Abstract
Heat shock protein 90 (Hsp90) is a prime target for antitumor therapies. The information obtained by MD simulations is combined with NMR data to provide a cross-validated atomic resolution model of the complementary interactions of Hsp90 with a peptidic (Shepherdin) and a non peptidic (5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside, AICAR) inhibitor, showing antiproliferative and proapoptotic activity in multiple tumor cell lines. This approach highlights the relevant role of imidazolic moiety in the interaction of both antagonist molecules. In AICAR bound state one conformation of those present in solution is selected, where imidazolic, H4 and H5 protons have a key role in defining a non polar region contacting Hsp90 surface. The dynamic equilibrium between N-type and S-type puckered forms of AICAR ribofuranoside moiety is shown to be functional to inhibitor binding. The first experimental structural data on these inhibitors are presented and discussed as hints for future design of improved molecules.
Keywords: Antitumor Agents, Drug Design, Molecular Dynamics, NMR, Hsp90
Heat shock protein 90 (Hsp90) is a molecular chaperone and is one of the most abundant proteins expressed in cells (1). It is a member of the heat shock protein family which is up regulated during stress response. These kinds of proteins protect the cell when exposed to elevated temperatures but their overexpression can be triggered also by other kinds of stress such as inflammation, hypoxia, exposure of cells to toxins and other pathological conditions like tumours. In order to respond to these insults and to ensure cancer cell survival in the face of an otherwise hostile environment, Hsp90 oversees the correct conformational development of a diverse ensemble of client proteins. Hsp90 is indeed the most abundant chaperone expressed in eukaryotic and prokaryotic cells. In mammalian cells, there are mainly two cytosolic Hsp90 isoforms and the human Hsp90α form shows 85% sequence identity with Hsp90β (2). Hsp90 consists of three structural domains: N-terminal domain (NT) of about 25KDa, the middle domain (M) of about 40KDa and the C-terminal domain (CT) of about 12KDa. The most studied and best-understood domain is the N-terminal. This domain contains the ATP binding site and shows high homology not only amongst members of Hsp family but also amongst members of the ATPase/kinase superfamily (3). In unstressed cells Hsp90 works as molecular chaperone assisting folding of newly synthesized proteins and regulates the trafficking of the denaturated proteins in cells under pathological and stressed conditions. Cancerous cells overexpress a number of proteins including P13K and AKT, whose inhibition causes apoptosis, and various steroid receptors such as p53, ErB2, Src, Abl, Raf which are all clients of Hsp90. It appears that all these Hsp90 clients are implicated in signal transduction and cell-cycle control; mutations or misregulation of these proteins cause uncontrolled development and proliferation of tumor cells. Hsp90 is also required for induction of vascular endothelial growth factor and nitric oxide synthase, both important for de novo angiogenesis. It also promotes the proliferation and invasion of metastasis by assisting the matrix metalloproteinase MMP2. Hsp90, with the help of its cochaperones, modulates tumour cell apoptosis trough effects on AKT (4), tumor necrosis factor receptors (TNFR) and nuclear factor-kB (NF-kB) function (5). Moreover Hsp90 stabilizes the mutants of these proteins involved in tumor growth and is involved in metastastasis and proliferation.
The important role that Hsp90 plays in tumor growth has made it a prime target for antitumor therapies. Most of the designed inhibitors act on Hsp90 competing for the ATP binding site at the N-terminal domain (6). The first known inhibitor of N-terminal domain of Hsp90 (Hsp90-NT) is the antibiotic geldanamycin which appeared to reduce tumor growth of 50% (7). In recent years a great interest has grown around the search for new Hsp90 inhibitors competing for the ATP binding site. X-Ray and NMR first, and in silico studies later, were used to design and optimize lead compounds able to inhibit Hsp90 (6, 8).
An example showing the utility of computational biology approaches in the discovery of new small molecule inhibitors of Hsp90-NT is represented by the use of molecular dynamics (MD) simulations and structural analysis to develop the peptidomimetic antagonist Shepherdin (9) into the small molecule 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) (10). Shepherdin is the minimal peptidic sequence from survivin, an essential regulator of cell proliferation, differentiation and apoptosis overexpressed in cancer (11), which is able to block the interaction of survivin with Hsp90 in vitro (12) and cause killing of tumour cells by apoptotic and nonapoptotic mechanisms (9). Starting from a model of the Shepherdin-Hsp90 complex (9), long timescale MD simulations were used to single out the most relevant functional groups required for the antagonist to productively bind the chaperone. This information was then translated into dynamic pharmacophore models taking the full protein and ligand flexibility into account. The use of the pharmacophore in a virtual screening effort allowed to identify AICAR as a novel, active Hsp90 inhibitor, able to compete with ATP for binding at the N terminal domain, destabilize various Hsp90-clients complex in vivo and inhibit cell proliferation in various tumor cell lines, while leaving normal cells unscathed (10). The 3D structural model of the AICAR-Hsp90 complex, obtained by computational studies(10), has to be validated experimentally to be really helpful in the context of rational drug design. NMR has recently emerged as a high-throughput experimental technique in drug discovery, in determining possible binding affinities and in determining the region of interaction.
In this paper we have set out to combine the information independently obtained by MD simulations and experimental NMR data to provide a cross-validated and cross-filtered atomic resolution model of the complementary interactions within the binding site. All-atom MD simulations allowing full flexibility of the ligand and the receptor generate diverse sets of configurations for the complex, whereby both the ligand and the binding site of receptor may visit different conformations available on the complex free energy landscape. In analogy with protein folding, it is important to discern native complex conformations, from near-native or non-native ones. In this context, native conformations (states) are the ones that are mostly accessed and populated in solution at equilibrium conditions. Long time scale MD simulations provide a general view of the different possible states. NMR analysis, as discussed herein, helps select and filter only those conformations that verify specific structural constraints obtained in solution at equilibrium, recapitulating ensemble properties that are specific only to selected molecular configurations. This combined approach may be particularly useful in the presence of large receptor proteins, not yet amenable for full NMR analysis or in the presence of highly flexible ligands, for which X-ray may fail to provide an atomic structure. Molecular dynamics simulations have been previously successfully applied to the optimisation of X-ray and NMR structures of macromolecules, significantly improving the efficiency of structure calculation and refinement, and allowing more and more challenging systems to be analyzed (13, 14). The efficiency of a similar in-silico-experimental approach was recently demonstrated for the development of new agents targeting angiogenic factors (15, 16).
In this study the first experimental structural data on Shepherdin and on the novel AICAR inhibitor in complex with Hsp90-NT are presented and discussed with clear implications for future design of improved inhibitors.
Materials and Methods
Hsp90-NT expression and inhibitor synthesis
Human Hsp90- NT, was cloned in pGex-4T3 and pFLAG-CMW 6c vector, expressed in E. Coli as GST fusion protein and purified from the GST frame by overnight thrombin (1 U/ml) cleavage as previously described(9, 12). Shepherdin peptide was synthesized by the W.M Keck Biotechnology Research Center at Yale University School of Medicine using solid phase tBoc chemistry followed by reverse-phase high-pressure liquid chromatography, mass spectrometry and solid phase purification. AICAR was purchased by Sigma.
Sample preparation
Shepherdin peptide (NH2-K1HSSGCAFL9-COOH) was dissolved at 1.2 mM peptide concentration in 30 mM phosphate buffer (95% H2O, 5% D2O), 6 mM DTT, 100 mM NaCl, pH 6.7. For NMR analysis of the interactions 1.6 mM shepherdin was studied in the presence of Hsp90-NT (50 μM). AICAR (5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside) was dissolved at 1.8 mM in the same buffer and interaction studies were performed in the presence of Hsp90-NT 60 μM. To evaluate the J12 and J34 of AICAR in the bound state, 0.1mM Hsp90 samples were titrated with the ligand from P:L ratios 1:1 to 1:3.
NMR
All NMR spectra were recorded on Bruker DMX spectrometer operating at 500 MHz equipped with a triple resonance probehead, incorporating gradients in the z-axis. All data were collected at 280 K on a spectral width of 6510 Hz. 2D-TOCSY (spin lock time of 80 ms), NOESY (mixing times of 100 and 250 ms) and ROESY (spin lock time of 250 ms) spectra were recorded using standard sequences (17), (18) on Shepherdin peptide in order to assign all the resonances at pH 6.8. Data were processed with NMRPipe(19) and visualized by NMRView(20). The proton assignments are reported in Table S1 of Supplemental Data. Longitudinal relaxation times T1 of Shepherdin and AICAR resonances were measured in the presence of Hsp90-NT with the standard inversion recovery method. Data points (32 K) were acquired to cover a sweep width of 10 ppm and a relaxation delay of 6 s was used. Data were analysed using Bruker Topspin software. Shepherdin T1 values were found to be rather constant along the peptide side-chains with values in the range 0.4-0.6 s. The resonance at 2.1 ppm, corresponding to an impurity present in Shepherdin samples, showed a longer T1 value (0.9 s) and strong STD effects in the spectra. It should be mentioned that, as previously reported in the literature (21), T1 values affect STD signal intensities and higher T1 gives rise to apparent stronger STD effects. AICAR T1 values were found to range between 0.4-0.9 s.
For the acquisition of NMR Saturation Transfer Difference (STD) experiments a 1D pulse sequence incorporating a T1ρ filter to remove disturbing protein signals was used (22) and the duration of the T1r filter was 80 ms. STD spectra were recorded with a spectral width of 6510 Hz and 32 K data points. On-resonance irradiations were performed at different frequencies in the methyl region and off resonance irradiation was performed at −20000 Hz, using a series of Gaussian pulses with a 1% truncation and 50 ms duration to give total saturation times of 0.25, 0.5, 1, 2, 3, 4, 6, 8 sec. STD NMR spectra were acquired with a total of 1024 transients in addition to 16 scans to allow the sample to come to equilibrium. The subtraction of the two FIDs was achieved by phase cycling.
1H chemical shift assignment of AICAR resonances was performed with standard methods. NOESY and ROESY spectra with mixing times of 100 and 250 ms were collected on AICAR alone and in the presence of Hsp90-NT at protein:ligand ratio 1:28 ratio. No purging spin lock period to remove the NMR signals of the macromolecule background was employed. To minimize the unwanted Hartmann-Hahn effects in the ROESY spectra a moderate spin lock pulse was used with a field strength of 2390 Hz and the transmitter frequency was set at 6 ppm (23). In order to compare NOESY and ROESY cross peaks volumes in the absence and in the presence of Hsp90-NT protein, taking into account cross-peak intensity loss due to T1 relaxation, fractional volumes were calculated by dividing cross-peak volumes by the average of two resolved diagonal peaks. The contribution of the bound form of the ligand to NOESY/ROESY cross-relaxation rate, observed for protons i and s on a ligand in fast exchange with a high molecular-mass receptor, is described by Equation (1) (24):
| (1) |
where pf and pb are the mole fractions and σisf and σisb are the cross-relaxation rates of the free and bound ligand respectively. The bound fraction was assumed to be 0.04 at 1:28 Hsp90-NT:AICAR ratio. The ROE volumes were corrected for the offset differences between the correlation peaks and the spin lock pulse offset (25).
A rough estimate of distances from NOE/ROE cross peaks volumes was calculated assuming an average distance between H4-H5 of 2.8 Å as deduced from MD simulations of free and bound AICAR (vide infra). The percentage of the two puckered forms S-type (C-2-endo, C-3-exo) and N-type (C-2-exo, C-3-endo) was estimated on the basis of the measured 1H J12 and J34 coupling constants in the free and bound state (Table S2 in Supplemental data). J couplings of the bound form (Jb) were deduced from Equation (2):
| (2) |
where Jobs and Jf are the J couplings observed in the spectra of the bound and free AICAR, respectively and pb and pf are the mole fractions of bound and free species, respectively.
MD Simulations and analysis
The docking procedure and simulations for the Shepherdin-Hsp90-NT and AICAR-Hsp90-NT complexes were already described in Meli et al 2006 (10).
Conformational cluster analysis of the trajectories was performed using the method described in Daura et al. (26): the count number of neighbours using a cut-off of 2 Å RMSD between the optimal backbone superposition of different structures was performed and the structure with the largest number of neighbors with all its neighbours was taken as cluster and eliminated from the pool. This procedure is repeated for the remaining structures in the pool. The three most populated clusters, representative of the most visited structures along the MD simulations, were selected. The central structure of each cluster, recapitulating the main structural properties of that cluster, was selected as representative for subsequent analysis and cross-validation through NMR experimental measurements.
The same procedure as outlined above was used for the conformational analysis of isolated AICAR free in solution.
The estimate of the population of the ribose ring puckered conformers (N or S-type) was performed by measuring the value of the C4-C3-C2-C1 dihedral angle over MD trajectory of free and bound ligand.
Pymol (DeLano, W.L. The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA, USA) was used for graphical representation of the results.
Results and Discussion
NMR spectroscopy is exploited here to increase the knowledge of structural and conformational features favouring Hsp90 recognition/inhibition by Shepherdin and AICAR, so far widely described by MD and docking studies (10).
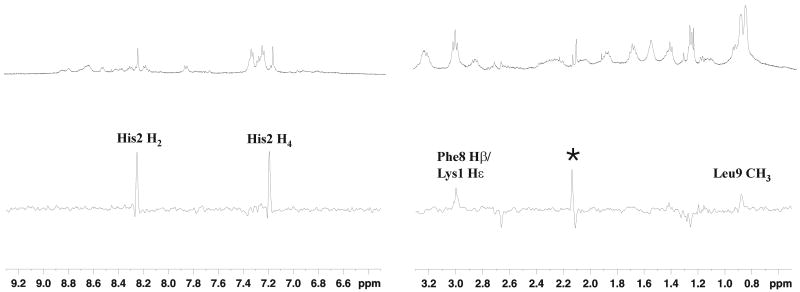
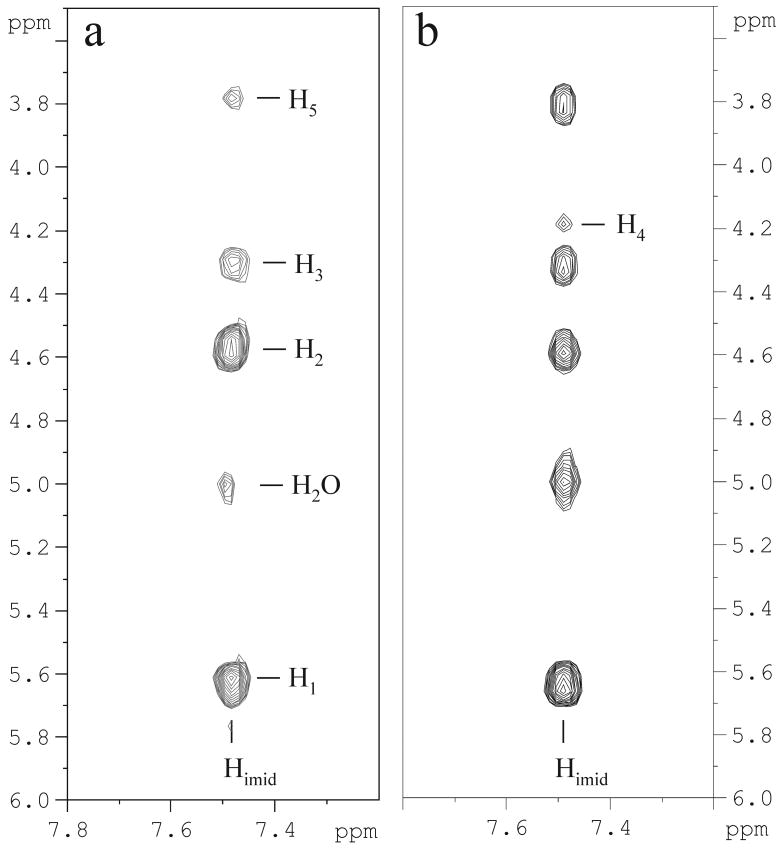
NMR interaction studies were performed through STD experiments for Shepherdin/Hsp90 system in order to map the peptide protons in more intimate contact with the protein. The addition of Hsp90-NT to Shepherdin sample in a 1:34 ratio induced a broadening of the peptide signals due to the presence of exchange between the bound and the free form of the peptide (27), thus clearly indicating the presence of binding interactions. STD NMR experiments were recorded with different saturation times and irradiating frequencies in the methyl spectral region of the protein and a selected spectrum is shown in Figure 1. NMR spectra suggest that Hsp90 recognition is strongly mediated by the aromatic ring of His2 (H2 and H4). In addition, STD signals indicate a smaller involvement of the side chains of Leu9, Phe8 and/or Lys1. These findings are in good agreement with the MD data according to which the His2 ring can satisfy H-bonding conditions with Hsp90 being involved in interactions both as an acceptor (Nε atom) and as a donor (NδH atom) (9). Indeed, the Shepherdin-based screening strategy suggested to map an imidazole ring moiety of two pharmacophoric models (PHARM1 and PHARM3) on the position of the His2 ring. PHARM1, used as query for a search of non-peptidic small molecules in the data base of NCI_3D yielded, among others, AICAR as a novel Hsp90 inhibitor (Figure 2). AICAR was shown to exhibit a high antiproliferative and proapoptotic activity in multiple tumour cell lines (10), and therefore represents a good starting point for further developing new anticancer drugs. Structural and conformational properties of the inhibitor in the free and bound state, together with the relevant ligand groups in close proximity to Hsp90 were therefore characterized by NMR. A clear indication of the binding of AICAR to Hsp90 came from the line broadening observed for the ligand resonances in the presence of a protein excess. The bound conformation of AICAR could be deduced from the comparison of Tr-NOESY experiments recorded in the presence of Hsp90 with NOESY data recorded on the free ligand. NOESY experiments on the free AICAR resulted in positive NOEs (opposite sign of diagonal) for all interproton vectors. Negative NOE cross-peaks were observed (same sign as the diagonal) in the presence of Hsp90 and this change in the sign corresponds to a slower tumbling molecule, as it is expected when the ligand is bound to the receptor (Figure 3). A new cross peak was observed between the imidazolic proton (Himid) and H4, suggesting the stabilization, upon binding, of an AICAR conformation characterized by a shorter distance between the two protons. In order to analyse the conformational changes, eventually affecting other inter-proton distances, a rough estimate of the contribution of the bound fraction of the ligand to the Tr-NOESY cross-peak intensities was calculated (see Experimental Procedures). The fractional NOESY intensities of the free and bound peptide showed a general increase of the NOESY intensities in the bound form, as expected when the ligand is bound to the protein. Assuming similar effective correlation times and spin diffusion pathways for the different proton pairs within the molecule in the same state (free or bound), the ratio of the NOE volumes was used to estimate changes in inter-proton distances. The largest changes in distance factors (Table 1) affected Himid-H5 and Himid-H4 pairs, suggesting that these inter-proton distances are approximately 1.3-1.4 times shorter than in the free state. These results showed that in the bound state one conformation of those present in solution is selected and is characterized by shorter distances between the imidazolic proton and H4 and H5 protons of the ribose ring. Tr-NOESY experiments could not be used for a quantitative deduction of inter-proton distances because the only proton pair separated by known distance are the geminal H5 protons, which are not resolved (isochronous) in the spectra. However a semi-quantitative picture from the available NMR data was achieved converting the NOE volumes in inter-proton distances assuming an average distance of 2.8 Å between H4 and H5/H5′protons, as suggested by the analysis of the main cluster of free and bound MD conformers (vide infra). The results obtained from NOESY experiments were cross checked by comparison with the distances derived from ROESY data (Table 1), since the artifacts which may appear in each of these two experiments are known to have different origins (28). The distances obtained from NOESY and ROESY showed a very good agreement suggesting that the cross peak volumes are not affected by artifactual contributions.
Figure 1.
STD NMR spectrum of Shepherdin peptide in the presence of Hsp90-NT. Upper panel: 1H NMR reference spectrum of 1.2 mM Shepherdin in the presence of 50 μM Hsp90-NT in 30 mM buffer phosphate (95% H2O, 5% D2O), 100 mM NaCl, 6 mM DTT, pH 6.7 recorded at 280K on a 500 MHz Bruker spectrometer. Lower panel: 1H NMR STD spectrum of the same sample. A saturation time of 2 sec at -602 Hz was used. The assignment of the observed STD signals is reported, an asterisk marks a signal due to an impurity (see Material and Methods).
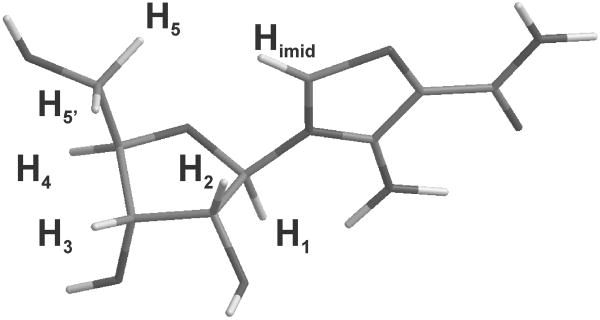
Figure 2.
Structure of AICAR inhibitor with labels for hydrogens mentioned in the paper.
Figure 3.
Selection of NOESY spectrum of free AICAR (panel a) and of Tr-NOESY of AICAR bound to Hsp90-NT (1:28 Hsp90-NT: AICAR ratio) (panel b) at 280 K on a 500 MHz Bruker spectrometer. A mixing time of 250 ms was employed. Cross peaks in panel a) have opposite sign with respect to diagonal peaks, while cross peaks in panel b) have the same sign as diagonal peaks. The assignment of the cross peaks is reported.
Table 1.
Comparison of proton distances in free and bound AICAR from NMR and MD calculationsa
| Cross peak | Distance factor from NOESY | NOE distance (Å) |
ROE distance (Å) |
MD distance (Å) |
||||
|---|---|---|---|---|---|---|---|---|
| Free | Bound | Free | Bound | Free | Bound | Free | Bound | |
| H1-H2 | 1.2 | 1.1 | 3.3 | 3.1 | 3.3 | 2.7 | 3.1 | 2.7 |
| H1-H4 | 1.3 | 1.2 | 3.8 | 3.4 | 3.5 | 3.0 | 3.4 | 3.5 |
| H1-H5* | > 1.7b | 1.7 | > 5b | 4.8 | > 5b | > 5b | 4.3, 5.0 | 4.4, 5.2 |
| H2-H3 | 1.0 | 0.9 | 2.8 | 2.6 | - | - | 2.4 | 2.4 |
| H3-H5* | 1.1 | 1.2 | 3.2 | 3.3 | 4.2 | 3.3 | 2.5, 2.9 | 2.5, 2.8 |
| H4-H5* | 1.0 | 1.0 | 2.8 | 2.8 | 2.8 | 2.8 | 2.5, 3.0 | 2.7, 3.0 |
| Himid-H1 | 1.0 | 0.9 | 2.9 | 2.7 | 2.7 | 2.8 | 3.6 | 3.6 |
| Himid-H2 | 1.0 | 1.0 | 2.8 | 2.9 | 2.7 | 3.2 | 3.6 | 4.3 |
| Himid-H3 | 1.3 | 1.2 | 3.7 | 3.3 | 3.7 | 3.0 | 5.2 | 3.2 |
| Himid-H4 | > 1.7b | 1.3 | > 5b | 3.8 | > 5b | 3.5 | 4.4 | 4.3 |
| Himid-H5* | 1.7 | 1.3 | 4.8 | 3.8 | 4.5 | 3.9 | 4.7, 3.2 | 3.8, 2.3 |
The average distance observed for H4-H5 pair from MD simulations in free and bound AICAR (2.8 Å) was used to deduce a rough estimate of distances from NOE/ROE cross peaks volumes.
the cross peak was not observed in the spectrum.
Possible conformational changes characterizing the ligand upon complex formation were investigated also with all atom MD simulations. Intramolecular inter-proton distances were calculated for both free and complexed AICAR on the representative structures of the three conformational clusters recapitulating more than 90% of all the conformations visited during the respective simulations. The comparison of theoretical and experimental distance values (Table S3 in Supplemental Data) suggests that for both free and bound simulations only structures belonging to the first and most populated MD clusters fit well with NMR data. Indeed structures belonging to second and third clusters show inter-proton distances between H5,5′ and Himid significantly higher than 5 Å, that would not give rise to an observable dipolar coupling, detectable through NOE measurements. The analysis of the representative structure of the first cluster indicated a general good agreement between computationally and experimentally determined inter-proton distances in the free inhibitor and in the complex with Hsp90 (Table 1), supporting the view that upon binding the preferred imidazolic ring conformation brings Himid closer to H4 and H5,5′protons (Figure S1 Supporting information).
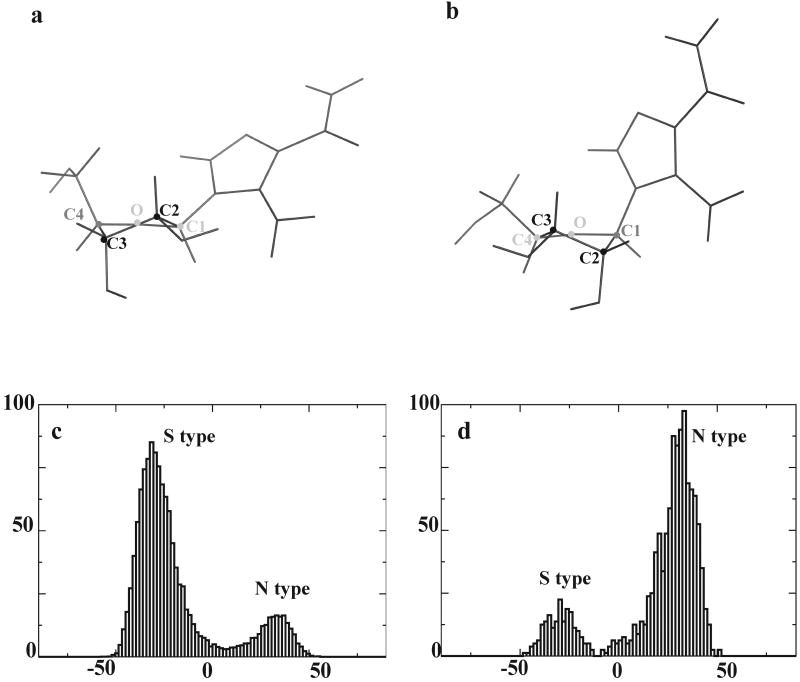
The analysis of NMR spectra also allowed to describe dynamic equilibrium of AICAR ribofuranoside moiety between the two puckered forms S-type (C-2-endo, C-3-exo) and N-type (C-2-exo, C-3-endo). Indeed the percentage of each conformer present in solution can be estimated on the basis of the measured 1H J12 and J34 coupling constants in the free and bound state (25). Interestingly the proportions of the S-type/N-type conformers change upon binding: in the free state the most populated state appeared to be the S-type, whereas the conformation preferred by bound AICAR was the N-type. In particular the N-type population, estimated from 1H J12 and J34 values (Table S2), increased from 35% in the free state to more than 50% when AICAR was bound to Hsp90. The experimental observation prompted us to deeply investigate whether the change of conformational equilibrium could be extrapolated from the MD trajectories of free and bound ligand. Figure 4 shows that the dihedral population defining the S or N-type puckered conformers undergoes a dramatic variation when passing from the free to the complexed ligand. The S-type population representing the preferred conformation for AICAR in solution, while the N-type appearing to be prevalently populated in the complex with Hsp90-NT. NMR and MD data revealed that the dynamic interconversion of AICAR ribofuranoside moiety is functional to binding. It is worth mentioning that the analysis of the reported X-Ray structures for the complex of Hsp90-NT with ADP-Mg (PDB ID: 1BYQ) and for the whole Hsp90 with an ATP analogue (PDB ID: 2CG9) showed that the ribose ring puckered conformer in the bound state was the N-type.
Figure 4.
AICAR representative structures obtained from MD simulations for free (panel a) and bound state (panel b). Distribution of the dihedral angle values defining the S- or N- type puckering for AICAR free in solution (panel c) and in complex with Hsp90-NT (panel d). The distributions have been calculated by analyzing all the structures visited during the respective MD trajectories.
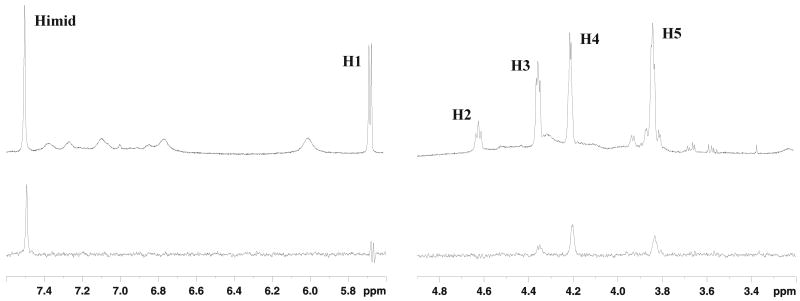
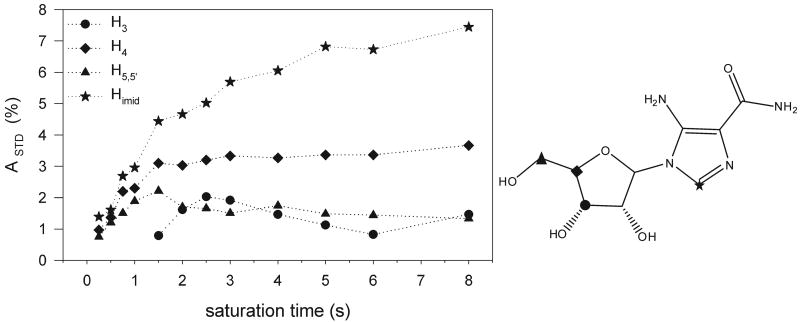
In order to collect experimental evidences of the kind of interactions established by AICAR with Hsp90, STD experiments were performed. The STD spectrum of AICAR (Figure 5) proved that Himid together with H3, H4 and H5 protons of the ribose moiety receives saturation transfer from the protein, giving rise to STD NMR signals. In an attempt to compare the relative contribution of each proton to the contact interface, STD experiments were performed at different saturation times and the build-up curves of the saturation degree (STD factor, ASTD) against the saturation time are shown in Figure 6. It has been previously reported that both the build–up rate and the height of STD factor plateau are strongly affected by T1 relaxation of the single protons(21), indeed at long saturation times higher STD factors are measured for protons with higher T1 values. However at saturation times shorter than half of the shortest T1 ASTD reflects the average proximity of the ligand protons to the protein surface. The sensitivity under short saturation conditions (saturation delay of 250 ms) is greatly reduced and the percentages of saturation transfer could be determined only for the imidazole proton (14%), for H4 (10%) and H5 (8%) sugar protons, giving the highest STD effects and providing the best contact interactions with the receptor. These results supported the proposed bound conformation where Himid, H4 and H5,5′ are close in space.
Figure 5.
STD NMR spectrum of AICAR at 280 K on a 500 MHz Bruker spectrometer. Upper panel: 1D NMR reference spectrum of 1.8 mM AICAR in the presence of 60 μM Hsp90-NT in 30 mM buffer phosphate (95% D2O, 5% H2O), 100 mM NaCl, pH 6.7. Lower panel: STD spectrum of the same sample. A saturation time of 3 sec at -456 Hz was used. The assignment of the AICAR proton resonances is reported.
Figure 6.
Plot of STD factors (ASDT) of the AICAR/Hsp90 system versus saturation time (left panel). AICAR was used with 28-fold molar excess over Hsp90-NT. ASTD values are plotted for Himid (star), H3 (circle), H4 (diamond) and H5,5′ (triangle). Protons showing STD effects are depicted on the chemical structure with the same symbols (right panel). T1 relaxation times measured for the investigated signals are: Himid (0.9 s), H3 (0.6s), H4 (0.7 s) and H5,5′ (0.4 s).
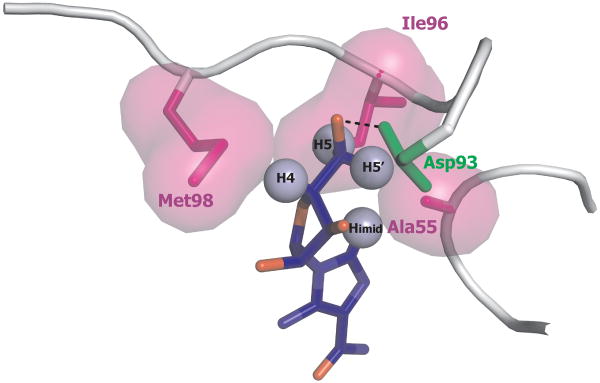
Analysis of MD trajectories indicated that AICAR binds into the ATP-binding pocket of Hsp90 (10), as observed for most common and well-studied inhibitors, such as geldanamycin, radicicol, and derivatives, whose therapeutic effect is via ATP competitive binding at NT domain(6, 29). The main structural features of the Hsp90-AICAR complexes, fit nicely with the results obtained from NMR-STD measurements. In particular, NMR-STD measurements showed the relevance of Himid together with H4 and H5,5′ protons in the stabilization of the complex. The analysis of the representative structure of the most populated cluster (cluster 1) shows that the mentioned protons are surrounded by a few sidechains, namely Ala55, Ile96, Met98 (Figure 7), which could stabilize the complex through hydrophobic interactions with the non-polar side of the bound conformation of AICAR. The hydroxyl group in position 5 of the ribose moiety is involved in H-bonds with the side-chain of Asp93, an important residue for binding and catalysis (6), and/or T184, thus not interfering with the hydrophobic interface. In agreement with NMR experimental determinations, MD pointed out also the relevant role of the carbohydrate ring of AICAR in binding being involved in direct hydrogen bonds with the side chain of Asp93, through the OH group in positions 5 of the ribose ring. STD NMR analysis could not detect any observable involvement in binding from H2. The analysis of the representative MD structure indicates that this proton is farther from the protein side-chains than imidazole, H4 and H5 protons, however an interaction with the protein cannot be excluded. This apparent inconsistency points to the dynamic character of binding of AICAR to Hsp90, where alternative inhibitor poses are possible in solution.
Figure 7.
AICAR/Hsp90 interface as deduced from NMR data and MD simulations. AICAR protons showing the highest saturation transfer from Hsp90-NT are labelled and represented as light blue spheres. Hsp90 residues Ala55, Ile96, Met98, making hydrophobic contacts with AICAR, are represented by their Van der Waals surface in magenta. The H-bond between the hydroxyl group in position 5 of AICAR ribose moiety with Asp93 side-chain, is depicted as a black dotted line.
MD results showed that the inhibitor contacts the charged region of the Hsp90 ATP binding site, defined by Asn51 and Asp54, establishing several hydrogen bonding interactions through the NH in position 3 and the carboxamide at position 4 of the imidazole ring. NMR cannot afford a clear indication of the role of the mentioned protons, which are hardly observable at the present experimental conditions due to their labile nature. However the high ASTD factor estimated for Himid strongly supports the relevant role of the imidazole ring in Hsp90 binding.
MD and NMR data proposed a binding mode slightly different with respect to ATP/Hsp90 complex observed in X-ray structure (PDB ID: 2CG9). It is however to be considered that in the present analysis we are considering the isolated N-terminal Hsp90 domain, while the X-ray structure containing the ATP molecule was obtained using full-length Hsp90. This determines non-negligible conformational changes especially in the two N-terminal domains, which form a tight interface and swap beta-strand 1 and helix 1 to stabilize the ATP complex. When the N-terminal domain of the full-length dimer is superimposed to the isolated N-terminal domain in complex with AICAR the positioning of ATP would determine a steric clash between the phosphate groups of the nucleotide and one of the loops protruding into the active site which can be avoided by allowing the ligand to move and adapt to the binding site, as it actually happens during MD simulations. Interestingly, the observed positioning of AICAR in the active site is more consistent with the structures of the Hsp90 N-terminal domain in complex with several small molecule inhibitors observed for instance by Wright et al (30) or by Immormino et al. (31). Indeed in the former case, it is possible to see that the imidazol ring of AICAR occupies the same space as the aromatic rings of the Vernalis compounds, while the ribose moiety traces the hydrogen-bonding functionalities of the substituents (Figure S2 Supporting Information). The comparison with the positioning of the inhibitors developed by the Chiosis group suggests that AICAR aromatic imidazole ring traces the position of the purine ring of the inhibitor (Figure S3 Supporting Information).
Conclusions
Obtaining three-dimensional macromolecular structures of protein/inhibitor complexes is a challenging and time-consuming task. Methods allowing a quick and reliable determination of the 3D structure of a complex are highly needed in the context of rational drug design. Experimental methods based on X-ray crystallography have been extremely successful, and are becoming a high-throughput method. However, it requires the easy availability of diffracting protein crystals, condition never met in the case of the Shepherdin/Hsp90 complex. Moreover, crystal structures may provide a static representation of the complex, in which internal dynamics and conformational equilibria may not be accounted for.
NMR has the potential to provide dynamic information on the adduct in solution, but the determination of the three-dimensional structure may involve the assignment of thousands of intra-protein nuclear Overhauser effects, while the only relevant ones are the few intermolecular cross-peaks between protein and ligand signals.
Computational predictions obtained through docking programs have reached a high level of sophistication and important results (32, 33), being extremely valuable in the early ligand design phases. One of the drawbacks is once more represented by the absence of dynamic information on the complexes generated.
In this paper, we have proposed a combination of computational biology and NMR approaches for relatively fast and reliable determination of protein–ligand structural models, keeping into explicit account the roles of conformational flexibility as well as the possibility for a certain bimolecular complex to visit alternative configurations available on the energy landscape. In order to fully exploit this information in drug-design efforts in the characterization of new complexes, one has to be able to select the native-states representatives (most populated configurations corresponding to free energy minima on the energy landscape) from non-native ones. The approach we propose relies on the validity of docking and MD simulations to generate ensembles of structures representative of the situation in solution, while the NMR addition allows to single out only the structures of the ligand in the complex that are significantly populated at equilibrium. All-atom MD simulations in explicit solvent have come of age, and it is now possible to run long reliable simulations on small clusters. NMR techniques such as the ones discussed here do not require the lengthy assignment of all protein resonances or the need for isotopically enriched, expensive protein samples. Importantly, this in silico-experimental approach, where the results obtained independently from the two methodologies are combined, sheds direct light on possible differences in the structure and dynamics of the ligand when in the free state and when complexed to the receptor.
In particular the MD-NMR approach was able to single out the relevant groups involved in interaction, with a clear indication of the importance of the imidazole moiety for both peptidic and non peptidic antagonist molecules. In addition to the strongly interaction mediated by the Shepherdin aromatic ring of His2, a hydrophobic contribution through the apolar side chains of Leu9, Phe8 and/or Lys1 was also suggested by NMR data for the recognition of Hsp90. These findings suggest that in the search of more promising pharmacophoric models, the addition of an apolar group mimicking the hydrophobic interactions could improve the inhibitor affinity.
Structural data on AICAR highlighted that, upon binding a conformation characterized by shorter H4-Himid and H5,5′-Himid distances is stabilized, defining a non polar region of the molecule. NMR indicated that H4, H5,5′ and Himid are the key AICAR atoms contacting Hsp90 surface and MD suggested that they are close to an hydrophobic region defined by Ala55, Ile96, Met98 side-chains (Figure 7).
Finally NMR and MD data revealed that a dynamic equilibrium between N-type and S-type puckered forms of AICAR ribofuranoside moiety is functional to inhibitor binding.
The data here presented will be used to improve pharmacophore models for the rational identification/design of new and more active Hsp90 inhibitors as new anticancer therapeutic candidates. The combined in silico-experimental approach outlined in this paper can be of general utility in drug design, allowing to select specific binding poses among the high number of possible structures of the complex, observed when taking the dynamics of both the ligand and the receptor into account.
Supplementary Material
Acknowledgments
This work was supported by grants from Italian Association for Cancer Research (code: 5890), National Institutes of Health: CA90917, CA118005 and CA78810 and Fondazione CARIPLO. We gratefully thanks Dr. Katiuscia Pagano for help with Pymol software.
References
- 1.Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79:129–68. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 2.Chen B, Zhong D, Monteiro A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 2006;7:156. doi: 10.1186/1471-2164-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prodromou C, Pearl LH. Structure and functional relationships of Hsp90. Curr Cancer Drug Targets. 2003;3:301–23. doi: 10.2174/1568009033481877. [DOI] [PubMed] [Google Scholar]
- 4.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10832–7. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 6.Sgobba M, Rastelli G. Structure-based and in silico design of Hsp90 inhibitors. ChemMedChem. 2009;4:1399–409. doi: 10.1002/cmdc.200900256. [DOI] [PubMed] [Google Scholar]
- 7.Goetz MP, Toft DO, Ames MM, Erlichman C. The Hsp90 chaperone complex as a novel target for cancer therapy. Ann Oncol. 2003;14:1169–76. doi: 10.1093/annonc/mdg316. [DOI] [PubMed] [Google Scholar]
- 8.Huth JR, Park C, Petros AM, Kunzer AR, Wendt MD, Wang X, et al. Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies. Chemical biology & drug design. 2007;70:1–12. doi: 10.1111/j.1747-0285.2007.00535.x. [DOI] [PubMed] [Google Scholar]
- 9.Plescia J, Salz W, Xia F, Pennati M, Zaffaroni N, Daidone MG, et al. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–68. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 10.Meli M, Pennati M, Curto M, Daidone MG, Plescia J, Toba S, et al. Small-molecule targeting of heat shock protein 90 chaperone function: rational identification of a new anticancer lead. Journal of medicinal chemistry. 2006;49:7721–30. doi: 10.1021/jm060836y. [DOI] [PubMed] [Google Scholar]
- 11.Altieri DC. Validating survivin as a cancer therapeutic target. Nature reviews. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 12.Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, et al. Regulation of survivin function by Hsp90. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13791–6. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunger AT, Adams PD, Rice LM. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–36. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 14.Brunger AT, Adams PD. Molecular dynamics applied to X-ray structure refinement. Accounts of chemical research. 2002;35:404–12. doi: 10.1021/ar010034r. [DOI] [PubMed] [Google Scholar]
- 15.Colombo G, Margosio B, Ragona L, Neves M, Bonifacio S, Annis DS, et al. Non-peptidic thrombospondin-1 mimics as fibroblast growth factor-2 inhibitors: an integrated strategy for the development of new antiangiogenic compounds. The Journal of biological chemistry. 2010;285:8733–42. doi: 10.1074/jbc.M109.085605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leali D, Bianchi R, Bugatti A, Nicoli S, Mitola S, Ragona L, et al. Fibroblast Growth Factor 2-antagonist Activity of a Long-Pentraxin 3-derived Antiangiogenic Pentapeptide. Journal of cellular and molecular medicine. 2009 doi: 10.1111/j.1582-4934.2009.00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bax A, D DG. Practical aspects of two-dimencional transverse NOE spectroscopy. Practical aspects of two-dimencional transverse NOE spectroscopy. 1985:207–13. [Google Scholar]
- 18.Hwang TL, Shaka AJ. water supression that works. Exitation sculpting using arbitrary wave-forms and pulsed field gradients. journal of magnetic resonance. 1995;112:275–9. [Google Scholar]
- 19.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 20.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology (Clifton, NJ. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 21.Yan J, Kline AD, Mo H, Shapiro MJ, Zartler ER. The effect of relaxation on the epitope mapping by saturation transfer difference NMR. J Magn Reson. 2003;163:270–6. doi: 10.1016/s1090-7807(03)00106-x. [DOI] [PubMed] [Google Scholar]
- 22.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–17. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 23.Bax A. Correction of cross-peak intensities in 2D spin locked NOE spectroscopy for offset and artman-Hahn effects. Journal of magnetic resonance. 1988;77:134–47. [Google Scholar]
- 24.Adams ER, D EA, Gizachew D, DeLeo FR, Yu L, Volpp BD, Vlases M, Jesaitis AJ, Quinn MT. interaction of human neutrophil flavocytochrome b with cytosolic proteins: transferred-NOESY NMR studies of a gp91phox C-terminal peptide bound to p47phox. Biochem J. 1997;325:249–57. doi: 10.1042/bj3250249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ammalahti E, B M, Cadet J, Molko D. NMR assignments and conformational studies of two diastereomeric oxidation products of 2′-deoxycytidine. Magnetic Resonance in Chemistry. 1998;36:363–70. [Google Scholar]
- 26.Daura X, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Reversible peptide folding in solution by molecular dynamics simulation. journal of Molecular Biology. 1998;280:925–32. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]
- 27.Burritt JB, Busse SC, Gizachew D, Siemsen DW, Quinn MT, Bond CW, et al. Antibody imprint of a membrane protein surface. Phagocyte flavocytochrome b. The Journal of biological chemistry. 1998;273:24847–52. doi: 10.1074/jbc.273.38.24847. [DOI] [PubMed] [Google Scholar]
- 28.Bazzo R, E CJ, Wormald MR, Rademacher TW, Dwek RA. Complete computer simulation of ROESY experiments, including Hartmann-Hahn effects. Chem Phys lett. 1990;174:313–7. [Google Scholar]
- 29.Dehner A, Furrer J, Richter K, Schuster I, Buchner J, Kessler H. NMR chemical shift perturbation study of the N-terminal domain of Hsp90 upon binding of ADP, AMP-PNP, geldanamycin, and radicicol. Chembiochem. 2003;4:870–7. doi: 10.1002/cbic.200300658. [DOI] [PubMed] [Google Scholar]
- 30.Wright L, Barril X, Dymock B, Sheridan L, Surgenor A, Beswick M, et al. Structure-activity relationships in purine-based inhibitor binding to HSP90 isoforms. Chemistry & biology. 2004;11:775–85. doi: 10.1016/j.chembiol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 31.Immormino RM, Kang Y, Chiosis G, Gewirth DT. Structural and quantum chemical studies of 8-aryl-sulfanyl adenine class Hsp90 inhibitors. Journal of medicinal chemistry. 2006;49:4953–60. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 32.Morra G, Verkhivker G, Colombo G. Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS computational biology. 2009;5:e1000323. doi: 10.1371/journal.pcbi.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, et al. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. The Journal of clinical investigation. 2009;119:454–64. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.