Abstract
Most MALDI instrumentation uses UV lasers. We have designed a MALDI–IM–oTOF–MS which employs both a Nd:YAG laser pumped optical parametric oscillator (OPOTEK, λ = 2.8–3.2 μm at 20 Hz) to perform IR–LDI or IR–MALDI and a Nd:YLF laser (Crystalaser, λ = 249 nm at 200 Hz) for the UV. Ion mobility (IM) gives a fast separation and analysis of biomolecules from complex mixtures in which ions of similar chemical type fall along well-defined “trend lines”. Our data shows that ion mobility allows multiply charged monomers and multimers to be resolved; thus, yielding pure spectra of the singly charged protein ion which are virtually devoid of chemical noise. In addition, we have demonstrated that IR–LDI produced similar results as IR–MALDI for the direct tissue analysis of phospholipids from rat brain.
Keywords: ion-mobility MALDI, UV lasers, IR lasers, adducts reduction
Introduction
Matrix-assisted laser desorption ionization (MALDI) is usually performed with ultraviolet (UV) lasers. However, infrared (IR) lasers have been used since the vibrational absorption of certain matrices (native OH) enables a softer desorption/ionization process.1,2 Until recently, the lack of versatile and inexpensive pulsed IR lasers has limited IR–MALDI to research instruments. The Er:YAG laser, with a wavelength of 2.94 μm is well suited for H atom vibrational absorption and has been used extensively by Hillenkamp et al.2 A pulsed CO2 gas laser operating at 10.6 μm was also used for MALDI.3 Recently, tunable infrared sources such as the free-electron laser4 and the tunable optical parametric oscillator (OPO)5–7 have been employed. The advent of smaller and less expensive OPO systems with higher pulse frequencies is making IR–MALDI mass spectrometry increasingly possible. In addition, the allure of a less invasive (matrix free) IR laser desorption/ionization (LDI) technique holds great promise for biological applications such as direct cell and tissue analysis (8–10).
The soft desorption/ionization provided by IR irradiation often results in reduced fragmentation when compared to UV. Recently, IR–MALDI using a glycerol matrix in an orthogonal TOF–MS was employed for the analysis of large proteins up to 150 kDa11, after extensive desalting of both the glycerol and protein solutions.
Desalting of protein/glycerol solutions is essential for reducing adduction in IR–MALDI. However desalting is often impractical for many proteomics applications and impossible for tissue analysis. To address this problem, we have combined IR–MALDI and IR–LDI with ion-mobility orthogonal time-of-flight mass spectrometry (IM–oTOF–MS). Our results are similar to the previously mentioned IR–MALDI study11 without the necessity of desalting the matrix or protein solution. Furthermore, the use of ion mobility allows multiply charged monomers and multimers to be resolved yielding pure singly charged protein ion spectra virtually devoid of chemical noise. In addition, we have demonstrated that IR–LDI and IR–MALDI produce similar results for the direct tissue analysis of phospholipids from rat brain tissue slices.
Experimental Section
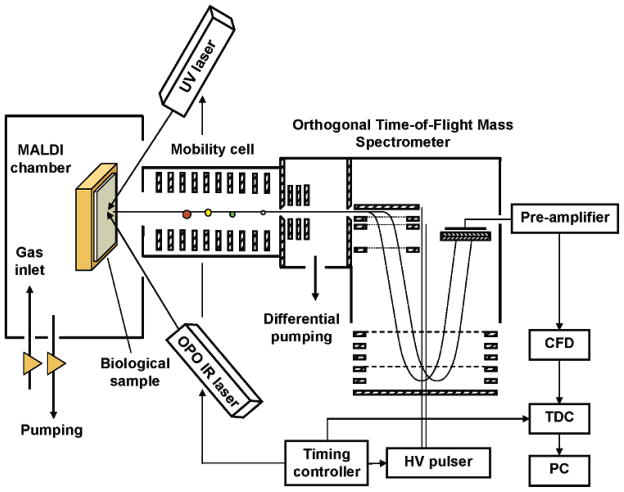
Figure 1 shows our MALDI–IM–oTOF–MS (Ionwerks Inc. Houston TX). A 14 cm long mobility cell is biased with a voltage of 1900 V and produces a mobility resolution of 30 at a He pressure of around 3 Torr. After the preseparated ions exit the mobility cell they pass through a differentially pumped region and enter an oTOF–MS, which achieves a resolution of 3000 for ions with m/z 1000. An X–Y sample stage provides 1 μm accuracy in beam positioning and sample scanning of a standard 96-well sample plate. The sample is at ground potential, which requires the application of an attractive high voltage bias to the entrance and exit end of the ion mobility cell to drag the ions through the He gas. Furthermore, the entire mass spectrometer must be floated on top of this bias voltage.
Figure 1.
Schematic of IR and UV–MALDI–IM–oTOF–MS.
A Nd:YAG laser pumped optical parametric oscillator (OPOTEK, λ = 2.8–3.2 μm at 20 Hz) is used to perform IR–LDI or IR–MALDI and a Nd:YLF laser (Crystalaser, λ = 249 nm at 200 Hz) for the UV. In contrast to high vacuum MALDI, the ablation plume is collisionally cooled within microseconds by interaction with the He drift gas. After the laser pulse, the ions drift to the end of the mobility cell which is biased by 1900 V applied to a resistive divider network connected between the sample plate and the exit of the mobility spectrometer. The mobility separated ions then pass through the skimmer into a differentially pumped orthogonal time-of-flight mass spectrometer where they are mass analyzed and the spectra recorded as a function of mobility drift time after the desorption laser pulse. The mobility drift times are up to several milliseconds while the flight times within the mass spectrometer are tens of microseconds. Therefore, several hundred mass spectra are acquired after every laser pulse at intervals of every 30 to 150 μs (depending on the mass range), and are each stored individually along with its associated mobility time. This process is repeated for several hundred laser shots, until each of the mass spectra contains sufficient intensity to construct plots, such as the ones shown in Figure 2, of the molecular ion mobility as a function of m/z. The data are presented as 2D false color contour plots of ion intensity as a function of ion mobility drift time and mass/charge.
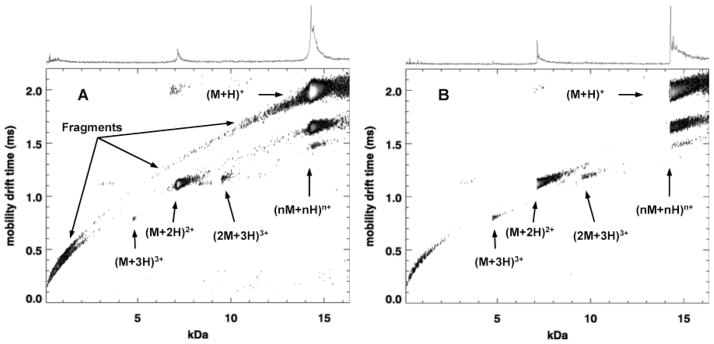
Figure 2.
MALDI–IM–TOF–MS spectra of lysozyme: UV (A) and IR (B). The singly charged fragments in A (which are absent in the IR data of B) form a peptide “trend line” which in the case of the pure Lysozyme must arise by in-source decay. This line has been explicitly added to Figure 3A,B (top panel) which shows an expanded mobility-m/z region around the [M + H]+.
Chicken lysozyme (MW 14313 Da) and equine heart cytochrome C (MW 12 384 Da) were dissolved in a saturated DHB solution (in 50% ethanol) for a final concentration of 100 pmol/μL. A 3-μL portion was deposited on the sample plate to form a large sample spot (~5 mm in diameter) and left to dry at room temperature. Both IR and UV spectra were acquired from different regions of the same sample spot and several replicate runs were conducted. Replicates were also obtained from different sample spots.
Brain tissue sections were prepared as previously described.12
Results and Discussion
Figure 2A,B shows typical UV and IR–MALDI–IM–oTOF–MS spectra of lysozyme in DHB. The IR spectrum (Figure 2b) is virtually devoid of singly charged fragments in contrast to the UV spectrum (Figure 2a). Similar IR–MALDI–IM–oTOF–MS results were also obtained from cytochrome C (Data not shown). On average, each spectrum was acquired until the peak intensity of the MH+ was around 100 ion count. The 1D mass spectrum is derived from the 2D data by adding all ion signals at each m/z irrespective of their mobility; this summed spectrum is shown along the top of each of the 2D–IM–oTOF–MS plots and is approximately what would be seen in a conventional mass spectrometer. The ion mobility separates the chemical noise and multiply charged monomers and multimers on trend lines below the MH+ in both the UV and
IR data. Hence, we can numerically isolate the pure MH+ and singly charged fragment spectra in both cases (Figure 3). The most striking observation is the lack of fragmentation of MH+ and MNa+ in the IR data. The low mass “in source” fragments from the IR desorption/ionization, are almost exclusively along the low mass region of the doubly charged trend line. By contrast in the UV spectrum, the singly charged in-source decay fragments extend prominently in a single trend line from the MH+ down to the lower mass range.
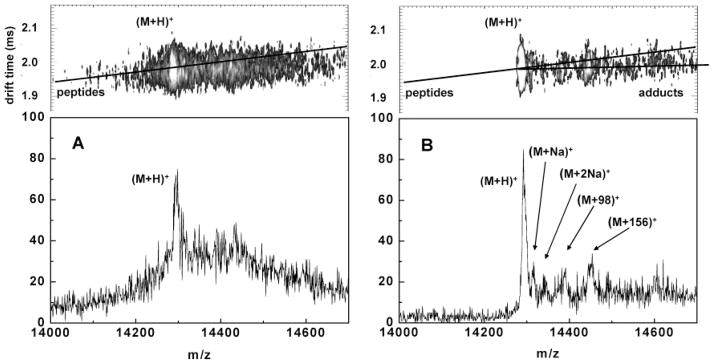
Figure 3.
Derived mass spectra (bottom panels) from the windowed regions (top panel) around [M + H]+ in the ion mobility-m/z data from Figure 2 – UV (A) and IR (B). The peptide trend line has been added from Figure 2A in both top panels and the near horizontal “Adducts” trend line is also shown in B.
The singly charged one-dimensional mass/charge spectra shown in Figure 3 are derived from the IR and UV data of Figure 2A,B from a region of the 2D IM-m/z data around their MH+ ions (shown in the top panel of Figure 3A,B). The chemical noise and fragment broadening of the UV MH+ (3A) are evident. The MH+ peak width for the IR and the UV were 12 and 18 amu, respectively. Because of reduced fragmentation in the IR spectra, the adduct peaks MNa+, [M+2Na]+, and [M+DHB]+are also well resolved. The adduct peaks are further identified by their near horizontal shift relative to the MH+ location shown in the top panel IM-m/z data. This is emphasized by a line labeled “adducts” in Figure 3B which guides the eye through the most prominent adduct peaks. We have also added a “peptide” trend line (both in 3A,B top panels) which is derived from a linear extrapolation of a line through fragment ions near the [M + H]+ in the UV data of 2A.
Increasing both the mobility and mass resolutions in future instruments should allow improved identification through two-dimensional numerical deconvolution iteratively applied along the m/z and IM axes. Increased separation of adducted peptide/protein ions from other coexisting isobaric [M + H]+ peptide/protein ions can be achieved with such a procedure. Improved detection of larger protein ions is planed in future redesigns of our existing instrument. Increasing the moderate intensity of [M + H]+ ions in Figures 2 and 3 is at present limited by detection efficiency, as only 4 keV ions collide with the MCP detector surface.
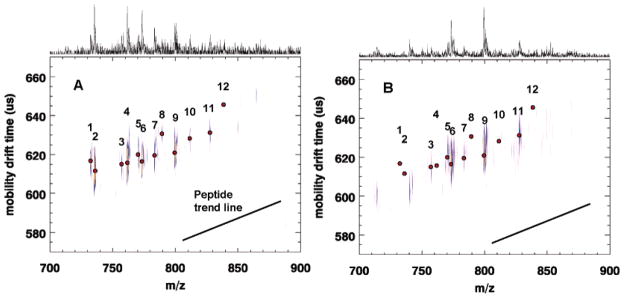
Additional experiments were conducted comparing IR–LDI with IR–MALDI for direct tissue analysis of phospholipids. Figure 4 shows the comparison of the 2D spectra of a rat cerebellum section: (A) IR–MALDI with DHB matrix and (B) IR–LDI. Assignments of major lipid peaks are given in Table 1 and are discussed elsewhere.12 The MH+ of lipids are almost totally absent in the IR–LDI spectrum (4B); this greatly simplifies the IR–LDI spectrum over the more complex IR–MALDI in panel (4A) in which MH+, MNa+, and MK+ are evident. The IR–MALDI (4B) and UV–MALDI (not shown) produce nearly identical lipid spectra (although the relative desorption efficiencies of lipid ions at the two wavelengths is not known). Similar results for direct lipid and peptide analysis in tissues by matrix free IR–LDI–TOF–MS have been seen recently by Dreisewerd et. al. (private communication), who previously used glycerol to successfully analyze gangliosides.13 In addition, IM may give some separation and identification of lipids based on the type of lipid headgroup, although more work is necessary to confirm this point.
Figure 4.
Comparison of (A) IR–MALDI–IM–oTOF–MS and (B) IR–LDI from brain tissue slices.
Table 1.
| 1 | [(SM18:0)+H]+ | 5 | [(SM18:0)+K]+ | 9 | [(PC34:1)+K]+ |
| 2 | [(PC32:0)+H]+ | 6 | [(PC32:0)+K]+ | 10 | [(PC36:1)+Na]+ |
| 3 | [(PC32:0)+Na]+ | 7 | [(PC34:1)+Na]+ | 11 | [(PC36:1)+K]+ |
| 4 | [(PC34:1)+H]+ | 8 | [(PC36:1)+H]+ | 12 | [(SM24:0)+Na]+ |
Conclusions
The combination of IR–MALDI and Ion Mobility–MS could become a useful tool for the identification of proteins in complex mixtures by separating different charge states and adduct ions, thus simultaneously reducing the need for desalting while providing protein spectra as a function of charge state. In particular, the parent ions of proteins remain remarkably intact when acquired by IR–MALDI benefiting important applications, such as biomarker identification, top down proteomics, and tissue imaging. Furthermore, the IR–LDI of lipids appears to be a robust technique for imaging analysis of this important class of biomolecules. Our results strongly argue that more efficient IR matrices should be identified. Nanoparticulates, which can localize in the near surface region, were shown to be effective UV–MALDI matrices for tissue probing14 and may now be considered as possible IR matrix candidates (along with any other material whose ions can be filtered from the bioanalytes by IMS).
Acknowledgments
Ionwerks is grateful for partial support from NIH for NIDA contracts N44DA-3-7727 and HHSN271200477384C, NRCC phase I Grant No. 1R43RR020238-01, and to Marjorie Schultz for providing personal funds for the remainder of our participation in this work. A.S.W. and J.A.S. appreciate Isabelle Fournier and Michel Salzet generously discussing their unpublished data on matrix free coaxial IR–LDI of lipids from tissue. This research was also funded by the NIH/NIDA IRP.
References
- 1.Berkenkamp S, Karas M, Hillenkamp F. Proc Natl Acad Sci USA. 1996;93:7003–7007. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkenkamp S, Menzel C, Karas M, Hillenkamp F. Rapid Commun Mass Spectrom. 1997;11:1399–1406. [Google Scholar]
- 3.Overberg A, Karas M, Hillenkamp F. Rapid Commun Mass Spectrom. 1991;5:128–131. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- 4.Cramer R, Hillenkamp F, Haglund RF. J Am Soc Mass Spectrom. 1996;7:1187–1193. doi: 10.1016/S1044-0305(96)00111-0. [DOI] [PubMed] [Google Scholar]
- 5.Sadeghi M, Olumee Z, Tang X, Vertes A, Jiang ZX, Henderson AJ, Lee HS, Prasad CR. Rapid Commun Mass Spectrom. 1997;11:393–397. doi: 10.1002/(SICI)1097-0231(19970228)11:4<393::AID-RCM868>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell KL, McGarity DR, Murray KK. J Mass Spectrom. 1997;32:1374–1377. [Google Scholar]
- 7.Menzel C, Dreisewerd K, Berkenkamp S, Hillenkamp F. Int J Mass Spectrom. 2001;207:73–96. doi: 10.1002/1096-9888(200011)35:11<1320::AID-JMS66>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya SH, Raiford TJ, Murray KK. Anal Chem. 2002;74:2228–2231. doi: 10.1021/ac0112972. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Little MW, Rousell DJ, Laboy JL, Murray KK. Anal Chem. 2004;76:1078–1082. doi: 10.1021/ac034879n. [DOI] [PubMed] [Google Scholar]
- 10.Rousell DJ, Dutta SM, Little MW, Murray KK. Anal Chem. 2004;39:1182–1189. doi: 10.1021/ac034879n. [DOI] [PubMed] [Google Scholar]
- 11.Berkenkamp S, Hillenkamp F. Infrared MALDI with an Orthogonal Ion Extraction TOF for high performance analysis of large proteins. Annual ASMS meeting; San Antonio, TX. 2005. [Google Scholar]
- 12.Jackson SN, Wang HYJ, Ugarov MV, Egan T, Schultz JA, Woods AS. Direct Tissue Analysis of Phospholipids in Rat Brain Using MALDI TOFMS and MALDI-Ion Mobility TOFMS. J Am Soc Mass Spectrom. 2005;16:133–138. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Dreisewerd K, Muthing J, Rohlfing A, Meisen I, Vukelic Z, Peter-Katalinic J, Hillenkamp F, Berkenkamp S. Analysis of gangliosides directly from thin-layer chromatography plates by infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry with a glycerol matrix. Anal Chem. 2005;77:4098–4107. doi: 10.1021/ac048373w. [DOI] [PubMed] [Google Scholar]
- 14.Tempez A, Ugarov M, Egan T, Schultz JA, Novikov A, Della-Negra S, Lebeyec Y, Pautrat M, Caroff M, Smentkowski VS, Wang HYJ, Jackson SN, Woods AS. Matrix implanted laser desorption ionization (MILDI) combined with ion mobility-mass spectrometry for bio-surface analysis. J Proteome Res. 2005;4:540–545. doi: 10.1021/pr0497879. [DOI] [PubMed] [Google Scholar]