Abstract
Allele-specific DNA methylation (ASM) and allele-specific gene expression (ASE) have long been studied in genomic imprinting and X chromosome inactivation. But these types of allelic asymmetries, along with allele-specific transcription factor binding (ASTF), have turned out to be far more pervasive—affecting many non-imprinted autosomal genes in normal human tissues. ASM, ASE and ASTF have now been mapped genome-wide by microarray-based methods and NextGen sequencing. Multiple studies agree that all three types of allelic asymmetries, as well as the related phenomena of expression and methylation quantitative trait loci, are mostly accounted for by cis-acting regulatory polymorphisms. The precise mechanisms by which this occurs are not yet understood, but there are some testable hypotheses and already a few direct clues. Future challenges include achieving higher resolution maps to locate the epicenters of cis-regulated ASM, using this information to test mechanistic models, and applying genome-wide maps of ASE/ASM/ASTF to pinpoint functional regulatory polymorphisms influencing disease susceptibility.
INTRODUCTION
Genome sequencing, expression profiling and now genome-wide mapping of epigenetic markings have been huge advances that have brought us into the so-called post-genomic era. Equally important, we now have saturating genome-wide maps of common DNA polymorphisms in humans, and the concept of haplotypes has been fully developed and widely applied. Going forward, the fields of genetics and epigenetics are starting to capitalize on this basic groundwork to explore allele-specific phenomena at unprecedented levels of detail. Realizing that this research area will continue to expand rapidly, I take this opportunity to survey the current landscape, particularly focusing on the role of cis-acting DNA polymorphisms in setting up allele-specific DNA methylation (ASM) and allele-specific gene expression (ASE).
ALLELIC ASYMMETRIES ARE STRONG AT IMPRINTED LOCI BUT IMPRINTED GENES ARE RARE
Genomic or parental imprinting produces strong ASM and ASE in a parent-of-origin-dependent manner. The imprint, which is an extremely potent dose-regulating mechanism, is purely epigenetic and, strikingly, is completely erased and reset each time the allele passes through the germline. While one can find more optimistic projections, the number of known imprinted genes appears to be reaching an asymptote at around 100, or <1% of the mammalian gene repertoire (1). Imprinting is a non-Mendelian phenomenon par excellence, and this relative rarity of imprinted genes is completely consistent with the overall success of Mendel's laws in human and mouse genetics. It is also consistent with classical experiments using mice carrying Robertsonian chromosomal translocations, which showed that only some, not all, whole chromosome uniparental disomies produce abnormal phenotypes. These elegant genetic studies suggested early on that some chromosomes may be devoid of imprinted genes (2). Nonetheless, imprinted genes are crucial for normal mammalian development (1,3), and mechanistic studies of imprinting have laid an important and impressive groundwork for understanding allele-specific gene regulation. Similarly, methods developed to study imprinting are now the workhorse tools for analyzing the types of non-imprinted allele-specific phenomena that are the main focus of this review. Examples include bisulfite conversion of DNA followed by PCR spanning an SNP and cloning of the products to reveal ASM, and comparing allelic representation in PCR products from cDNA versus gDNA to score ASE. Genome-wide scanning methods like methylation analysis on SNP arrays (MSNP) were initially developed with the hope of finding additional imprinted genes, but instead uncovered the novel and more widespread phenomenon of non-imprinted ASM (4). Other new approaches that have been published for analyzing imprinted domains, such as chromatin immunoprecipitation (ChIP)–chip or ChIP–Seq, to search for ‘overlapping’ activating (methylated H3K4) and repressive (methylated H3K9) histone modifications that in fact represent the two oppositely poised (active/inactive) parental alleles (5), may also prove useful for finding loci with non-imprinted allelic asymmetries. As I discuss more in a later section, prior work on the mechanisms of genomic imprinting will also likely be relevant for understanding the mechanisms that produce non-imprinted allelic asymmetries.
CIS-REGULATED ALLELIC ASYMMETRIES ARE COMMON
Allelic asymmetries are now recognized as very common at non-imprinted loci. Here I consider three related classes of asymmetry affecting non-imprinted genes: allele-specific expression of mRNAs and non-coding RNAs (ASE), ASM, allele-specific chromatin modifications and transcription factor binding (ASTF). A fourth class, random monoallelic expression and DNA methylation, is also important for gene regulation (6), but it is not covered here. ASE refers to asymmetric mRNA or non-coding RNA expression from the two alleles; alternative abbreviations in the literature include AE and AI (allelic imbalance). ASE is scored in heterozygous samples by comparing the representation of the two alleles of a given SNP in genomic DNA (by definition 50:50) to their representation in the corresponding mRNA or non-coding RNA, generally assayed as cDNA. This type of gDNA/cDNA comparison has now been carried out genome-wide by many laboratories, first using SNP arrays and more recently massively parallel NextGen sequencing. A clear conclusion from all studies is that ASE is quite frequent across the human genome, and it usually reflects the presence of a cis-acting regulatory polymorphism or regulatory haplotypes near or encompassing the gene (references in Table 1). In contrast, these studies, and expression quantitative trait locus (eQTL) screens discussed below, have shown that ASE due to trans-acting genetic or epigenetic mechanisms is relatively rare. ASE due to cis-acting regulatory polymorphisms is typically a quantitative phenomenon; it does not produce ‘all or none’ monoallelic expression but instead results in a bias in the ratio of transcripts from the two alleles. ASE due to cis-effects at a given locus is therefore a continuous variable, and thoughtful statistical approaches are essential for setting cutoffs and making meaningful statements about its frequency (7). In contrast, when ASE is produced by other mechanisms, such as genomic imprinting or, even more rarely, heterozygous germline mutations causing nonsense-mediated mRNA decay (7,8), the allelic bias can be very strong, with close to monoallelic expression.
Table 1.
Methods and conclusions from screens for ASE at non-imprinted loci
| Tissues or cell types (n) | Primary screening methods and validations | Findings and conclusions | References |
|---|---|---|---|
| LCL (81) | Illumina ASE Cancer Panel (1380 genes) with cDNA versus gDNA probes; validations by Sanger sequencing (cDNA versus gDNA) | Using criteria of 1.5-fold differential expression, ASE is widespread, affecting the expression of 20% of genes in LCLs. There can be discrepancies between lists of cis-acting polymorphisms from direct ASE measurements versus eQTL approaches | (53) |
| LCL (100) | Illumina Golden Gate assays for 2117 SNPs; comparison of gDNA versus cDNA; validations by Sanger sequencing | Cis-regulated ASE is widespread by lenient criteria, but is less frequent when criteria are a strong allelic bias and consistency among individuals. Rare extreme ASE can be due to mutations causing nonsense-mediated mRNA decay (e.g. BRCA1). | (7); see also (8) |
| LCL (53), osteoblasts (2), fibroblasts (3) | Illumina 1M BeadArrays with cDNA versus gDNA probes; validations by Sanger sequencing | SNPs linked to cis-acting variants explain 45% of loci with ASE in LCLs; imprinted genes account for only 0.1% of ASE. Significant but imperfect correlations found between direct ASE measurements and eQTL data. Several loci associated with autoimmune diseases (e.g. C8orf13-BLK) show cis-regulated ASE in LCLs. | (16) |
| LCL (55) and osteoblasts (55); sample pooling strategy | RNA-Seq (cDNA/gDNA) from pooled samples; validations by Sanger sequencing of gDNA versus cDNA in individual samples | Positive loci from direct analysis of ASE show a significant, but not complete, overlap with positive loci from eQTL screens. Multiple genes previously implicated as disease-associated in GWAS show ASE in LCLs | (12) |
| LCL (1), fibroblasts (2), keratinocytes (1), human embryonal stem cells (hESC) (4) | RNA-Seq (cDNA/gDNA) of padlock probe amplicons (27 000 SNPs in 10 345 genes), from multiple cell lines derived from single donors | Much of the observed inter-sample variation in ASE was accounted for by genetic background, presumably reflecting cis-effects of regulatory polymorphisms. Tissue-specific variations in ASE were also found. ASE was stable across independently cloned hESC lines. | (54) |
| T-cells (four pairs of samples; basal and activated by anti-CD3/CD28) | RNA-Seq (cDNA/gDNA); validations by quantitative cloning and sequencing and direct Sanger sequencing | Approximately 5% of HapMap-validated exonic SNPs tag transcripts with ASE in T-cells. Indel polymorphisms can be a confounder in ASE assays. Several genes associated with autoimmune diseases from GWAS data show ASE in T-cells (e.g. CD6, IL21R). | (19) |
| LCL (63 and 69 individuals from HapMap; two ethnicities) | RNA-Seq (cDNA/gDNA); similar approaches taken in these two studies | Compared with microarray-based approaches in the same cell type, RNA-Seq at sufficient depth is more sensitive in detecting ASE. SNPs affecting allele-specific mRNA splicing can be evaluated | (10,11) |
Examples from the recent literature are shown. The table is not comprehensive; additional studies, including earlier work, are cited in other reviews (42,51,52). All experiments include internal statistical validations of the microarray data; secondary validations refer to downstream assays by independent methods. The listed cells and tissues are of human origin.
The term eQTL is related to ASE, but not synonymous. The typical strategy for mapping eQTLs is to correlate SNP genotypes with separate data from mRNA expression profiling in large numbers of individuals. Standard microarray-based methods are easily adapted for this purpose and lend themselves to high genomic coverage and high sample throughput (Table 2). Homozygotes for the minor and major alleles at each SNP are highly informative for this type of eQTL mapping, while they are not informative for direct measurements of ASE, in which the allelic expression bias can only be examined in heterozygotes. There has been some discussion of the relative sensitivity and accuracy of these two approaches. As a tool for finding and validating regulatory SNPs measuring ASE has the major advantage of being internally controlled. It directly compares expression of the two alleles within one individual, rather than measuring associations of SNP genotypes with net expression of the gene across subjects, which can suffer from the limited precision of microarray assays and unpredictable effects of environmental influences in each individual. But both approaches are valid and assessing correlations of SNPs and haplotypes with net transcript levels gets more directly at the biologically relevant outcome—namely net gene expression. Further, for technical reasons, mapping eQTLs has allowed certain types of questions to be answered faster than mapping ASE directly. For example, Stranger and colleagues used transcriptome profiling in lymphoblastoid cell lines (LCLs) from individuals included in HapMap to sort out the relative contributions of SNPs versus DNA copy number variants (CNVs) to inter-individual differences in gene expression. They found that, while SNPs and CNVs both contributed, the majority of genotype-dependent expression variation (84%) in these cells was attributable to SNPs, which were not acting as surrogates for the CNVs (9). With available methods, the lists of cis-regulated genes obtained by ASE mapping versus eQTL analyses are significantly but not perfectly overlapping (references in Tables 1 and 2). One assumes that the overlap will improve as the methods are further refined, and a recent study using NextGen sequencing provided some support for this notion (10). NextGen sequencing is already increasing the information content of genome-wide studies dealing with ASE. In addition to possibly giving more linear estimates of the abundance of major transcripts, mapping ASE by RNA-Seq has revealed cis-acting SNPs in splice donor and acceptor sequences that affect exon usage in alternatively spliced transcripts (10,11), and it can facilitate the analysis of intronic SNPs in primary RNA transcripts, which substantially increases the number of informative biological samples (12).
Table 2.
Methods and conclusions from screens for cis-acting eQTLs
| Tissues or cell types (n) | Primary screening methods and validations | Findings and conclusions | References |
|---|---|---|---|
| LCL [60; European Caucasian (CEU) individuals from HapMap] | RNA profiling on custom Illumina BeadArrays (630 genes); correlations with HapMap SNP genotypes | From 374 informative genes interrogated in 60 individuals, genetic variants with significant cis-effects on transcript levels were found for 10–40 genes. Trans-acting effects are rare. Groups of multiple SNPs that are cis-associated with eQTLs are often clustered in haplotype blocks | (55) |
| LCL (210; HapMap individuals) | Illumina Sentrix Human-6 Expression BeadChip (14 925 transcripts); SNP genotypes from HapMap and CNVs from bacterial artificial chromosome (BAC) array CGH | Large-scale (typically >100 kb) CNVs are associated with about 10–25% as many gene expression phenotypes as captured by ∼700 000 SNPs. The cis-effects of large CNVs on eQTLs seem to extend over greater distances on average than the cis-effects from SNPs. Influences of smaller CNVs remain to be studied | (9) |
| LCL (HapMap samples; 60 CEU, 41 Chinese from Beijing and 41 Japanese from Tokyo) | Expression profiling on Affy Genome Focus Array; genotypes from HapMap | Expression QTLs differ between European- and Asian-derived populations by t-tests for ∼25% of genes. With a more stringent cutoff of 2-fold differential expression, the frequency is ∼1% of genes. Most of the variation is due to frequency differences at cis-linked regulatory polymorphisms, or more rarely CNVs. | (56); see also (57) |
| PBMC (1240) from 30 extended families | Expression profiling (Illumina WG-6); microsatellite genotyping to test heritability of eQTLs by cis- versus trans-mechanisms | Proportion of genes that are cis-regulated by nearby polymorphisms (eQTLs strongly linked to the closest microsatellite marker) is ∼7% in PBMC. Trans-regulation is rare. In an example of a cis-regulated gene, VNN1, the promoter contains an SNP that alters GTF (Sp1) binding | (58) |
| PBL (1002); adipose tissue (673) | Expression profiling on custom 23K microarrays; microsatellite genotyping for 1732 markers in three-generation pedigrees | By tracking inheritance, the authors found cis-acting eQTLs for 9.4% of loci in blood cells, and 5.8% in adipose tissue. About half of the eQTLs found in PBL were shared by adipose tissue | (59) |
| Three cell types (LCL, fibroblasts and T-cells) from each of 75 individuals. | RNA expression profiled on WG-6 v3 BeadChips (Illumina); SNP genotyping on Illumina 550K SNP arrays; validations of ASE by allele-specific MS (Sequenom) | From 69 to 80% of regulatory variants were found to act in a cell-type-specific manner; cell-type-specific eQTLs were found at larger distances from genes and at lower effect size, compared with universal eQTLs, suggesting that distant regulatory elements such as enhancers or insulators might be involved | (17) |
| Liver (427) | RNA profiling on Agilent array (34 266 genes including non-coding RNAs). Genotyping on Affy 500K and Illumina 650Y SNP arrays | Many eQTLs (∼10% of genes surveyed) identified as linked to cis-acting polymorphisms in the liver. The results support RPS26 and not ERBB3 as a susceptibility gene for type 1 diabetes on Hsa12 and identify SORT1 and CELSR2 as cis-regulated susceptibility genes for coronary artery disease and plasma LDL cholesterol levels | (20) |
| Monocytes (1490) | mRNA profiled on Illumina HT-12 BeadChips; genotyping on Affy 6.0 SNP arrays | This study had a very large sample size. Among 12 808 genes expressed in monocytes, 2745 eQTLs were detected (21%), the large majority (90%) being cis-modulated. For several loci, there was evidence for an gene–environment interaction of smoking history with eQTL strength | (21) |
| LCL (109; CEU individuals from HapMap) | Statistical approach to extract causal relationships in overlays of GWAS signals with eQTLs | Regulatory trait concordance (RTC) accounts for local LD structure and integrates eQTLs and GWAS results to reveal disease associations that are due to cis-acting variants | (14) |
Examples from the recent literature are shown. This is a very active research area and the table is not comprehensive. All experiments include internal statistical validations of the microarray data; secondary validations refer to downstream assays by independent methods. Cells and tissues are of human origin.
CIS-REGULATED ASE IS TISSUE-SPECIFIC AND INDIVIDUAL-SPECIFIC
All of the allele-specific phenomena discussed here are tissue-specific, so choosing the appropriate tissues and cell types is crucial for getting useful information. Among more than 16 recent large-scale studies of ASE, either by direct measurements or by eQTL analyses, about half utilized exclusively LCLs (Tables 1 and 2). This reliance on a renewable source of DNA and RNA is understandable in the methods development phase, and it allowed several groups to rapidly take advantage of available dense SNP genotyping data for these immortalized cell lines from CEPH and the HapMap (now 1000 Genomes) project. Efficient methodology developed using LCLs as the source of RNA included not just microarray-based and NextGen sequencing protocols, but also the essential statistical methods for dealing with the data, including methods for overlaying eQTL and ASE maps with GWAS data to extract functional conclusions (13,14). However, there are well-documented problems with clonal selection in established LCLs (15). Methods to monitor and correct for this problem have been developed (16), but it is still gratifying to see that all groups working in this area are now analyzing primary human cell types. As listed in Tables 1 and 2, some impressive studies of ASE or eQTLs have now been published using osteoblasts, non-transformed fibroblasts, keratinocytes, human ES cells or induced pluripotent stem cells (iPS), primary peripheral blood mononuclear cells, resting and PHA-stimulated T-lymphocytes, monocytes, adipose tissues and normal liver samples. Some studies are finding substantial overlap (up to 30% of eQTLs) between different types, including LCLs, suggesting the existence of a category of ‘universal eQTLs’, but more than half of all eQTLs seem to be private to specific tissues (17). Data from these pioneering studies on LCLs and primary cells and tissues are all in public repositories and the resulting maps of eQTLs and ASE will be a valuable adjunct to studies of human gene regulation and genetic variation for years to come.
The necessity of using well-chosen cells and tissues is not just academic; from the studies to date, it is already clear that the overlap of cis-regulated genes with GWAS signals will make sense only in a tissue-specific context. For example, data from analyzing LCLs and T-cells have shown overlap mainly with GWAS signals for autoimmune diseases, while data from liver samples have shown overlap with GWAS signals for lipid profiles, Type II diabetes and coronary artery heart disease (16,18–20).
So, how many genes show ASE and/or eQTLs in specific types of human cells or tissues? This number can be a moving target, as it depends on cell type and the stringency of the cutoffs for defining a significant allelic bias. For example, Chakravarti and colleagues developed an unbiased statistical approach to establish the most lenient cutoff for calling an observation ASE (7). As defined by their approach, ASE was found to be quite widespread in LCLs: 19.6% of heterozygotes at 78% of SNPs at 84% of genes demonstrated ASE in these immortalized cell lines, with a mean allelic bias of 1.6-fold. As listed in Tables 1 and 2, other studies of LCLs have come to similar figures, even up to 30% of genes surveyed (16). Estimates in primary tissues have tended to be somewhat lower, but still of the order of 10–20% of genes are affected by this phenomenon (Tables 1 and 2). In evaluating the frequency of ASE, it is crucial to carry out validations of the microarray or NextGen sequencing data using independent gene-specific molecular assays, as has been done in some, but not all, of the published studies. Without independent validations, it is impossible to know the false-positive rates of the initial screens. Lastly regarding the frequency and strength of ASE and eQTLs in the human genome, it is clear that the extent of the allelic bias can vary substantially among individuals, some of whom are expected to share the same genotype (7,21). Some of this variation may be influenced by trans-acting loci or by the environment. Most environmental effects on ASE are probably not major when considered singly, but it has been shown that single types of exposures that have very strong biological impacts, e.g. cigaret smoking, can produce quantitative effects on ASE that are detectable when the epidemiological study is sufficiently powered (21). All of these themes are also relevant to ASM, which I consider next.
ASM IS ALSO COMMONLY DUE TO CIS-EFFECTS OF GENETIC POLYMORPHISMS
In 2008, Kerkel et al. (4) used the MSNP method, pre-digestion of genomic DNA by methylation-sensitive restriction enzyme(s) followed by probe synthesis and hybridization of SNP arrays (Fig. 1), to examine ASM in several human tissues, including peripheral blood leucocytes (PBL), hematopoietic stem cells and placenta. Their study was designed to detect new examples of imprinted genes, but they found only old examples of such genes, instead identifying numerous examples of previously unsuspected ASM at loci outside of imprinted regions. Most of these examples of ASM outside of imprinted genes showed a strong correlation with local SNP genotypes, indicating cis-regulation of the phenomenon. The observed ASM, which was validated by pre-digestion PCR/RFLP assays and bisulfite sequencing, was found to be tissue-specific, and for a given positive locus, it was seen in 40–95% of heterozygotes. That paper was quickly followed by other reports (Table 3), including one by Zhang et al. (22), who used bisulfite sequencing of PBL DNA to document SNP-dependent ASM in CpG-rich sequences in or near four genes on human chromosome 21, and larger genome-wide studies by Schalkwyk et al. (23) who used MSNP on high-density Affymetrix 6.0 SNP arrays to profile ASM in blood leukocytes and buccal cells, validating their results by bisulfite conversion of genomic DNA followed by SNaPshot assays. Hellman and Chess (24), who had previously published a method similar to MSNP to study DNA methylation on human X-chromosomes, went on to use 500K SNP arrays to study autosomal loci and found that ∼10% of SNP-tagged regions have genotype-dependent ASM in LCLs (25). Extending these types of observations to a well-controlled mouse model system, Schilling et al. (26) did a genome-wide analysis in macrophages from two common laboratory strains (C57BL/6 and BALB/c) and in F1 hybrid offspring. They found that ASM was frequent and widely distributed across the genome, and that the allelic asymmetry in DNA methylation was largely attributable to cis-acting polymorphisms. In another study, Lee and coworkers (27) carried out 500K MSNP on human LCLs and esophageal tissues (normal and cancerous) and observed that methylation profiles are individual-specific as well as tissue-specific, suggesting an effect of genetic background on CpG methylation at many loci. ASM per se was not scored in their study but might be extractable from their primary data, which has been deposited at NCBI. From all of these studies, we know that when ASM is found, it can vary from a highly localized asymmetry in methylation affecting only one or several CpGs to examples in which a large number of contiguous CpGs are coordinately affected. Most examples of ASM affect DNA sequences outside of CpG islands, but there are rare examples in which even large CpG-dense islands can show this phenomenon (22). The region shown in Figure 1 is a typical example of cis-regulated ASM affecting a moderate-sized cluster of CpG sites in an intergenic CG-rich region that is not long enough or CG-dense enough to qualify as an island.
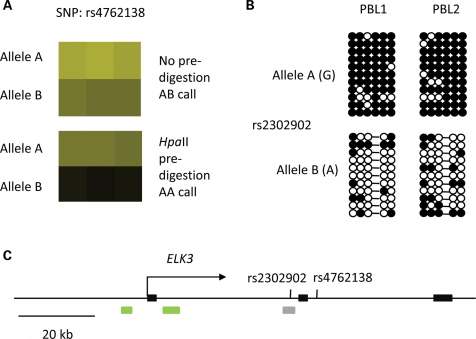
Figure 1.
Example of primary data and validations showing non-imprinted ASM. (A) Primary data from MSNP on Affymetrix 6.0 arrays. Pre-digestion of the genomic DNA (PBL sample) with the methylation-sensitive restriction enzyme HpaII prior to linker ligation and PCR for probe synthesis leads to a dropout of the B-allele, suggesting that this allele is relatively hypomethylated. (B) Validation by bisulfite conversion of genomic DNA followed by PCR, cloning and sequencing of multiple clones. Black circles are methylated CpG dinucleotides; white circles are unmethylated CpGs; the dash indicates a CpG SNP in one individual. The two alleles are distinguished in the group of clones by an SNP (rs2302902) that is not destroyed by the bisulfite conversion. Using pre-digestion/PCR assays, the hypermethylated allele was the A-allele at rs4762138 in each of 11 informative PBL samples, consistent with cis-regulation of DNA methylation. (C) Map of the ELK3 gene, containing the intragenic index SNP on the microarrays (rs4762138) and the nearby SNP (rs2302902) that was utilized for distinguishing the alleles in the bisulfite sequencing. The region with ASM subjected to sequencing (gray bar) is CG-rich but is not a CpG island; two CpG islands are near the gene promoter (green bars). Black rectangles are exons 1 and 2. The functional role of the allelic asymmetry at this position in the ELK3 gene is under investigation. Unpublished data are from K. Kerkel and B.T.
Table 3.
Methods and conclusions from screens for ASM and mQTLs at non-imprinted loci
| Tissues or cell types (n) | Primary screening methods and validations | Findings and conclusions | References |
|---|---|---|---|
| PBL (6); placenta (3); other normal tissues (7) | MSNP Affy 50K/250K; validations by pre-digestion/RFLP assays and bisulfite-Seq in larger series of individuals; ASE tested for selected loci | ASM at non-imprinted loci is a recurrent phenomenon in humans. At 12/16 validated examples (75%), the ASM was strongly correlated to SNP genotypes. ASM was found to be tissue-specific and for a given locus, it was seen in 40–95% of heterozygotes. For some genes, the ASM is associated with ASE (e.g. VNN1, CYP2A7) | (4) |
| Macrophages from B6 ×BALB/c F1 mice | methylation-dependent immunoprecipitation (MeDIP) on tiling arrays; validations by bisulfite-MS and bisulfite-Seq | Non-imprinted genes with ASM can be identified in F1 hybrid mice, and the majority (five of six examples validated by bisulfite-Seq) are regulated in cis by strain-specific genetic polymorphisms | (26) |
| PBL (20) | Targeted bisulfite-Seq (16 amplicons on Hsa21); validations by HpaII pre-digestion/Seq | ASM was found in several CGIs (∼10%) on Hsa21. For one example (C21orf81), the ASM was associated with ASE. ASM was not seen in all heterozygous individuals (95–13% depending on locus) | (22) |
| PBL (30); esophageal carcinoma (30); normal esophagus (30) | MSNP Affy 500K | Methylation profiles are individual-specific as well as tissue-specific, suggesting an effect of genetic background on CpG methylation at many loci | (27) |
| LCL (12); PBL (2) | MSNP Affy 500K, with MSRE digestions; validations by bisulfite-Seq | ∼10% of SNP-tagged regions have ASM in LCLs; mostly genotype-dependent. There is a weak but detectable short-range effect of CpG SNPs on ASM at adjacent non-polymorphic CpGs | (25) |
| Cell lines; ES, iPS and fibroblast (16) | Targeted bisulfite-Seq (padlock probes); ∼2500 promoter regions | ∼30% of SNP-tagged regions have ASM in clonal cell lines, using a lenient cutoff of >0.1 methylation frequency difference. There is an increased frequency of CpG SNPs near (non-polymorphic) CpGs with ASM | (30) |
| PBL (10); buccal cells (10) | MSNP Affy 6.0 (1 M) arrays; validations by bisulfite-Seq in index cases and by MS-SNuPE in a larger series of individuals. ASE was also investigated. | Non-imprinted ASM, regulated in cis by nearby DNA polymorphisms, is widespread (∼3.5% of SNP-tagged regions using a lenient cutoff of >0.1 methylation frequency difference). At some loci, the genotype-dependent ASM affects only a few closely spaced CpGs. For some genes, the ASM is strongly correlated with ASE | (23) |
| Cerebellar cortex (153) | Methylation on Illumina Infinium 27K; fractional methylation cross-tabulated to SNPs and mRNA expression; validations by bisulfite-Pyroseq | Cis-correlations of SNPs with CpG methylation in promoter regions can be identified by an mQTL approach and occur over distances equivalent to haplotype blocks (up to 100 kb). ∼13% of SNPs that are highly correlated with CpG methylation are also associated with genotype-dependent mRNA expression | (28) |
| Brains (150 individuals; four regions assayed from each brain) | Methylation on Illumina 27K Infinium arrays; SNP genotyping on Illumina HumanHap550 BeadChips; RNA expression on Illumina HumanRef-8 BeadChips | Both eQTLs and mQTLs were found to be mostly cis-regulated. There was substantial sharing of eQTLs and mQTLs across the four brain regions. A low but significant proportion, about 5% of genes examined, showed correlations between mQTLs and eQTLs | (29) |
Examples from the recent literature are shown. Additional studies are discussed in the text and in another review (42). All experiments include internal statistical validations of the microarray data; secondary validations refer to downstream assays by independent methods. Cells and tissues are of human origin except where otherwise stated.
A methylation QTL (mQTL) approach, analogous in design to prior studies of eQTLs, has also been quite successful, with two groups applying this strategy to human brain regions and finding strong evidence for widespread cis-regulation of DNA methylation patterns (28,29). NextGen sequencing of genomic DNA after bisulfite conversion can also be useful for analyzing ASM, as recently shown in a proof-of-principle study by Shoemaker et al. (30), and as per-sample costs go down, this approach can be expected to eventually supersede microarray-based methods.
In all of these studies in human cells and tissues, when sequence-dependent ASM was found at a given locus, its dependance on the genotype at closely adjacent SNP(s) was close to absolute. ASM is linked to ASE at some but not all loci; in the Schalkwyk et al. study, more than 150 ASM-associated SNPs, distributed across each of the human chromosomes, were found to be significantly associated with the expression of nearby genes. The frequency with which ASM is associated with ASE will likely depend on the tissue being examined and the methods and platforms used; the two mQTL studies in human brain tissues gave estimates of 5 and 13% of mQTLs associating strongly with eQTLs (28,29). Whether these estimates might be somewhat on the low side due to cell-type heterogeneity in brain tissues will no doubt be answered by future studies using purified neurons and glial cells. Along these same lines, the theme of individual specificity from studies of ASE and eQTLs also pertains, quite strikingly, to ASM and mQTLs (4,23). Is individual specificity of ASM a type of incomplete penetrance due to unpredictable trans-acting and environmental influences, or does it reflect our incomplete knowledge of the precise haplotypes surrounding each index SNP in each individual? The answers might emerge from genetic epidemiological studies including exposure information, and improvements in methods for ascertaining long-range haplotypes with direct information on phasing of SNPs and other DNA polymorphisms in each individual.
POSSIBLE MECHANISMS OF CIS-REGULATED ASE AND ASM
Polymorphisms affecting transcription factor binding
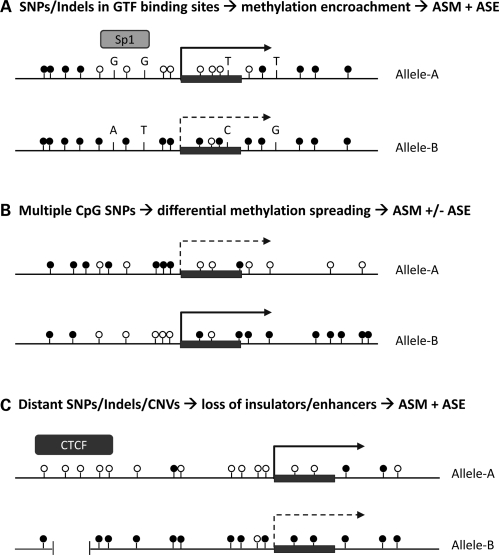
It seems a safe bet a priori that many of these genetic effects on ASE and ASM will in the end prove to be due to allele-specific affinity of DNA-binding proteins (transcription factors in the broad sense) for critical polymorphic cis-acting regulatory elements. As perhaps the most obvious example, starting with early work done by the Cedar and Turker laboratories and now nicely followed up by others, general transcription factors (GTFs) that bind to gene promoters have long been studied as candidates for playing an active role in protecting unmethylated CpG islands from a poorly understood but experimentally measureable process of ‘methylation encroachment’ from their highly methylated flanking sequences (31–35). According to the model in Figure 2A, SNPs or indels in the recognition sequences for GTFs like Sp1 could in principle lead to altered GTF binding, followed by methylation encroachment on one of the two alleles. Importantly, this model does not require that the GTF itself have methylation-dependent binding, simply that it protects the promoter from methylation encroachment. In fact, Boumber et al. (36) recently showed that a 12 bp indel polymorphism in a Sp1 GTF-binding site in the promoter of the RIL gene affects the propensity of this gene to become methylated in human leukemias. This does not necessarily imply that such a mechanism accounts for instances of ASM in normal cells, but it offers a good rationale for now performing systematic genome-wide search for ASM around polymorphic GTF-binding sites.
Figure 2.
Possible mechanisms of cis-regulated ASE and ASM. The cis-acting effects of sequence polymorphisms can be either short-range (A and B) or long-range (C). Details of each model are discussed in the text. Working out the relevance of each mechanism will depend on developing efficient methods for high-resolution mapping of allelic asymmetries and for determining long-range haplotypes in large collections of primary tissues. Black circles, methylated CpG dinucleotides; white circles, unmethylated CpGs; dark gray rectangle and arrow, first exon of the gene.
Polymorphisms affecting CpG dinucleotides
Essentially, all DNA methylation in humans occurs in CpG dinucleotides. By definition, ASM is measured at non-polymorphic CpG sites, but the methylation status of such sites can be influenced, at least in theory, by allele-specific differences in the density of CpGs in the surrounding local DNA sequence. Thus, another mechanistic model has SNPs that create or delete CpGs (‘CpG SNPs’) influencing the propensity of neighboring non-polymorphic CpGs to become cytosine-methylated. In regions of the DNA where overall CpG density is high and CpG SNPs are abundant, one can imagine more efficient spreading of methylation through the allele that has a higher preservation of CpGs, perhaps by more efficient cooperative binding (37) of the enzymes and cofactors in the methylation machinery (Fig. 2B). Alternatively, a subset of CpG SNPs could influence the binding of specific transcription factors, either positively or negatively, similar to the model in Figure 2A. Intriguingly, two recent genome-wide surveys of ASM have noted a small but statistically significant excess of CpG SNPs near loci with ASM (25,30).
Polymorphisms affecting insulators and long-range chromosome structure
The roles of insulator elements have been well studied at classical model loci like the globin genes, and also at several different imprinted loci where they show parent-of-origin-dependent ASM (1,38,39). There are already precedents for naturally occurring genetic lesions in such sequences within the human genome, for example, micro-deletions in the insulator located upstream of the imprinted H19 gene, which lead to over-expression of IGF2 and cause some cases of the Beckwith–Wiedemann overgrowth syndrome (40). As shown in Figure 2C, qualitative or quantitative alterations in insulator function due to SNPs or indels could lead to ASE and ASM. Lastly, given that large-scale CNVs are fairly common in human genomes, a gross alteration in chromosome structure secondary to CNVs is another a priori possibility that could account for some instances of ASE and ASM.
CIS-REGULATED ASTF IS WIDESPREAD AND LINKED TO ASE
All of the above mechanistic models can begin to be addressed by combining ASE/ASM mapping with genome-wide mapping of ASTF. Here the abbreviation ASTF broadly includes allele-specific affinities of insulator-binding proteins and allele-specific chromatin modifications. In fact, an impressive initial group of papers have already appeared on this topic, using ChIP with downstream analysis either by probe synthesis from the IP DNA and hybridization to SNP arrays or, more recently, NextGen sequencing to query SNP representation in the IP DNA (references in Table 4). Taken together, these studies provide proof-of-principle for mapping not only allele-specific histone modifications (a method which has also been extensively used by laboratories working on imprinted chromosomal domains) but also allele-specific DNA occupancy of RNA PolII, allele-specific binding of the transcription factor NF-κB and the insulator-binding protein/transcription factor CTCF and, as a surrogate for open chromatin, mapping of allele-specific DNase I hypersensitive sites. In each of these studies, allele specificity has been found at large numbers of SNP-tagged loci. Encouragingly, in the study by McDaniell et al. (41) at least some of the allele specificity of CTCF binding could be accounted for by SNPs located within CTCF consensus-binding motifs. This observation provides some experimental support for the mechanistic model in Figure 2C.
Table 4.
Methods and conclusions from screens for allele-specific chromatin and ASTF at non-imprinted loci
| Tissues or cell types (n) | Primary screening methods and validations | Findings and conclusions | References |
|---|---|---|---|
| LCL (12; from 2 CEPH families) | RNA PolII and histone modification ChIP–Chip on Affy 10K SNP arrays; ChIP antibodies validated by Q-PCR of IPs at imprinted loci | ASTF as measured by PolII occupancy and chromatin modifications (H3K4, H3K9, H3K27 methylation state) behaves as a genetically determined trait in families | (60) |
| IMR90 fibroblast line (1) | RNA PolII ChIP-SNP on Illumina Hap300 BeadChips; validations by PCR sequencing of IPs; ASE by cDNA/gDNA comparison using Illumina GoldenGate assays | Of 11 9821 heterozygous SNPS, 466 (239 RefSeq genes and 18 small RNAs) showed allele-specific enrichment in PolII IPs with an average fold change of 3.98. For 20 examples among these genes, the allele-specific RNA PolII binding was shown to correlate with ASE in the IMR90 cells. | (61); see also (62) |
| LCL (10) | RNA PolII and NF-κB ChIP-Seq data overlaid to SNP genotypes from HapMap and CNVs | SNPs and structural variants are frequently associated with RNA PolII and NF-κB binding differences. The fraction of binding differences coinciding with genetic variations can be very high: 35% for NF-κB and 26% for PolII in these LCLs | (63) |
| LCL (6; 4 parents and 2 children from HapMap project) | DNase I HS site (DNase-Seq) mapping and ChIP-Seq for CTCF–binding sites; validations by PCR/MS (Sequenom) on ChIP material | Transmission of SNP alleles suggests a heritable genetic basis for a large proportion of the allele-specific binding of CTCF. At sites where CTCF showed ASTF, the binding motif score tended to be higher for the favored allele, whereas at sites lacking differences in CTCF binding, motif scores were similar | (41) |
All experiments include internal statistical validations of the microarray data; secondary validations refer to downstream assays by independent methods. Cells and tissues are of human origin.
APPLICATIONS OF ALLELE-SPECIFIC MAPPING
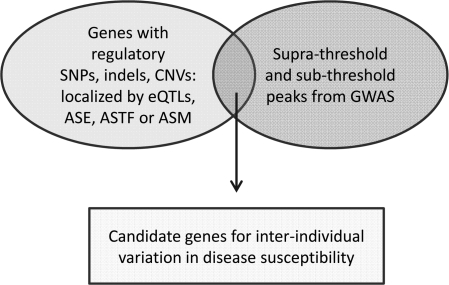
As introduced by the above discussion, one practical use of ASE/ASM mapping is to help extract maximum information from GWAS. Here I paraphrase from my recent commentary focusing on this application (42). There is some debate now on the relative merits of the ‘common disease–common variant’ versus ‘multiple rare variant’ hypotheses for explaining complex disorders. Nonetheless, as predicted by the common variant model, GWAS have in fact identified many well replicated and biologically credible loci for disease susceptibility. In the process, these types of studies have come up against two technical roadblocks: First, most (∼90%) of the supra-threshold disease association signals are at non-coding SNPs (43–45). Among these statistical signals which ones are due to bona fide functional regulatory SNPs, and how can these SNPs be identified? Second, because of multiple comparisons, the threshold for significance needs to be set high, at P < 10−7 or P < 5 × 10−8, so there are numerous sub-threshold peaks that are difficult to interpret. Are some of these signals true-positives that should not be discarded? Independent lines of evidence are needed, and a promising direct approach is to combine statistical evidence from GWAS with functional evidence for the presence of cis-acting regulatory SNPs, indels or CNVs from mapping of eQTLs, ASE and ASM (Fig. 3). Statistical methods for carrying out such overlaps have recently been published [for example (14, 21)].
Figure 3.
Allele-specific mapping for maximizing information from GWAS. This figure is adapted from Tycko (42). It shows the general strategy for overlaying information from genome-wide maps of allelic asymmetries with data from GWAS. Mapping ASE, ASTF and ASM can help to pinpoint bona fide regulatory polymorphisms, which reveal their presence by conferring the allelic asymmetries in the relevant disease target tissue. Supra-threshold and sub-threshold GWAS peaks that co-localize with these regulatory polymorphisms warrant attention as true-positive signals.
Beyond providing evidence for rSNPs being near a gene of interest, mapping ASE can help to close in on the precise positions of functional SNPs. Forton et al. (46) used ASE and haplotypes analysis to map cis-regulatory elements in chromosome band 5q31, thereby pinpointing the location of cis-acting DNA sequences that regulate the IL13 gene from a distance of 250 Kb upstream. Other examples in this fast-moving area are discussed in my previous review (42) and listed here in Tables 1 and 2. In the future, it will be interesting to see whether the methods developed for overlapping GWAS with ASE and eQTL data in these papers can also work using ASM as the marker for nearby regulatory polymorphisms. To use this strategy, it will first be necessary to develop more efficient methods for complete ASM profiling over megabase regions of DNA to define the epicenters of the allelic asymmetry.
CONCLUSIONS AND REMAINING QUESTIONS
As this field matures over the next decade, there will likely be two lines of important descriptive work, with continuing genome-wide analyses moving forward in parallel with more focused ‘fine-mapping’ studies of genes and chromosomal regions of interest, to more tightly pin down the identities of regulatory polymorphisms and haplotypes. Doing so will be necessary both for testing mechanistic hypotheses and for completing the tasks started by GWAS—namely to fully understand the etiologies of complex genetic diseases. Even the au courant research strategy of searching for rare genetic variants to explain complex diseases could benefit from incorporating ASE, ASM and ASTF mapping. In particular, not all pathogenic variants, even if rare, will be non-synonymous coding changes, so the functional significance of rare non-coding variants will still need to be grappled with. Fine-mapping ASM to find the true epicenters of allelic asymmetry will be essential for testing mechanistic models. None of the available datasets yet provides this type of information. In the near future, microarrays with custom designs will be useful for achieving greater coverage of SNPs in CpG-rich sequences while continuing to achieve high sample throughput at reasonable costs. NextGen bisulfite sequencing with reduced genomic representation and padlock probe methods (30,47), and high-throughput bisulfite PCR on new microfluidic and microdroplet instruments (48,49), will also be essential in the near term for achieving regional genomic coverage at single base-pair resolution. Ultimately the ‘$1000 epigenome’ will become a reality. But this will not be enough—all questions in this field come back to understanding the functions of cis-acting variants in the DNA of a given chromosome homolog over both short and long distances. So it will also be important to continue using samples from multi-generation families, and to develop direct methods for establishing the phase of SNPs, indels and CNVs, in other words, their physical linkage over long stretches of the DNA (50).
FUNDING
This work was supported by grants R21 CA125461-02, R01 AG036040-01, and R01 AG035020-01 from the NIH and by grants from the March of Dimes and the Douglas Kroll Foundation of the Leukemia and Lymphoma Society.
ACKNOWLEDGEMENTS
I thank Barbara Stranger and Andrew Chess for their helpful comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Weaver J.R., Susiarjo M., Bartolomei M.S. Imprinting and epigenetic changes in the early embryo. Mamm. Genome. 2009;20:532–543. doi: 10.1007/s00335-009-9225-2. doi:10.1007/s00335-009-9225-2. [DOI] [PubMed] [Google Scholar]
- 2.Cattanach B.M., Beechey C.V., Peters J. Interactions between imprinting effects: summary and review. Cytogenet. Genome Res. 2006;113:17–23. doi: 10.1159/000090810. doi:10.1159/000090810. [DOI] [PubMed] [Google Scholar]
- 3.Tycko B., John R. Imprinted genes and placental growth: implications for the developmental origins of health and disease. In: Burton J., Barker D., Moffett A., Thornburg K., editors. The Placenta and Human Developmental Programming. Cambridge, UK: Cambridge University Press; 2010. pp. 57–69. [Google Scholar]
- 4.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E., et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. doi:10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 5.Dindot S.V., Person R., Strivens M., Garcia R., Beaudet A.L. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–1383. doi: 10.1101/gr.089185.108. doi:10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. doi:10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 7.Tan A.C., Fan J.B., Karikari C., Bibikova M., Garcia E.W., Zhou L., Barker D., Serre D., Feldmann G., Hruban R.H., et al. Allele-specific expression in the germline of patients with familial pancreatic cancer: an unbiased approach to cancer gene discovery. Cancer Biol. Ther. 2008;7:135–144. doi: 10.4161/cbt.7.1.5199. doi:10.4161/cbt.7.1.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Weaver J., Bove B.A., Vanderveer L.A., Weil S.C., Miron A., Daly M.B., Godwin A.K. Allelic imbalance in BRCA1 and BRCA2 gene expression is associated with an increased breast cancer risk. Hum. Mol. Genet. 2008;17:1336–1348. doi: 10.1093/hmg/ddn022. doi:10.1093/hmg/ddn022. [DOI] [PubMed] [Google Scholar]
- 9.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. doi:10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery S.B., Sammeth M., Gutierrez-Arcelus M., Lach R.P., Ingle C., Nisbett J., Guigo R., Dermitzakis E.T. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. doi:10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. doi:10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verlaan D.J., Ge B., Grundberg E., Hoberman R., Lam K.C., Koka V., Dias J., Gurd S., Martin N.W., Mallmin H., et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–127. doi: 10.1101/gr.084798.108. doi:10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. doi:10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. doi:10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plagnol V., Uz E., Wallace C., Stevens H., Clayton D., Ozcelik T., Todd J.A. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS ONE. 2008;3:e2966. doi: 10.1371/journal.pone.0002966. doi:10.1371/journal.pone.0002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C., Gagne V., et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. doi:10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 17.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. doi:10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong H., Beaulaurier J., Lum P.Y., Molony C., Yang X., Macneil D.J., Weingarth D.T., Zhang B., Greenawalt D., Dobrin R., et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932.. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heap G.A., Yang J.H., Downes K., Healy B.C., Hunt K.A., Bockett N., Franke L., Dubois P.C., Mein C.A., Dobson R.J., et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. doi:10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. doi:10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. doi:10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Rohde C., Reinhardt R., Voelcker-Rehage C., Jeltsch A. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol. 2009;10:R138. doi: 10.1186/gb-2009-10-12-r138. doi:10.1186/gb-2009-10-12-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N., Plomin R., Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. doi:10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. doi:10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 25.Hellman A., Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. doi:10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling E., El Chartouni C., Rehli M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009;19:2028–2035. doi: 10.1101/gr.095562.109. doi:10.1101/gr.095562.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H.H., Hu N., Wang C., Ding T., Dunn B.K., Goldstein A.M., Taylor P.R., Lee M.P. Influence of genetic background and tissue types on global DNA methylation patterns. PLoS ONE. 2010;5:e9355. doi: 10.1371/journal.pone.0009355. doi:10.1371/journal.pone.0009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D., Cheng L., Badner J.A., Chen C., Chen Q., Luo W., Craig D.W., Redman M., Gershon E.S., Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am. J. Hum. Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. doi:10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. doi:10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoemaker R., Deng J., Wang W., Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. doi:10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mummaneni P., Yates P., Simpson J., Rose J., Turker M.S. The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res. 1998;26:5163–5169. doi: 10.1093/nar/26.22.5163. doi:10.1093/nar/26.22.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandeis M., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. doi:10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 33.Han L., Lin I.G., Hsieh C.L. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. doi:10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senigl F., Plachy J., Hejnar J. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 2008;82:7818–7827. doi: 10.1128/JVI.00419-08. doi:10.1128/JVI.00419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebhard C., Benner C., Ehrich M., Schwarzfischer L., Schilling E., Klug M., Dietmaier W., Thiede C., Holler E., Andreesen R., et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. doi:10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 36.Boumber Y.A., Kondo Y., Chen X., Shen L., Guo Y., Tellez C., Estecio M.R., Ahmed S., Issa J.P. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4:e1000162. doi: 10.1371/journal.pgen.1000162. doi:10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh R.P., Horowitz-Scherer R.A., Nikitina T., Shlyakhtenko L.S., Woodcock C.L. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell Biol. 2010 doi: 10.1128/MCB.00379-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reik W., Murrell A., Lewis A., Mitsuya K., Umlauf D., Dean W., Higgins M., Feil R. Chromosome loops, insulators, and histone methylation: new insights into regulation of imprinting in clusters. Cold Spring Harb. Symp. Quant. Biol. 2004;69:29–37. doi: 10.1101/sqb.2004.69.29. [DOI] [PubMed] [Google Scholar]
- 39.Fitzpatrick G.V., Pugacheva E.M., Shin J.Y., Abdullaev Z., Yang Y., Khatod K., Lobanenkov V.V., Higgins M.J. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell Biol. 2007;27:2636–2647. doi: 10.1128/MCB.02036-06. doi:10.1128/MCB.02036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparago A., Cerrato F., Vernucci M., Ferrero G.B., Silengo M.C., Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann syndrome. Nat. Genet. 2004;36:958–960. doi: 10.1038/ng1410. doi:10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 41.McDaniell R., Lee B.K., Song L., Liu Z., Boyle A.P., Erdos M.R., Scott L.J., Morken M.A., Kucera K.S., Battenhouse A., et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. doi:10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tycko B. Mapping allele-specific DNA methylation: a new tool for maximizing information from GWAS. Am. J. Hum. Genet. 2010;86:109–112. doi: 10.1016/j.ajhg.2010.01.021. doi:10.1016/j.ajhg.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nica A.C., Dermitzakis E.T. Using gene expression to investigate the genetic basis of complex disorders. Hum. Mol. Genet. 2008;17:R129–134. doi: 10.1093/hmg/ddn285. doi:10.1093/hmg/ddn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Easton D.F., Eeles R.A. Genome-wide association studies in cancer. Hum. Mol. Genet. 2008;17:R109–115. doi: 10.1093/hmg/ddn287. doi:10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 45.Lettre G., Rioux J.D. Autoimmune diseases: insights from genome-wide association studies. Hum. Mol. Genet. 2008;17:R116–121. doi: 10.1093/hmg/ddn246. doi:10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forton J.T., Udalova I.A., Campino S., Rockett K.A., Hull J., Kwiatkowski D.P. Localization of a long-range cis-regulatory element of IL13 by allelic transcript ratio mapping. Genome Res. 2007;17:82–87. doi: 10.1101/gr.5663007. doi:10.1101/gr.5663007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmermann B.G., Grill S., Holzgreve W., Zhong X.Y., Jackson L.G., Hahn S. Digital PCR: a powerful new tool for noninvasive prenatal diagnosis? Prenat. Diagn. 2008;28:1087–1093. doi: 10.1002/pd.2150. [DOI] [PubMed] [Google Scholar]
- 49.Tewhey R., Warner J.B., Nakano M., Libby B., Medkova M., David P.H., Kotsopoulos S.K., Samuels M.L., Hutchison J.B., Larson J.W., et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. doi:10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao M., Wan E., Chu C., Hsueh W.C., Cao Y., Kwok P.Y. Direct determination of haplotypes from single DNA molecules. Nat. Methods. 2009;6:199–201. doi: 10.1038/nmeth.1301. doi:10.1038/nmeth.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dermitzakis E.T., Stranger B.E. Genetic variation in human gene expression. Mamm. Genome. 2006;17:503–508. doi: 10.1007/s00335-006-0005-y. doi:10.1007/s00335-006-0005-y. [DOI] [PubMed] [Google Scholar]
- 52.Pastinen T., Hudson T.J. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. doi:10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 53.Serre D., Gurd S., Ge B., Sladek R., Sinnett D., Harmsen E., Bibikova M., Chudin E., Barker D.L., Dickinson T., et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. doi:10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K., Li J.B., Gao Y., Egli D., Xie B., Deng J., Li Z., Lee J.H., Aach J., Leproust E.M., et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat. Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. doi:10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. doi:10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spielman R.S., Bastone L.A., Burdick J.T., Morley M., Ewens W.J., Cheung V.G. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39:226–231. doi: 10.1038/ng1955. doi:10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storey J.D., Madeoy J., Strout J.L., Wurfel M., Ronald J., Akey J.M. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007;80:502–509. doi: 10.1086/512017. doi:10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. doi:10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 59.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi:10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 60.Kadota M., Yang H.H., Hu N., Wang C., Hu Y., Taylor P.R., Buetow K.H., Lee M.P. Allele-specific chromatin immunoprecipitation studies show genetic influence on chromatin state in human genome. PLoS Genet. 2007;3:e81. doi: 10.1371/journal.pgen.0030081. doi:10.1371/journal.pgen.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maynard N.D., Chen J., Stuart R.K., Fan J.B., Ren B. Genome-wide mapping of allele-specific protein–DNA interactions in human cells. Nat. Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 62.Knight J.C., Keating B.J., Rockett K.A., Kwiatkowski D.P. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat. Genet. 2003;33:469–475. doi: 10.1038/ng1124. doi:10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 63.Kasowski M., Grubert F., Heffelfinger C., Hariharan M., Asabere A., Waszak S.M., Habegger L., Rozowsky J., Shi M., Urban A.E., et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. doi:10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]