rather is regulated by external factors. Specifically, based on our current understanding, we believe that these three systems are regulated in response to the growth phase of the cell (Fig. 2). Why then is cross talk employed?
If we consider the logic of this cross talk, then a simple pattern emerges (Fig. 8). Specifically, cross talk appears to make the expression of these three systems mutually exclusive, though only to the degree to which they are expressed during the infection cycle. In this regard, it helps to reinforce the transcriptional hierarchy. For example, the expression of the flagellar genes represses the expression of the type 1 fimbrial genes and vice versa. This mutual repression is perfectly logical when considering that Salmonella cannot move and adhere/persist at the same time. Similarly, the expression of the flagellar genes enhances SPI1 gene expression, whereas the expression of the SPI1 gene represses flagellar gene expression. Regulation in this case would suggest that only actively motile cells try to invade. As a corollary, persisting cells do not invade, consistent with the fact that the expression of the type 1 fimbrial genes represses the expression of the SPI1 genes. Lastly, once the cells decide to invade, logically then motility is no longer required.
FIG. 8.
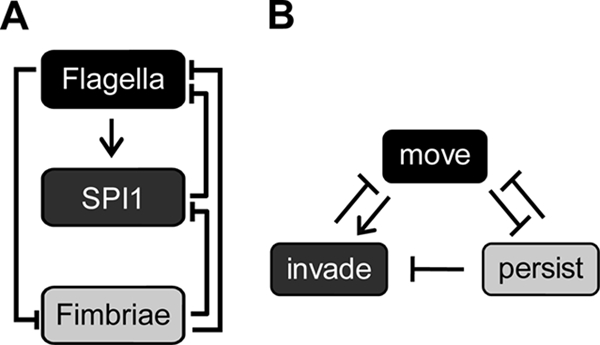
Salmonella invasion program. (A) Diagram of transcriptional cross talk between the flagellar, SPI1, and type 1 fimbrial gene systems. (B) Inferred logic of transcriptional cross talk, where the decision to “move” results from flagellar gene expression, the decision to “invade” results from SPI1 gene expression, and the decision to “persist” results from type 1 fimbrial gene expression.
One limitation of this model is that it does not account for all the other systems involved in the infection cycle. For example, Salmonella has at least 13 distinct fimbrial systems (68) whose expression might also be coordinated with the expression of flagellar, SPI1, and type 1 fimbrial genes. Salmonella also possesses a nonfimbrial adhesin encoded in Salmonella pathogenicity island 4 (SPI4) (28). Previously, we demonstrated that the expression of the SPI4 adhesin is regulated by SprB, a SPI1-encoded regulator (73). Aside from fimbriae and adhesins, Salmonella also possesses a second T3SS encoded within Salmonella pathogenicity island 2 (SPI2) (34). The SPI2 T3SS is used to survive and replicate within host cells during systemic phases of infection. The SPI2 genes also play a role in inducing intestinal inflammation and are known to be regulated by HilD, a SPI1 regulator (7). However, chemical and environmental cues are required to activate SPI2 gene expression, most notably low Mg2+ concentrations (16) and acidic pH (6); HilD is not required for SPI2 gene expression during systemic infection (20). How the cells coordinate the expression of SPI1 and SPI2 genes is still not well understood.
Clearly, this model of the coordinate expression of Salmonella virulence genes (Fig. 8) is still incomplete, as it considers only a small subset of the systems involved in the infection cycle. In addition, it is based on just one mode of growth. Likely, cross talk is more significant when growth is irregular and the environment is variable, as opposed to our in vitro experiments where growth is uninterrupted and the environment is fixed. Further investigations are necessary to fully characterize the role of regulatory cross talk in coordinating gene expression during invasion. The significance of this study is that it is the first to systemically study the effect of regulatory cross talk on the expression dynamics of flagellar, SPI1, and type 1 fimbrial genes.
Acknowledgments
This work was partially supported by National Science Foundation grant CBET 0644744 to C.V.R., Public Health Service grants GM083601 and GM054365 to C.V.R. and AI63230 to J.M.S., and BBSRC grant no. BB/D015855/1 awarded to P.D.A. S.S. was supported by a Drickamer Fellowship, awarded by the Department of Chemical and Biomolecular Engineering at the University of Illinois at Urbana-Champaign.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Althouse, C., S. Patterson, P. Fedorka-Cray, and R. E. Isaacson. 2003. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71:6446-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslanzadeh, J., and L. J. Paulissen. 1992. Role of type 1 and type 3 fimbriae on the adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol. Immunol. 36:351-359. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 73:1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante, V. H., L. C. Martinez, F. J. Santana, L. A. Knodler, O. Steele-Mortimer, and J. L. Puente. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591-14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 12.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 17.Desai, T. A., and C. V. Rao. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 76:1524-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145(Pt. 5):1023-1031. [DOI] [PubMed] [Google Scholar]
- 19.Duguid, J. P., M. R. Darekar, and D. W. Wheater. 1976. Fimbriae and infectivity in Salmonella typhimurium. J. Med. Microbiol. 9:459-473. [DOI] [PubMed] [Google Scholar]
- 20.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691-705. [DOI] [PubMed] [Google Scholar]
- 21.Ellermeier, C. D., and J. M. Slauch. 2006. The genus Salmonella. Springer, New York, NY.
- 22.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 24.Ernst, R. K., D. M. Dombroski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewen, S. W., P. J. Naughton, G. Grant, M. Sojka, E. Allen-Vercoe, S. Bardocz, C. J. Thorns, and A. Pusztai. 1997. Salmonella enterica var Typhimurium and Salmonella enterica var Enteritidis express type 1 fimbriae in the rat in vivo. FEMS Immunol. Med. Microbiol. 18:185-192. [DOI] [PubMed] [Google Scholar]
- 26.Franchi, L., A. Amer, M. Body-Malapel, T. D. Kanneganti, N. Ozoren, R. Jagirdar, N. Inohara, P. Vandenabeele, J. Bertin, A. Coyle, E. P. Grant, and G. Nunez. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576-582. [DOI] [PubMed] [Google Scholar]
- 27.Francis, C. L., T. A. Ryan, B. D. Jones, S. J. Smith, and S. Falkow. 1993. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364:639-642. [DOI] [PubMed] [Google Scholar]
- 28.Gerlach, R. G., D. Jackel, B. Stecher, C. Wagner, A. Lupas, W. D. Hardt, and M. Hensel. 2007. Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834-1850. [DOI] [PubMed] [Google Scholar]
- 29.Ginocchio, C. C., S. B. Olmsted, C. L. Wells, and J. E. Galan. 1994. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell 76:717-724. [DOI] [PubMed] [Google Scholar]
- 30.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, A., M. A. Lasaro, J. C. Sirard, J. P. Kraehenbuhl, and D. M. Schifferli. 2007. Adhesin-dependent binding and uptake of Salmonella enterica serovar Typhimurium by dendritic cells. Microbiology 153:1059-1069. [DOI] [PubMed] [Google Scholar]
- 32.Hakkila, K., M. Maksimow, M. Karp, and M. Virta. 2002. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 301:235-242. [DOI] [PubMed] [Google Scholar]
- 33.Hayward, R. D., and V. Koronakis. 2002. Direct modulation of the host cell cytoskeleton by Salmonella actin-binding proteins. Trends Cell Biol. 12:15-20. [DOI] [PubMed] [Google Scholar]
- 34.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 35.Horiuchi, S., Y. Inagaki, N. Okamura, R. Nakaya, and N. Yamamoto. 1992. Type 1 pili enhance the invasion of Salmonella braenderup and Salmonella typhimurium to HeLa cells. Microbiol. Immunol. 36:593-602. [DOI] [PubMed] [Google Scholar]
- 36.Humphries, A. D., S. M. Townsend, R. A. Kingsley, T. L. Nicholson, R. M. Tsolis, and A. J. Baumler. 2001. Role of fimbriae as antigens and intestinal colonization factors of Salmonella serovars. FEMS Microbiol. Lett. 201:121-125. [DOI] [PubMed] [Google Scholar]
- 37.Ikebe, T., S. Iyoda, and K. Kutsukake. 1999. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 74:179-183. [DOI] [PubMed] [Google Scholar]
- 38.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 39.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones, G. W., and L. A. Richardson. 1981. The attachment to, and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose-resistant haemagglutinating activities. J. Gen. Microbiol. 127:361-370. [DOI] [PubMed] [Google Scholar]
- 41.Kage, H., A. Takaya, M. Ohya, and T. Yamamoto. 2008. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J. Bacteriol. 190:2470-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080-2083. [DOI] [PubMed] [Google Scholar]
- 43.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlinsey, J. E. 2007. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 421:199-209. [DOI] [PubMed] [Google Scholar]
- 45.Khoramian-Falsafi, T., S. Harayama, K. Kutsukake, and J. C. Pechere. 1990. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9:47-53. [DOI] [PubMed] [Google Scholar]
- 46.Kimbrough, T. G., and S. I. Miller. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75-82. [DOI] [PubMed] [Google Scholar]
- 47.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 49.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ledeboer, N. A., J. G. Frye, M. McClelland, and B. D. Jones. 2006. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74:3156-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. U. S. A. 89:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. U. S. A. 97:12283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, D., C. V. Rao, and J. M. Slauch. 2008. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J. Bacteriol. 190:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindquist, B. L., E. Lebenthal, P. C. Lee, M. W. Stinson, and J. M. Merrick. 1987. Adherence of Salmonella typhimurium to small-intestinal enterocytes of the rat. Infect. Immun. 55:3044-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, S. L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of P(prgH) from Salmonella pathogenicity island 1. J. Bacteriol. 183:4876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCormick, B. A., C. A. Parkos, S. P. Colgan, D. K. Carnes, and J. L. Madara. 1998. Apical secretion of a pathogen-elicited epithelial chemoattractant activity in response to surface colonization of intestinal epithelia by Salmonella typhimurium. J. Immunol. 160:455-466. [PubMed] [Google Scholar]
- 60.Miao, E. A., C. M. Alpuche-Aranda, M. Dors, A. E. Clark, M. W. Bader, S. I. Miller, and A. Aderem. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569-575. [DOI] [PubMed] [Google Scholar]
- 61.Miao, E. A., E. Andersen-Nissen, S. E. Warren, and A. Aderem. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29:275-288. [DOI] [PubMed] [Google Scholar]
- 62.Miller, S. I., and P. A. Pegues. 2000. Salmonella species, including Salmonella typhi. Churchill Livingstone, Philadelphia, PA.
- 63.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15:749-759. [DOI] [PubMed] [Google Scholar]
- 64.Monod, J. 1966. From enzymatic adaptation to allosteric transitions. Science 154:475-483. [DOI] [PubMed] [Google Scholar]
- 65.Nambu, T., and K. Kutsukake. 2000. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology 146(Pt. 5):1171-1178. [DOI] [PubMed] [Google Scholar]
- 66.Ogunniyi, A. D., I. Kotlarski, R. Morona, and P. A. Manning. 1997. Role of SefA subunit protein of SEF14 fimbriae in the pathogenesis of Salmonella enterica serovar Enteritidis. Infect. Immun. 65:708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Old, D. C., I. Corneil, L. F. Gibson, A. D. Thomson, and J. P. Duguid. 1968. Fimbriation, pellicle formation and the amount of growth of salmonellas in broth. J. Gen. Microbiol. 51:1-16. [DOI] [PubMed] [Google Scholar]
- 68.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 69.Purcell, B. K., J. Pruckler, and S. Clegg. 1987. Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J. Bacteriol. 169:5831-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossolini, G. M., P. Muscas, A. Chiesurin, and G. Satta. 1993. Analysis of the Salmonella fim gene cluster: identification of a new gene (fimI) encoding a fimbrin-like protein and located downstream from the fimA gene. FEMS Microbiol. Lett. 114:259-265. [DOI] [PubMed] [Google Scholar]
- 71.Saini, S., J. D. Brown, P. D. Aldridge, and C. V. Rao. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J. Bacteriol. 190:4979-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saini, S., J. A. Pearl, and C. V. Rao. 2009. Role of FimW, FimY, and FimZ in regulating the expression of type I fimbriae in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3003-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saini, S., and C. V. Rao. 2010. SprB is the molecular link between Salmonella pathogenicity island 1 (SPI1) and SPI4. J. Bacteriol. 192:2459-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 75.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun, Y. H., H. G. Rolan, and R. M. Tsolis. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 282:33897-33901. [DOI] [PubMed] [Google Scholar]
- 78.Tavendale, A., C. K. Jardine, D. C. Old, and J. P. Duguid. 1983. Haemagglutinins and adhesion of Salmonella typhimurium to HEp2 and HeLa cells. J. Med. Microbiol. 16:371-380. [DOI] [PubMed] [Google Scholar]
- 79.Thijs, I. M., S. C. De Keersmaecker, A. Fadda, K. Engelen, H. Zhao, M. McClelland, K. Marchal, and J. Vanderleyden. 2007. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J. Bacteriol. 189:4587-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tinker, J. K., and S. Clegg. 2000. Characterization of FimY as a coactivator of type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 68:3305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tinker, J. K., L. S. Hancox, and S. Clegg. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Immerseel, F., V. Eeckhaut, F. Boyen, F. Pasmans, F. Haesebrouck, and R. Ducatelle. 2008. Mutations influencing expression of the Salmonella enterica serovar Enteritidis pathogenicity island I key regulator hilA. Antonie Van Leeuwenhoek 94:455-461. [DOI] [PubMed] [Google Scholar]
- 83.Vestby, L. K., T. Moretro, S. Langsrud, E. Heir, and L. L. Nesse. 2009. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vijay-Kumar, M., H. Wu, R. Jones, G. Grant, B. Babbin, T. P. King, D. Kelly, A. T. Gewirtz, and A. S. Neish. 2006. Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am. J. Pathol. 169:1686-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang, S., R. T. Fleming, E. M. Westbrook, P. Matsumura, and D. B. McKay. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798-808. [DOI] [PubMed] [Google Scholar]
- 86.Weening, E. H., J. D. Barker, M. C. Laarakker, A. D. Humphries, R. M. Tsolis, and A. J. Baumler. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilson, R. L., J. Elthon, S. Clegg, and B. D. Jones. 2000. Salmonella enterica serovars Gallinarum and Pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68:4782-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 90.Yeh, K. S., L. S. Hancox, and S. Clegg. 1995. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J. Bacteriol. 177:6861-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yeh, K. S., J. K. Tinker, and S. Clegg. 2002. FimZ binds the Salmonella typhimurium fimA promoter region and may regulate its own expression with FimY. Microbiol. Immunol. 46:1-10. [DOI] [PubMed] [Google Scholar]
- 92.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]