Abstract
Genetic variants predicted to seriously disrupt the function of human protein-coding genes—so-called loss-of-function (LOF) variants—have traditionally been viewed in the context of severe Mendelian disease. However, recent large-scale sequencing and genotyping projects have revealed a surprisingly large number of these variants in the genomes of apparently healthy individuals—at least 100 per genome, including more than 30 in a homozygous state—suggesting a previously unappreciated level of variation in functional gene content between humans. These variants are mostly found at low frequency, suggesting that they are enriched for mildly deleterious polymorphisms suppressed by negative natural selection, and thus represent an attractive set of candidate variants for complex disease susceptibility. However, they are also enriched for sequencing and annotation artefacts, so overall present serious challenges for clinical sequencing projects seeking to identify severe disease genes amidst the ‘noise’ of technical error and benign genetic polymorphism. Systematic, high-quality catalogues of LOF variants present in the genomes of healthy individuals, built from the output of large-scale sequencing studies such as the 1000 Genomes Project, will help to distinguish between benign and disease-causing LOF variants, and will provide valuable resources for clinical genomics.
INTRODUCTION
Rapid advances in DNA sequencing technology are now making large-scale clinical genomics a reality. Currently, groups around the world are sequencing all of the protein-coding genes, or even the entire genomes, of thousands of patients to search for disease-causing mutations. Such studies have already yielded novel disease genes (1), and seem poised to identify many more over the next few years. Building on studies of copy number variation (2,3) and single-base substitutions that introduce new stop codons (nonsense SNPs) (4), they have also revealed an unexpected feature of human genomes: the existence of many dozens of genetic variants predicted to severely disrupt protein-coding genes in every human genome, even those from healthy individuals. The existence of these variants in such high numbers raises intriguing questions about recent human evolutionary history, and poses a major challenge for clinical geneticists: how can we find true disease-causing mutations amidst this sea of gene-disrupting, but apparently benign, variants?
In this review, we will discuss recent genome-scale findings about the prevalence of these loss-of-function (LOF) variants. In theory, LOF variants can act by disrupting any essential genetic element, including non-coding regulatory motifs, but we will focus on disruptions to protein-coding genes. In addition, although severe loss of function can result from seemingly mild perturbations (such as substitutions of a single amino acid in an active site), we will restrict our discussion to variants that substantially truncate or entirely eliminate protein-coding transcripts, because in these cases a functional impact can be assigned from examination of the sequence with greater confidence.
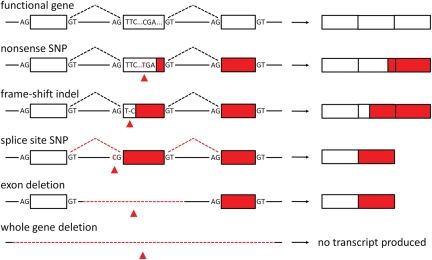
Figure 1 illustrates the variety of LOF variants that can arise within protein-coding genes, ranging from single-base substitutions such as nonsense SNPs or splice site disruptions, through small insertions/deletions (indels) that change the reading frame or remove a splice site, to larger deletions that remove either crucial exons or entire genes.
Figure 1.
Classes of LOF variant affecting protein-coding regions. A model three-exon gene is shown both intact (top) and following the introduction of various types of LOF variant (red triangles). Effects on the transcript produced by the gene are shown at the right. LOF variants typically result in a loss of protein-coding functionality downstream of the variant (red boxes).
LOF variants vary considerably in their effects on human phenotype. Most obviously, they can represent severely deleterious disease-causing mutations which, in healthy individuals, should be restricted to recessive alleles present in a heterozygous state. However, they will also include mildly deleterious variants with small effects on fitness, and neutral variants disrupting the function of non-essential genes. We further demonstrate that they include additional and perhaps less obvious categories: advantageous variants, and (importantly) sequencing and annotation errors. Developing strategies to distinguish between these categories is a key challenge for the field.
As we write this review, the trickle of genome sequences is about to become a flood, and we will focus particularly on the steps needed to achieve the clinical geneticist's goal of identifying a causal mutation in an entire genome sequence.
HISTORICAL CONTEXT
Clinical genetic studies have traditionally relied on an implicit assumption that variants that severely disrupt gene function are likely to be disease-causing. This intuitive approach is understandable, and can be powerful when there are other lines of evidence supporting the pathogenicity of that variant (for instance, in vitro or in vivo functional studies, or strong linkage signals). However, such intuition can also lead investigators astray: for example, protein deficiency resulting from a common, benign nonsense SNP in the ACTN3 gene (described below) was initially considered a possible causal candidate for severe muscle disease (5).
In fact, examples of benign LOF variation were observed as early as 1900 with the discovery of the first variable blood-group markers, the ABO antigens (6): the O allele is an LOF variant due to a single-base deletion and has arisen independently on several occasions (7). Pharmacogenetic studies have revealed extensive variation in drug-metabolizing capacity between individuals, frequently caused by LOF variants of drug-metabolizing enzymes: for example, an LOF allele of CYP2C19 arises from an SNP creating a novel splice acceptor site, present at ∼30% in Han Chinese (8). Other drug-metabolizing variation proved to arise from larger rearrangements (including complete deletion) of genes, such as CYP2D6 (9).
Subsequently, a series of large-scale surveys revealed that LOF variants are surprisingly common in healthy individuals. For example, a systematic survey of 805 reported nonsense SNPs in 1151 individuals from 56 worldwide populations (4) found that 169 were variable within the genotyped individuals, with each individual carrying on average 32 of them (14 in a homozygous state); 99 genes were homozygously inactivated in one or more individuals. More recently, a high-resolution, genome-wide survey of common copy-number variations identified 213 complete deletions of RefSeq genes and 34 deletions of whole exons leading to frameshifts in HapMap samples (2).
Given that many of these variants are present at relatively high frequencies (and seen in a homozygous state) in healthy individuals, it seems clear that gene loss is often benign. More surprisingly, several cases have been identified—either by comparing the phenotypes of those with and without the LOF variant, or by examining the pattern of variation in the surrounding DNA in the population for signatures indicative of rapid evolutionary spread or positive selection (10)—where gene disruption actually appears to be beneficial. Examples include CASP12, where LOF due to a nonsense SNP decreases the chance of developing severe sepsis and increases survival in modern hospital surroundings (11) and has conferred a long-term evolutionary advantage (12); ACTN3, where a nonsense SNP that results in complete protein deficiency is associated with altered human muscle function and athletic performance (reviewed in 13) and shows evidence for recent positive selection in non-African populations (14); and UGT2B17, where LOF due to complete deletion of the gene shows a signal of positive selection in East Asians (15). Indeed, some have speculated that LOF might frequently have been advantageous in human evolution: the ‘less-is-more’ hypothesis (16). A fuller understanding of the LOF variants segregating in the current population and those fixed on the human lineage (17) will allow the importance of this mode of evolution to be evaluated more fully.
Observations such as those above demonstrate the presence of abundant LOF variants in healthy people and raise a number of questions: how many variants of this kind are carried by each individual, how many genes can show heterozygous or homozygous LOF without severe consequences and what is the spectrum of disadvantage and advantage associated with LOF variants?
CHALLENGES IN IDENTIFYING LOF VARIANTS
It may intuitively seem that it should be straightforward to identify LOF variants belonging to the clear categories discussed in this review (deletions that remove or truncate a gene, frameshifting indels and SNPs that create or destroy a canonical splice site, or introduce a stop codon). However, this is far from being the case. Even in the high-quality, intensively studied human reference sequence, gene annotation is incomplete and imperfect (18). Consequently, LOF variants in genes or exons that are not annotated, or are annotated as pseudogenes, will not be recognized; conversely, pseudogenes that are annotated as genes, or intronic regions that are mistakenly included in the gene model, may appear to carry LOF variants, but these will be false positives. For example, the ACTN3 gene discussed above is currently annotated in the Ensembl reference gene set as a ‘polymorphic pseudogene’ and thus excluded from many genome-wide analyses.
Even when annotation is accurate, it can often be difficult to determine whether or not a coding variant is truly LOF. For example, many LOF variants will directly or indirectly (via a frame shift) generate a premature stop codon in the affected transcript; in some cases, this will result in the degradation of the entire transcript via nonsense-mediated decay (19), but in other cases it may result in the production of a truncated but nonetheless functional protein.
In addition, alternative splicing is a common characteristic of human genes (20), and many LOF variants affect only a fraction of the transcripts, with other transcripts skipping the affected exon and potentially rescuing the biological function of the gene. For example, a nonsense SNP in the MOBKL2C gene truncates the predicted protein product of one known splice variant by >90% but leaves two alternative transcripts intact, whereas a nonsense SNP in the ASCC1 gene is predicted to truncate two of the gene's transcripts by 80%, but leaves another six splice variants intact (4).
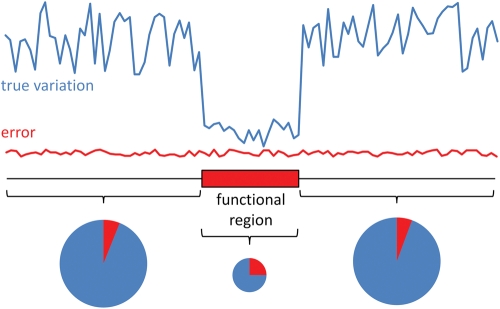
All sequence data—particularly when generated by next-generation, short-read sequencing technologies—contain errors arising from base mis-calls and read mis-mapping (21). These will be encountered among apparent LOF variants to a much greater degree than many other classes of variation for the reason illustrated in Figure 2: a lower expected rate of polymorphism at LOF sites, coupled with broadly uniform error rates across all functional classes of variant, is expected to lead to a substantially higher false-positive rate for LOF variants. These effects mean that LOF variants will typically be enriched for all of the various sources of error that plague large-scale genomic studies in general, all of which will be familiar to readers engaged in clinical sequencing studies.
Figure 2.
Functional regions are enriched for sequencing errors and other artefacts. Top shows a schematic plot of levels of true sequence variation (blue) and sequencing error (red). Bottom shows pie charts with circle size proportional to total observed variation, and red and blue sections proportional to error and true variation, respectively. In functional regions, true variation is suppressed by natural selection, but error remains approximately uniform. This results in fewer observed variants in functional regions, but a higher error rate in those observed.
Finally, there can be a question of whether an LOF variant has been inherited, or has arisen somatically. This problem is particularly acute when cell-line DNA (which can accumulate de novo mutations during cell divisions in culture) is sequenced (22) and a de novo heterozygous causal variant is expected in a patient. When germ-line inheritance is expected, this can be tested; when it is not, follow-up might be carried out in multiple patient tissues, or using model systems.
LOF VARIANTS IN THE SEQUENCING ERA
Despite difficulties in recognizing true LOF variants, applications of high-throughput DNA sequencing technologies over the last 2 years have provided an increasingly high-resolution view of the patterns of variation throughout the human genome, including insights into the extent of LOF polymorphism in healthy humans.
In a resequencing study of 718 X-chromosomal genes in 208 families with X-linked mental retardation, 30 genes were found to contain a truncating variant, but more than half of these were either present in controls or did not segregate with the disease phenotype in families, suggesting that they were not associated with the disorder (23). This high number is particularly striking since selection against deleterious LOF variants is expected to be strong for X-chromosomal genes due to hemizygosity in males.
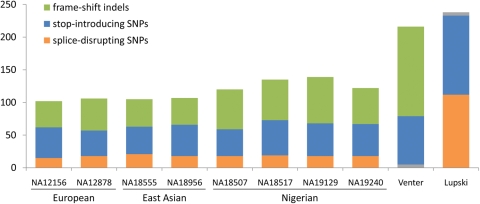
Insight into the wider distribution of LOF variation within the genome has come from individual whole-genome sequences generated using a variety of technologies. The first published individual genome sequence—and by most standards still the highest quality—was that of Venter, generated using ‘first-generation’ capillary sequencing technology (24). A subsequent analysis of variants in the protein-coding regions of Venter's genome (25) identified 74 stop SNPs and 137 frame-shift indel variants (Figure 3). However, the authors also noted the challenges of interpreting these variants: nearly half of the nonsense SNPs were found in hypothetical genes, and frame-shift indels clustered significantly towards the boundaries of genes and coding exons, making it more likely that functional transcripts could be generated from alternative splicing of the putative LOF allele.
Figure 3.
Reported numbers of LOF variants per individual genome in several published large-scale sequencing studies. Individuals labelled European, East Asian or Nigerian are HapMap individuals from reference (27). Numbers for Venter and Lupski are from references (25) and (28), respectively. Small grey segments in Venter and Lupski histograms indicate unreported numbers for splice-disrupting SNPs and frame-shift indels, respectively.
Since the publication of Venter's genome, several dozen complete genomes and exomes (targeted sequencing of the protein-coding regions of the genome) have been generated using ‘second-generation’ sequencing technologies (26). Unfortunately, few of the resulting publications have provided systematic counts of observed LOF variants. Even where such numbers are provided, differences between studies in terms of the sequencing technology, read-mapping and variant-calling algorithms and annotation sets make it difficult to compare studies.
These differences, and the problems with sequencing and annotation artefacts raised above, mean that there is as yet no clear consensus on the number of LOF variants present in an individual genome. To illustrate the extent of the discrepancy, one recent exome sequencing study (27) reported an average of 45 nonsense SNPs, 16 splice-disrupting SNPs and 46 frameshift indels per genome in individuals of non-African ancestry, whereas a whole-genome sequence of a European male (28) reported 121 stop SNPs and 112 splice-disrupting SNPs but did not report frame-shift indels (Figure 3). At least some of this discrepancy relates to the relatively conservative consensus coding sequence (CCDS) gene set (29) targeted by the exome sequencing study, but differences in sequencing technology and variant calling thresholds likely also played a role.
Several lessons can be drawn from the data generated so far. First, the current catalogue of human LOF variants is incomplete—especially for insertion/deletion variants, which are still more difficult than SNPs to ascertain from short-read sequence data due to their frequently repetitive sequence context—and also likely contains many sequencing and annotation artefacts. Second, although there is wide variation in per-individual numbers between studies (Figure 3), LOF variants are certainly more prevalent within human genomes than most observers would have predicted prior to the genomic era; using even the more conservative estimates thus far from large-scale sequencing (27), each individual carries at least 100 of these variants, and at least 30 in the homozygous state.
Third, the distribution of allele frequencies of LOF variants suggests—as might be expected—that they as a class tend to be evolutionarily deleterious (4), in turn suggesting that they may provide a rich source of potentially causal variation for complex diseases. Finally, however, the sheer number of these variants present within each human genome also poses major analytical challenges for clinical genome sequencing studies seeking to identify a single disease-causing mutation, especially in cases where there is little or no additional information (such as linkage data) available to filter out spurious variants.
MOVING FORWARD
As mentioned above, comparison of published genomic data sets is challenging due to heterogeneity in the technologies and analysis techniques employed, the annotation sets used and the degree of filtering and validation of variants. In order to better understand the full spectrum of LOF variation in the human genome, it will be necessary to take a more systematic approach to the analysis of genome-wide sequence data.
We and others are currently performing this type of analysis as part of the 1000 Genomes Project, an international collaboration generating low-coverage whole-genome and high-coverage exome sequence data from 2500 individuals from 27 diverse populations (http://www.1000genomes.org/page.php); the results of a pilot study are expected to be published around the same time as this review. The project is stimulating improvements in the annotation of both gene models and disease-associated variants in databases such as the Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php). It is expected to provide a catalogue of most coding LOF variants present at ≥5% frequency in the populations studied by the end of 2010 and ≥0.1% when the project is complete.
Simply collecting observed LOF variants from a high-throughput survey, however, is not sufficient to provide a useful resource, since we expect a significant minority of these variants to be false positives for the reasons outlined above. Therefore, we are subjecting many of the LOF variants collected by the project to both experimental validation and careful manual reannotation of the surrounding gene structure, thus providing a core set of true LOF variants to serve as the basis for interpretation of these variants in other sequencing studies. In the long term, this catalogue will prove most useful in conjunction with experimental functional annotation. A first step in this direction will be the incorporation of data from transcriptome sequencing of multiple individuals across multiple tissues with whole-genome sequences, allowing direct assessment of the effects of putative LOF variants on transcript level and structure.
One key challenge moving forward will be in identifying LOF variants in non-coding regions—mutations affecting distant regulatory elements that may have effects on gene expression every bit as profound as changes in coding sequences (30). Adding to the complication of assessing functionality of non-coding variants will be the fact that in some cases the resulting effects will be tissue-specific. For example, the classic Duffy O allele is an LOF variant in which the DARC protein is absent specifically from the red blood cell membrane in many Africans, a benign variation conferring resistance to vivax malaria. The DARC coding region is intact and the protein is expressed in other tissues, but an SNP 46 bp upstream of the transcription start site disrupts a binding site for the erythroid transcription factor GATA1 and abolishes red cell expression (31).
We expect systematic catalogues of LOF variants to prove useful to clinical geneticists in a number of ways. Most simply, such catalogues will be available for researchers to match against LOF variants observed in patient samples, allowing them to quickly determine whether the variant has been previously observed in healthy individuals, and if so whether heterozygous or homozygous, and at what frequency. Second, it will be possible to see whether a gene that contains a potentially causal variant has previously been observed to contain homozygous LOF variants in healthy individuals, thus making it an unlikely candidate for a disease-causing mutation. Finally, the generation of a high-quality catalogue of LOF variation will make it possible to compare the functional and evolutionary properties of LOF-tolerant genes with genes implicated in severe disease, potentially creating a signature of LOF tolerance that will allow researchers to prioritize the downstream analysis of novel LOF-containing genes according to a predicted probability of disease causation.
CONCLUSIONS
Our understanding of the normal pattern of LOF variation will expand as more and more high-quality complete genomes are generated from a wider variety of human populations. A crucial stepping stone will be large-scale exome sequencing, an affordable alternative to whole-genome sequencing that focuses on the protein-coding regions where functional variants are enriched. However, care must be taken to ensure that the targeted regions are drawn from a comprehensive gene set; it is likely that the use of the conservative CCDS annotation (29) in the design of many current exome-targeting chips (e.g. that used in reference 27) will result in true coding variation being missed. The more comprehensive GENCODE annotation (18) provides an alternative already being adopted by some companies.
A key lesson for clinical geneticists is that the implicit assumption that LOF variants (and indeed other changes predicted to be damaging to the protein) are necessarily deleterious to human health is a dangerous one, especially when such an assumption is used to infer disease causality for a novel variant. In fact, the studies reviewed above demonstrate that healthy humans carry many dozens of LOF variants, most of which have little or no effect on health (at least in the heterozygous state).
As we enter the era of large-scale sequencing, it will become increasingly easier to identify the full spectrum of gene-disrupting mutations present in a patient's genome—but determining which of those variants, if any, is actually responsible for causing disease will remain a non-trivial challenge. Comprehensive catalogues of the location, frequency and properties of the full spectrum of human variation will provide an important resource for such investigations.
Conflict of Interest statement. None declared.
FUNDING
Our work is funded by The Wellcome Trust. D.G.M. is funded by an Overseas Biomedical Fellowship from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. doi:10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. doi:10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. doi:10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yngvadottir B., Xue Y., Searle S., Hunt S., Delgado M., Morrison J., Whittaker P., Deloukas P., Tyler-Smith C. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am. J. Hum. Genet. 2009;84:224–234. doi: 10.1016/j.ajhg.2009.01.008. doi:10.1016/j.ajhg.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.North K.N., Beggs A.H. Deficiency of a skeletal muscle isoform of alpha-actinin (alpha-actinin-3) in merosin-positive congenital muscular dystrophy. Neuromuscul. Disord. 1996;6:229–235. doi: 10.1016/0960-8966(96)00361-6. doi:10.1016/0960-8966(96)00361-6. [DOI] [PubMed] [Google Scholar]
- 6.Landsteiner K. Zur Kenntnis der antifermentativen, lytischen und agglutinierenden Wirkungen des Blutserums und der Lymphe. Zentral. Bakteriol. 1900;27:357–362. [Google Scholar]
- 7.Calafell F., Roubinet F., Ramirez-Soriano A., Saitou N., Bertranpetit J., Blancher A. Evolutionary dynamics of the human ABO gene. Hum. Genet. 2008;124:123–135. doi: 10.1007/s00439-008-0530-8. doi:10.1007/s00439-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 8.de Morais S.M., Wilkinson G.R., Blaisdell J., Nakamura K., Meyer U.A., Goldstein J.A. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 9.Gaedigk A., Blum M., Gaedigk R., Eichelbaum M., Meyer U.A. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am. J. Hum. Genet. 1991;48:943–950. [PMC free article] [PubMed] [Google Scholar]
- 10.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 11.Saleh M., Vaillancourt J.P., Graham R.K., Huyck M., Srinivasula S.M., Alnemri E.S., Steinberg M.H., Nolan V., Baldwin C.T., Hotchkiss R.S., et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. doi:10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y., Daly A., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am. J. Hum. Genet. 2006;78:659–670. doi: 10.1086/503116. doi:10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur D.G., North K.N. ACTN3: a genetic influence on muscle function and athletic performance. Exerc. Sci. Sport Sci. Rev. 2007;35:30–34. doi: 10.1097/JES.0b013e31802d8874. doi:10.1097/JES.0b013e31802d8874. [DOI] [PubMed] [Google Scholar]
- 14.MacArthur D.G., Seto J.T., Raftery J.M., Quinlan K.G., Huttley G.A., Hook J.W., Lemckert F.A., Kee A.J., Edwards M.R., Berman Y., et al. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 2007;39:1261–1265. doi: 10.1038/ng2122. doi:10.1038/ng2122. [DOI] [PubMed] [Google Scholar]
- 15.Xue Y., Sun D., Daly A., Yang F., Zhou X., Zhao M., Huang N., Zerjal T., Lee C., Carter N.P., et al. Adaptive evolution of UGT2B17 copy-number variation. Am. J. Hum. Genet. 2008;83:337–346. doi: 10.1016/j.ajhg.2008.08.004. doi:10.1016/j.ajhg.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson M.V. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. doi:10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z.D., Frankish A., Hunt T., Harrow J., Gerstein M. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol. 2010;11:R26. doi: 10.1186/gb-2010-11-3-r26. doi:10.1186/gb-2010-11-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):1–9. doi: 10.1186/gb-2006-7-s1-s4. S4 doi:10.1186/gb-2006-7-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. doi:10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 20.Keren H., Lev-Maor G., Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. doi:10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 21.Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. doi:10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 22.Xue Y., Wang Q., Long Q., Ng B.L., Swerdlow H., Burton J., Skuce C., Taylor R., Abdellah Z., Zhao Y., et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. doi:10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarpey P.S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O'Meara S., Latimer C., Dicks E., Menzies A., et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 2009;41:535–543. doi: 10.1038/ng.367. doi:10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G., et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. doi:10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng P.C., Levy S., Huang J., Stockwell T.B., Walenz B.P., Li K., Axelrod N., Busam D.A., Strausberg R.L., Venter J.C. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. doi:10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yngvadottir B., Macarthur D.G., Jin H., Tyler-Smith C. The promise and reality of personal genomics. Genome Biol. 2009;10:237. doi: 10.1186/gb-2009-10-9-237. doi:10.1186/gb-2009-10-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. doi:10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A., et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. doi:10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. doi:10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. doi:10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tournamille C., Colin Y., Cartron J.P., Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. doi:10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]