Abstract
The human D4 dopamine receptor is a synaptic neurotransmitter receptor responsible for neuronal signaling in the mesolimbic system of the brain, an area of the brain that regulates emotion and complex behavior. Its structure makes it a very unusual and interesting G protein-coupled receptor (GPCR) as it has several polymorphic variants of its gene in the region encoding the third intracellular loop (IL3). This region contains from two to seven or more similar 48 base pair repeats. These repeats cause this protein to have a very high disorder index and this, in turn, makes it very interactive with other proteins. Among GPCRs in general, the unusually proline-rich IL3 is unique to the D4 receptor (D4R). We believe that, as in the D2R, this region of the receptor plays a role in it’s interaction with other receptors.
Keywords: Dopamine D4 Receptor, Disordered proteins, AKAP, Heteromers, Adenosine A2A Receptor
Introduction
The neurotransmitter dopamine interacts with two dopamine receptors’ type, the D1 and D2. The D1 has one subtype (D5), while the D2 has two subtypes the D3 and D4. However, D1 and D2 have the largest expression in the brain. The D2, D3 and D4 receptors can inhibit adenylate cyclase, while D1 and D5 activate it (1–3). The D4 receptor (D4R) has structural, functional, and pharmacological characteristics closely related to the dopamine D2 receptor. According to Van Tol, both genomic structure (i.e. intron-exon organization) and primary sequence show very high similarity to the D2 receptor (4–7). We have previously demonstrated that the D2 receptor (D2R; P14416) has an arginine (Arg) rich epitope 217RRRRKRVNpTKRpS228 at the amino-terminus of the third intracellular loop (IL3) and that this epitope is involved in heteromer formation with an adenosine A2A receptor’s (A2A R) [P29274] epitope located in its carboxy terminus and containing a phosphorylated serine residue [Ser374] from a casein kinase (CK) consensus site (Figure 1A and Table 1) (8–12). Downstream from that Arg-rich area are Thr225 and Ser228 which have consensus sites amenable to phosphorylation with protein kinase A/protein kinase C (PKA/PKC). However, in all D4R subtypes the Arg-rich domain is located in the carboxy terminus of IL3 while the phosphorylatable residues, two Thr, are located in a PKA/PKC consensus sites, one downstream and another upstream, flank the Arg-rich epitope sequence, QpTRRRRRAKIpT. (Figure 1B).
Figure 1.
(A) Location of Arg-rich epitope. (B) AKAP anchoring kinases and phosphatases.
Table 1.
Dopamine D2, D3 and D4 conserved basic epitope.
| IL3 | # aa | Receptor | Arg-rich epitope sequence |
|---|---|---|---|
| 211–373 | 163 | Dopamine D2 | VLRRRRKRVNpTKRpSSR |
| 210–329 | 120 | Dopamine D3 | KQRRRKRILpTRQNSQC |
| 214–346 | 133 | Dopamine D4 | QpTRRRRRAKIpTGRERK |
IL3, third intracellular loop.
The D4 receptors presynaptic localization is in glutamatergic terminals while the postsynaptic localization is in the dendrites of the GABAergic efferent neurons. The (A2A R) is mostly localized in the striatum in glutamatergic terminals and in GABAergic efferent neurons, the striatopallidal neurons. Their co-localization suggests the possibility of heteromer formation between A2A R and D4 R (11,13,14). The dopamine D4 has several subtypes that differ in the number of polymorphic repeat sequence of 2–10 repeated units of 16 amino acids that occur in the IL3. D4 receptor genes encoding receptors with two (D4.2), four (D4.4) [Q8NGM5-1], or seven (D4.7) [P21917] repeat units are the most abundant (4–7). The D4 sequence is also unusually rich in its content of the amino acid proline. Src homology 3 binding domains (SH3) domains are recognized by proline-rich proteins.
Disordered proteins
Many proteins or segments of proteins lack three-dimensional structure. These unstructured regions are called ‘disordered regions’. According to Dyson and Wright’s excellent review (15), these proteins are most often then not highly conserved between species in both composition and sequence. This is the case for the A2AR, D2R, D3R, and D4R (16) and, contrary to the traditional view that functional proteins always have a stable three-dimensional structure (some do not), nevertheless disordered regions are often functional. They are rich in disorder promoting amino acids (A, R, G, Q, S, P, E, and K) and have a low content of order-promoting amino acids (W, C, F, I, Y, V, L, and N) (16). Thus, they are characterized by an overall high net charge and low hydrophobicity, which supports our findings that epitopes involved in the initiation of receptor heteromerization do it through salt bridge formation driven by an electrostatic interaction not a hydrophobic one (9–12). Usually disordered segments of a protein fold on binding to their biological targets, which gives them the conformation that a disorder protein lacks. The disorder to order transitions, which take place during binding interactions of the disordered regions, makes the interaction highly reversible and at the same time highly specific. D2, D3, D4, and A2A receptors also qualify for the title of ‘hub proteins’ (15–18), which are defined as proteins with greater than five interaction partners. Bioinformatics analysis of the A2AR and D2 R using various programs (16–18) clearly shows that the IL3 of the D2R and the carboxyl terminus of the A2AR have both a high disorder index. This explains their propensity to interact with each other and with other receptors (17–22). It also emphasizes the importance of the electrostatic interaction between the D2R, D3R, and D4R IL3′ and the A2AR carboxyl terminus epitope and other receptors similar epitope (12), as indeed the first and necessary step in heteromer formation.
Heteromer formation
To test for the possibility of heteromer formation between the D4R Arg-rich epitope and the adenosine A2A receptor epitope SAQEpSQGNT that interacts with the Arg-rich regions of D2R, D3R, and D4 R, synthetic peptides representing the phosphorylated and non-phosphorylated D4 epitope [QpTRRRRRAKIpT and QTRRRRRAKIT] and the A2AR epitope were made and an equimolar solution of all three peptides was analyzed by MALDI mass spectrometry in positive ion mode. As in the case of the D2R-A2A interaction, the D4R and A2AR interaction seen in the MALDI mass spectrum in Figure 2, shows formation of non-covalent complexes (NCXs) between the epitopes of the D4R and A2AR (relative abundance 6 and 25% respectively). The normalized relative abundance of the MH+ of the NCXs of SAQEpSQGNT with the epitopes of the D4 R QpTRRRRRAKIpT and QTRRRRRAKIT was 11.0% and 25.0%, respectively. Normalization was obtained by dividing the relative abundance of the molecular ion [MH+] of the NCXs by the relative abundance of the MH+ of the corresponding phosphorylated and non-phosphorylated epitopes of the D4 receptor. We previously demonstrated that heteromerization of adenosine A2A with dopamine D2 receptors and glutamate N-methyl-d-aspartate (NMDA) receptors (NR1-1 subunit) with dopamine D1 receptors depends on epitope–epitope electrostatic interactions (9,10) and that this is a general mechanism for receptor heteromerization (12). We found striking similarities between the epitopes involved in the A2A-D2 and NMDA-D1 receptor heteromerization. Similarities were seen in an (1) Arg-rich epitope, localized in the N-terminal portion of the IL3 of the D2 receptor, and the C-terminus of the NR1-1 subunit of the NMDA receptor (2); on the opposing epitopes of the C-termini of both A2A and D1 receptors, a serine (Ser) residue susceptible of phosphorylation by CK1 (pSer374 and pSer397 respectively) (3); a PKA consensus site adjacent to the Arg-rich epitope of the D2 receptor and the NR1-1 subunit of the NMDA receptor. The same parameters are found with the D4R-A2A interaction.
Figure 2.
Mass spectrum showing the non-covalent complex formed between the phosphorylated and non-phosphorylated the epitopes of the D4R and A2AR.
The role of proline
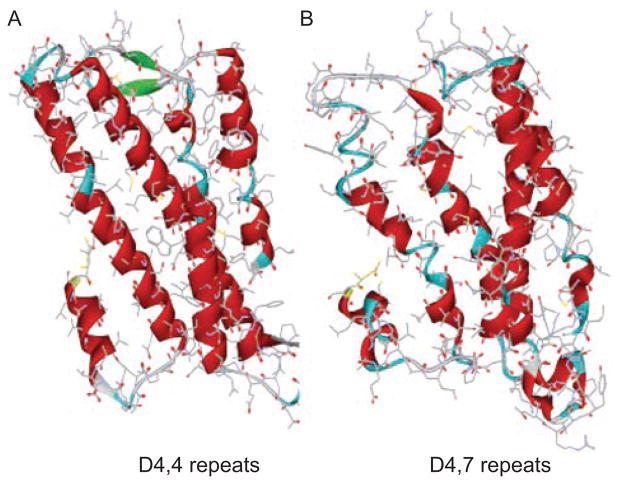
Amino acid composition and modifications determine the structure and function of a protein. When perusing the amino acid sequence of the D4R subtypes, one immediately notices the overwhelming presence of the amino acid residue proline which is the only cyclic amino acid. Its cyclic structure strongly influences the secondary structures of proteins, as its amino group is a secondary amine, so formation of a peptide bond yields an amide in which there is no N-H hydrogen bond donor function. Due to its rigidity, proline is frequently located in turns or bends, which are often on the surface of a protein (23). Thus the kinking of protein strands generated by proline results in destabilized α-helix formation. This is clearly seen in Figure 3 where a model of the D4-4 Rpts-IL3 has more α-helices than the D4-7 Rpts-IL3. Proline-rich segments found in the IL3 of the D4 receptor in all species allow it to serve as a docking site for signaling proteins that become targets for phospholipase methylation [phospholipid methylation (PLM)]-based modulation.
Figure 3.
T model of the D4,4 repeats (A) third intracellular loop (IL3) has more α-helices than the D4,7 repeats-IL3 (B).
The frequency of proline residues in most proteins is 5%. However, as seen in Table 2 it is around 12 and 15% in D4-4 Rpts and D4-7 Rpts, respectively and 27 and 30% in their IL3, while it is somewhat elevated in D2R and D3 R IL3 (8 and 9% respectively; Table 2). This is an unusually high proline content for mammalian proteins, as high proline concentration is usually seen in thermophiles’ proteins. In humans and other primates, the D4 receptor possesses anywhere from 2 to 11 additional proline-rich repeat segments (Figure 3), and a higher number of repeats (i.e. seven) brings an increased risk of attention-deficit hyperactivity disorder (ADHD) (5).
Table 2.
Percentage of prolines in the D2, D3 and D4 receptors and in their third intracellular loop.
| Receptor | # aa Res | # Of prolines | % Of total aa Res |
|---|---|---|---|
| D4-7 Rpts | 467 | 69 | 14.8% |
| D4-7 Rpts-IL3 | 181 | 54 | 29.3% |
| D4-4 Rpts | 419 | 50 | 11.9% |
| D4-4 Rpts-IL3 | 133 | 35 | 26.5% |
| D2 | 443 | 27 | 6.1% |
| D2-IL3 | 163 | 15 | 9.2% |
| D3 | 400 | 20 | 5.0% |
| D3-IL3 | 120 | 10 | 8.3% |
IL3, third intracellular loop.
A-kinase anchoring proteins
A-kinase anchoring proteins (AKAPs) are scaffold proteins which coordinate the subcellular localization of second-messenger-regulated enzymes such as: PKA or PKC and protein phosphatases. They also have domains that interact with G protein-coupled receptors (GPCRs). AKAPs facilitate and localize dopamine-initiated signal cascades, leading to activation of protein kinases, as well as the turning-off of such signals. Second messengers regulate synaptic plasticity by influencing the balance between kinase and phosphatase activity. AKAP interacts with PKA and a calcium-dependent phosphatase PP2B. AKAPs (24) are signal organizing molecules that compartmentalize kinases and phosphatases that are regulated by second messengers. D4R IL3 contains several SH3 in its polymorphic repeat sequence. SH3 domains are found in numerous proteins involved with signal transduction pathways, and form β-barrel like structures that bind proline-rich sequences. Therefore the presence of multiple repeats greatly increases the # of prolines and subsequently the # of SH3 recognition sites. Thus, proteins such as AKAP, which contain SH3 domains recognized by the proline-rich D4 IL3, can tether the kinases PKA and PKC, and phosphatases making phosphorylation/dephosphorylation of the D4,7 repeats more likely than phosphorylation of the D4,4 repeats variants.
The SH3 domains β-barrel shape consisting of five β-strands is arranged as two tightly packed anti-parallel β sheets (25,26). The SH3-type fold is found in both eukaryotes and prokaryotes. SH3 domains are usually found in disordered proteins that have the propensity to interact with other proteins thus mediating the assembly of certain protein complexes, by binding to proline-rich motifs in certain proteins. SH3 domains are mostly found in intracellular proteins domain which is the case for the D4R IL3.
SH3-binding epitopes usually have the consensus sequence:
| -X | -P | -p | -X | -P- |
| 1 | 2 | 3 | 4 | 5 |
Residues 1 and 4 are usually aliphatic, 2 and 5 are always proline, while 3 is also sometimes a proline. The protein AKAP contains SH3 domains recognized by the proline-rich D4 IL3.
In conclusion, what might drive the function of the D4R is the unusual predominance of prolines in its sequence, leading to anchoring of several AKAPs on its IL3. These AKAPs anchor kinases and phosphatases in the proximity of the basic epitope located at the carboxy terminus of IL3, thereby locating all the needed enzymes for phosphorylation and dephosphorylation in the proximity of the important epitope. When phosphorylated, [QpTRRRRRAKIpT] is less likely to interact with phosphorylated epitopes of the A2AR (11%) and when not phosphorylated [QTRRRRRAKIT] the interaction likelihood doubles to 25%. The human D4 dopamine receptor is a synaptic neurotransmitter receptor responsible for neuronal signaling in the mesolimbic system of the brain, a part of the brain that regulates emotion and complex behavior. Phosphorylation/dephosphorylation has always been considered to be an ON/OFF switch. The D4R structure makes it a very efficient switch and is a particularly interesting GPCR because the large number of polymorphic variants of the receptor gene in humans. This results in variations that affect the efficiency of this receptor as a switch. It seems that the more repeats the more efficient it probably becomes. The gene varies in the nucleotide sequence of the region encoding IL3, which contains from two to seven or more similar 48 base pair repeats. (These nucleotide repeats code for more than 18 different sequences.) Among GPCR, the proline-rich IL3 is unique to the D4R. We believe that as in the D2R, the role of this region of the receptor is to interact with other receptors (12,27).
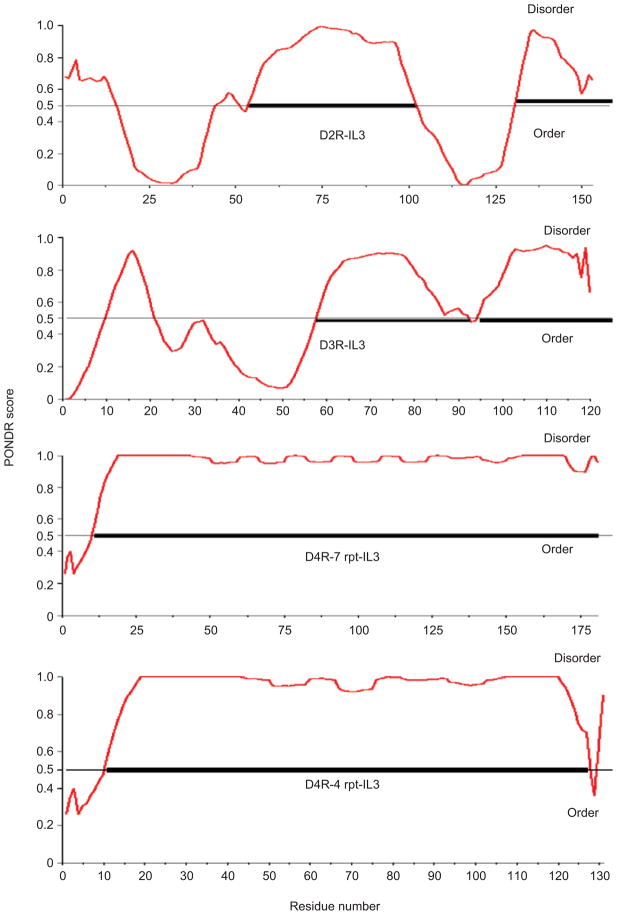
In addition, proline-rich segments found in the IL3 of the D4 receptor in all species allow it to serve as a docking site for signaling proteins that become targets for PLM-based modulation. In humans and other primates, the D4R proline-rich segments (Figure 4), and a higher number of repeats (i.e. seven) shows maximum disorder (28) 90 and 97%, respectively for the D4R-4 rpts and D4R-7Rpts (Table 3) and is accompanied by an increased risk of ADHD (5).
Figure 4.
Shows graphs of the disorder index of the D2, D3 and D4 (four and seven repeat) third intracellular loop The D4-7 is twice as disordered as the D2 or D3, therefore it is more interactive.
Table 3.
% Disorder in IL3 of dopamine receptor.
| Receptor | % Disorder |
|---|---|
| D4R-7 rpts | 97 |
| D4R-4 rpts | 90 |
| D2R | 26 |
| D3R | 22 |
D2, D3, D4 receptor; IL3, third intracellular loop.
The D4 R SH3-binding sites are within a region involved in the control of receptor internalization. Through heteromerization, receptors establish new functional interactions, and acquire new functional properties that differ from those of either receptor unit when not part of a receptor heteromer unit.
We would like to emphasize that SH3 are protein domains found in many proteins involved with signal transduction pathways. SH3 domains form β-barrel like structures that bind proline-rich sequences. Hence the presence of multiple repeats greatly increases the # of prolines and therefore the # of SH3 recognition sites. This fact results in proteins such as AKAP, which contain SH3 domains, to be recognized by the proline-rich D4 IL3. This can, in turn, tether the kinases PKA and PKC and phosphatases thus making phosphorylation/dephosphorylation of the D4,7 repeats more likely than phosphorylation of the D4,4 repeats or other variants containing less prolines. One cannot emphasize enough the role of AKAPs as scaffold proteins which coordinates the subcellular localization of second-messenger-regulated enzymes such as: PKA or PKC and protein phosphatases. AKAPs facilitate and localize dopamine-initiated signal cascades, leading to activation of protein kinases, as well as the turning-off of such signals. Second messengers regulate synaptic plasticity by influencing the balance between kinase and phosphatase activity. AKAP interacts with PKA and a calcium-dependent phosphatase PP2B.
Footnotes
Declaration of interest
This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, NIH. The authors thank the Office of National Drug Control Policy (ONDCP) for instrumentation funding, without which this and other projects could not have been accomplished.
References
- 1.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 2.Aizman O, Brismar H, Uhlén P, Zettergren E, Levey AI, Forssberg H, Greengard P, Aperia A. Anatomical and physiological evidence for D1 and D2 dopamine receptor co-localization in neostriatal neurons. Nat Neurosci. 2000;3:226–230. doi: 10.1038/72929. [DOI] [PubMed] [Google Scholar]
- 3.Surmeier DJ, Song WJ, Yan Z. Coordinated expression of dopamine receptors in neostriatal medium spiny neurons. J Neurosci. 1996;16:6579–6591. doi: 10.1523/JNEUROSCI.16-20-06579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Tol HH, Wu CM, Guan HC, Ohara K, Bunzow JR, Civelli O, Kennedy J, Seeman P, Niznik HB, Jovanovic V. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358:149–152. doi: 10.1038/358149a0. [DOI] [PubMed] [Google Scholar]
- 5.Oak JN, Oldenhof J, Van Tol HH. The dopamine D(4) receptor: one decade of research. Eur J Pharmacol. 2000;405:303–327. doi: 10.1016/s0014-2999(00)00562-8. [DOI] [PubMed] [Google Scholar]
- 6.Livak KJ, Rogers J, Lichter JB. Variability of dopamine D4 receptor (DRD4) gene sequence within and among nonhuman primate species. Proc Natl Acad Sci USA. 1995;92:427–431. doi: 10.1073/pnas.92.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oldenhof J, Vickery R, Anafi M, Oak J, Ray A, Schoots O, Pawson T, von Zastrow M, Van Tol HH. SH3 binding domains in the dopamine D4 receptor. Biochemistry. 1998;37:15726–15736. doi: 10.1021/bi981634+. [DOI] [PubMed] [Google Scholar]
- 8.Ciruela F, Burgueño J, Casadó V, Canals M, Marcellino D, Goldberg SR, Bader M, Fuxe K, Agnati LF, Lluis C, Franco R, Ferré S, Woods AS. Combining mass spectrometry and pulldown techniques for the study of receptor heteromerization. Direct epitope-epitope electrostatic interactions between adenosine A2A and dopamine D2 receptors. Anal Chem. 2004;76:5354–5363. doi: 10.1021/ac049295f. [DOI] [PubMed] [Google Scholar]
- 9.Woods AS, Ciruela F, Fuxe K, Agnati LF, Lluis C, Franco R, Ferré S. Role of electrostatic interaction in receptor-receptor heteromerization. J Mol Neurosci. 2005;26:125–132. doi: 10.1385/JMN:26:2-3:125. [DOI] [PubMed] [Google Scholar]
- 10.Woods AS, Ferré S. Amazing stability of the arginine-phosphate electrostatic interaction. J Proteome Res. 2005;4:1397–1402. doi: 10.1021/pr050077s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuxe K, Marcellino D, Guidolin D, Woods AS, Agnati LF. Heterodimers and receptor mosaics of different types of g-protein- coupled receptors. Physiology. 2008;23:322–332. doi: 10.1152/physiol.00028.2008. [DOI] [PubMed] [Google Scholar]
- 12.Woods AS, et al. Electrostatic interactions as key determinants of the quaternary structure of receptor heteromers. JBC. 2010 doi: 10.1074/jbc.M110.115634. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanganelli S, Sandager Nielsen K, Ferraro L, Antonelli T, Kehr J, Franco R, Ferré S, Agnati LF, Fuxe K, Scheel-Krüger J. Striatal plasticity at the network level. Focus on adenosine A2A and D2 interactions in models of Parkinson’s Disease. Parkinsonism Relat Disord. 2004;10:273–280. doi: 10.1016/j.parkreldis.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Fuxe K, Canals M, Torvinen M, Marcellino D, Terasmaa A, Genedani S, Leo G, Guidolin D, Diaz-Cabiale Z, Rivera A, Lundstrom L, Langel U, Narvaez J, Tanganelli S, Lluis C, Ferré S, Woods A, Franco R, Agnati LF. Intramembrane receptor-receptor interactions: a novel principle in molecular medicine. J Neural Transm. 2007;114:49–75. doi: 10.1007/s00702-006-0589-0. [DOI] [PubMed] [Google Scholar]
- 15.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 16.Agnati LF, Fuxe K, Woods A, Genedani S, Guidolin D. Theoretical considerations on the topological organization of receptor mosaics. Curr Protein Pept Sci. 2009;10:559–569. doi: 10.2174/138920309789630606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 19.Romero P, Obradovic Z, Dunker AK. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 20.Linding R, Jensen F, Diella P, Bork T, Gibson J, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Vullo A, Bortolami O, Pollastri G, Tosatto SCE. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34:W164–W168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creighton TE. Proteins Structure and Molecular properties. 2. New-York: W.H. Freeman and Company; 1993. Chapter 3: amino acid residues; pp. 6–20. [Google Scholar]
- 24.Malbon CC, Tao J, Hsien-yu Wang HY. AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem J. 2004;379:1–9. doi: 10.1042/BJ20031648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawson T, Schlessingert J. SH2 and SH3 domains. Curr Biol. 1993;3:434–42. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 26.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 27.Ferré S, Baler R, Bouvier M, Caron MG, Devi LA, Durroux T, Fuxe K, George SR, Javitch JA, Lohse MJ, Mackie K, Milligan G, Pfleger KD, Pin JP, Volkow ND, Waldhoer M, Woods AS, Franco R. Building a new conceptual framework for receptor heteromers. Nat Chem Biol. 2009;5:131–134. doi: 10.1038/nchembio0309-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radivojac P, Obradovic Z, Brown CJ, Bunker AK. Prediction of boundaries between intrinsically ordered and disordered protein regions. Pac Symp Biocomput. 2003;8:216–227. [PubMed] [Google Scholar]