Abstract
While maintaining anatomical integrity, matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS) has allowed researchers to directly probe tissue, map the distribution of analytes and elucidate molecular structure with minimal preparation. MALDI-ion mobility (IM)-orthogonal time-of-flight mass spectrometry (oTOFMS) provides an advantage by initially separating different classes of biomolecules such as lipids, peptides, and nucleotides by their IM drift times prior to mass analysis. In the present work the distribution of phosphatidlycholine and cerebroside species was mapped from 16 μm thick coronal rat brain sections using MALDI-IM-oTOFMS. Furthermore, the use of gold nanoparticles as a matrix enables detection of cerebrosides, which although highly concentrated in brain tissue, are not easily observed as positive ions because of intense signals from lipids such as phosphatidlycholines and sphingomyelins.
Keywords: ion mobility MS, lipid imaging, brain tissue, phosphatidylcholines, cerebrosides
INTRODUCTION
Tissue profiling and imaging by matrix assisted laser desorption/ionization (MALDI) mass spectrometry has allowed the direct analysis and localization of biomolecules (proteins, peptides, lipids) while maintaining the anatomical integrity of the tissue.1–3 In this method, the matrix is added directly to the tissue section which is then mass analyzed with no additional preparation. Successful applications of this technique have led to the profiling and mapping of proteins,4,5 peptides,6–9 lipids,10–14 and drugs15–17 in both normal and pathological tissues. Owing to the in situ nature of this method, purification and chromatographic separation steps are not possible since the spatial location of the bioanalyte is paramount. Therefore, without the power of chromatography, the mass spectra acquired can be extremely complex and difficult to interpret. This is particularly true at m/z less than 2 kDa where lipid, peptide, and matrix peaks overlap in MALDI-MS.
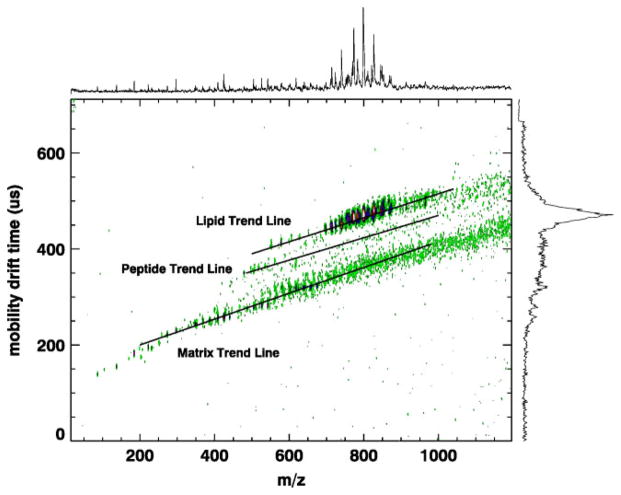
The advantages of MALDI-ion mobility (IM)-orthogonal time-of-flight mass spectrometry (oTOFMS) over MALDI-MS have been demonstrated by comparing spot profiles from standard samples as well as tissue.18–23 MALDI-IM-oTOFMS offers the potential for real-time separations within the several 100 ms time interval between successive focused laser desorption pulses. The IM separation is based on the gas-phase ion density and is done prior to mass analysis. This enables sorting biological families such as lipids, peptides, and nucleotides within the 2D space defined by the ions’ mobility drift time and m/z.20–23 An example of such a plot is shown in Fig. 1, where familial ‘trend lines’ comprising lipids, peptides, and matrix are separated from each other by differences in their IM drift times of about 10–15%. Moreover, random experimental noise is distributed over the entire 2D IM–m/z space instead of within the one-dimensional m/z spectra obtained with MALDI-MS. In addition, IM separates multiple charge states, which enable partitioning of ‘chemical noise’ into useful signals. This is particularly helpful for enhancing the information content available from protein and peptide ions since mass spectra of each purified charge state can be numerically reconstructed. For example, the distribution of exclusively singly charged molecular ions (MH)+ of peptides or proteins can easily be obtained.24 The advantages of MALDI-IM-oTOFMS over MALDI-MS which have been previously demonstrated for tissue spot profiling are expected to also transfer to the use of MALDI-IM-oTOFMS in tissue imaging.
Figure 1.
MALDI-IM 2D plot of a rat brain tissue section with 2,5-dihydroxy benzoic acid (DHB) matrix in positive ion mode. Many of the peaks in the trend line identified as ‘matrix’ can be assigned to DHB clusters or DHB clusters + potassium. The rectangular region of interest, 16 ms wide (mobility drift time) and 100 amu long (centered at 800 m/z), was defined in software for subsequent use during acquisition of the data in Fig. 2.
The present study focuses on mapping lipid distribution in brain tissue sections since they are the most common biomolecules found in the brain (12%). They comprise 50% of the dry weight of the brain and play an important role in its anatomy, physiology, and pathophysiology. Furthermore, direct tissue analysis by MALDI of the lipid component of the rat brain has been well studied.12,14,25–27 The proof of the principle of imaging by MALDI-IM-TOFMS is best done on a known system; moreover, the lipid images should contain a much reduced background noise which are usually due to random signals and isobaric interferences (e.g. peptides and matrix) by analogy to the spot profile comparisons previously mentioned.20–23 In addition, the use of gold nanoparticles as matrix has allowed us to detect and image cerebrosides, which are not typically seen in positive ion mode when using an organic matrix such as 2,5-dihydroxy benzoic acid (DHB).
EXPERIMENTAL
Mass spectrometer
Data was acquired with a periodic focusing MALDI-IM-TOFMS instrument in positive ion mode (Ionwerks Inc., Houston, TX). A mobility resolution of 30 (half-width of the peak of mobility/drift time) and a mass resolution of 3000 for m/z 1000 using an oTOFMS are routinely achieved. The length of the mobility cell is 15 cm. It is operated at 1700 V with 3.5 Torr He pressure. An X–Y sample stage (National Aperture Inc, folded micro-stage, model MM-3M-F-2) provides 1 μm accuracy in beam positioning and sample scanning. A Nd : YLF UV laser (Crystalaser, λ = 349 nm at 200 Hz) is used to generate ions in the source at the operating pressure of the mobility cell. After the laser shot, the ions float to the end of the mobility cell under the force of a high voltage field applied between the sample plate and successive electrode rings within the mobility spectrometer. The mobility-separated ions then pass through the skimmer into a differentially pumped oTOFMS where they are mass analyzed and the spectra recorded as a function of mobility drift time after the desorption laser pulse. The mobility drift times are up to several milliseconds while the flight times within the mass spectrometer are tens of microseconds. Therefore, several mass spectra can be obtained after every laser pulse. After the laser pulse, individual mass spectra are acquired at intervals of 30–150 ms (depending on the mass range) and are stored individually along with the mobility time at which they were acquired.
The images presented in this study were acquired with a 200 μm spatial resolution and a 3 s acquisition time per step. The laser spot size is less than 50 microns and the angle of incidence is 60° (tilt measured from the surface normal). Thus within the 3 s acquisition, a swath of 100 μm or less is cut across the pixel. The area within each pixel that is interrogated by the laser is maximally estimated from postimaging micrographs of these erosion tracks as being 250 × 100 μm for the first pixel in each line scan and 200 × 100 μm for each pixel thereafter. The surface area of available material interrogated is thus estimated to be 50% or less of the total pixel area depending on the actual profile of the laser beam. A specific region of interest in both mass and mobility dimensions is preselected within the software. Offsets in both mass and mobility data acquisition parameters limit incoming data to a narrow mass range of approximately 100 Da and a mobility drift time range spanning 16 μs (at m/z 800). The region of interest mass bins and corresponding intensities are saved to a file along with the positional coordinates.
Tissue sectioning and handling
All the animal use and handling in this work abides by the Guide for the Care and Use of Laboratory Animals (NIH). Male Sprague–Dawley rats (Harlan Industries, Indianapolis, IN) between 300 and 420 g were used. Rats were euthanized with isoflurane and were decapitated upon cessation of respiration. The brains were quickly removed from the skull and frozen in dry ice-chilled isopentane for 15 s, prior to storage at −80 °C. The frozen rat brains were transferred from −80 °C to the cryostat chamber (CM 3050 S; Leica Microsystems Nussloch GmbH, Nussloch, Germany) at −20 °C where they were allowed to thermally equilibrate for 45 min. The tissue samples were attached to the cryostat sample stages using ice slush made from distilled water. This procedure has been described in detail previously.25 The brains were cut into 16 μm sections and placed onto the MALDI sample targets. The plates were stored at −80 °C until analysis.
Matrix deposition
Two matrices were used in this study: DHB (Fluka, Switzerland) and a PSG gold colloid (5.5 nm, 1.2 × 1014 particles/ml, nanoComposix, San Diego, CA). DHB was prepared in 90% ethanol at 100 mg/ml concentration, while the gold colloid was diluted 1 : 4 (v/v) in ethanol. The matrix solutions were sprayed on the tissue sections with an artistic airbrush14 (Aztek A470/80 Airbrush System, the Testors Corporation, Rockford, IL). Each target insert was sprayed with 5–6 spray cycles. A 2 min wait between spray cycles was observed to ensure sufficient evaporation of organic solvents from the target insert surface. One spray cycle is defined as an organized rastering of spraying, which results in complete coverage of the target insert surface by the spraying mist. The distance between the nozzle of the airbrush and the target plate was kept between 15 and 20 cm. The matrix was perpendicularly sprayed onto the target plate.
RESULTS AND DISCUSSIONS
Figure 1 illustrates a typical MALDI-IM-TOF MS spectrum of rat brain tissue using DHB matrix in positive ion mode. In this figure, the data is presented as a 2D contour plot of ion intensity as a function of IM drift time (y-axis) and mass (m/z, x-axis). Furthermore, the composite 1D IM spectrum (right y-axis) and 1D mass spectrum (top x-axis) are included in the figure. In this spectrum, the major biomolecules detected are the phospholipid species, in particular, phosphatidylcholine (PC) and sphingomyelin (SM). These lipids were detected in positive ion mode from rat brain sections in earlier studies using MALDI-IM-TOFMS and MALDI-TOFMS.25,26 In a previous study22 using standards, it was demonstrated that different classes of biomolecules such as lipids, oligonucleotides, and peptides form distinct familial trend lines on a 2D plot of mobility drift times versus m/z. Furthermore, it was shown in this study that lipid ion species were consistently around 12% slower than isobaric peptides. This result was also observed in direct tissue analysis using MALDI-IM-TOFMS.25 As can be seen in Fig. 1, there is a distinct lipid trend line which is separated from other biomolecules, and it is this 2D range of mobility time and mass that was used to acquire the lipid images in this study.
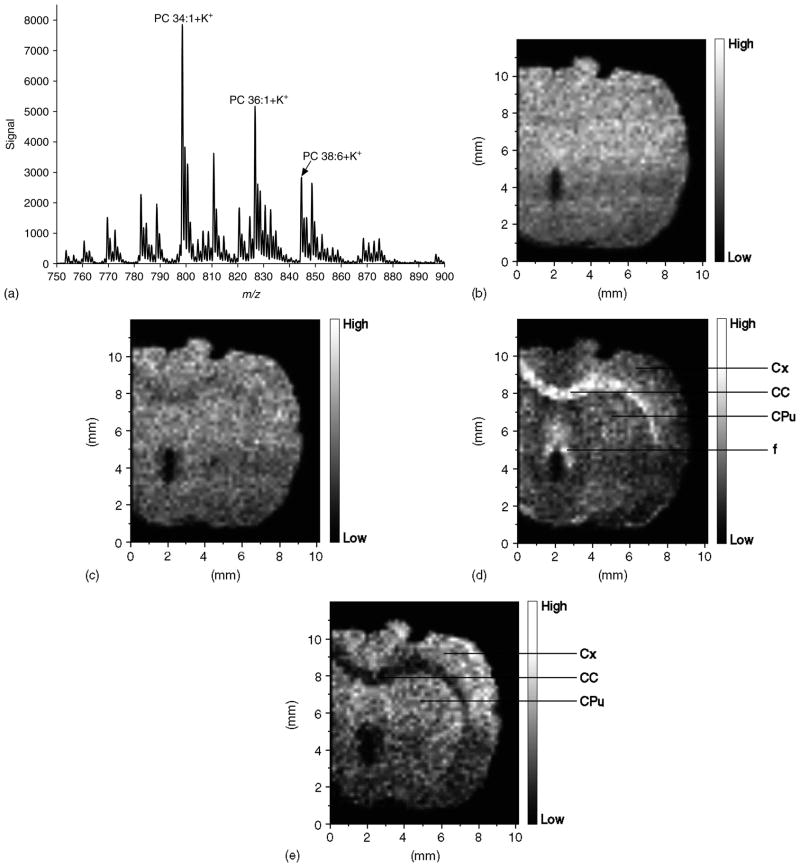
Figure 2 shows the results of a MALDI-IM imaging run obtained with DHB matrix from a rat cerebrum in positive ion mode. A mass range of m/z 750–900 and a mobility drift time of 476 ms were used to acquire the data in Fig. 2. Figure 2(a) represents the total 1D mass spectrum for the run, while Fig. 2(b) is the image produced using the total ion count. The image in Fig. 2(b) shows the shape of the section; however, no clear anatomical regions are observed. Figure 2(c) illustrates the distribution of m/z 798.6 (PC 34 : 1 + K), which is the most abundant PC species in rat brain and is widely distributed throughout the brain. Figure 2(d) shows the distribution of m/z 826.6 (PC 36 : 1 + K), while Fig. 2(e) illustrates the distribution of m/z 844.6 (PC 38 : 6 + K). Anatomically distinct regions are detected in Fig. 2(d), with the highest intensities observed in white matter regions including the corpus callosum and fornix, while in Fig. 2(e) the highest intensities are observed in the cerebral cortex and caudate putamen, both gray matter regions. These results agree with a previous study using MALDI-TOFMS for direct profiling of rat brain, in which PC 36 : 1 was shown to be more abundant in white matter while PC 38 : 6 was shown to be more abundant in gray matter.26
Figure 2.
MALDI-IM images using DHB matrix in positive ion mode. (a) 1D mass spectrum obtained from section. (b) Image of total ion count in Fig. 2(a). (c) Image of mass peak at m/z 798.6. (d) Image of mass peak at m/z 826.6. (e) Image of mass peak at m/z 844.6. Images are 51 × 60 pixels. Abbreviations: Cx – cerebral cortex; CC – corpus callosum; CPu – caudate putamen; f – fornix.
Previous studies have used implanted gold clusters28,29 and gold nanoparticles30 as novel MALDI matrices. In two of these studies,28,29 the gold clusters were implanted directly on rat brain tissue sections. Mass spectra containing strong peaks from cationized cerebroside species were obtained in positive ion mode. This is in contrast to the mass spectra obtained from rat brain tissues using traditional organic acid matrices in which PC species are the dominant signals in this lipid mass range in positive ion mode. MALDI-IM-oTOFMS is an enabling technology for using gold nanoparticulates as matrices since it removes a significant interference from ionized gold clusters and their attachments and allows spot profile analysis of weak signals from peptides and minority lipid species in the presence of the dominant majority lipid signals.28,29
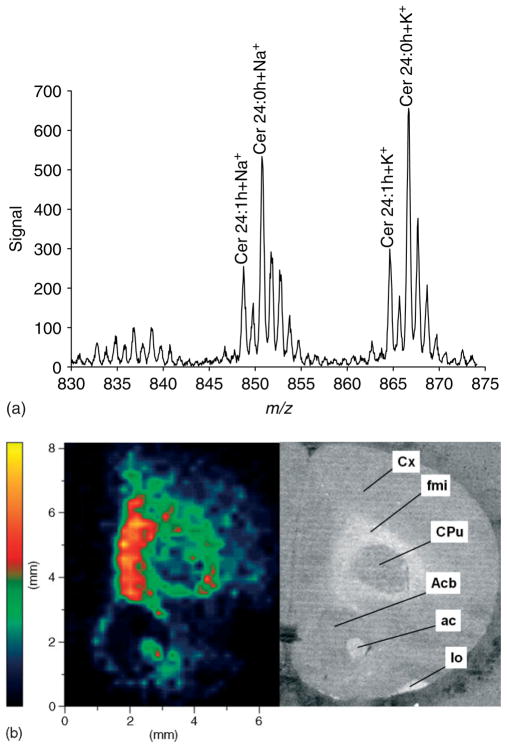
Motivated by these prior results28,29 we have used 5.5 nm gold nanoparticles in this study to activate and measure the spatial distribution of cerebroside species in rat brain sections. Figure 3 illustrates the results of a MALDI-IM-oTOFMS image obtained using 5.5 nm gold nanoparticles as a matrix for a rat brain cerebrum coronal section in positive ion mode. A mass range of m/z 830–875 and a mobility drift time of 578 ms were used to acquire the data in Fig. 3. Figure 3(a) represents the total 1D mass spectrum obtained by summing all ions within this IM–m/z region of interest irrespective of spatial location. Major mass peaks detected in this spectrum correspond to the sodiated and potassiated cerebroside 24 : 0 OH at m/z 850.7 and 866.8 and the sodiated and potassiated cerebroside 24 : 1 OH at m/z 848.7 and 864.8. Figure 3(b) is a spatial image, tracking only the distribution of sodiated cerebroside 24 : 0 OH at m/z 850.7. In the Fig. 3(b) image, the highest intensities are observed in white matter regions corresponding to the forceps minor of the corpus callosum and anterior commissure as labeled in the adjacent frame showing a photo of the tissue section. Images analogous to Fig. 3(b) (data not shown) were derived from the region of interest using the total ion count data from potassiated cerebroside 24 : 0 OH, sodiated cerebroside 24 : 1 OH, or potassiated cerebroside 24 : 1 OH. These images show that the highest intensities were recorded in white matter regions. This distribution of cerebrosides in brain tissue is expected since cerebrosides are known to be highly concentrated in white matter regions, especially in myelin.31 Furthermore, electrospray-MS of lipid extracts from rat brain32 and secondary-ion mass spectrometry imaging of rat brain sections33 have also recorded higher abundance of cerebrosides in white matter regions.
Figure 3.
MALDI-IM images using 5.5 nm Au particles in positive ion mode. (a) 1D mass spectrum recorded from the tissue section. (b) Image of sodiated cerebroside 24 : 0 OH at m/z 850.7. The image is a 40 × 40 pixel. The adjacent frame is a photograph of the rat brain section. Abbreviations: Cx – cortex; fmi – forceps minor of the corpus callosum; Cpu – caudate putamen (striatum); Acb – nucleus accumbens; ac – anterior commissure; lo – lateral olfactory tract.
This study suggests that IM coupled to MALDI-MS has the potential to be a useful tool for spot profiling and spatial imaging of tissue due to its ability to enhance image details by numerically subtracting noise and matrix-derived signals as well as separation of isobaric biomolecules that may be present in adjacent familial trend lines. Future studies will include the analysis and mapping of peptides and drugs in tissue, in which the selection of the mobility time will be used to filter out interfering signals from the more abundant lipid or matrix ions in the m/z. In addition, in this work the use of gold nanoparticles as a matrix has permitted the detection and mapping of the neutral cerebrosides, which although present in high concentration, regularly go undetected in positive ion mode when using organic acid matrices owing to the suppression by positively charged lipid species such as PCs and SMs.
At present, during data acquisition we exclude all ions except those with the prescribed mobility and mass region of interest. This is done to reduce the data file size and is not a fundamental limitation. In future, improvement in software and hardware instrumentation will permit a complete IM–m/z representation of every ion within each pixel, which will be acquired and saved. Thus, it will be possible to post-process the stored data to create images of any desired ion type with an adequate number of the desired ions from which to establish a high dynamic range image.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, NIH. The authors thank the Office of National Drug Control Policy (ONDCP) for instrumentation funding, without which this and other projects could not have been accomplished. This project has also been funded in part at Ionwerks with Federal funds from the National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services, under contracts No. HHSN271200677593C, HHSN271200677563C, and N44DA-3-7727.
References
- 1.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Current Opinion in Chemical Biology. 2007;11:1. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell RL, Caprioloi RM. Tissue profiling by mass spectrometry: a review of methodology and applications. Molecular and Cellular Proteomics. 2005;4:394. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Chaurand P, Schwartz SA, Reyzer ML, Caprioli RM. Imaging mass spectrometry: principles and potentials. Toxicologic Pathology. 2005;33:92. doi: 10.1080/01926230590881862. [DOI] [PubMed] [Google Scholar]
- 4.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. Journal of Proteome Research. 2006;5:2889. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 5.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. Journal of Mass Spectrometry. 2007;42:254. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 6.Fournier I, Day R, Salzet M. Direct analysis of neuropeptides by in situ MALDI-TOF mass spectrometry in the rat brain. Neuro Endocrinology Letters. 2003;24:9. [PubMed] [Google Scholar]
- 7.Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Analytical Chemistry. 2006;78:7267. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoeckli M, Knochenmuss R, McCombie G, Mueller D, Rohner T, Staab D, Wiederhold K-H. MALDI MS imaging of amyloid. Methods in Enzymology. 2006;412:94. doi: 10.1016/S0076-6879(06)12007-8. [DOI] [PubMed] [Google Scholar]
- 9.Taban IM, Altelaar AFM, van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, Baykut G. Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. Journal of the American Society for Mass Spectrometry. 2007;18:145. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Touboul D, Piednoel H, Voisin V, De La Porte S, Brunelle A, Halgand F, Laprevote O. Changes in phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry imaging. European Journal of Mass Spectrometry. 2004;10:657. doi: 10.1255/ejms.671. [DOI] [PubMed] [Google Scholar]
- 11.Rujoi M, Estrada R, Yappert MC. In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Analytical Chemistry. 2004;76:1657. doi: 10.1021/ac0349680. [DOI] [PubMed] [Google Scholar]
- 12.Woods AS, Jackson SN. Brain tissue lipidomics: direct probing using MALDI MS. The AAPS Journal. 2006;8:E39. doi: 10.1007/BF02854910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones JJ, Borgmann S, Wilkins CL, O’Brein RM. Characterizing the phospholipid profiles in mammalian tissues by MALDI FTMS. Analytical Chemistry. 2006;78:3062. doi: 10.1021/ac0600858. [DOI] [PubMed] [Google Scholar]
- 14.Garrett TJ, Prieto-Conaway MC, Kovtoun V, Bui H, Izgarian N, Stafford G, Yost RA. Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. International Journal of Mass Spectrometry. 2007;260:166. [Google Scholar]
- 15.Wang H-YJ, Jackson SN, McEuen J, Woods AS. Localization and analyses of small drug molecules in rat brain tissue sections. Analytical Chemistry. 2005;77:6682. doi: 10.1021/ac050868d. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh Y, Casale R, Fukuda E, Chen J, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Communications in Mass Spectrometry. 2006;20:965. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 17.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Analytical Chemistry. 2006;78:6448. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 18.Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, Gonin M, Schultz JA. Coupling high-pressure MALDI with ion mobility/orthogonal time-of-flight mass spectrometry. Analytical Chemistry. 2000;72:3965. doi: 10.1021/ac0005619. [DOI] [PubMed] [Google Scholar]
- 19.Von Helden G, Wyttenbach T, Bowers MT. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science. 1995;267:1483. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 20.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Ion mobility-mass Spectrometry: a new paradigm for proteomics. International Journal of Mass Spectrometry. 2005;240:301. [Google Scholar]
- 21.Ruotolo BT, Gillig KJ, Stone EG, Russell DH. Capacity of ion mobility mass spectrometry: separation of peptides in helium buffer gas. Journal of Chromatography B. 2002;782:385. doi: 10.1016/s1570-0232(02)00566-4. [DOI] [PubMed] [Google Scholar]
- 22.Woods AS, Fuhrer K, Egan T, Ugarov M, Koomen J, Gonin M, Gillig KJ, Schultz JA. Lipid/peptide/nucleotide separation with MALDI-ion mobility-TOFMS. Analytical Chemistry. 2004;76:2187. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- 23.McLean JA, Russell DH. New vistas for mass spectrometry-based proteomics and biotechnology: rapid two-dimensional separations using gas-phase electrophoresis/ion mobility-mass spectrometry. American Biotechnology Laboratory. 2005;23:18. [Google Scholar]
- 24.Woods AS, Ugarov M, Jackson SN, Egan T, Wang H-YJ, Murray KK, Schultz JA. IR-MALDI-LDI combined with ion mobility orthogonal time-of-flight mass spectrometry. Journal of Proteome Research. 2006;5:1484. doi: 10.1021/pr060055l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson SN, Wang H-YJ, Woods AS, Ugarov M, Egan T, Schultz JA. Direct tissue analysis of phospholipids in rat brain using MALDI-TOFMS and MALDI-ion mobility-TOFMS. Journal of the American Society for Mass Spectrometry. 2005;16:133. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Jackson SN, Wang H-YJ, Woods AS. Direct profiling of lipid distribution in brain tissue using MALDI-TOF MS. Analytical Chemistry. 2005;77:4523. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- 27.Jackson SN, Wang H-YJ, Woods AS. In situ structural characterization of glycerophospholipids and sulfatides in brain tissue using MALDI-MS/MS. Journal of the American Society for Mass Spectrometry. 2007;18:17. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Novikov A, Caroff M, Della-Negra S, Lebeyec Y, Pautrat M, Schultz JA, Tempez A, Wang H-YJ, Jackson SN, Woods AS. Matrix-implanted laser desorption/ionization mass spectrometry. Analytical Chemistry. 2004;76:7288. doi: 10.1021/ac049123i. [DOI] [PubMed] [Google Scholar]
- 29.Tempez A, Ugarov M, Egan T, Schultz JA, Novikov A, Della-Negra S, Lebeyec Y, Pautrat M, Caroff M, Smentkowski VS, Wang H-YJ, Jackson SN, Woods AS. Matrix implanted laser desorption ionization (MILDI) combined with ion mobility-mass spectrometry for bio-surface analysis. Journal of Proteome Research. 2005;4:540. doi: 10.1021/pr0497879. [DOI] [PubMed] [Google Scholar]
- 30.McLean JA, Stumpo KA, Russell DH. Size-selected (2–10nm) gold nanoparticles for matrix assisted laser desorption ionization of peptides. Journal of the American Chemical Society. 2005;127:5304. doi: 10.1021/ja043907w. [DOI] [PubMed] [Google Scholar]
- 31.Agranoff BW, Benjamins JA, Hajra AK. In: Basic Neurochemistry Molecular, Cellular and Medical Aspects. 6. Siegel GJ, Agranoff BW, Albers RW, Fisher SK, Uhler MD, editors. Lippincott Williams & Wilkins; Philadelphia: 1999. p. 47. [Google Scholar]
- 32.Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. Journal of Lipid Research. 2005;46:163. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Borner K, Nygren H, Hagenhoff B, Malmberg P, Tallarek E, Mansson J-E. Distribution of cholesterol and galactosylceramide in rat cerebellar white matter. Biochimica et Biophysica Acta. 2006;1761:335. doi: 10.1016/j.bbalip.2006.02.021. [DOI] [PubMed] [Google Scholar]