Abstract
Genomic tiling arrays, cDNA sequencing and, more recently, RNA-Seq have provided initial insights into the extent and depth of transcribed sequence across human and other genomes. These methods have led to greatly improved annotations of protein-coding genes, but have also identified transcription outside of annotated exons. One resultant issue that has aroused dispute is the balance of transcription of known exons against transcription outside of known exons. While non-genic ‘dark matter’ transcription was found by tiling arrays to be pervasive, it was seen to contribute only a small percentage of the polyadenylated transcriptome in some RNA-Seq experiments. This apparent contradiction has been compounded by a lack of clarity about what exactly constitutes a protein-coding gene. It remains unclear, for example, whether or not all transcripts that overlap on either strand within a genomic locus should be assigned to a single gene locus, including those that fail to share promoters, exons and splice junctions. The inability of tiling arrays and RNA-Seq to count transcripts, rather than exons or exon pairs, adds to these difficulties. While there is agreement that thousands of apparently non-coding loci are present outside of protein-coding genes in the human genome, there is vigorous debate of what constitutes evidence for their functionality. These issues will only be resolved upon the demonstration, or otherwise, that organismal or cellular phenotypes frequently result when non-coding RNA loci are disrupted.
INTRODUCTION
The low protein-coding gene count has been, perhaps, one of the two most surprising findings since the human draft genome sequence was announced 10 years ago. The number of human protein-coding genes has recently settled at approximately 20 000, having been estimated in the mid-1990s as being over 60 000 (1). These early estimates were based on cDNA and expressed sequence tag (EST) data, and their wide margins of error reflect in part that there was a substantial quantity of human transcribed sequence that was wrongly thought to encode protein. Indeed, we have still to determine how many entirely non-coding transcripts are expressed from the human genome and what functions they possess. Evolutionary studies produced the second surprise from the human genome sequence; that is, there is a large amount of apparently functional, yet non-coding, DNA contained in the human genome, estimated as being at least 4-fold the amount of protein-coding sequence (reviewed in 2). When such evolutionarily constrained DNA sequence is also transcribed, it immediately becomes a good candidate for being a functional non-protein-coding RNA (ncRNA) locus.
Defining a transcript, or its locus, as being non-coding is unsatisfactory simply because of its contrariety. Human genes commonly possess both coding and non-coding transcripts and, without detailed experimental studies, it is difficult to accurately distinguish non-coding from coding sequence in short transcripts (3,4). Labelling a transcript as being ‘intergenic’ (meaning that it is transcribed entirely from sequence intervening between two adjacent coding genes) is likewise fraught with difficulties. These stem from inaccurate or incomplete gene models, and from the unrealistic premise that coding gene loci and intergenic non-coding loci are always distinct, overlapping in neither chromosomal nor exonic sequence.
It is with these difficulties in mind that the central issue of this review is now introduced: what fraction of all intergenic sequence in the human genome is transcribed into stable non-coding RNA products? In line with recent publications, we shall refer to this transcribed and intergenic sequence as ‘dark matter’, although the initial definition of this term covered all intergenic sequence, irrespective of functionality or expression (5). A census of functional and transcribed dark matter in the human genome will be important not just for completing its functional repertoire, but also for revealing new mechanisms of transcriptional regulation and other novel functions. Looking beyond the identification of non-coding transcripts, it will also be important to know their full-length sequences and their expression patterns across diverse cell types and tissues at multiple developmental stages in different species. While recent developments in short-read sequencing technologies are helping to reveal these expression patterns, full-length sequence information remains largely inaccessible. This is due to ncRNAs' low expression levels and to positional information of short reads within a transcript being mostly lost upon sequencing. While computational approaches have shown modest success in reconstructing full-length transcripts (6,7), this issue will only be resolved when longer read sequencing technologies become available.
cDNA SANGER SEQUENCING
Should transcribed intergenic sequences be added to the gene count, thereby producing a full census not only of protein-coding genes but non-coding genes too? In 2002, the FANTOM2 consortium reported 33 409 ‘transcriptional units' (TUs) in the mouse, of which 11 665 appeared not to be protein coding (8). They defined a TU as a cluster of one or more transcripts that share at least one base of exonic sequence on the same strand. Advantages of this definition are that it is simple and unambiguous, although one disadvantage is that it will merge two otherwise separate loci when they are bridged by a single rare, perhaps artefactual, transcript (see below). The FANTOM projects revealed how non-coding TU transcripts differ greatly from their coding counterparts. While most coding transcripts are multiexonic and are well conserved between human and mouse, the majority of FANTOM2 non-coding transcripts contain only a single exon and most show insufficient similarity to be alignable to transcribed human sequence. Importantly, most also exhibit very low levels of expression relative to protein-coding transcripts. In the subsequent FANTOM3 project, over 3500 apparently non-coding transcripts were defined, each with support from other sequence sources (9), yet again tending to be rare, poorly conserved and unspliced. One surprising indication that the recruitment of transcriptional machinery to these locations is ultimately functional is that their promoter regions tend to be better conserved than promoters of protein-coding genes (9).
Overall, such differences have not persuaded many that thousands of non-coding TUs should be added to the gene counts of mouse or human. If ncRNA transcripts are of low abundance, with their sequences showing only a low level of evolutionary constraint, and if only a low fraction are spliced, then might these ncRNAs instead be without function (10,11)? Experimental approaches that enrich for lowly expressed transcripts, for example, may have inadvertently identified many transcripts that are biological artefacts, whose expression results from a fortuitous assembly of the transcriptional machinery on neutrally evolving sequence (11).
TILING ARRAYS
Rather than diminishing in size, transcribed dark matter was found by tiling microarrays to be even more abundant in human, mouse and other genomes (reviewed in 11–13). This technology detects transcription using probes that are regularly spaced on the genome. Consequently, although tiling arrays can detect transcription, they cannot determine the complete extent of transcripts. Results from these arrays revealed that transcribed sequence from human cytosolic or nuclear polyadenylated RNA was roughly equally distributed between known protein-coding and unknown apparently non-coding transcripts (14). Furthermore, RNA transcripts lacking polyadenylation represented approximately half of the transcriptome, and a third of all transcribed sequences were detected in both poly A− and poly A+ forms (14). In a small 1% sample of the human genome, 93% of bases appeared to be transcribed, three-quarters of which could be verified by an alternative technique (15).
Nevertheless, tiling arrays have not definitively shown widespread transcription outside of known genes. This is because results, in the main, have not been faithfully reproduced in separate studies using different samples and microarray platforms (13). Consequently, it has been concluded that most of the observed dark matter transcription is either specific to a platform and thus may reflect experimental artefacts, or else is tissue specific (13). There are indications that intergenic transcripts are, indeed, often tissue specific. This is because not only is their expression often low, but compared with protein-coding transcripts their promoters are less often of the ‘housekeeping gene’ CpG type (16).
RNA-SEQ
Genomic tiling microarrays are prone to artefacts arising from cross-hybridization (17). Massively parallel whole transcriptome sequencing (‘RNA-Seq’), on the other hand, is largely free of such effects (17) and, moreover, can detect transcript expression levels over a wider dynamic range (18). An added benefit is that, with paired-end sequencing, pairs of RNA regions separated by 200–500 bp within the same transcript can be sequenced. Although this provides some connectivity information between exons, it still does not always allow the full extent of a transcript's sequence to be determined directly. Typically, tens of millions of fragment reads are mapped to a reference genome sequence and intersected with existing or novel transcript and gene annotations. In theory, by mapping reads that fail to intersect with exons or introns from known transcripts, both the proportion of reads that contribute to transcribed dark matter (‘dark matter mass’) and the fraction of the genome sequence covered by such reads (‘dark matter coverage’) can be calculated.
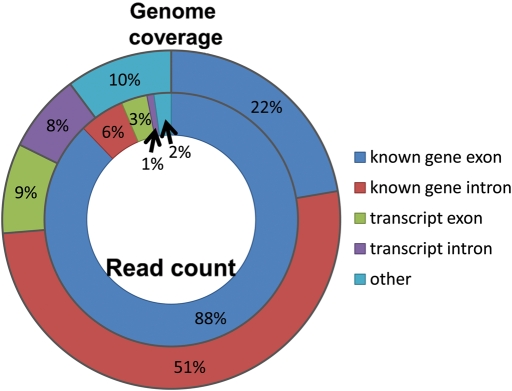
RNA-Seq experiments have been performed for many species, for a variety of different tissues or cells, and for polyA+ or total RNA (reviewed in 11). Although proportions vary considerably, for illustrative purposes, we shall here consider data sets from human brain tissue and cell lines recently described by van Bakel et al. (19). In this experiment, 94% of reads mapped to exons (88%) or introns (6%) of known protein-coding genes, leaving only 4% overlapping other transcripts (from ESTs, cDNAs etc.) and a further 2% mapping to other genomic regions previously lacking evidence of transcription (Fig. 1). Dark matter mass is thus relatively low, consistent with previous observations from cDNA sequencing and tiling arrays that ncRNAs, with some notable exceptions (19), tend to be expressed at low levels. Dark matter coverage, on the other hand, is relatively high with over a quarter of all transcribed regions not overlapping known genes (Fig. 1). In summary, dark matter transcription, at low levels, appears to be widespread across the human genome.
Figure 1.
Exons from known genes are associated with 88% of uniquely mapping short reads, but provide 22% of genomic sequence that is transcribed [human data from van Bakel et al. (19)]. On the other hand, only 6% of uniquely mapping reads are in intergenic sequence, but these lowly expressing regions cover about one quarter of all transcribed genomic sequence.
The proportions shown in Figure 1, however, are far from definitive. On the one hand, the extent of transcriptional dark matter coverage will be overestimated because our catalogues of ‘known’ protein-coding genes, transcripts and exons remain far from complete. For instance, even a 43 exon 2 Mb human gene (RP25) was until recently unknown (20), and 7395 novel splice junctions in known mouse genes were revealed by a recent RNA-Seq experiment (6). The incompleteness of the protein-coding transcriptome will thus account for some reads currently being mapped to intronic and intergenic sequence.
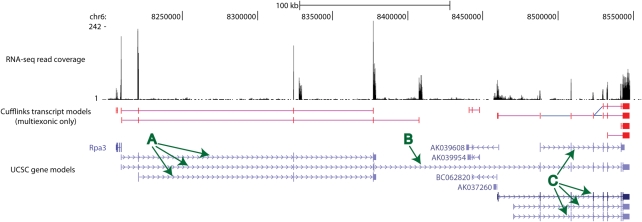
On the other hand, for three reasons, dark matter mass will currently be underestimated. First, any read cluster containing a read whose sequence overlaps, even by a single base, with an exon or intron of a known gene will be assigned to that gene model, even if large numbers of this cluster's reads do not overlap with this gene. Ambiguous gene annotations in mouse chromosome 6qA1 will serve to illustrate this and other issues (Fig. 2). Here, two protein-coding gene loci Gig18 (transcripts labelled A) and Glcci1 (labelled C) are adjacent on mouse chromosome 6qA1, with four apparently non-coding antisense transcripts (AK039608, BC062820, AK037260 and AK039954) being expressed from intervening sequence (Fig. 2). Nevertheless, a single cDNA (AF374476; labelled B) has been sequenced that spans both loci, thereby encompassing all exons from the four ncRNA loci within one of its introns. Mapped reads (coverage indicated in Fig. 2) that fall within these four ncRNA loci thus are assigned not as intergenic reads, but instead as reads that are intronic to a protein-coding gene. Figure 2 also serves as an illustration of the second reason why dark matter transcription could be underestimated. The four ncRNAs are transcribed on the opposite strand to both Gig18 and Glcci1; yet because most published paired-end RNA-Seq experiments cannot directly resolve strandedness, many antisense transcript reads will be misclassified. This is a considerable problem since in one experiment, 11% of the reads were antisense to annotated genes (21). Moreover, although strandedness is commonly inferred from splicing dinucleotide (GT-AG) motifs, this will be in error for those antisense ncRNAs that map exactly to an intron/exon junction, yet do so in the antisense orientation (22). The third contributing factor to dark matter mass underestimates arises when transcriptomes are constructed only from sequence reads that map uniquely, or near uniquely, within the reference genome assembly. Reads from highly repetitive DNA, such as transposable element (TE) sequence, often can be mapped to many sites. Such sequence is rare in protein-coding exons, but frequent in non-coding exons, leading to the preferential fragmentation of non-coding TE-containing transcript models and the underestimation of their expression levels.
Figure 2.
Cufflinks (6) multiexonic transcript models from short reads mapped to mouse chromosome 6 (assembly mm9, bases 8 200 479–8 555 654) and viewed using the UCSC Genome Browser (56). Short reads (coverage represented at top) map not just to known exons of coding and non-coding loci, but also to introns and gene termini. Multiexonic cufflinks transcript models consistent with known gene annotations (shown in blue, based on RefSeq, UniProt, GenBank, CCDS and Comparative Genomics) are shown as red exons joined by pink lines (blue joining lines indicate introns not supported by the data). Three protein-coding loci (Rpa3, Gig18 and Glcci1) and four FANTOM ncRNA loci (AK039608, AK039954, BC062820 and AK037260) are represented. Transcripts discussed in the main text are labelled A–C. Mouse brain data were generated using the Illumina genome analyzer (unpublished data).
TRANSCRIPTIONAL NOISE
Sequence reads that map outside of exons from known gene models may thus represent new protein-coding or non-coding genes, antisense transcription and alternative transcripts, including 5′ and 3′ extensions to genes involving additional promoters and polyadenylation sites (13). However, such reads may also reflect experimental artefacts such as genomic DNA contaminants, or they may reflect biological artefacts. Many intronic reads, for example, are likely to represent unprocessed or partially processed RNAs (19). Intergenic reads, on the other hand, may result from the random initiation and elongation of RNA polymerase II-mediated transcription (23). If noisy transcription predominates, then an even spread of RNA-Seq reads in intergenic sequence is expected. Nevertheless, intergenic reads, even those observed only once in a sample, are spread non-randomly across the genome (19). It thus appears that random transcriptional noise cannot account for all intergenic expression. Some noise, however, might be non-random. This is because when the transcriptional machinery is recruited to a bona fide gene promoter, transcriptional activity can ‘ripple out’ within a ∼100 kb radius resulting in ‘illegitimate’ transcription from coding and non-coding loci (24).
INTERGENIC TRANSCRIPTS AND THEIR FUNCTIONS
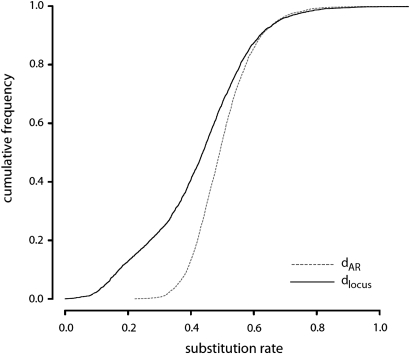
Together cDNA sequencing, tiling arrays and RNA-Seq approaches have identified thousands of long (>200 bp) intergenic ncRNA (lincRNA) loci in human and mouse genomes. While functions have been assigned to only a few of these lincRNA loci (reviewed in 25–27), four lines of indirect evidence support their functionality more generally. First, many loci harbour chromatin signatures that typically mark promoter regions (histone-3 Lys4 trimethylation; H3K4me3) and transcribed regions (histone-3 Lys36 trimethylation; H3K36me3) (28), and many overlap regions of open chromatin (19). Second, they tend to show differential expression, for example, in response to retinoic acid or lipopolysaccharide, or among different tissues (29,30). Third, they accumulate fewer substitutions within their promoters, their transcribed sequences and their dinucleotide splicing motifs than neutrally evolving sequence, all of which are indicators of sustained purifying selection (7,9,31,32). For example, nucleotide substitutions in 3390 human lincRNAs (from data presented in Fig. 1) when aligned to mouse tend to occur 10% less frequently than in putatively neutrally evolving sequence (Fig. 3). This indicates that a minority of nucleotide changes, specifically those that are deleterious, have been purged from functional sequence. Lastly, lincRNAs are enriched in predicted secondary structures which would indicate their involvement in trans-acting mechanisms (16,32).
Figure 3.
Nucleotide substitution rates tend to be slower in lincRNA sequence (solid line) than in putatively neutral sequence (dotted line). Cumulative frequency histogram of nucleotide substitution rates for 3390 human lincRNA sequence aligned to mouse (solid line) compared with aligned putatively neutral sequence (dotted line). Neutrally evolving sequence has been acquired from genomically adjacent TE sequence inferred to have been present in the last common ancestor of human and mouse (‘ancestral repeats’, ‘ARs’). Of 16 268 human intergenic seqfrags obtained from a RNA-Seq experiment (19), 3390 were chosen since they contain at least 100 bp of sequence aligned to mouse. Nucleotide substitution rates in lincRNA loci (dlocus) and ancestral repeats (dAR) were calculated using a method that accounts for GC content (32). LincRNA loci tend to have evolved significantly and 10% more slowly than neighbouring neutral sequence (median dlocus/dAR = 0.902, median dlocus = 0.438, median dAR = 0.488; two-sided Mann–Whitney test, P<10−15). Further sets of lincRNA loci derived from cDNA sequencing and chromatin signatures show very similar degrees of evolutionary constraint (dlocus/dAR = 0.887 and 0.904, respectively) (32).
It has been noted that long- and short-ncRNA loci, whether defined using cDNA (31) or tiling arrays (33), or RNA-Seq (19), tend to lie close to known protein-coding genes. This has led to the hypothesis that in their expression, these transcripts are ‘linked’ in some manner to protein-coding genes (19). This link could take the form of alternative promoters and polyadenylation sites, or could reflect the by-products of abortive initiation or transcriptional pausing (11,34,35). On the other hand, that long RNAs transcribed close to protein-coding genes are functional is suggested by their unusual concentration in the vicinity of a particular class of genes, namely those encoding transcription factors (16,28). Many long and short RNAs have also been observed to interact with PRC2 (polycomb repressive complex-2, a H3K27 trimethylase) and appear to cause repression in cis of polycomb target genes (28,36–39). In contrast, transcription of RNAs at intragenic or intergenic enhancer elements appears to activate in cis the expression of protein-coding genes (40,41). Thus, the generality of these explanations, whether most ncRNAs associated with protein-coding genes contribute to the spatiotemporal control of gene expression or else result from ‘leaky’ transcription, remains unresolved.
DARK MATTER AND THE NUMBER OF GENES
Between 1200 and 2200 dark matter, long intergenic ncRNA loci have been identified in cDNA sequencing and RNA-Seq experiments for mammals (7,19,32,42). Should these thousands of newly identified ncRNA loci be considered genes, thus adding to our current list of ∼20 000 functional protein-coding genes? In a recent article, van Bakel and Hughes (11) argue that they should not, even when their transcripts are differentially expressed, are conserved across species and are localized within cells and interact with proteins. They argue that established techniques, such as gene knockout, knockdown mutagenesis or over-expression, which frequently are successful in confirming the functionality of protein-coding loci, should equally be employed to assay the functionality of putative non-coding loci. In the interests of fairness, it should be said that if these stringent criteria for functionality were to be applied to all predicted protein-coding genes, then their number would fall far short of 20 000. This is because, for example, only 6000 protein-coding genes, when disrupted in mouse, are associated with an overt phenotype (43). Nevertheless, it is clear that substantial direct evidence of functionality, including knockout phenotypes or disease association, will be required to convince many that the status of an ncRNA locus should be promoted to that of a gene. To date, such evidence has been forthcoming for only a handful of non-coding RNAs (44–48).
Yet, some preconceived ideas of what properties exclude ncRNA loci from being bona fide genes, drawn as they are from decades of fruitful research into proteins, are perhaps wide of the mark. Protein-coding genes are typically long and multiexonic, and their mature transcripts highly expressed and highly conserved, with their sequences lacking TEs. In contrast, even those ncRNAs whose mechanism has been established are relatively short, with one or few exons, they are often lowly expressed and poorly conserved, and TEs have frequently inserted into their sequence. One assumption that may yet be found wanting is that very low abundance transcripts typically lack function. For example, any mechanism involving a high-affinity interaction of RNA with DNA sites, such as at a single gene locus, centromere or telomere, whose cellular copy number is low, might proceed with only a few transcripts per cell. As in X-chromosome inactivation, low abundant lincRNAs may act in cis as ‘guides and tethers’ attaching themselves to their sites of transcription (49). In contrast, those lincRNAs with stable secondary structures and that act in trans perhaps are likely to be more abundant. Some lincRNAs will do both, acting in trans by binding a transcriptional cofactor, and in cis, binding within this complex to a physically linked enhancer, just as has been observed for a mouse lincRNA, Evf-2 (50).
As ncRNA transcripts increasingly become the focus of investigation, it is likely that their contribution to the human gene count will rise. The rise will be least if we aggregate a lincRNA transcript locus with its adjacent protein-coding gene whose expression it regulates, and greatest if we count such cis-acting loci separately. It is because of such arbitrariness that the count of functional human (coding and non-coding) transcripts will increasingly be seen as a more meaningful quantity than the number of human genes (51). For as the true transcriptional complexity of loci is revealed, with sense, antisense, and enhancer-, promoter- or UTR- associated transcripts, the concept of a ‘gene’ (52) will increasingly appear incomplete and overly simplistic. Moreover, when third-generation sequencing technologies (53,54) allow the full extent of all transcripts' sequences to be determined, the entire transcriptional repertoires of different species and different cells will finally be revealed. Only then, when we have these data and the results of detailed mechanistic studies at hand, will dark matter transcription be revealed as either ‘sound and fury, signifying nothing’ [as it has recently been described (55)] or else as functional elements that are crucial to the biology of our species.
FUNDING
This work was supported by the BBSRC (grant BB/F007590/1 to C.P.P.); by the European Research Council (DARCGENs to C.P.P.); by a Medical Research Council Programme grant (to C.P.P.); by a Marshall Scholarship (to T.G.B.); by the NIH Oxford Cambridge Scholars Program (to T.G.B.); and by New College, Oxford (to T.G.B.).
ACKNOWLEDGEMENTS
We are very grateful to Dr Ana C. Marques for many insightful discussions, and for the results presented in Figure 3.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pertea M., Salzberg S.L. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 11:206. doi: 10.1186/gb-2010-11-5-206. doi:10.1186/gb-2010-11-5-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponting C.P. The functional repertoires of metazoan genomes. Nat. Rev. Genet. 2008;9:689–698. doi: 10.1038/nrg2413. doi:10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- 3.Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. doi:10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frith M.C., Forrest A.R., Nourbakhsh E., Pang K.C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T.L., Grimmond S.M. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. doi:10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. doi:10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 6.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. doi:10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., Fan L., Koziol M.J., Gnirke A., Nusbaum C., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. doi:10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. doi:10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 9.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K. Mouse transcriptome: neutral evolution of 'non-coding' complementary DNAs. Nature. 2004;431 1 p following 757; discussion following 757. [PubMed] [Google Scholar]
- 11.van Bakel H., Hughes T.R. Establishing legitimacy and function in the new transcriptome. Brief. Funct. Genomic. Proteomic. 2009;8:424–436. doi: 10.1093/bfgp/elp037. doi:10.1093/bfgp/elp037. [DOI] [PubMed] [Google Scholar]
- 12.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. doi:10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 13.Johnson J.M., Edwards S., Shoemaker D., Schadt E.E. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. doi:10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. doi:10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 15.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponjavic J., Oliver P.L., Lunter G., Ponting C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. doi:10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal A., Koppstein D., Rozowsky J., Sboner A., Habegger L., Hillier L.W., Sasidharan R., Reinke V., Waterston R.H., Gerstein M. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics. 2010;11:383. doi: 10.1186/1471-2164-11-383. doi:10.1186/1471-2164-11-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. doi:10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Bakel H., Nislow C., Blencowe B.J., Hughes T.R. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. doi:10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abd El-Aziz M.M., Barragan I., O'Driscoll C.A., Goodstadt L., Prigmore E., Borrego S., Mena M., Pieras J.I., El-Ashry M.F., Safieh L.A., et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat. Genet. 2008;40:1285–1287. doi: 10.1038/ng.241. doi:10.1038/ng.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. doi:10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rederstorff M., Bernhart S.H., Tanzer A., Zywicki M., Perfler K., Lukasser M., Hofacker I.L., Huttenhofer A. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38:e113. doi: 10.1093/nar/gkq057. doi:10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. doi:10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 24.Ebisuya M., Yamamoto T., Nakajima M., Nishida E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. doi:10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 25.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. doi:10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. doi:10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. doi:10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 28.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. doi:10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. doi:10.1016/S0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 30.Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. doi:10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. doi:10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marques A.C., Ponting C.P. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. doi:10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. doi:10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 34.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. doi:10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponjavic J., Ponting C.P. The long and the short of RNA maps. Bioessays. 2007;29:1077–1080. doi: 10.1002/bies.20669. doi:10.1002/bies.20669. [DOI] [PubMed] [Google Scholar]
- 36.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. doi:10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondal T., Rasmussen M., Pandey G.K., Isaksson A., Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. doi:10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanhere A., Viiri K., Araujo C.C., Rasaiyaah J., Bouwman R.D., Whyte W.A., Pereira C.F., Brookes E., Walker K., Bell G.W., et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. doi:10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szutorisz H., Dillon N., Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. doi:10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. doi:10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia H., Osak M., Bogu G.K., Stanton L.W., Johnson R., Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Eppig J.T. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. doi:10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewejohann L., Skryabin B.V., Sachser N., Prehn C., Heiduschka P., Thanos S., Jordan U., Dell'Omo G., Vyssotski A.L., Pleskacheva M.G., et al. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 2004;154:273–289. doi: 10.1016/j.bbr.2004.02.015. doi:10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Heinen T.J., Staubach F., Haming D., Tautz D. Emergence of a new gene from an intergenic region. Curr. Biol. 2009;19:1527–1531. doi: 10.1016/j.cub.2009.07.049. doi:10.1016/j.cub.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 46.Bond A.M., Vangompel M.J., Sametsky E.A., Clark M.F., Savage J.C., Disterhoft J.F., Kohtz J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. doi:10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon F.E., Nutt C.L., Cheunsuchon P., Nakayama Y., Provencher K.A., Rice K.A., Zhou Y., Zhang X., Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443–2452. doi: 10.1210/en.2009-1151. doi:10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuster-Gossler K., Bilinski P., Sado T., Ferguson-Smith A., Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. doi:10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Lee J.T. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. doi:10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. doi:10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gingeras T.R. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. doi:10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 52.Gerstein M.B., Bruce C., Rozowsky J.S., Zheng D., Du J., Korbel J.O., Emanuelsson O., Zhang Z.D., Weissman S., Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. doi:10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 53.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. doi:10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 54.Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. doi:10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 55.Robinson R. Dark matter transcripts: sound and fury, signifying nothing? PLoS Biol. 2010;8:e1000370. doi: 10.1371/journal.pbio.1000370. doi:10.1371/journal.pbio.1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J., et al. The UCSC Genome Browser database: update. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. doi:10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]