Abstract
With the delivery of millions of sequence reads in a single experiment, next-generation sequencing (NGS) is currently revolutionizing surveys of microorganism diversity. In particular, when applied to Eukaryotes, we are still lacking a rigorous comparison of morphological and NGS-based diversity estimates. In this report, we studied the diversity and the seasonal community turnover of alveolates (Ciliophora and Dinophyceae) in an oligotrophic freshwater lake by SSU amplicon sequencing with NGS as well as by classical morphological analysis. We complemented the morphological analysis by single-cell PCR followed by Sanger sequencing to provide an unambiguous link to the NGS data. We show that NGS and morphological analyses generally capture frequency shifts of abundant taxa over our seasonal samples. The observed incongruencies are probably largely due to rDNA copy number variation among taxa and heterogeneity in the efficiency of cell lysis. Overall, NGS-based amplicon sequencing was superior in detecting rare species. We propose that in the absence of other nuclear markers less susceptible to copy number variation, rDNA-based diversity studies need to be adjusted for confounding effects of copy number variation.
Keywords: alveolata, ciliphora, dinophyceae, microorganism diversity, next-generation sequencing
Introduction
Phagotrophic protists are a major component in any ecosystem and are responsible for the majority of eukaryotic respiration and production (Foissner 1987; Boenigk & Arndt 2002) whereas their autotrophic counterparts are important primary producers (Caron et al. 2009). They are ubiquitous and abundant in all types of habitats and channel carbon and nutrient flow from one of the largest standing stocks of living biomass, i.e. bacteria and archaea (Cho & Azam 1990; Caron et al. 1995), to higher trophic levels. Thus, with respect to global biogeochemical cycles, protists are a major force that shapes the movement and fate of microbial biomass in aquatic ecosystems (Fenchel 1987; Sherr & Sherr 1994).
The diversity of protists is remarkable compared to that of the metazoa and embryophytes (Moreira & Lopez-Garcia 2002; Stoeck & Epstein 2003; Bass & Cavalier-Smith 2004; Countway et al. 2005). Recent studies demonstrated that protist diversity is vastly underestimated, potentially by orders of magnitude – less than 10% of the sequences discovered in cultivation-independent molecular surveys were previously known (Leander & Keeling 2003; Slapeta et al. 2005; Boenigk et al. 2006).
Historically, protist diversity studies largely relied on morphological surveys (Beaver & Crisman 1989; Gaedke & Wickham 2004) but more recently cultivation-independent molecular surveys have been used (e.g. Moreira & Lopez-Garcia 2002; Richards et al. 2005; Slapeta et al. 2005). Irrespective of the method used, the number of individuals studied is rather low, i.e. up to several hundred individuals per sample or less in morphological surveys (e.g. Auer & Arndt 2001; Weitere & Arndt 2003) or several hundred gene copies in molecular surveys (Diez et al. 2001; Moreira & Lopez-Garcia 2002; Stoeck & Epstein 2003; Massana et al. 2004). In the light of the high species diversity of protists, it is obvious that fundamental disputes in microbial ecology, such as the ‘everything is everywhere’ debate, cannot be satisfactorily addressed with the currently available data. Furthermore, molecular and morphological surveys found important differences in the community composition. Within the limits of selective amplification due to primer specificity (Jeon et al. 2008), specific taxonomic groups seem systematically to be over- or underrepresented: for instance, whereas alveolates often dominate in molecular surveys (e.g. Behnke et al. 2006), these taxa usually account for only a few per cent in morphological surveys (Laybourn-Parry et al. 1997; Finlay & Esteban 1998; Savin et al. 2004).
The large amount of sequence reads provided by NGS methods provide a unique opportunity to resolve the discrepancy between morphological and molecular studies. Furthermore, due to the large amount of sequencing data produced, it is anticipated that it will be possible to address the species richness of protists. Using the 454 technology, we show the limitations of SSU sequences for measuring species abundances and the strength of NGS for estimating species richness.
Materials and methods
Sampling and sample preparation
Between March 2007 and October 2007 we collected approximately every third week samples from Lake Fuschlsee (10 samples). The oligotrophic Lake Fuschlsee is located in the district of the Salzkammergut (Austria) (47°48′10″N, 13°16′20″E, maximum depth = 66 m). Integrated samples covering the upper 10 m of the water column were collected within the pelagic zone with a sampling tube. Three integrated samples were pooled prior to further processing. Subsamples were immediately transferred into 100-mL nontransparent flasks and preserved with Lugol's iodine solution (2% final concentration) for morphological analysis and single-cell PCR. A second set of subsamples of 100 mL each were filtered onto 0.2 μm polycarbonate filters for NGS. Filters were air-dried and frozen at −80 °C until further processing.
Microscopical analysis
We counted the abundance of protists in Sedgwick Rafter counting cells using an inverted microscope (Nikon Eclipse) at 200x magnification. Species composition of ciliates was determined following Utermöhl (1958). Briefly, aliquots of 10 mL were settled in the chambers onto coverslips. The complete coverslip area was scanned at 200x magnification. Each individual encountered was determined to species level at 1000X magnification. The Ciliophora were identified according to the key of Foissner et al. (1999). The small oligotrichs Halteria spp. and Pelagohalteria spp. were not clearly distinguishable and therefore not differentiated in our analysis. Except for the clearly distinguishable species Pelagostrombidium fallax, we also grouped the two genera Limnostrombidium and Pelagostrombidium as strombidiid ciliates, as some species of these genera cannot be separated quantitatively (Müller et al. 2002).
Single-cell PCR
We applied single-cell PCR to link sequences to morphospecies. As some morphospecies were represented by several distinct sequences, we analysed up to 10 cells of a given morphospecies in order to check for sequence heterogeneity. Single cells were isolated following to the protocol of Auinger et al. (2008). Briefly, the plankton samples (preserved with Lugol's iodine solution) were repeatedly washed with sterile preserved medium to remove freely dissolved DNA. Subsequently, one millilitre of the preserved sample was transferred to a cover slide and inspected for target cells at a total magnification of ×200 (Zeiss Axiovert 200). One cell (corresponding to 5 μL) was transferred to a 400 μL drop of Lugol's iodine working solution on a separate slide. From this final drop, the cells were picked and (in a drop of ~10 μL) transferred to a PCR tube, which already contained 10 μL of the thiosulfate working solution (corresponding to 390 μg Na2S2O3/mL). Ten microlitres of the remaining fluid was transferred to a second PCR tube as a negative control for freely dissolved DNA and further processed in the same way. The PCR tubes were subsequently heated at 95 °C for 5 min and immediately afterwards shock frozen at −24 °C to break cells and denature proteins.
DNA amplification and sequence handling
The SSU rRNA gene was amplified with the broad eukaryotic SSU targeting forward primer EK82f (5′-GA-AACTGCGAATGGCTC-3′) and the reverse primer Proto5r (5′-GACGGGCGGTGTGTAC-3′), which have been successfully applied for the direct amplification of the SSU rRNA gene from single cells (Auinger et al. 2008). PCR conditions are given in Auinger et al. (2008). If nested PCR was necessary, the following primers were used: forward Primer Sogin2f (5′-AGGGTTCGATTCCGGAG-3′) and reverse primer Nu-SSU-898r (5′-TCCAAGAATTTCACCTC-3′).
The PCR products were commercially purified and sequenced (Cogenics). Success rate of the protocol was >90%. The sequences were processed as previously described in (Auinger et al. 2008), and deposited in the NCBI database (accession numbers GQ281552–GQ28157).
Sample preparation for NGS
For each temporal sample we extracted genomic DNA using the DNeasy Tissue kit (QIAGEN) according to the supplier's instructions. We used HPLC-purified PCR primers, which carry sequences specific for the SSU (fw: ATTAGGGTTCGATTCCGGAGAGG, rv: CTGGAATTACCGCGGSTGCTG) and a 5′-tail for the 454 sequencing (adapter A: GCCTCCCTCGCGCCATCAG, adapter B: GCCTTGCCAGCCCGCTCAG). The forward primer also contained a 4-bp tag for each of the temporal samples inserted between the 454-adapter A and the SSU-specific part. The forward primer was based on the established broad eukaryotic primer Sogin2f (Auinger et al. 2008) whereas the reverse primer was newly designed. For primer design, we aligned sequences from representative taxa covering all major freshwater protists. PCR primers were designed in conserved regions of the SSU rRNA gene to target eukaryotes and primer specificity was tested by analysing clone libraries based on Sanger sequencing and later by analysis of the NGS data set. The primers target all eukaryotic supergroups; some lineages may, however, not be targeted as it is generally known for broad eukaryotic primer sets (Stoeck et al. 2006). The primers amplify a 180–200 bp fragment of the SSU rRNA gene including the V3 region. PCR was carried out using 0.4U Phusion High-Fidelity DNA Polymerase (Finnzymes Oy), 200 μm dNTPs and 0.25 pmol of each primer. The cycling profile consisted of 1′ denaturation at 98 °C, followed 22 PCR cycles (10″ 98 °C, 15″ 65 °C, 20″ 72 °C) with a final extension step of 7′ 72 °C.
In order to minimize recombinant PCR products, we only performed 22 PCR cycles and compensated for the lower yield by pooling the products of 10 PCRs. The pooled PCR products of each temporal sample were gel-purified using QIAquick Gel Extraction Kit (QIAGEN) and quantified on an agarose gel. Approximately equal amounts from each temporal sample were combined into one DNA sample. The pooled DNA was sequenced using a 454 FLX sequencer.
Bioinformatic analyses
Primer clipping
We developed a special primer clipping software, which is specifically designed to account for sequencing artefacts, which add or delete nucleotides during the 454 sequencing process. Reads for which an indel was identified at the transition to the sequencing primer were also removed. The detailed clipping algorithm will be described elsewhere (R. Vinay Pandex, V. Nolte, C. Schlotterer, unpublished). The primer clipping is included in the software package CANGS, which can be downloaded from: http://i122server.vu_wien.ac.at/pop/software.html
Quality filtering
In addition to primer clipping, we removed all sequences that did not fit the following criteria: (i) no Ns; (ii) quality score >24, when averaged across the read after trimming adapters and primers; (iii) no sequencing error in the PCR primers; (iv) minimum sequence length of 200 bp (including PCR primers); and (v) at least two copies of the read present in the entire data set before clipping primers. These procedures eliminated ~37% of all sequences.
Taxonomic classification
In order to minimize the computational burden for genetic distance calculation and blast analysis, we first identified all non-redundant sequences. In the non-redundant sequence data set each sequence variant is only represented once, thus it does not contain redundant sequences. We blasted all non-redundant sequences against the NCBI database and retrieved the taxonomic classification of the best hit. In the case of multiple hits with identical E-values, we selected the most detailed taxonomic classification.
Linking single-cell sequence data with NGS
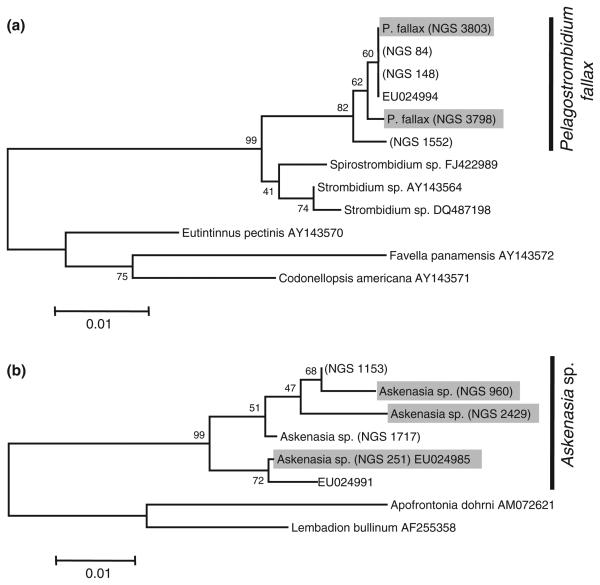
All Sanger sequences obtained from single-cell analysis were used to identify identical sequences in our 454 sequence library. At least three individual cells were analysed per species [Askenasia sp. (5 cells), Pelagostrombidium fallax (6 cells), Ceratium hirundinella (3 cells), Balanion planktonicum (3 cells), Dinobryon spp. (20 cells)]. For Askenasia sp. and P. fallax multiple Sanger sequences were obtained from different cells identified as the same morphospecies (Fig. 1). In these cases, we used phylogenetic analysis based on sequences from the NGS library, the NCBI database and the single-cell sequences to confirm that these sequences cluster together and do not originate from diverged species with a similar morphology. For the phylogenetic analysis we included all sequences obtained from single-cell PCR, the most similar sequences from our NGS database as well as from the NCBI database (using blast). Furthermore, we included at least two more distantly related species as outgroup. Phylogenetic analyses were performed using the software package MEGA4. For subsequent analyses we combined all NGS of the cluster (Fig. 1), as we consider them to originate from the same species.
Fig. 1.
NJ trees (bootstrap 1000 replicates; gaps complete deletion; nucleotide maximum composite likelihood) for (a) Pelagostrombidium fallax and (b) Askenasia sp. Strain codes starting with next-generation sequencing (NGS) indicate the sequence abundance in our NGS library. Grey background indicates that this sequence has been verified by single cell PCR.
Rarefaction analysis
After calculating all pairwise distances of the non-redundant reads with mafft_distance (version 6.608beta; Katho et al. 2009), we constructed a distance matrix, which accounted for the frequency of each sequence in the sample (i.e.: if one sequence occurs twice, this results in two entries in the data matrix). This distance matrix was used to run DOTUR using the default settings (Schloss & Handelsman 2005).
Results and discussion
Comparison of NGS data with morphological analysis
Community composition
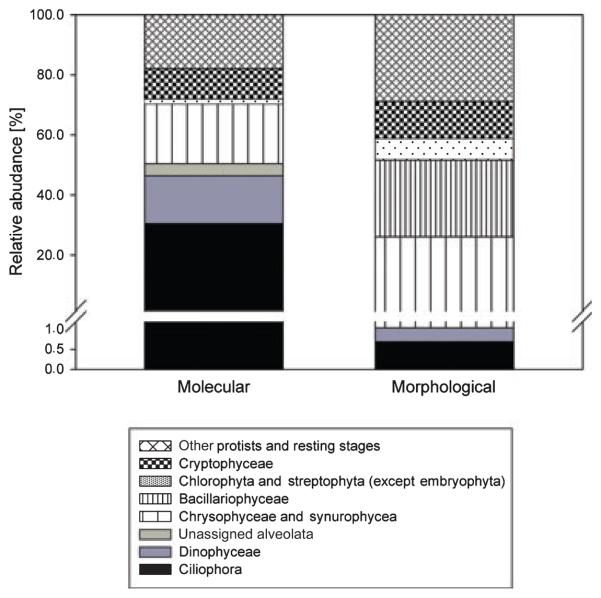
Using FLX sequencing we obtained 447 910 reads of which 279 336 (62.4%) remained after our stringent quality filtering. Based on these rDNA sequence reads, Ciliophora is the most abundant group of protists (30.5%), followed by chrysophytes (including synurophytes) (19.9%), dinoflagellates (15.9%) and cryptophytes (10.2%). This abundance pattern is sharply contrasted by the morphological analysis. The most frequent group were diatoms (25.5%), which are represented by a few sequence reads. Another large discrepancy was noted for Alveolates, with Ciliophora and dinoflagellates contributing 0.69% and 0.35% respectively in the morphological analysis. The abundance of chrysophytes (including synurophytes) (25.0%), Cryptophytes (12.3%) and green algae (7.2%) approximately matched the molecular analysis (Fig. 2).
Fig. 2.
Community composition based on molecular and morphological data. Values correspond to the average across all samples.
The underrepresentation of diatoms in the NGS data is best explained by the difficulties of breaking their shells during DNA extraction. The over-representation of Alveolates could be explained by the well-described high copy number of the SSU rRNA genes (Dyal et al. 1995; Zhu et al. 2005), which is contrasted by a presumably low copy number in most flagellates (cf. Zhu et al. 2005). Furthermore, it is conceivable that cell loss in preserved samples has also contributed to the observed discrepancy between morphological and NGS measurement of species abundance. Sime-Ngando & Grolieère (1991) demonstrated that 10–30% of the cells are lost in samples preserved for morphological analyses. If the cell losses are taxon-specific and differ between species they can bias the relative taxon abundances in the morphological samples.
Species abundance
We observed large discrepancies between morphological and molecular analysis in our comparison of taxonomic groups. Hence, we were interested whether this phenomenon is observed to the same extent for all individual species. We focused on taxa, which are sufficiently abundant in the morphological and molecular analysis and allowed for unambiguous taxon identification. Interestingly, our analysis confirmed the general overestimation of alveolates by NGS (Figs 2 and 3). Nevertheless, the overestimation varied substantially among the species analysed. The most pronounced difference was observed for Askenasia sp., with a ratio of sequence to cell abundance of 203, followed by Pelagostrombidium fallax (39.4), Balanion planktonicum (5.2) and Ceratium hirundinella (4.5).
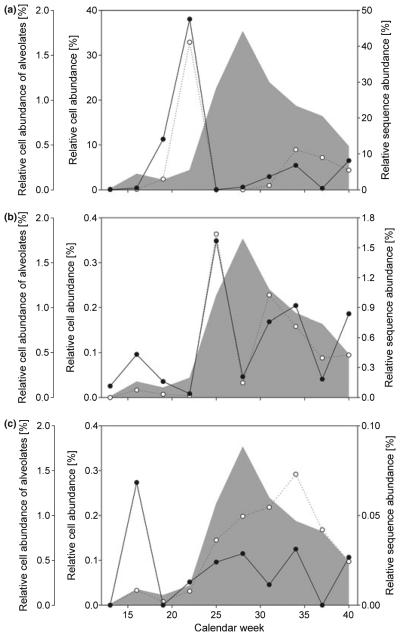
Fig. 3.
Relative abundance of species and their respective sequences. With an increasing correlation between species cell abundance (white dots) and alveolate cell abundance (grey shaded area) seasonal changes are less efficiently captured by sequence abundances (black dots). See text and Table 1 for explanation. (a) Dinobryon divergens and Dinobryon bavaricum; (b) Balanion planktonicum; (c) Ceratium hirundinella.
While our results suggest that NGS of rDNA genes is not well suited to determine absolute species abundances, we were interested to test if NGS accurately measures abundance shifts. Hence, we compared the species abundance over the sampling period based on morphological and sequence data. We found that the change in seasonal abundance was equally well recognized by molecular and morphological data for some species, e.g. a common ciliate (B. planktonicum) and the chrysophyte Dinobryon divergens (Fig. 3) which resulted in a high correlation between molecular (NGS) and morphological abundance measurements (B. planktonicum: r = 0.913, P = 0.000229; D. divergens: r = 0.932; P < 0.0001; Table 1). However, for other taxa the abundance shifts inferred by NGS and morphology were not very similar (e.g. C. hirundinella: r = 0.0961; P = 0.792; P. fallax r = −0.236, P = 0.511).
Table 1.
Correlation (Pearson's product moment) of cell abundances of different taxa with the total cell abundance of alveolates and with the abundance of the affiliated sequence(s)
| Correlation of species abundance with alveolate cell abundance |
Correlation between cell abundance and sequence abundance |
|||
|---|---|---|---|---|
| r | P | r | P | |
| Dinobryon divergens/Dinobryon bavaricum | −0.261 | 0.467 | 0.932 | <0.0001 |
| Balanion planktonicum | 0.548 | 0.101 | 0.913 | 0.0003 |
| Askenasia sp. | 0.645 | 0.0442 | −0.131 | 0.719 |
| Ceratium hirundinella | 0.858 | 0.0015 | 0.0961 | 0.792 |
| Pelagostrombidium fallax | 0.900 | 0.0004 | −0.236 | 0.511 |
Significant correlations are given in bold.
We reasoned that the non-concordant results may be attributed to the use of relative frequencies of the NGS data. As alveolate sequences constitute the largest fraction of sequence reads we focused on their seasonal abundance pattern and noted a substantial change in abundance with the highest relative abundances in week 28 (Fig. 3). Interestingly, the total abundance of alveolate cells has a pronounced effect on the goodness of the correlation between cell and sequence abundance: When the abundance of a given species (e.g. D. divergens, B. planktonicum) is uncorrelated to alveolate abundance, the seasonal abundance shifts were highly consistent between morphological estimates and NGS data (Fig. 3, Table 1). In contrast, if the morphological abundance of the target species (e.g. Ceratium hirundinella) is correlated with alveolate abundance pattern, we did not observe a good correlation between molecular and morphological data (Fig. 3c, Table 1).
Although the change in alveolate abundance is only moderate when measured based on morphological data (~10-fold), the high SSU rRNA gene copy number of alveolates amplifies the abundance changes observed in NGS data. As an increase of alveolate abundance results in disproportionately more reads in a seasonal sample other species, even if their abundance does not change, will be represented by fewer reads. We propose that non-alveolate abundance changes could be more accurately measured if seasonal libraries are analysed without alveolates. Nevertheless, it is also clear that alveolate abundance is not the only factor potentially influencing a discrepancy between NGS and morphological analysis.
Resting stages contribute to NGS-based diversity estimates
Resting stages are typically not determined in morphological surveys, as their taxonomic identification is challenging. Nevertheless, they contribute to NGS-based abundance estimates, as their cell walls may be dissolved by typical DNA extraction protocols. In our samples we noted an abundance of resting stages, but the lack of morphological resolution prevented a more detailed analysis linking the abundance of resting stages to NGS data.
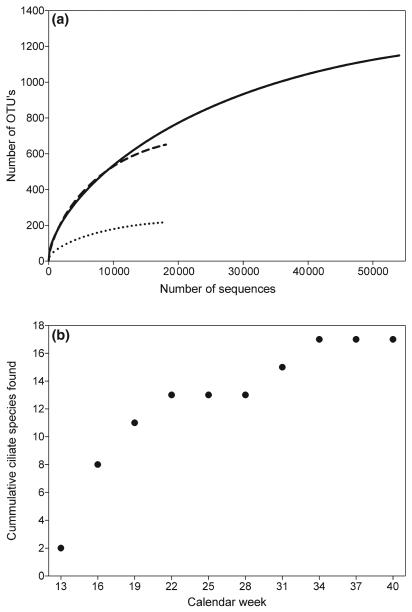
NGS methods are superior in detecting taxon richness
The morphological analysis suggested low species richness. For instance, we found 17 ciliate species and a maximum abundance of 8352 Ind/L (>1000 individual cells were analysed), which lies well in the range of taxa for oligotrophic lakes determined based on morphology (Simek et al. 1995; Foissner et al. 1999; Felip et al. 2002). The NGS approach yielded 216 ciliate and 651 dinophyte OTUs (Alveolata: 1149 OTU's; Fig. 4). Consistent with earlier molecular studies using Sanger sequencing (Richards et al. 2005; Slapeta et al. 2005), we found that NGS-based diversity estimates largely exceed those based on morphological surveys. Typically several individuals need to be inspected for unambiguous species identification in morphological surveys. As protist communities are often dominated by few abundant taxa, rare taxa may easily be overlooked or mistakenly pooled with a similar more abundant taxon.
Fig. 4.
Species richness estimates. (a) Rarefaction analysis for next-generation sequencing using the pooled samples set for Alveolata (straight line), Ciliophora (dotted line) and Dinophyceae (dashed line). OTUs with up to 2% sequence divergence were treated as a single species B) cumulative number of ciliates found in the morohological sample.
The high rate of sequencing errors results in the potential problem of misclassifying reads with sequencing errors as new species. In our analyses, we attempted to minimize the problem by excluding all reads that occurred only once in the data set. Furthermore, we accounted for the high frequency of indel mutations in 454 reads, by not considering indels as an informative character.
Given that we know from Sanger sequences that our fragment does contain true indels, we will on the other hand likely miss some species.
In the recent years, NGS approaches have been gaining popularity for estimating biodiversity in bacteria. In this report, we have extended the same approach to single-celled eukaryotes. Through the combination of single-cell sequencing with morphological analyses, we were able to link NGS data with classic morphology-based diversity estimates.
Our data clearly indicate that rDNA-based NGS data should not be used without additional controls for the estimates of protist biodiversity. We identified three problems; first quantitative DNA extraction could be compromised by heterogeneity in cell wall stability. Second, rDNA copy number variation affects the relative abundance estimates of all species in the sample. Third, we noted that the number of resting stages varies among samples.
Although improved DNA extraction protocols could help addressing the difficulty to break some cell walls efficiently, the variation in copy number poses a more severe problem. In order to compare samples, it is mandatory to standardize NGS data, mostly by the total number of reads.
We showed that variation in abundance of species with different rDNA repeat counts could severely affect the abundance estimates of all species in the library. It may be possible to ameliorate this problem by using either group-specific PCR primers or by restricting the analysis to species with similar rDNA repeat numbers. Nevertheless, we do not think that this lineage-specific analysis of rDNA sequences is the best strategy for future NGS projects. Rather, we strongly advocate for a community effort to develop single copy markers that could replace rDNA sequences for protist research. Environmental genomics projects, such as the ones performed for bacteria, will be very helpful in the identification of suitable sequences. With increasing read lengths of the NGS technologies, we anticipate that single copy markers will eventually become available that combine sufficient phylogenetic resolution over a broad taxonomic range in order to link barcoding efforts in protists with diversity studies. Once such data will be available, it will be possible to evaluate to what extent PCR biases (e.g. preferential amplification of some genotypes) affect the species representation.
Acknowledgement
We thank the Austrian Science Fund for financial support (Project FWF P19706).
Footnotes
Conflicts of interest
The authors have no conflict of interest to declare and note that the sponsors of the issue had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Auer B, Arndt H. Taxonomic composition and biomass of heterotrophic flagellates in relation to lake trophy and season. Freshwater Biology. 2001;46:959–972. [Google Scholar]
- Auinger B, Pfandl K, Boenigk J. Improved methodology for identification of protists and microalgae from plankton samples preserved in Lugol's iodine solution: combining microscopic analysis with single-cell PCR. Applied and Environmental Microbiology. 2008;74:2505–2510. doi: 10.1128/AEM.01803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass D, Cavalier-Smith T. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa) International Journal of Systematic and Evolutionary Microbiology. 2004;54:2393–2404. doi: 10.1099/ijs.0.63229-0. [DOI] [PubMed] [Google Scholar]
- Beaver JR, Crisman TL. The role of ciliated protozoa in pelagic freshwater ecosystems. Microbial Ecology. 1989;17:111–136. doi: 10.1007/BF02011847. [DOI] [PubMed] [Google Scholar]
- Behnke A, Bunge J, Barger K, et al. Microeukaryote community patterns along an O-2/H2S gradient in a supersulfidic anoxic Fjord (Framvaren, Norway) Applied and Environmental Microbiology. 2006;72:3626–3636. doi: 10.1128/AEM.72.5.3626-3636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boenigk J, Arndt H. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie van Leeuwenhoek. 2002;81:465–480. doi: 10.1023/a:1020509305868. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Pfandl K, Garstecki T, et al. Evidence for geographic isolation and signs of endemism within a protistan morphospecies. Applied and Environmental Microbiology. 2006;72:5159–5164. doi: 10.1128/AEM.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA, Dam HG, Kremer P, et al. The contribution of microorganisms to particulate carbon and nitrogen in surface waters of the Sargasso Sea near Bermuda. Deep-Sea Research I. 1995;42:943–972. [Google Scholar]
- Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: a perspective. ISME Journal. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- Cho B, Azam F. Biogeochemical significance of bacterial biomass in the ocean's euphotic zone. Marine Ecology Progress Series. 1990;63:253–259. [Google Scholar]
- Countway PD, Gast RJ, Savai P, et al. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. Journal of Eukaryotic Microbiology. 2005;52:95–106. doi: 10.1111/j.1550-7408.2005.05202006.x. [DOI] [PubMed] [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study ofgenetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Applied and Environmental Microbiology. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyal PL, Hope S, Roberts DM, Embley TM. Use of the PCR and fluorescent-probes to recover SSU ribosomal-RNA sequences from single cells of the ciliate protozoan Sphatitium. Molecular Ecology. 1995;4:499–503. doi: 10.1111/j.1365-294x.1995.tb00244.x. [DOI] [PubMed] [Google Scholar]
- Felip M, Wille A, Sattler B, et al. Microbial communities in the winter cover and the water column of an alpine lake: system connectivity and uncoupling. Aquatic Microbial Ecology. 2002;29:123–134. [Google Scholar]
- Fenchel T. Ecology of Protozoa. Springer-Verlag; Berlin: 1987. [Google Scholar]
- Finlay BJ, Esteban GF. Freshwater protozoa: biodiversity and ecological function. Biodervisity and Conservation. 1998;7:1163–1186. [Google Scholar]
- Foissner W. Soil protozoa: fundamental problems, ecological significance, adaptations in ciliates and testaceans, bioindicators, and guide to the literature. Progress in Protistology. 1987;2:69–212. [Google Scholar]
- Foissner W, Berger H, Schaumburg H. Identification and ecology of limnetic plankton ciliates. Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft. 1999;3:1–793. [Google Scholar]
- Gaedke U, Wickham S. Ciliate dynamics in response to changing biotic and abiotic conditions in a large, deep lake (L Constance) Aquatic Microbial Ecology. 2004;34:247–261. [Google Scholar]
- Jeon S, Bunge J, Leslin C, et al. Environmental rRNA inventories miss over half of protistan diversity. BMC Microbiology. 2008;8:222. doi: 10.1186/1471-2180-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katho K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Laybourn-Parry J, James M, McKnight D, et al. The microbial plankton of Lake Fryxell, Southern Victoria Land, Antarctica, during the summers of 1992 and 1994. Polar Biology. 1997;17:54–61. [Google Scholar]
- Leander BS, Keeling PJ. Morphostasis in alveolate evolution. Trends in Ecology & Evolution. 2003;18:395–402. [Google Scholar]
- Massana R, Castresana J, Balagué V, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Applied and Environmental Microbiology. 2004;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends in Microbiology. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Müller H, Stadler P, Weisse T. Seasonal dynamics of cyst formation of strombidiid ciliates in alpine Lake Mondsee, Austria. Aquatic Microbial Ecology. 2002;29:181–188. [Google Scholar]
- Richards TA, Vepritskiy AA, Gouliamova DE, et al. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environmental Microbiology. 2005;7:1413–1425. doi: 10.1111/j.1462-2920.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- Savin MC, Martin JL, LeGresley M, et al. Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microbial Ecology. 2004;48:51–65. doi: 10.1007/s00248-003-1033-8. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microbial Ecology. 1994;28:223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- Šimek K, Bobková J, Macek M, et al. Ciliate grazing on picoplankton in a eutrophic reservoir during the summer phytoplankton maximum: a study at the species and community level. Limnology and Oceanography. 1995;40:1077–1090. [Google Scholar]
- Sime-Ngando T, Grolière CA. Effets quantitatifs des fixateurs sur le stockage des ciliés d'eaux douces. Archiv für Protistenkunde. 1991;140:109–120. [Google Scholar]
- Slapeta J, Moreira D, Lopez-Garcia P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proceedings of the Royal Society of London, Series B: Biological Sciences. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Epstein S. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Applied and Environmental Microbiology. 2003;69:2657–2663. doi: 10.1128/AEM.69.5.2657-2663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Utermöhl H. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitteilungen. Internationale Vereinigung Für Theoretische und Angewandte Limnologie. 1958;5:567–596. [Google Scholar]
- Weitere M, Arndt H. Structure of the heterotrophic flagellate community in the water column of the River Rhine (Germany) European Journal of Protistology. 2003;39:287–300. [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiology Ecology. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]