Abstract
The duplication architecture of the human genome predisposes our species to recurrent copy number variation and disease. Emerging data suggest that this mechanism of mutation contributes to both common and rare diseases. Two features regarding this form of mutation have emerged. First, common structural polymorphisms create susceptible and protective chromosomal architectures. These structural polymorphisms occur at varying frequencies in populations, leading to different susceptibility and ethnic predilection. Second, a subset of rearrangements shows extreme variability in expressivity. We propose that two types of genomic disorders may be distinguished: syndromic forms where the phenotypic features are largely invariant and those where the same molecular lesion associates with a diverse set of diagnoses including epilepsy, schizophrenia, autism, intellectual disability and congenital malformations. Copy number variation analyses of patient genomes reveal that disease type and severity may be explained by the occurrence of additional rare events and their inheritance within families. We propose that the overall burden of copy number variants creates differing sensitized backgrounds during development leading to different thresholds and disease outcomes. We suggest that the accumulation of multiple high-penetrant alleles of low frequency may serve as a more general model for complex genetic diseases, posing a significant challenge for diagnostics and disease management.
INTRODUCTION
Genomic disorders were originally described as large deletions and duplications that are highly penetrant, mostly de novo in origin, and typically identified in affected individuals with intellectual disability/multiple congenital malformations. Some examples include Smith–Magenis syndrome (MIM: 182290), DiGeorge/velocardiofacial syndrome (MIM: 188400, 192430) and Williams–Beuren syndrome (MIM: 194050). These classical genomic disorders have been well characterized in the past two decades with genotype–phenotype correlation studies implicating causative genes, mouse models recapitulating the human clinical features, and standardized management protocols and support groups established.
Application of higher definition molecular techniques, including single-nucleotide polymorphism microarrays or array comparative genomic hybridization (CGH), has allowed genotyping of larger disease cohorts and controls. Two major principles have emerged from these more recent studies: (i) common copy number polymorphism predisposes certain chromosomes to recurrent deletions and duplications and (ii) association of the same recurrent genomic lesion with apparently very diverse phenotypes. The latter has begun to illuminate common neurodevelopmental pathways and helps to explain the comorbidity of diverse neurological manifestations within the same families. The distinction between variability of expressivity and reduced penetrance depending on the diagnosis has become an important consideration for these rare mutational events. We will explore the mechanisms, models and implications underlying these two different aspects.
GENOMIC SUSCEPTIBILITY TO RECURRENT DELETIONS AND DUPLICATIONS
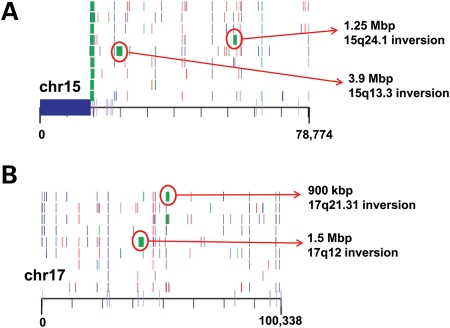
Seminal work on Charcot–Marie–Tooth disease (1,2) and hereditary neuropathy with liability to pressure palsies (HNPP) (3) directly implicated low-copy repeats or segmental duplications as substrates for unequal crossover or non-allelic homologous recombination (NAHR) resulting in duplications or deletions (4). Discovery of genomic disorders enriched within chromosomal regions flanked by segmental duplications further strengthened the NAHR model (5,6), and predisposition to such events was attributed to the length (≥10 kb) and percentage identity (≥95%) of participating segmental duplication blocks (5). It was also recognized that sites flanking copy number variants (CNVs) were particularly polymorphic with increased copy numbers of segmental duplications associating with recurrent rearrangements (7,8). For example, increased copies of segmental duplications flanking the chromosome 7q11.23 region have been linked to an elevated risk for rearrangements causing Williams syndrome (9). An intimate association between inversion polymorphisms and deletions and duplications was recognized early on for genomic disorders such as Williams syndrome (10), 8p23.1 microdeletion (11,12) and 15q11 microdeletions (13). Recently, fosmid paired-end sequencing followed by fluorescent in situ hybridization also identified six human disease-associated inversion polymorphisms in three HapMap populations for 3q29, 15q24.1, 15q13.3, 17q21.31, 8p23 and 17q12 (14,15) (Fig. 1). However, the molecular basis of these associations was not clear until recently.
Figure 1.
Rare CNVs and inversion polymorphisms. A structural variation map for chromosome 15 (A) and chromosome 17 (B) is shown (15). Of relevance to this review, inversion polymorphisms (green blocks) associated with genomic disorders for both the chromosomes are shown. Of note, these inversions likely harbor the directly orientated segmental duplication and such inversions are usually identified in the parents of probands carrying a CNV for the intervening region. This image is modified from Kidd et al. (15).
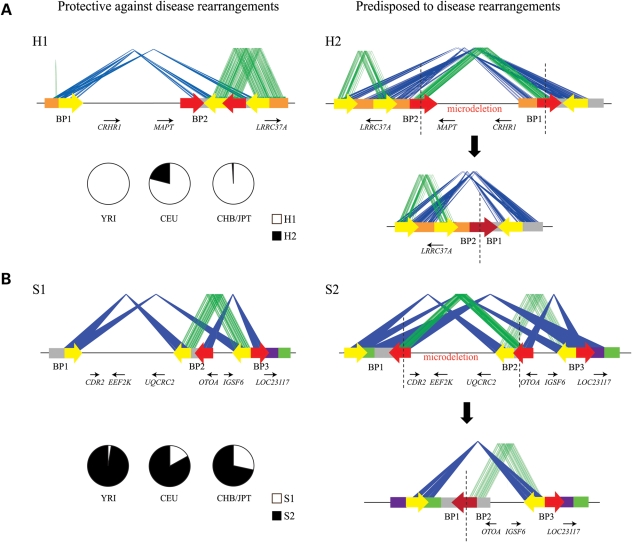
Stefansson et al. (16) reported one such structurally complex region on chromosome 17q21.31 of the human genome. Two ancient haplotypes map to this region: the directly orientated (or H1 based on the build36 version of the human genome assembly) and the inverted orientation (H2) (Fig. 2A). Interestingly, no recombination occurs between the two haplotypes and, thus, there is a large extended stretch of linkage disequilibrium in this region. Only the H2 haplotype harbors two highly identical (38 kb, ∼98%) blocks of segmental duplications in direct orientation facilitating NAHR events (6,17). This structure predisposes ∼20% of Europeans to a recurrent 480 kb microdeletion (6,17–19). Notably, the H2 haplotype occurs at an extremely low frequency in the Asian and African populations (16). Among the San population of Southern Africa, the H2 allele has been discovered at low frequency (20) yet without the predisposing segmental duplication that promotes unequal crossover. The available data suggest that the duplication-associated inversion and the 17q21.31 recurrent microdeletion are largely restricted to individuals of European and Mediterranean descent.
Figure 2.
Genomic predisposition to disease rearrangements. The schematic depicts the common structural configurations for 17q21.31 (A) and 16p12.1 (B), with block arrows depicting the orientation of the segmental duplication blocks. The blue lines connect the identical segmental duplications in the inverted orientation, and green lines connect those in the direct orientation. The red arrows are the segmental duplications that participate in the NAHR events resulting in the microdeletion with the fusion of the two segmental duplication blocks (dark red arrow). The genes lying within these regions are also shown, with small arrows depicting the transcriptional orientation. H1 represents the direct configuration wherein the predisposing duplications are not in the same orientation. H2 represents a vulnerable structure with directly oriented duplication blocks. S1 is the protective structure and S2 has the predisposing structure. Note the orientation of both S1 and S2 is correct but inverted compared with the current version (build36) of the human genome (see Antonacci et al. (22) for more details). Also shown is the frequency of H1 and H2 (for 17q21.31) and S1 and S2 (for 16p12.1) in the HapMap populations.
Similarly, investigation into segmental duplication architecture of the more recently described 16p12.1 microdeletion (21) revealed two common structural configurations with worldwide frequencies of 17.6% (S1) and 82.4% (S2) (22) (Fig. 2B). Of note, the two human haplotypes differ by 333 kb of additional duplicated sequence present in S2 but not in S1. Again, similar to the 17q21.31 region, the S2 configuration directly orients the specific duplication blocks (68 kb, >99%), thereby predisposing to disease rearrangement. Although the S1 structure is protective against the disease-associated rearrangement, S2 is the most common structure with frequencies of 97.5% in Africans (YRI), 83.1% in Europeans (CEU) and 71.6% in Asian populations (CHB/JPT). Similar analyses of other hotspot regions of the genome will likely identify other specific predilections to specific disease rearrangements.
ADDITIONAL PHENOTYPES FOR KNOWN GENOMIC DISORDERS
The dichotomy between syndromic genomic disorders and those with more variable phenotypes is longstanding. For example, microdeletion on chromosome 22q11.2 presented with a variety of phenotypes, including velocardiofacial syndrome (23,24), DiGeorge syndrome (25–27), isolated cardiac outflow tract defects (28,29) and schizophrenia (30). In contrast, specific phenotypes aiding relatively straightforward clinical diagnosis were noted for Williams syndrome resulting from a 7q11.23 microdeletion (31) and Angelman syndrome (32) or Prader–Willi syndrome (33) resulting from maternal (34) or paternal deletion (35), respectively, of the imprinted loci on chromosome 15q11.2q13 (36,37). Nearly 20 new genomic disorders have been reported recently, almost twice the number compared with what was known in the past two decades. These novel disorders are recurrent and map within genomic hotspots flanked by segmental duplications. Despite initial claims, most of these disorders are not specific and phenotypes have been associated with, but not limited to, intellectual disability/congenital malformations, epilepsy, schizophrenia, autism, cardiac and renal anomalies and obesity (Table 1).
Table 1.
Variable clinical features associated with genomic disorders
| Genomic loci | Position | Size | Candidate gene(s) | Major phenotypes | CNVs in cases | Incidence (%) | CNVs in controls | Incidence (%) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1q21.1 deletion | chr1: 145.0–146.35Mb | 1.35 Mb | GJA5, GJA8, CHD1L, HYDIN2 | Learning disability, congenital anomaly, microcephaly, cataracts | 52/21 775 | 0.24 | 0/4737 | 0 | (95,104) |
| Schizophrenia | 17/7918 | 0.21 | 11/46 502 | 0.02 | (76,77) | ||||
| Tetralogy of Fallot | 1/512 | 0.20 | 0/2265 | 0 | (105) | ||||
| Congenital heart disease | 3/505 | 0.59 | 0/520 | 0 | (106) | ||||
| 1q21.1 duplication | chr1: 145.0–146.35 Mb | 1.35 Mb | GJA5, GJA8, CHD1L, HYDIN2 | Learning disability, autism spectrum disorder, macrocephaly, behavioral features | 26/21 775 | 0.12 | 0/4737 | 0 | (95,104) |
| Tetralogy of Fallot | 4/512 | 0.78 | 0/2265 | 0 | (105) | ||||
| 3q29 deletion | chr3: 197.4–198.9 Mb | 1.5 Mb | PAK2, DLG1 | Mild-to-moderate learning disability, mild dysmorphic features, autism, bipolar disorder | 14/14 698 | 0.10 | NA | — | (107–110) |
| 3q29 duplication | chr3: 197.4–198.9 Mb | 1.5 Mb | PAK2, DLG1 | Mild-to-moderate learning disability, microcephaly, obesity | 19/14 698 | 0.13 | NA | — | (107,111) |
| 15q11.2 deletion | chr15: 20.30–20.80Mb | 500 kb | NIPA1, NIPA2, CYFIP1 | Idiopathic generalized epilepsy | 12/1234 | 0.97 | 2/3022 | 0.07 | (54) |
| Schizophrenia | 49/7918 | 0.62 | 103/46 497 | 0.22 | (49,76,77) | ||||
| Learning disability | 8/1010 | 0.79 | 3/2493 | 0.12 | (44) | ||||
| Behavioral problems, developmental delay, autism spectrum disorders, craniofacial features | 9/1576 | 0.57 | NA | — | (112) | ||||
| 15q13.3 deletion | chr15: 28.70–30.20Mb | 1.5 Mb | CHRNA7 | Idiopathic generalized epilepsy | 12/1223 | 0.98 | 0/3699 | 0 | (74) |
| Learning disability, seizures | 22/8706 | 0.25 | 0/2962 | 0 | (72,113) | ||||
| Cognitive impairment, expressive language deficits, autism spectrum disorder, behavioral features, no epilepsy | 5/1445 | 0.35 | NA | — | (114) | ||||
| Autism spectrum disorder | NA | — | NA | — | (75) | ||||
| Schizophrenia | 17/7918 | 0.21 | 8/45 103 | 0.02 | (49,76,77) | ||||
| Rage/aggressive behaviors, autism, learning disability | 14/8200 | 0.17 | NA | — | (78) | ||||
| 15q13.3 duplication | chr15: 28.70–30.20Mb | 1.5 Mb | CHRNA7 | Behavioral features, depression, schizophrenia, learning disability | 8/15 456 | 0.05 | 23/3699 | 0.62 | (74,113,115) |
| Autism, language delay, no epilepsy | 3/1445 | 0.21 | NA | — | (114) | ||||
| 16p11.2 deletion | chr16: 29.50–30.10Mb | 600 kb | SEZ6L2, ALDOA, TBX6, QPRT | Autism, learning disability | 13/2252 | 0.58 | 5/23 502 | 0.02 | (39) |
| Autism | 8/1139 | 0.70 | 0/2489 | 0 | (38,41) | ||||
| Developmental delay, speech delay, behavioral problems, no autism | 74/15 067 | 0.49 | 0/2393 | 0 | (40,44,45) | ||||
| Speech/language delay, congenital anomaly, seizures, macrocephaly, autism | 27/7400 | 0.36 | NA | — | (116) | ||||
| Autism, learning disability | 17/2172 | 0.78 | NA | — | (117) | ||||
| Obesity | 50/20 312 | 0.25 | 1/7434 | 0.01 | (46) | ||||
| 16p11.2 duplication | chr16: 29.50–30.10Mb | 600 kb | SEZ6L2, ALDOA, TBX6, QPRT | Autism, learning disability | 7/2252 | 0.31 | 7/23 502 | 0.03 | (39) |
| Motor delay, congenital anomaly, behavioral features, and microcephaly | 18/7400 | 0.24 | NA | — | (116) | ||||
| Schizophrenia, microcephaly | 26/8590 | 0.30 | 8/28 406 | 0.03 | (117) | ||||
| Learning disability, speech and language delay | 32/9773 | 0.33 | 1/2393 | — | (45) | ||||
| 16p11.2 deletion | chr16:20.50–20.90Mb | 400 kb | SH2B1, ATXN2L, ATP2A1 | Obesity | 5/300 | 1.67 | 2/7366 | 0.03 | (47) |
| Mental retardation | 31/23 084 | 0.13 | 1/7700 | 0.12 | (48) | ||||
| 16p12.1 deletion | chr16: 21.85–22.37Mb | 520 kb | EEF2K, CDR2, POLR3E | Learning disability/multiple congenital anomaly | 42/21 127 | 0.20 | 8/14 839 | 0.05 | (21) |
| 16p13.11 deletion | chr16: 15.4–16.4Mb | 1 Mb | NDE1, MYH11, ABCC1 | Learning disability/multiple congenital anomaly | 5/1027 | 0.49 | 0/2014 | 0 | (53) |
| Autism, learning disability | 3/182 | 1.65 | 0/600 | 0 | (52) | ||||
| Sporadic epilepsy syndromes | 23/3812 | 0.60 | 0/1299 | 0 | (55) | ||||
| Idiopathic generalized epilepsy | 6/1234 | 0.49 | 2/3022 | 0.07 | (54) | ||||
| 16p13.11 duplication | chr16: 15.4–16.4Mb | 1 Mb | NDE1, MYH11, ABCC1 | Schizophrenia | 16/4816 | 0.33 | 38/37 871 | 0.10 | (49,56) |
| Autism, learning disability | 3/182 | 1.65 | 0/600 | 0 | (52) | ||||
| Learning disability | 11/1010 | 1.09 | 2/2493 | 0.08 | (44) | ||||
| 17p12 deletion | chr17:14.0–15.5Mb | 1.5 Mb | PMP22 | Hereditary neuropathy with liability to pressure palsies | NA | — | NA | — | (3) |
| Schizophrenia | 8/5089 | 0.16 | 6/38 884 | 0.02 | (49) | ||||
| 17q12 deletion | chr17: 31.80–33.30Mb | 1.5 Mb | TCF2, LHX1, ACACA,HNF1β | Renal cysts, diabetes, seizures | NA | — | NA | — | (50) |
| Learning disability, congenital anomaly, renal cysts, seizures | NA | — | NA | — | (51) | ||||
| 17q12 duplication | chr17: 31.80–33.30Mb | 1.5 Mb | TCF2, LHX1, ACACA,HNF1β | Learning disability and seizures | NA | — | NA | — | (50) |
| Learning disability, behavioral abnormalities, no seizures | NA | — | NA | — | (51) | ||||
| 17p11.2 duplication | chr17: 16.6–20.2Mb | 3.6 Mb | RAI1 | Autism, cardiac anomalies, developmental delay, failure to thrive | NA | — | NA | — | (58) |
| No autism, speech and language impairment, learning disability | NA | — | NA | — | (118) | ||||
| 22q11.2 deletion | chr22: 17.0–20.0Mb | 3 Mb | TBX1, COMT | Cardiac outflow tract defects, learning disability, facial dysmorphology, velopharyngeal insufficiency, cleft palate, thymic hypoplasia, parathyroid hypoplasia | NA | — | NA | — | (24,25,27,119) |
| Schizophrenia | 18/7038 | 0.26 | 0/44 602 | 0 | (30,49,76,77) | ||||
| 22q11.2 duplication | chr22: 17.0–20.0Mb | 3 Mb | TBX1, COMT | Learning disability, congenital malformation, behavioral features | NA | — | NA | — | (120,121) |
| Phenotypically normal parents | NA | — | NA | — | (122) | ||||
| 22q11.2 distal deletion | chr22: 20.2–21.9Mb | 1.7 Mb | UBE2L2, GNAZ, MAPK1 | Growth retardation, developmental delay, craniofacial/skeletal features, cardiac defects, behavioral features | 6/8000 | 0.08 | NA | — | (123) |
| 22q11.2 distal duplication | chr22: 20.2–21.9Mb | 1.7 Mb | UBE2L2, GNAZ, MAPK1 | Normal-to-profound developmental delay, micro- or macrocephaly, normal tone or profound hypotonia | 18/22 096 | 0.08 | NA | — | (124) |
NA, data not available.
For example, microdeletions in 16p11.2 containing the TBX6 gene was initially reported to be associated in individuals with autism (38) and autism spectrum disorder (39), whereas the same events were identified in cases ascertained for developmental delay and congenital malformations (40). Although further studies validated the association of this microdeletion to both autism (41–43) and idiopathic mental retardation (44,45), using high-density array CGH, Walters et al. (46) also documented an increased incidence of obesity in cases with microdeletion of this locus.
There are now numerous examples of this widening spectrum of clinical diagnoses (Table 1). Microdeletion on 16p11.2 containing the SH2B1 gene was initially reported as an obesity locus (47); however, recent studies have also shown a strong association with developmental delay (48). These studies have also associated a previously unreported phenotype to a well-characterized canonical syndrome mainly due to ascertainment bias. For example, the microdeletion on chromosome on 17p12 is classically known to be causal for HNPP (3). Interestingly, a large, multi-center study also implicated microdeletion on 17p12 in schizophrenia susceptibility (49). Initially, del17q12 was discovered in cases with cystic renal disease and maturity-onset diabetes of the young but without cognitive impairment (50). Nagamani et al. (51) expanded the phenotypic spectrum of this disorder to include cognitive impairment, seizures and brain malformations. Deletions and duplications in 16p13.11 were initially reported in cases with intellectual disability (44,52) and autism (52,53); subsequent studies then also linked these CNVs to epilepsy (54,55) and schizophrenia (56).
Thus, certain challenges surround the discovery of these newly described CNVs: (i) many of these novel CNVs have been found in the control populations as well as in unaffected family members and (ii) lack of detailed clinical information and standardized phenotyping poses a challenge for diagnosis, counseling and management of such disorders. In light of the effect size of some of these variants in large case–control studies, the discovery of the same lesion in the general population likely reflects variability of expressivity as opposed to reduced penetrance of the variant. Studying these deletions and duplications within the context of a well-phenotyped family would likely reveal that most of these larger events have phenotypic consequence although the effect in some cases may be more subtle depending on other genetic and non-genetic modifiers.
VARIABLE EXPRESSIVITY IN GENOMIC DISORDERS
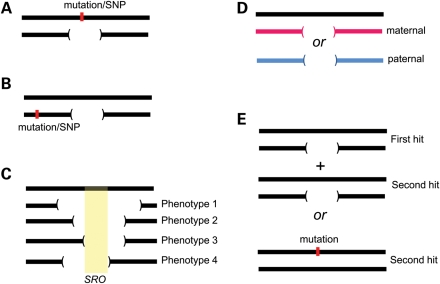
There are several explanations for variable expressivity and clinical heterogeneity in genomic disorders (Fig. 3). First, the breakpoints of the events may not be identical. Atypical deletions and duplications involving contiguous dosage-sensitive genes within the region often explained the observed clinical variability in many genomic disorders. Examples include Smith–Magenis syndrome (57), Potocki–Lupski syndrome (58), 15q24.3 microdeletion (59,60) and Williams syndrome (61). Many of these atypical CNVs tend to result from alternate segmental duplications (62) or Alu sequences (63) acting as recombination substrates leading to non-recurrent rearrangements. Mapping of these atypical CNVs has further enabled the delineation of a disease-associated critical interval, for example, in Alagille syndrome (JAG1) (64,65), Smith–Magenis syndrome (RAI1) (66) and demarcation of the phenotype-associated gene in Williams syndrome [GTF2IRD1 and GTF2I for craniofacial abnormalities and visuospatial defects (61,67) and NCF1 for hypertension (68)]. Similarly, by correlating the patient phenotypes to the non-recurrent atypical 17p13.3 deletions, the PAFAH1B1 or LIS1 and YWHAE genes were attributed to a majority of clinical features associated with lissencephaly or Miller–Dieker syndrome, respectively (69–71). Although detection of smaller or atypical deletions has pinpointed genes related to specific phenotypes, for some genomic disorders the CNV size (examples include del1q21.1, del16p13.11 and dup16p13.11) did not correlate with the phenotypes observed, indicating other mechanisms for the clinical heterogeneity.
Figure 3.
Phenotypic variability in genomic disorders. Possible causes for phenotypic variability are shown: (A) unmasking of recessive mutations or functional polymorphism within the CNV region, (B) a single-nucleotide change in close proximity to the CNV altering the expression pattern of the genes in the region, and (C) different sizes of the CNV leads to different phenotypes, as in Williams syndrome (61). Sometimes, a ‘core’ gene is responsible for a majority of the phenotypes, whereas other genes within the region contribute to the variability and severity, as in Smith–Magenis syndrome (57). (D) Parent-of-origin effects or imprinting of the paternal or maternal derived allele/genes as in Prader-Willi or Angelman syndrome and (E) the two-hit model wherein the severity and variability are due to alteration (CNV or a point mutation) of multiple functionally relevant genes.
A major challenge for the newly reported genomic disorders has been the pathological association of diverse neurological phenotypes with CNVs mapping to segmental duplications. For these CNVs, the breakpoints have not been refined to a specific basepair position due to the complexity of the duplicated sequences. However, such areas are enriched for genes of unknown function, so it is possible that subtle differences within the breakpoints themselves can account for the variability. A good example is the 15q13.3 microdeletion that maps between two large identical blocks of segmental duplications, distal to the Prader–Willi/Angelman locus. Originally, these microdeletions were described in nine cases with developmental delay/multiple congenital anomalies and seizures (72). The same event was later identified to be the most prevalent risk factor for common epilepsies; in fact, accounting for ∼1% of the sporadic cases as well as familial cases with idiopathic generalized epilepsy (73,74) and sporadic epilepsy syndromes (55). As more cases with a broad spectrum of developmental and neuropsychiatric disorders and control samples were analyzed, it became clear that the 15q13.3 microdeletion was associated with a wide range of outcomes as well being reported among normal individuals. Whereas the same microdeletion was enriched in cases with autism (75) and individuals with schizophrenia (76,77), Ben-Shachar et al. (78) also reported aggressive and possible antisocial behaviors in carrier parents. Shinawi et al. (79) were further able to reduce the disease-associated critical region when they genotyped four cases with variable neurodevelopmental phenotypes and identified a 680 kb region encompassing the CHRNA7 and OTUD7 genes. Le Pichon et al. (80) identified a patient with severe neurodevelopmental deficits and a homozygous 15q13.3 microdeletion suggesting dosage sensitivity for several genes within this region including TRIPM1 and CHRNA7. Although accumulating evidence suggests that CHRNA7 is the causal gene for these neurological phenotypes, it is likely that heterozygous deletions result in neuropsychiatric and/or epilepsy phenotypes, dependent on other CNV hits and genetic modifiers, and that the absence of CHRNA7 would result in a more severe cognitive manifestation.
Haploinsufficiency or dosage sensitivity for one or more genes within the genomic region has been the commonly proposed functional impact of a CNV. Genes associated with autosomal dominant disorders are also likely to be exposed due to the hemizygosity of the particular gene-containing interval; examples include Williams syndrome [ELN for supravalvular aortic stenosis (81)], Smith–Magenis syndrome [RAI1 for mental retardation, sleep disturbance, self-injurious behaviors (66)] and del1q21.1 [GJA5 for congenital cardiac disease (82,83) and GJA8 for cataracts (84)]. Mutations in NIPA1 within the 15q11.2 (BP1–BP2) region have been reported in autosomal dominant hereditary spastic paraplegia (85). Interestingly, 15q11.2 deletion, resulting in hemizygosity of NIPA2, has been linked with an increased risk for diverse neuropsychiatric features (Table 1). The MYH11 gene, previously implicated in aortic aneurysms and dissections (86), maps within the 16p13.11 CNV region; however, probably due to ascertainment bias, aortic defects have not yet been reported in studies on 16p13.11 CNVs.
Furthermore, recessive genes reside within the CNV regions, and the chances of finding a recessive mutation along with a microdeletion are rare (frequency of spontaneous mutation × frequency of the deletion event), but plausible (Fig. 3). Profound sensorineural hearing loss has been reported in patients with Smith–Magenis syndrome whose deletions unmask the recessive mutations in the myosin (MYO15A) gene located within the 17p11.2 region (87). Functional polymorphisms within COMT and FXII, unmasked by hemizygous deletions, have also been reported to result in cognitive decline and psychosis in patients with del22q11.2 and reduced activity of coagulation factor 12 in Sotos syndrome, respectively (88,89). Chronic granulomatous disease resulting from unmasking of NCF1 recessive mutation has been reported in relation to the Williams syndrome deletion (90). Similarly, metachromatic leukodystrophy was reported in a case with 22q13 deletion and a hemizygous mutation in the arylsulfatase A (ARSA) gene (91). Oculocutaneous albinism was reported in an individual with Prader–Willi syndrome deletion and a recessive mutation in the OCA2 gene (92). A combination of symptoms of Wolfram syndrome and Wolf–Hirschhorn syndrome was detected in a case with a WFS1 mutation unmasked by a chromosome 4p deletion (93). Kumar et al. (94) performed sequencing analysis on eight genes within the 16p11.2 autism-associated region in five autism cases with the microdeletion and approximately 100 cases without the microdeletion and 100 ethnicity-matched controls. Although single-nucleotide variants in the coding exons were detected in SEZ6L2 in the initial set of autism cohorts, the authors were not able to replicate this finding in a replication set. However, sequencing efforts to identify recessive alleles in genes within the 1q21.1 (95,96), 16p13.11 (53) and 16p12.1 (21) regions have not yet met with much success. Targeted sequencing of genes within the pathogenic CNV region using the newly available technologies such as molecular inversion probe assays (97) would be useful to find potential candidate genes.
A TWO-HIT MODEL FOR PHENOTYPIC VARIABILITY
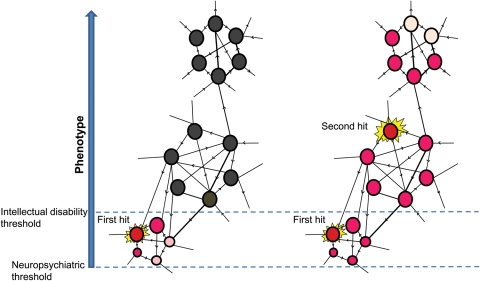
Most studies on previously discovered microdeletions and microduplications associated with neurodevelopmental phenotypes were focused on the impact of genes within a specific region. Recently, a 520 kb microdeletion on the 16p12.1 was identified to be non-syndromic, i.e. associated with variable phenotypes and was inherited from a parent in 95% of the cases (21), in stark contrast to other genomic disorders where most mutations are de novo. Further follow-up on the families of probands showed that the carrier parents were, in fact, suffering from subclinical manifestations of mild neuropsychiatric illness including depression, bipolar disorder, mild learning disability or seizures. What is the cause for the phenotypic variability between the probands and the carrier parents? It was observed that ∼25% of the probands also carried another large deletion or duplication second hit, elsewhere in the genome. This represented a 40-fold increase for two or more CNVs > 500 kb in length when compared with the general population. The clinical features in probands with two hits were different from those associated with just the second hit. It is evident that two large CNVs in a single individual would increase or decrease the dosage for multiple genes, thereby creating a sensitized genomic background. It is likely that one hit is sufficient to reach a threshold just enough to induce some neuropsychiatric features but a second hit pushes one toward a more severe phenotype with intellectual disability and developmental delay (Fig. 4).
Figure 4.
A model to explain the functional impact of two hits. The schematic depicts a network of genes in a phenotype-associated pathway. A single hit disrupts the pathway, which is just sufficient to reach a threshold to induce neuropsychiatric features (left). A second hit pushes the threshold of the resultant phenotype toward intellectual disability (right). Two hits can therefore generate a more severe phenotype, a distinct phenotype or an additional phenotype (comorbidity) compared with the outcome from the either the first or second hit.
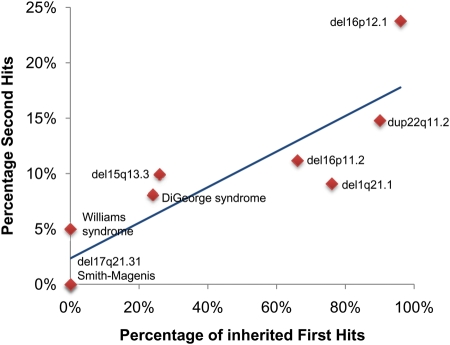
The two-hit model was initially applied to explain the variable expressivity in 16p12.1 microdeletion. Further evaluation of a handful of other genomic disorders associated with syndromic or variable phenotypes showed a clear clustering of two hits in CNVs with variable phenotypes when compared with syndromic mutational events. Further, a strong correlation was observed between the proportion of inherited first hits and the frequency of second hits (Fig. 5). This relationship reflects as a function of fitness wherein genomic disorders associated with severe syndromes are essentially under strong purifying selection and therefore can arise only through de novo events. However, others often present with milder phenotypes are subjected to strong but reduced selection, and are therefore passed on from parents to offspring for a few generations until the variant crosses paths with a second germline hit and then manifests as a severe phenotype. Although the two-hit model was initially applied to large CNVs, the second hit could also, in principle, be a smaller CNV or a single-nucleotide change affecting a phenotypically related gene or a risk allele derived from a parent. Assuming that nosologically distinct diseases such as epilepsy, intellectual disability, schizophrenia and autism share common neurodevelopmental pathways, the disease outcome may differ depending on the overall burden of haplosensitive genes in an individual and the pathways that are affected (Fig. 4). Ultimately, such mutations are removed from the population but the overall genetic load and high frequency of large, recurrent CNVs lead to a significant contribution to disease. The two-hit model is attractive in the sense that it can explain the often-documented comorbidity between intellectual disability and other neurological phenotypes (98,99) within families.
Figure 5.
The two-hit model for variable expressivity and comorbidity. The graph plots the percentage of second hits and the frequency of inherited first hits for nine genomic disorders. Regression analysis suggests a significant correlation (r = 0.87, P < 0.01). See text for more details.
CONCLUSION
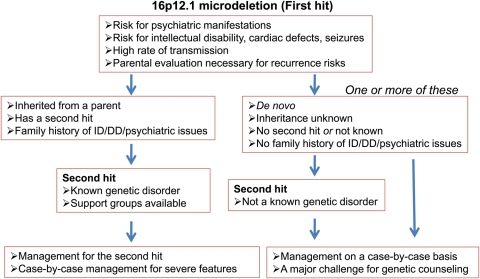
Standard management protocols have been developed for several genomic disorders including DiGeorge, Smith–Magenis and Williams syndromes. As many of the new ‘first-hit’ CNVs are inherited from a parent, management of cases with two hits is challenging, especially when the pathogenicity of the second hit is not yet well established. In addition to the assessment of recurrence risks, management of cases with two hits has to rely on a case-by-case workup or follow the standard protocols in case either hit is a known genomic disorder (Fig. 6). Detailed multi-disciplinary assessment of many cases is warranted for a definitive diagnosis and for teasing out the genomic contribution for neurodevelopmental phenotypes. Although collectively quite common, the low prevalence of any single event justifies the screening of hundreds of thousands of cases to provide evidence-based criteria for the assessment of pathogenicity and diagnosis. The development of an international consortium—the ISCA (100)—and associated databases—dbVAR, ECARUCA (101) and DECIPHER (102)—is an important first step in this regard.
Figure 6.
Management of genomic disorders with two hits. An example management flow chart for 16p12.1 microdeletion is shown. It is important to note that there is a high risk for recurrence given that several genomic disorders associated with two hits are also highly inherited.
Given the rapid progress in the field of genomic sequencing, it is evident that the microarrays are going to be replaced by exome and whole-genome sequencing in the near future (103). These will ultimately become the standard for testing copy number variation. Such rapid advancements will uncover a deluge of potentially disruptive mutations including small indels and point mutations, further challenging genetic diagnosis and management. In such situations wherein diseases seem to behave in a non-Mendelian fashion, compounding rare variants of functional significance will be relevant. A more tractable solution would be to perform a detailed focus study of individuals sharing a particular lesion using a genotype-first approach. Such projects have already been proposed and are some of the most tangible deliverables of the genome era. Finally, it is evident that copy number and inversion polymorphisms can predispose some populations to disease and make others immune (Europeans predisposed and Asians protected against 17q21.31 lesions). Therefore, CNV studies for rare structural variants (0.001–0.1%) could lead to false associations without disease significance and should contain properly matched cases and controls.
FUNDING
This work was supported by NIH grant HD043569 (E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
ACKNOWLEDGEMENTS
We thank Luis Perez-Jurado for useful comments.
Conflict of Interest statement. E.E.E. is a scientific advisory board member for Pacific Biosciences.
REFERENCES
- 1.Raeymaekers P., Timmerman V., Nelis E., De Jonghe P., Hoogendijk J.E., Baas F., Barker D.F., Martin J.J., De Visser M., Bolhuis P.A., et al. Duplication in chromosome 17p11.2 in Charcot–Marie–Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group . Neuromuscul. Disord. 1991;1:93–97. doi: 10.1016/0960-8966(91)90055-w. doi:10.1016/0960-8966(91)90055-W. [DOI] [PubMed] [Google Scholar]
- 2.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A., et al. DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. doi:10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 3.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. doi:10.1016/0092-8674(93)90058-X. [DOI] [PubMed] [Google Scholar]
- 4.Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. doi:10.1016/S0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 5.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. doi:10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 6.Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. doi:10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 7.Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. doi:10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. doi:10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cusco I., Corominas R., Bayes M., Flores R., Rivera-Brugues N., Campuzano V., Perez-Jurado L.A. Copy number variation at the 7q11.23 segmental duplications is a susceptibility factor for the Williams–Beuren syndrome deletion. Genome Res. 2008;18:683–694. doi: 10.1101/gr.073197.107. doi:10.1101/gr.073197.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne L.R., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., et al. A 1.5 million-base pair inversion polymorphism in families with Williams–Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. doi:10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giglio S., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. doi:10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giglio S., Calvari V., Gregato G., Gimelli G., Camanini S., Giorda R., Ragusa A., Guerneri S., Selicorni A., Stumm M., et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 2002;71:276–285. doi: 10.1086/341610. doi:10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimelli G., Pujana M.A., Patricelli M.G., Russo S., Giardino D., Larizza L., Cheung J., Armengol L., Schinzel A., Estivill X., et al. Genomic inversions of human chromosome 15q11–q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum. Mol. Genet. 2003;12:849–858. doi: 10.1093/hmg/ddg101. doi:10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- 14.Antonacci F., Kidd J.M., Marques-Bonet T., Ventura M., Siswara P., Jiang Z., Eichler E.E. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 2009;18:2555–2566. doi: 10.1093/hmg/ddp187. doi:10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. doi:10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefansson H., Helgason A., Thorleifsson G., Steinthorsdottir V., Masson G., Barnard J., Baker A., Jonasdottir A., Ingason A., Gudnadottir V.G., et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. doi:10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 17.Zody M.C., Jiang Z., Fung H.C., Antonacci F., Hillier L.W., Cardone M.F., Graves T.A., Kidd J.M., Cheng Z., Abouelleil A., et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. doi:10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. doi:10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 19.Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. doi:10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 20.Schuster S.C., Miller W., Ratan A., Tomsho L.P., Giardine B., Kasson L.R., Harris R.S., Petersen D.C., Zhao F., Qi J., et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. doi:10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C., et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. doi:10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonacci F., Kidd J.M., Marques-Bonet T., Teague B., Ventura M., Girirajan S., Alkan C., Campbell C.D., Vives L., Malig M., et al. A large, complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat. Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burn J., Takao A., Wilson D., Cross I., Momma K., Wadey R., Scambler P., Goodship J. Conotruncal anomaly face syndrome is associated with a deletion within chromosome 22q11. J. Med. Genet. 1993;30:822–824. doi: 10.1136/jmg.30.10.822. doi:10.1136/jmg.30.10.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shprintzen R.J., Goldberg R.B., Lewin M.L., Sidoti E.J., Berkman M.D., Argamaso R.V., Young D. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J. 1978;15:56–62. [PubMed] [Google Scholar]
- 25.Driscoll D.A., Budarf M.L., Emanuel B.S. A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am. J. Hum. Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- 26.Scambler P.J., Kelly D., Lindsay E., Williamson R., Goldberg R., Shprintzen R., Wilson D.I., Goodship J.A., Cross I.E., Burn J. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339:1138–1139. doi: 10.1016/0140-6736(92)90734-k. doi:10.1016/0140-6736(92)90734-K. [DOI] [PubMed] [Google Scholar]
- 27.Wilson D.I., Cross I.E., Goodship J.A., Brown J., Scambler P.J., Bain H.H., Taylor J.F., Walsh K., Bankier A., Burn J., et al. A prospective cytogenetic study of 36 cases of DiGeorge syndrome. Am. J. Hum. Genet. 1992;51:957–963. [PMC free article] [PubMed] [Google Scholar]
- 28.Goldmuntz E., Driscoll D., Budarf M.L., Zackai E.H., McDonald-McGinn D.M., Biegel J.A., Emanuel B.S. Microdeletions of chromosomal region 22q11 in patients with congenital conotruncal cardiac defects. J. Med. Genet. 1993;30:807–812. doi: 10.1136/jmg.30.10.807. doi:10.1136/jmg.30.10.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson D.I., Goodship J.A., Burn J., Cross I.E., Scambler P.J. Deletions within chromosome 22q11 in familial congenital heart disease. Lancet. 1992;340:573–575. doi: 10.1016/0140-6736(92)92107-q. doi:10.1016/0140-6736(92)92107-Q. [DOI] [PubMed] [Google Scholar]
- 30.Karayiorgou M., Morris M.A., Morrow B., Shprintzen R.J., Goldberg R., Borrow J., Gos A., Nestadt G., Wolyniec P.S., Lasseter V.K., et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl Acad. Sci. USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. doi:10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tassabehji M. Williams–Beuren syndrome: a challenge for genotype–phenotype correlations. Hum. Mol. Genet. 2003;12((Spec no. 2)):R229–R237. doi: 10.1093/hmg/ddg299. doi:10.1093/hmg/ddg299. [DOI] [PubMed] [Google Scholar]
- 32.Magenis R.E., Brown M.G., Lacy D.A., Budden S., LaFranchi S. Is Angelman syndrome an alternate result of del(15)(q11q13)? Am. J. Med. Genet. 1987;28:829–838. doi: 10.1002/ajmg.1320280407. doi:10.1002/ajmg.1320280407. [DOI] [PubMed] [Google Scholar]
- 33.Ledbetter D.H., Riccardi V.M., Airhart S.D., Strobel R.J., Keenan B.S., Crawford J.D. Deletions of chromosome 15 as a cause of the Prader–Willi syndrome. N. Engl. J. Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. doi:10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- 34.Williams C.A., Zori R.T., Stone J.W., Gray B.A., Cantu E.S., Ostrer H. Maternal origin of 15q11–13 deletions in Angelman syndrome suggests a role for genomic imprinting. Am. J. Med. Genet. 1990;35:350–353. doi: 10.1002/ajmg.1320350308. doi:10.1002/ajmg.1320350308. [DOI] [PubMed] [Google Scholar]
- 35.Butler M.G., Palmer C.G. Parental origin of chromosome 15 deletion in Prader–Willi syndrome. Lancet. 1983;1:1285–1286. doi: 10.1016/s0140-6736(83)92745-9. doi:10.1016/S0140-6736(83)92745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horsthemke B., Wagstaff J. Mechanisms of imprinting of the Prader–Willi/Angelman region. Am. J. Med. Genet. A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. doi:10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 37.Hulten M., Armstrong S., Challinor P., Gould C., Hardy G., Leedham P., Lee T., McKeown C. Genomic imprinting in an Angelman and Prader–Willi translocation family. Lancet. 1991;338:638–639. doi: 10.1016/0140-6736(91)90652-6. doi:10.1016/0140-6736(91)90652-6. [DOI] [PubMed] [Google Scholar]
- 38.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr, Dobyns W.B., et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. doi:10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 39.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. doi:10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 40.Bijlsma E.K., Gijsbers A.C., Schuurs-Hoeijmakers J.H., van Haeringen A., Fransen van de Putte D.E., Anderlid B.M., Lundin J., Lapunzina P., Perez Jurado L.A., Delle Chiaie B., et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur. J. Med. Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. doi:10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. doi:10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. doi:10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mefford H.C., Cooper G.M., Zerr T., Smith J.D., Baker C., Shafer N., Thorland E.C., Skinner C., Schwartz C.E., Nickerson D.A., et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenfeld J.A., Coppinger J., Bejjani B.A., Girirajan S., Eichler E.E., Shaffer L.G., Ballif B.C. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J. Neurodevelop. Disord. 2010;2:26–38. doi: 10.1007/s11689-009-9037-4. doi:10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S., et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. doi:10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K., Saeed S., Hamilton-Shield J., Clayton-Smith J., O'Rahilly S., et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. doi:10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann-Gagescu R., Mefford H.C., Cowan C., Glew G.M., Hing A.V., Wallace S., Bader P.I., Hamati A., Reitnauer P.J., Smith R., et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet. Med. 2010 doi: 10.1097/GIM.0b013e3181ef4286. (in press) [DOI] [PubMed] [Google Scholar]
- 49.Kirov G., Grozeva D., Norton N., Ivanov D., Mantripragada K.K., Holmans P., Craddock N., Owen M.J., O'Donovan M.C. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. doi:10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mefford H.C., Clauin S., Sharp A.J., Moller R.S., Ullmann R., Kapur R., Pinkel D., Cooper G.M., Ventura M., Ropers H.H., et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am. J. Hum. Genet. 2007;81:1057–1069. doi: 10.1086/522591. doi:10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagamani S.C., Erez A., Shen J., Li C., Roeder E., Cox S., Karaviti L., Pearson M., Kang S.H., Sahoo T., et al. Clinical spectrum associated with recurrent genomic rearrangements in chromosome 17q12. Eur. J. Hum. Genet. 2010;18:278–284. doi: 10.1038/ejhg.2009.174. doi:10.1038/ejhg.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ullmann R., Turner G., Kirchhoff M., Chen W., Tonge B., Rosenberg C., Field M., Vianna-Morgante A.M., Christie L., Krepischi-Santos A.C., et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum. Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. doi:10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 53.Hannes F.D., Sharp A.J., Mefford H.C., de Ravel T., Ruivenkamp C.A., Breuning M.H., Fryns J.P., Devriendt K., Van Buggenhout G., Vogels A., et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J. Med. Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. doi:10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Kovel C.G., Trucks H., Helbig I., Mefford H.C., Baker C., Leu C., Kluck C., Muhle H., von Spiczak S., Ostertag P., et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. doi:10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinzen E.L., Radtke R.A., Urban T.J., Cavalleri G.L., Depondt C., Need A.C., Walley N.M., Nicoletti P., Ge D., Catarino C.B., et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am. J. Hum. Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. doi:10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingason A., Rujescu D., Cichon S., Sigurdsson E., Sigmundsson T., Pietilainen O.P., Buizer-Voskamp J.E., Strengman E., Francks C., Muglia P., et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol. Psychiatry. 2009 doi: 10.1038/mp.2009.101. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Girirajan S., Vlangos C.N., Szomju B.B., Edelman E., Trevors C.D., Dupuis L., Nezarati M., Bunyan D.J., Elsea S.H. Genotype–phenotype correlation in Smith–Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet. Med. 2006;8:417–427. doi: 10.1097/01.gim.0000228215.32110.89. doi:10.1097/01.gim.0000228215.32110.89. [DOI] [PubMed] [Google Scholar]
- 58.Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., et al. Characterization of Potocki–Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. doi:10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrieux J., Dubourg C., Rio M., Attie-Bitach T., Delaby E., Mathieu M., Journel H., Copin H., Blondeel E., Doco-Fenzy M., et al. Genotype–phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am. J. Med. Genet. A. 2009;149A:2813–2819. doi: 10.1002/ajmg.a.33097. doi:10.1002/ajmg.a.33097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharp A.J., Selzer R.R., Veltman J.A., Gimelli S., Gimelli G., Striano P., Coppola A., Regan R., Price S.M., Knoers N.V., et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. doi:10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 61.Tassabehji M., Hammond P., Karmiloff-Smith A., Thompson P., Thorgeirsson S.S., Durkin M.E., Popescu N.C., Hutton T., Metcalfe K., Rucka A., et al. GTF2IRD1 in craniofacial development of humans and mice. Science. 2005;310:1184–1187. doi: 10.1126/science.1116142. doi:10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- 62.Shaw C.J., Withers M.A., Lupski J.R. Uncommon deletions of the Smith–Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am. J. Hum. Genet. 2004;75:75–81. doi: 10.1086/422016. doi:10.1086/422016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babcock M., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. doi:10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L., Krantz I.D., Deng Y., Genin A., Banta A.B., Collins C.C., Qi M., Trask B.J., Kuo W.L., Cochran J., et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. doi:10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 65.Oda T., Elkahloun A.G., Pike B.L., Okajima K., Krantz I.D., Genin A., Piccoli D.A., Meltzer P.S., Spinner N.B., Collins F.S., et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. doi:10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 66.Slager R.E., Newton T.L., Vlangos C.N., Finucane B., Elsea S.H. Mutations in RAI1 associated with Smith–Magenis syndrome. Nat. Genet. 2003;33:466–468. doi: 10.1038/ng1126. doi:10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 67.Antonell A., Del Campo M., Magano L.F., Kaufmann L., de la Iglesia J.M., Gallastegui F., Flores R., Schweigmann U., Fauth C., Kotzot D., et al. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams–Beuren syndrome neurocognitive profile. J. Med. Genet. 2009;47:312–320. doi: 10.1136/jmg.2009.071712. doi:10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- 68.Del Campo M., Antonell A., Magano L.F., Munoz F.J., Flores R., Bayes M., Perez Jurado L.A. Hemizygosity at the NCF1 gene in patients with Williams–Beuren syndrome decreases their risk of hypertension. Am. J. Hum. Genet. 2006;78:533–542. doi: 10.1086/501073. doi:10.1086/501073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruno D.L., Anderlid B.M., Lindstrand A., van Ravenswaaij-Arts C., Ganesamoorthy D., Lundin J., Martin C.L., Douglas J., Nowak C., Adam M.P., et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J. Med. Genet. 2010;47:299–311. doi: 10.1136/jmg.2009.069906. doi:10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- 70.Cardoso C., Leventer R.J., Ward H.L., Toyo-Oka K., Chung J., Gross A., Martin C.L., Allanson J., Pilz D.T., Olney A.H., et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller–Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 2003;72:918–930. doi: 10.1086/374320. doi:10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagamani S.C., Zhang F., Shchelochkov O.A., Bi W., Ou Z., Scaglia F., Probst F.J., Shinawi M., Eng C., Hunter J.V., et al. Microdeletions including YWHAE in the Miller–Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J. Med. Genet. 2009;46:825–833. doi: 10.1136/jmg.2009.067637. doi:10.1136/jmg.2009.067637. [DOI] [PubMed] [Google Scholar]
- 72.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. doi:10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dibbens L.M., Mullen S., Helbig I., Mefford H.C., Bayly M.A., Bellows S., Leu C., Trucks H., Obermeier T., Wittig M., et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum. Mol. Genet. 2009;18:3626–3631. doi: 10.1093/hmg/ddp311. doi:10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Helbig I., Mefford H.C., Sharp A.J., Guipponi M., Fichera M., Franke A., Muhle H., de Kovel C., Baker C., von Spiczak S., et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. doi:10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pagnamenta A.T., Wing K., Sadighi Akha E., Knight S.J., Bolte S., Schmotzer G., Duketis E., Poustka F., Klauck S.M., Poustka A., et al. A 15q13.3 microdeletion segregating with autism. Eur. J. Hum. Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. doi:10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. doi:10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stefansson H., Rujescu D., Cichon S., Pietilainen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. doi:10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ben-Shachar S., Lanpher B., German J.R., Qasaymeh M., Potocki L., Nagamani S.C., Franco L.M., Malphrus A., Bottenfield G.W., Spence J.E., et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J. Med. Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. doi:10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shinawi M., Schaaf C.P., Bhatt S.S., Xia Z., Patel A., Cheung S.W., Lanpher B., Nagl S., Herding H.S., Nevinny-Stickel C., et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. doi:10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le Pichon J.B., Bittel D.C., Graf W.D., Yu S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am. J. Med. Genet. A. 2010;152A:1300–1304. doi: 10.1002/ajmg.a.33374. doi:10.1002/ajmg.a.33374. [DOI] [PubMed] [Google Scholar]
- 81.Ewart A.K., Morris C.A., Atkinson D., Jin W., Sternes K., Spallone P., Stock A.D., Leppert M., Keating M.T. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat. Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. doi:10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 82.Gollob M.H., Jones D.L., Krahn A.D., Danis L., Gong X.Q., Shao Q., Liu X., Veinot J.P., Tang A.S., Stewart A.F., et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N. Engl. J. Med. 2006;354:2677–2688. doi: 10.1056/NEJMoa052800. doi:10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 83.Gu H., Smith F.C., Taffet S.M., Delmar M. High incidence of cardiac malformations in connexin40-deficient mice. Circ. Res. 2003;93:201–206. doi: 10.1161/01.RES.0000084852.65396.70. doi:10.1161/01.RES.0000084852.65396.70. [DOI] [PubMed] [Google Scholar]
- 84.Shiels A., Mackay D., Ionides A., Berry V., Moore A., Bhattacharya S. A missense mutation in the human connexin50 gene (GJA8) underlies autosomal dominant ‘zonular pulverulent' cataract, on chromosome 1q. Am. J. Hum. Genet. 1998;62:526–532. doi: 10.1086/301762. doi:10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rainier S., Chai J.H., Tokarz D., Nicholls R.D., Fink J.K. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am. J. Hum. Genet. 2003;73:967–971. doi: 10.1086/378817. doi:10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu L., Vranckx R., Khau Van Kien P., Lalande A., Boisset N., Mathieu F., Wegman M., Glancy L., Gasc J.M., Brunotte F., et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat. Genet. 2006;38:343–349. doi: 10.1038/ng1721. doi:10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 87.Liburd N., Ghosh M., Riazuddin S., Naz S., Khan S., Ahmed Z., Riazuddin S., Liang Y., Menon P.S., Smith T., et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith–Magenis syndrome. Hum. Genet. 2001;109:535–541. doi: 10.1007/s004390100604. doi:10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- 88.Gothelf D., Eliez S., Thompson T., Hinard C., Penniman L., Feinstein C., Kwon H., Jin S., Jo B., Antonarakis S.E., et al. COMT genotype predicts longitudinal cognitive decline and psychosis in 22q11.2 deletion syndrome. Nat. Neurosci. 2005;8:1500–1502. doi: 10.1038/nn1572. doi:10.1038/nn1572. [DOI] [PubMed] [Google Scholar]
- 89.Kurotaki N., Shen J.J., Touyama M., Kondoh T., Visser R., Ozaki T., Nishimoto J., Shiihara T., Uetake K., Makita Y., et al. Phenotypic consequences of genetic variation at hemizygous alleles: Sotos syndrome is a contiguous gene syndrome incorporating coagulation factor twelve (FXII) deficiency. Genet. Med. 2005;7:479–483. doi: 10.1097/01.gim.0000177419.43309.37. doi:10.1097/01.GIM.0000177419.43309.37. [DOI] [PubMed] [Google Scholar]
- 90.Kabuki T., Kawai T., Kin Y., Joh K., Ohashi H., Kosho T., Yachie A., Kanegane H., Miyawaki T., Oh-ishi T. A case of Williams syndrome with p47-phox-deficient chronic granulomatous disease. Nihon Rinsho Meneki Gakkai Kaishi. 2003;26:299–303. doi: 10.2177/jsci.26.299. [DOI] [PubMed] [Google Scholar]
- 91.Bisgaard A.M., Kirchhoff M., Nielsen J.E., Kibaek M., Lund A., Schwartz M., Christensen E. Chromosomal deletion unmasking a recessive disease: 22q13 deletion syndrome and metachromatic leukodystrophy. Clin. Genet. 2009;75:175–179. doi: 10.1111/j.1399-0004.2008.01113.x. doi:10.1111/j.1399-0004.2008.01113.x. [DOI] [PubMed] [Google Scholar]
- 92.Lee S.T., Nicholls R.D., Bundey S., Laxova R., Musarella M., Spritz R.A. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader–Willi syndrome plus albinism. N. Engl. J. Med. 1994;330:529–534. doi: 10.1056/NEJM199402243300803. doi:10.1056/NEJM199402243300803. [DOI] [PubMed] [Google Scholar]
- 93.Flipsen-ten Berg K., van Hasselt P.M., Eleveld M.J., van der Wijst S.E., Hol F.A., de Vroede M.A., Beemer F.A., Hochstenbach P.F., Poot M. Unmasking of a hemizygous WFS1 gene mutation by a chromosome 4p deletion of 8.3 Mb in a patient with Wolf–Hirschhorn syndrome. Eur. J. Hum. Genet. 2007;15:1132–1138. doi: 10.1038/sj.ejhg.5201899. doi:10.1038/sj.ejhg.5201899. [DOI] [PubMed] [Google Scholar]
- 94.Kumar R.A., Marshall C.R., Badner J.A., Babatz T.D., Mukamel Z., Aldinger K.A., Sudi J., Brune C.W., Goh G., Karamohamed S., et al. Association and mutation analyses of 16p11.2 autism candidate genes. PLoS ONE. 2009;4:e4582. doi: 10.1371/journal.pone.0004582. doi:10.1371/journal.pone.0004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D., et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. doi:10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klopocki E., Schulze H., Strauss G., Ott C.E., Hall J., Trotier F., Fleischhauer S., Greenhalgh L., Newbury-Ecob R.A., Neumann L.M., et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am. J. Hum. Genet. 2007;80:232–240. doi: 10.1086/510919. doi:10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Igartua C., Turner E.H., Ng S.B., Hodges E., Hannon G.J., Bhattacharjee A., Rieder M.J., Nickerson D.A., Shendure J. Targeted enrichment of specific regions in the human genome by array hybridization. Curr. Protoc. Hum. Genet., 2010 doi: 10.1002/0471142905.hg1803s66. Chapter 18, Unit 18.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanderson T.L., Best J.J., Doody G.A., Owens D.G., Johnstone E.C. Neuroanatomy of comorbid schizophrenia and learning disability: a controlled study. Lancet. 1999;354:1867–1871. doi: 10.1016/s0140-6736(99)01049-1. doi:10.1016/S0140-6736(99)01049-1. [DOI] [PubMed] [Google Scholar]
- 99.Woodberry K.A., Giuliano A.J., Seidman L.J. Premorbid IQ in schizophrenia: a meta-analytic review. Am. J. Psychiatry. 2008;165:579–587. doi: 10.1176/appi.ajp.2008.07081242. doi:10.1176/appi.ajp.2008.07081242. [DOI] [PubMed] [Google Scholar]
- 100.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. doi:10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feenstra I., Fang J., Koolen D.A., Siezen A., Evans C., Winter R.M., Lees M.M., Riegel M., de Vries B.B., Van Ravenswaaij C.M., et al. European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations (ECARUCA): an online database for rare chromosome abnormalities. Eur. J. Med. Genet. 2006;49:279–291. doi: 10.1016/j.ejmg.2005.10.131. doi:10.1016/j.ejmg.2005.10.131. [DOI] [PubMed] [Google Scholar]
- 102.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. doi:10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alkan C., Kidd J.M., Marques-Bonet T., Aksay G., Antonacci F., Hormozdiari F., Kitzman J.O., Baker C., Malig M., Mutlu O., et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. doi:10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brunetti-Pierri N., Berg J.S., Scaglia F., Belmont J., Bacino C.A., Sahoo T., Lalani S.R., Graham B., Lee B., Shinawi M., et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. doi:10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greenway S.C., Pereira A.C., Lin J.C., DePalma S.R., Israel S.J., Mesquita S.M., Ergul E., Conta J.H., Korn J.M., McCarroll S.A., et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat. Genet. 2009;41:931–935. doi: 10.1038/ng.415. doi:10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Christiansen J., Dyck J.D., Elyas B.G., Lilley M., Bamforth J.S., Hicks M., Sprysak K.A., Tomaszewski R., Haase S.M., Vicen-Wyhony L.M., et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ. Res. 2004;94:1429–1435. doi: 10.1161/01.RES.0000130528.72330.5c. doi:10.1161/01.RES.0000130528.72330.5c. [DOI] [PubMed] [Google Scholar]
- 107.Ballif B.C., Theisen A., Coppinger J., Gowans G.C., Hersh J.H., Madan-Khetarpal S., Schmidt K.R., Tervo R., Escobar L.F., Friedrich C.A., et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol. Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. doi:10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clayton-Smith J., Giblin C., Smith R.A., Dunn C., Willatt L. Familial 3q29 microdeletion syndrome providing further evidence of involvement of the 3q29 region in bipolar disorder. Clin. Dysmorphol. 2010;19:128–132. doi: 10.1097/MCD.0b013e32833a1e3c. doi:10.1097/MCD.0b013e32833a1e3c. [DOI] [PubMed] [Google Scholar]
- 109.Digilio M.C., Bernardini L., Mingarelli R., Capolino R., Capalbo A., Giuffrida M.G., Versacci P., Novelli A., Dallapiccola B. 3q29 microdeletion: a mental retardation disorder unassociated with a recognizable phenotype in two mother-daughter pairs. Am. J. Med. Genet. A. 2009;149A:1777–1781. doi: 10.1002/ajmg.a.32965. doi:10.1002/ajmg.a.32965. [DOI] [PubMed] [Google Scholar]
- 110.Willatt L., Cox J., Barber J., Cabanas E.D., Collins A., Donnai D., FitzPatrick D.R., Maher E., Martin H., Parnau J., et al. 3q29 microdeletion syndrome: clinical and molecular characterization of a new syndrome. Am. J. Hum. Genet. 2005;77:154–160. doi: 10.1086/431653. doi:10.1086/431653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lisi E.C., Hamosh A., Doheny K.F., Squibb E., Jackson B., Galczynski R., Thomas G.H., Batista D.A. 3q29 interstitial microduplication: a new syndrome in a three-generation family. Am. J. Med. Genet. A. 2008;146A:601–609. doi: 10.1002/ajmg.a.32190. doi:10.1002/ajmg.a.32190. [DOI] [PubMed] [Google Scholar]
- 112.Doornbos M., Sikkema-Raddatz B., Ruijvenkamp C.A., Dijkhuizen T., Bijlsma E.K., Gijsbers A.C., Hilhorst-Hofstee Y., Hordijk R., Verbruggen K.T., Kerstjens-Frederikse W.S., et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader–Willi critical region, possibly associated with behavioural disturbances. Eur. J. Med. Genet. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. doi:10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 113.van Bon B.W., Mefford H.C., Menten B., Koolen D.A., Sharp A.J., Nillesen W.M., Innis J.W., de Ravel T.J., Mercer C.L., Fichera M., et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J. Med. Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. doi:10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J., et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. doi:10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szafranski P., Schaaf C.P., Person R.E., Gibson I.B., Xia Z., Mahadevan S., Wiszniewska J., Bacino C.A., Lalani S., Potocki L., et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum. Mutat. 2010;31:840–850. doi: 10.1002/humu.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shinawi M., Liu P., Kang S.H., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A., et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. doi:10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCarthy S.E., Makarov V., Kirov G., Addington A.M., McClellan J., Yoon S., Perkins D.O., Dickel D.E., Kusenda M., Krastoshevsky O., et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. doi:10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Greco D., Romano C., Reitano S., Barone C., Benedetto D.D., Castiglia L., Fichera M., Galesi O., Zingale M., Buono S., et al. Three new patients with dup(17)(p11.2p11.2) without autism. Clin. Genet. 2008;73:294–296. doi: 10.1111/j.1399-0004.2007.00959.x. doi:10.1111/j.1399-0004.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 119.Shprintzen R.J. Velocardiofacial syndrome and DiGeorge sequence. J. Med. Genet. 1994;31:423–424. doi: 10.1136/jmg.31.5.423-b. doi:10.1136/jmg.31.5.423-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ensenauer R.E., Adeyinka A., Flynn H.C., Michels V.V., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am. J. Hum. Genet. 2003;73:1027–1040. doi: 10.1086/378818. doi:10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ou Z., Berg J.S., Yonath H., Enciso V.B., Miller D.T., Picker J., Lenzi T., Keegan C.E., Sutton V.R., Belmont J., et al. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet. Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. doi:10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- 122.Yobb T.M., Somerville M.J., Willatt L., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am. J. Hum. Genet. 2005;76:865–876. doi: 10.1086/429841. doi:10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ben-Shachar S., Ou Z., Shaw C.A., Belmont J.W., Patel M.S., Hummel M., Amato S., Tartaglia N., Berg J., Sutton V.R., et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am. J. Hum. Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. doi:10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coppinger J., McDonald-McGinn D., Zackai E., Shane K., Atkin J.F., Asamoah A., Leland R., Weaver D.D., Lansky-Shafer S., Schmidt K., et al. Identification of familial and de novo microduplications of 22q11.21–q11.23 distal to the 22q11.21 microdeletion syndrome region. Hum. Mol. Genet. 2009;18:1377–1383. doi: 10.1093/hmg/ddp042. doi:10.1093/hmg/ddp042. [DOI] [PMC free article] [PubMed] [Google Scholar]