Abstract
The development of massively parallel sequencing technologies, coupled with new massively parallel DNA enrichment technologies (genomic capture), has allowed the sequencing of targeted regions of the human genome in rapidly increasing numbers of samples. Genomic capture can target specific areas in the genome, including genes of interest and linkage regions, but this limits the study to what is already known. Exome capture allows an unbiased investigation of the complete protein-coding regions in the genome. Researchers can use exome capture to focus on a critical part of the human genome, allowing larger numbers of samples than are currently practical with whole-genome sequencing. In this review, we briefly describe some of the methodologies currently used for genomic and exome capture and highlight recent applications of this technology.
INTRODUCTION
The introduction and widespread use of massively parallel sequencing has made it possible for individual laboratories to sequence a whole human genome. However, the cost and capacity required are still significant, especially considering that the function of much of the genome is still largely unknown. Before massively parallel sequencing, specific regions of the genome were targeted using PCR, followed by capillary sequencing. This approach was effective at narrowing the scope of investigation, but required a tightly defined guess as to which region should be targeted. Larger-scale studies have used this method [X-chromosome exons (1), human exome (2)], but this remains a major undertaking that is not feasible for many research groups. Recent studies have described new methods to target much larger regions of the human genome (up to ∼3 Mb) in a more cost- and time-efficient manner (reviewed in 3–6). Such methods, described as genome capture, genome partitioning, genome enrichment etc., are well suited to current massively parallel sequencing platforms, as they produce a pool of desired molecules that are separated by the parallel nature of the sequencing technologies themselves. Although these methods can cover more of the human genome in a shorter amount of time at reduced cost compared with PCR, they also require an educated guess as to which regions or genes may be interesting. Several of these methods have been extended to capture the human exome, eliminating the need to choose a subset of genes for interrogation and focussing on the best understood 1% of the genome, the protein-coding exons.
CAPTURE METHODS
Solid-phase hybridization
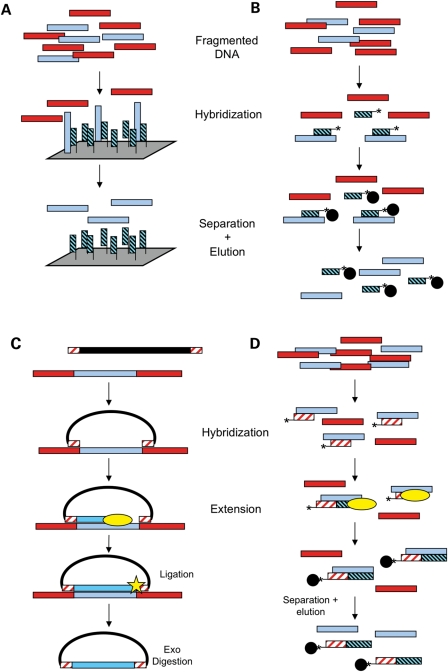
Solid-phase hybridization methods generally utilize probes complimentary to the sequences of interest affixed to a solid support, such as microarrays (7–11) (Fig. 1A) or filters (12). The total DNA is applied to the probes, where the desired fragments hybridize. The non-targeted fragments are subsequently washed away, and the enriched DNA is eluted for sequencing. Recently, these methods have been improved using multiple enrichment cycles (13,14). Agilent, Roche/Nimblegen and Febit offer commercial kits implementing these methods.
Figure 1.
Illustration of different capture methods. Light blue bars represent desired genomic sequence, red bars represent unwanted sequence. (A) Solid-phase hybridization. Bait probes (light blue and black) complementary to the desired sequence are synthesized on a microarray. Fragmented genomic DNA is applied, and the desired fragments hybridize. The array is washed, and desired fragments are eluted. (B) Liquid-phase hybridization. Bait probes (light blue and black) complementary to the desired regions are synthesized, often using microarray technology. The probes are generally biotinylated (asterisk). The bait probes are mixed with fragmented genomic DNA, and the desired fragments hybridize to baits in solution. Streptavidin beads (black circles) are added to allow physical separation. The bead-bait complexes are washed, and desired DNA is eluted. (C) MIP. Single-stranded probes composed of a universal linker backbone (black line) and arms complementary to the sequence flanking desired regions (red and white) are synthesized, often using microarray or microfluidics technology. The probes are added to genomic DNA and hybridize in an inverted manner. A polymerase (yellow oval) fills in the gap between the two arms. A ligase (yellow star) seals the nick, resulting in a closed single-strand circle. Genomic DNA is digested with exonucleases, and the captured DNA is amplified using sequences in the universal backbone. (D) PEC. Biotinylated primers (red and white) are added to fragmented genomic DNA, where they hybridize to the desired sequence. A polymerase (yellow oval) extends the primer, creating a tighter interaction. Streptavidin beads (black circles) are added and are used to physically separate the desired DNA from the unwanted DNA. The desired DNA is then eluted.
Liquid-phase hybridization
Liquid-phase hybridization is similar to solid phase; the probes in this method are not attached to a solid matrix, but instead are biotinylated (Fig. 1B). Following hybridization, the biotinylated probes (with the complementary desired genomic DNA) are bound to magnetic streptavidin beads and are separated from the undesired DNA by washing. After elution, enriched DNA can be sequenced. Initial reports on this method used biotinylated RNA probes (15) (commercially available from Agilent), and recent methods use DNA probes (commercially available from Roche/Nimblegen).
Polymerase-mediated capture
Although all capture methods use polymerases to amplify captured fragments, these methods use polymerases in a more integral way. Padlock probe technology has been extended to develop Molecular Inversion Probes (MIP) and Spacer Multiplex Amplification ReacTion (SMART), in which a single probe acts as both a primer to start elongation and a receiver to end elongation and allow ligation (Fig. 1C). Subsequent digestion of linear DNA leaves only the closed circular extension/ligation products with the desired sequence [MIP (16–19), SMART (20)]. Primer extension capture (PEC) was developed with small amounts of DNA in mind (Fig. 1D). This method uses a biotinylated primer with complimentary sequence to the DNA of interest. After annealing, the primer is extended, effectively generating a hybridization probe to capture the sequence of interest like other hybridization methods (21). Highly parallel PCR has been an effective method to prepare samples for capillary sequencing, and recent work has extended this idea using microfluidics. Instead of using plates with hundreds of wells, aqueous microdroplets can segregate thousands of individual reactions in the same tube, allowing for a much more highly parallel use of PCR (22) (commercially available from Raindance). Another commercially available kit uses restriction enzymes to fragment DNA; probes specific to the ends of desired fragments are used to amplify the desired sequence (Olink Genomics).
Regional capture
Other methods exist to isolate larger sections of the genome. Chromosome sorting (reviewed in 23) has long been useful for genomics. Massively parallel sequencing is well suited to sequence libraries generated by fragmenting flow sorted chromosomes and offers a way to sequence a single chromosome. When odd chromosomal structures are present, or DNA is only available from a handful of molecules, microdissection of metaphase chromosomes followed by sequencing has been reported (24). Although these methods require highly specialized instruments, they do offer a powerful approach for unique cases.
EXOME CAPTURE
Although many different methods for targeted capture have been described, only few have been extended to target the human exome. These methods belong to the hybridization type and include array-based hybridization (9,25,26) and liquid-based hybridization (27) [products available from Agilent Technologies (SureSelect), RocheNimbleGen (SeqCap/SeqCap EZ)]. In the future, other methods may also be able to scale up as well.
The term ‘whole human exome’ can be defined in many different ways. Two companies offer commercial kits for exome capture and have targeted the human consensus coding sequence regions (28), which cover ∼29 Mb of the genome. This is a more conservative set of genes and includes only protein-coding sequence. It covers ∼83% of the RefSeq coding exon bases. Both companies also target selected miRNAs, and extra regions can sometimes be added (Agilent). Although still a subset of the genome, exome capture allows the investigation of a more complete set of human genes with the cost and time advantages of genome capture.
APPLICATIONS
Following initial method descriptions, current research is applying genome capture methods to a variety of questions. From disease causation and diagnosis to evolutionary comparison of ancient genomes, genome capture and massively parallel sequencing is a powerful investigative tool.
Medical sequencing
One of the more common exome capture experiments will be the search for genetic variation underlying a particular disease. For some diseases, causative genes have been identified, and researchers can use custom captures to examine those genes for known and novel variants in their samples. For other diseases, whole exome capture is suitable, as the causative gene is unknown, or many different genes may contribute. Several recent studies have captured and sequenced different regions of individual genomes with known causative variants or genes. These proof of principle experiments demonstrate the utility, as well as some shortcomings, of capture followed by massively parallel sequencing. Ng et al. (26) have used array-based hybridization to sequence 12 human exomes (∼28 Mb). The study included four unrelated individuals with Freeman–Sheldon syndrome, a dominantly inherited rare Mendelian disorder. The investigators were able to identify variants in the known causative gene in each sample. Interestingly, the known causative gene was the only candidate following the application of numerous filters, including requiring a gene to have a novel variant in each sample. In their study of neurofibromatosis type 1, Chou et al. (29) used custom array capture and pyrosequencing to target the 280 kb region containing the NF1 gene, which is known to harbor causal dominant mutations. The authors captured DNA from two different samples with known genotypes, but were initially only able to recover a known single-base deletion. The other known variant, an Alu sequence insertion, was only observed after de novo assembly of unmapped reads. Additionally, the authors found many positions at which the captured genotypes did not agree with Sanger sequencing confirmations. They found that while some discrepancies were due to pyrosequencing errors, others were misalignments from the numerous pseudogenes of NF1, illustrating one of the potential pitfalls of the method. Hoischen et al. (30) also used array-based capture (∼2 Mb) and pyrosequencing to re-identify known variants in five individuals with autosomal recessive ataxia. They were able to initially identify 6/7 known variants investigated; the seventh variant was visible only after adding three times more sequence, although at a low number of reads (2/9 reads contained the mutation). A known variant trinucleotide repeat was not included in the design, due to the repetitive nature of these variants, and therefore not recovered. Raca et al. (31) searched for two known variants causative for Papillorenal syndrome using array-based capture targeting the causative gene, PAX2, as well as >100 candidate genes for other ocular disorders (370 kb), followed by pyrosequencing. They were able to identify a known substitution using the provided sequencing analysis software, but did not recover the known single-base deletion in a homo-polymer run, despite seeing reads containing the variant. The authors concluded the vendor provided software was conservative when dealing with insertions/deletions in homo-polymer runs, as pyrosequencing has a higher error rate with this type of sequence. Other analysis packages were able to identify the variant.
Although these studies were not designed to identify novel variants causative for disease, much can be learned from them. Importantly, not every known variant was recovered. This was due to low sequence depth at the variant position, as well as issues relating to repeat regions and alignment. One study estimated that the probability of detecting a causative variant in any given gene is ∼86%, although this ignores non-coding and structural variants (26). In order to ensure sufficient allele sampling, as well as to prevent sequencing errors from appearing to be actual variants, all four studies use or recommend a minimum sequence depth threshold, ranging from 8- to 30-fold depth of coverage. These recommendations will affect the amount of sequencing required for a given capture size and will therefore affect the cost of the experiment.
Targeted capture has also been used to identify novel genes that cause hereditary disorders. Novel, putative causative variants have recently been discovered for a variety of disorders [sensory/motor neuropathy with ataxia (32), Clericuzio-type poikiloderma with neutropenia (33), familial exudative vitreoretinopathy (34), recessive non-syndromic hearing loss (35), talipes equinovarus, atrial septal defect, robin sequence, persistent left superior vena cava (36)] using genome capture to target linkage regions from the affected families. The identified variants were almost all non-synonymous substitutions, but follow-up studies on additional unrelated samples using Sanger sequencing also identified insertions/deletions in the same genes (33,35). Volpi et al. (33) identified a substitution that disrupted a splice site, resulting in an exon skip and a frameshift. Interestingly, Johnston et al. (36) were able to identify variants in two different families (one non-sense, one frameshifting insertion) without sequencing the probands, for which DNA was not available. These studies demonstrate the ability of genomic capture to discover different types of novel variants important for human disease.
In addition to custom capture studies, two whole exome studies have been recently reported. In the first, Choi et al. identified a novel coding variant in a consanguineous region of an affected individual. The variant was a homozygous missense substitution in SLC26A3, a gene in which mutations are known to cause congenital chloride-losing diarrhea (25). This genetic finding allowed the researchers to correct an earlier diagnosis of the patient's disorder. Variants in the same gene were present in other individuals, allowing the corrected diagnosis for them as well. In the second study, Ng et al. used exome capture to search for variants causing Miller syndrome in three unrelated families. They identify variants in DHODH in all three families, using filters for novel variants that fit inheritance models. These studies both showed that exome capture is an effective way to discover causative variants and genes and to correctly diagnose heritable disorders caused by variants in known genes.
Human evolution
Recent advances in the sequencing of ancient DNA have also benefited from targeted capture. Researchers used PEC to specifically target mitochondrial DNA from five Neandertal samples (21). The PEC method allowed complete coverage of the Neandertal mtDNA, using only 5–50 ng of amplified pyrosequencing library template. More recently, researchers used array-based capture to target, in Neandertal DNA, non-synonymous substitutions that have been fixed in humans since the divergence from the human/chimpanzee ancestor (37). Although the array-based capture did not have the low DNA requirements of PEC, the method allowed sequencing of a Neandertal sample containing 99.8% contaminating microbial DNA. Owing to the high contamination, this sample was unsuitable for shotgun sequencing, but targeted capture allowed recovery for almost all of the Neandertal sequence at the desired positions. The authors were able to then identify 88 substitutions that have become fixed in humans since the split from Neandertal, giving insight into what distinguishes us at the genetic, and perhaps molecular level.
Exome capture has been used to investigate more recent variation as well. Researchers used whole exome capture to identify changes in allele frequency between high-altitude populations (Tibetans) and low altitude populations (Han Chinese and Danes) (38). They were able to identify a number of genes likely to have been selected for as a part of adaptation to a high-altitude environment. Several of these genes were identified in other studies using microarray genotyping (39,40). This suggests that exome capture techniques are accurate and useful for these types of allelic frequency studies and would be especially useful for rarer SNPs that may not be included on the microarray platforms. Both recent and ancient genetic differences have been investigated using exome capture, allowing us to see a more complete view of our evolutionary history.
Biological
Basic biology questions are also being investigated on a much greater scale than previously possible using genome capture. Although the genetic information in DNA is frequently the initial focus of genome studies, epigenetic modification of the DNA also plays an important role in the biological function of an organism. Two groups used genome capture with padlock probes (19) or array-based capture (41) to investigate DNA methylation using bisulfite sequencing. Both studies found this to be very accurate when compared with the standard capillary methods. The latter study also showed that sensitivity using array-based capture was high: 86–91% of targeted bases were covered by 10 or more reads. An additional study focussed not on methylation status, but on genetic variation at CpG sites, which are subject to a higher mutation rate via 5-methylcytosine deamination (17). Using padlock probes, the researchers were able to determine genotypes for ∼65% of targeted bases. The accuracy was very high when compared with an independent genotype assessment. These CpG region studies show that capture is useful to focus on the desired regions and is effective, even on difficult (high GC content) regions.
Copy number variation (CNV) is another source of genetic variation implicated in disease. The detection of copy number changes is often performed using low-resolution methods, such as array-comparative genomic hybridization and single nucleotide polymorphism (SNP) microarrays. Conrad et al. (42) have used targeted sequencing to capture breakpoint regions and identify the actual breaks with a high resolution. They were able to identify breakpoints for a number of known CNVs and were then able to classify the breaks into likely repair mechanisms used. The authors point out that this method is useful for CNVs in simpler regions, as repeat elements and complex genomic regions present challenges both for capture and post-sequence alignment.
Capture is not only limited to genomic DNA. Several studies have used targeted sequencing to investigate RNA as well. One group used padlock probes to target regions containing known RNA-editing sites (43). They were able to identify sites in 10 of 13 known edited genes, by comparing captures of genomic DNA and cDNA from various tissues. The authors chose 18 editing sites at random and confirmed 15 with capillary sequencing. This research showed that padlock capture techniques work with cDNA and can be used to identify sites of RNA editing. Hybridization capture was also shown to capture cDNA (44,45). In (44), the authors capture both cDNA and genomic DNA with an array-based method. They then determine allele-specific expression using both data sets. In (45), the authors use solution hybridization to focus on enriching cDNA from a set of genes of interest. They were able to effectively enrich these genes, suggesting that genes of low abundance could be detected without huge increases in total sequencing. Interestingly, they were also able to identify gene fusions, including fusions in which one gene was not targeted. Applying targeted sequencing to cDNA is another way to focus on specific questions, even without whole-genome sequence.
FUTURE
One of the main reasons for performing a capture experiment is the significantly increased cost and time required for whole-genome sequencing. However, the constant improvements to massively parallel sequencing technologies and the impending massively parallel single-molecule sequencing technologies will certainly reduce these cost and time barriers. One may wonder what role capture will play as whole-genome sequencing is no longer impractical. Although capture has inherent costs independent of sequencing, capture experiments focus on subsets of the whole genome and will therefore always require less sequencing. Thus, more capture experiments can be performed given a set amount of sequencing capacity. Higher sample numbers result in higher power to detect variation, a key metric for discovering causative variants, especially for more common disorders. An argument in favor of whole-genome sequencing is that it is unwise to limit the data by doing capture experiments; it may be worth the additional cost to sequence ‘everything’. While this may be true, if researchers are confident that the desired genome subset (linkage regions, CpG islands, genes of interest etc.) is all they need to look at, more samples can be examined, and the data are limited to what is of interest. Data fatigue from attempting to interpret whole-genome sequence is not insignificant. Will an investigator be able to pick out the important variants out of a list of millions of positions? Although capture data can also contain large numbers of variants, the number is nearly two orders of magnitude lower than that from whole-genome sequence, making secondary analyses much less onerous. This is particularly important when bioinformatics personnel and resources are limiting (annotating lists of hundreds of variants is possible to accomplish by hand; doing so for tens of thousands variants is not). Therefore, it seems likely that targeted sequencing will be useful along side of whole-genome sequencing. Researchers will need to consider all aspects of a given project before deciding on whether to proceed with whole genome or targeted sequencing. Fortunately, ever decreasing sequencing costs may allow mixed approaches. Targeted sequencing has been shown to be a robust, effective technique that leverages the unique aspects of massively parallel sequencing and has already yielded many exciting new discoveries.
FUNDING
The authors are supported by the Intramural Research Program of the National Human Genome Research Institute. Funding to pay the Open Access Charge was provided by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
ACKNOWLEDGEMENTS
Conflict of Interest statement. None declared.
REFERENCES
- 1.Tarpey P.S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O'Meara S., Latimer C., Dicks E., Menzies A., et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat. Genet. 2009;41:535–543. doi: 10.1038/ng.367. doi:10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. doi:10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garber K. Fixing the front end. Nat. Biotechnol. 2008;26:1101–1104. doi: 10.1038/nbt1008-1101. doi:10.1038/nbt1008-1101. [DOI] [PubMed] [Google Scholar]
- 4.Summerer D. Enabling technologies of genomic-scale sequence enrichment for targeted high-throughput sequencing. Genomics. 2009;94:363–368. doi: 10.1016/j.ygeno.2009.08.012. doi:10.1016/j.ygeno.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Turner E.H., Ng S.B., Nickerson D.A., Shendure J. Methods for genomic partitioning. Annu. Rev. Genomics Hum. Genet. 2009;10:263–284. doi: 10.1146/annurev-genom-082908-150112. doi:10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]
- 6.Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. doi:10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 7.Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. doi:10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 8.Okou D.T., Steinberg K.M., Middle C., Cutler D.J., Albert T.J., Zwick M.E. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. doi:10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 9.Hodges E., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. doi:10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 10.Hodges E., Rooks M., Xuan Z., Bhattacharjee A., Benjamin Gordon D., Brizuela L., Richard McCombie W., Hannon G.J. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 2009;4:960–974. doi: 10.1038/nprot.2009.68. doi:10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bau S., Schracke N., Kranzle M., Wu H., Stahler P.F., Hoheisel J.D., Beier M., Summerer D. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal. Bioanal. Chem. 2009;393:171–175. doi: 10.1007/s00216-008-2460-7. doi:10.1007/s00216-008-2460-7. [DOI] [PubMed] [Google Scholar]
- 12.Herman D.S., Hovingh G.K., Iartchouk O., Rehm H.L., Kucherlapati R., Seidman J.G., Seidman C.E. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat. Methods. 2009;6:507–510. doi: 10.1038/nmeth.1343. doi:10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summerer D., Wu H., Haase B., Cheng Y., Schracke N., Stahler C.F., Chee M.S., Stahler P.F., Beier M. Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing. Genome Res. 2009;19:1616–1621. doi: 10.1101/gr.091942.109. doi:10.1101/gr.091942.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H., O'Connor B.D., Merriman B., Funari V.A., Homer N., Chen Z., Cohn D.H., Nelson S.F. Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing. BMC Genomics. 2009;10:646. doi: 10.1186/1471-2164-10-646. doi:10.1186/1471-2164-10-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W., Fennell T., Giannoukos G., Fisher S., Russ C., et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. doi:10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porreca G.J., Zhang K., Li J.B., Xie B., Austin D., Vassallo S.L., LeProust E.M., Peck B.J., Emig C.J., Dahl F., et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. doi:10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 17.Li J.B., Gao Y., Aach J., Zhang K., Kryukov G.V., Xie B., Ahlford A., Yoon J.K., Rosenbaum A.M., Zaranek A.W., et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 2009;19:1606–1615. doi: 10.1101/gr.092213.109. doi:10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner E.H., Lee C., Ng S.B., Nickerson D.A., Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat. Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. doi:10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng J., Shoemaker R., Xie B., Gore A., LeProust E.M., Antosiewicz-Bourget J., Egli D., Maherali N., Park I.H., Yu J., et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. doi:10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnakumar S., Zheng J., Wilhelmy J., Faham M., Mindrinos M., Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc. Natl Acad. Sci. USA. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. doi:10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs A.W., Good J.M., Green R.E., Krause J., Maricic T., Stenzel U., Lalueza-Fox C., Rudan P., Brajkovic D., Kucan Z., et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. doi:10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 22.Tewhey R., Warner J.B., Nakano M., Libby B., Medkova M., David P.H., Kotsopoulos S.K., Samuels M.L., Hutchison J.B., Larson J.W., et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. doi:10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim S.F., van den Engh G. High-speed chromosome sorting. Chromosome Res. 2004;12:5–14. doi: 10.1023/b:chro.0000009328.96958.a6. doi:10.1023/B:CHRO.0000009328.96958.a6. [DOI] [PubMed] [Google Scholar]
- 24.Weise A., Timmermann B., Grabherr M., Werber M., Heyn P., Kosyakova N., Liehr T., Neitzel H., Konrat K., Bommer C., et al. High-throughput sequencing of microdissected chromosomal regions. Eur. J. Hum. Genet. 2010;18:457–462. doi: 10.1038/ejhg.2009.196. doi:10.1038/ejhg.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. doi:10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. doi:10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bainbridge M.N., Wang M., Burgess D.L., Kovar C., Rodesch M.J., D'Ascenzo M., Kitzman J., Wu Y.Q., Newsham I., Richmond T.A., et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. doi:10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. doi:10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou L.S., Liu C.S., Boese B., Zhang X., Mao R. DNA sequence capture and enrichment by microarray followed by next-generation sequencing for targeted resequencing: neurofibromatosis type 1 gene as a model. Clin. Chem. 2010;56:62–72. doi: 10.1373/clinchem.2009.132639. doi:10.1373/clinchem.2009.132639. [DOI] [PubMed] [Google Scholar]
- 30.Hoischen A., Gilissen C., Arts P., Wieskamp N., van der Vliet W., Vermeer S., Steehouwer M., de Vries P., Meijer R., Seiqueros J., et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum. Mutat. 2010;31:494–499. doi: 10.1002/humu.21221. doi:10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- 31.Raca G., Jackson C., Warman B., Bair T., Schimmenti L.A. Next generation sequencing in research and diagnostics of ocular birth defects. Mol. Genet. Metab., 2010;100:184–192. doi: 10.1016/j.ymgme.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brkanac Z., Spencer D., Shendure J., Robertson P.D., Matsushita M., Vu T., Bird T.D., Olson M.V., Raskind W.H. IFRD1 is a candidate gene for SMNA on chromosome 7q22–q23. Am. J. Hum. Genet. 2009;84:692–697. doi: 10.1016/j.ajhg.2009.04.008. doi:10.1016/j.ajhg.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volpi L., Roversi G., Colombo E.A., Leijsten N., Concolino D., Calabria A., Mencarelli M.A., Fimiani M., Macciardi F., Pfundt R., et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. doi:10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikopoulos K., Gilissen C., Hoischen A., van Nouhuys C.E., Boonstra F.N., Blokland E.A., Arts P., Wieskamp N., Strom T.M., Ayuso C., et al. Next-generation sequencing of a 40 Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am. J. Hum. Genet. 2010;86:240–247. doi: 10.1016/j.ajhg.2009.12.016. doi:10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehman A.U., Morell R.J., Belyantseva I.A., Khan S.Y., Boger E.T., Shahzad M., Ahmed Z.M., Riazuddin S., Khan S.N., Friedman T.B. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am. J. Hum. Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. doi:10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston J.J., Teer J.K., Cherukuri P.F., Hansen N.F., Loftus S.K., Chong K., Mullikin J.C., Biesecker L.G. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. doi:10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burbano H.A., Hodges E., Green R.E., Briggs A.W., Krause J., Meyer M., Good J.M., Maricic T., Johnson P.L., Xuan Z., et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. doi:10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X.P., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S., et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. doi:10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M., et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. doi:10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B., et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. doi:10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 41.Hodges E., Smith A.D., Kendall J., Xuan Z., Ravi K., Rooks M., Zhang M.Q., Ye K., Bhattacharjee A., Brizuela L., et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. doi:10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conrad D.F., Bird C., Blackburne B., Lindsay S., Mamanova L., Lee C., Turner D.J., Hurles M.E. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010;42:385–391. doi: 10.1038/ng.564. doi:10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. doi:10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 44.Heap G.A., Yang J.H., Downes K., Healy B.C., Hunt K.A., Bockett N., Franke L., Dubois P.C., Mein C.A., Dobson R.J., et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. doi:10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin J.Z., Berger M.F., Adiconis X., Rogov P., Melnikov A., Fennell T., Nusbaum C., Garraway L.A., Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. doi:10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]