Abstract
SIM1 (single-minded 1) haploinsufficiency is responsible for obesity in both humans and mice, but the contribution of frequent DNA variation to polygenic obesity is unknown. Sequencing of all exons, exon/intron boundaries, 870 base pairs (bp) of the putative promoter, and 1,095 bp of the 3′UTR of SIM1 gene in 143 obese children and 24 control adults identified 13 common variants. After analysis of the linkage disequilibrium (LD) structure, association study of eight variants was performed in 1,275 obese children and severely obese adults, in 1,395 control subjects, and in 578 obesity-selected pedigrees. A nominal evidence of association was found for the nonsynonymous variant P352T C/A (rs3734354) (P = 0.01, OR = 0.81 (0.70–0.95)), the +2,004 TGA −/insT SNP (rs35180395) (P = 0.02, OR = 1.21 (1.02–1.43)), the +2,215A/G TGA SNP (rs9386126) (P = 0.002, OR = 0.81 (0.71–0.93)), and pooled childhood/adult obesity. Even though transmission disequilibrium test (TDT) further supported the association of P352T and +2,004 −/inst T with obesity, none of these nominal associations remained significant after a multiple testing Bonferroni correction. Therefore, our study excludes a major contribution of SIM1 common variants in exons, 5′ and 3′ UTR regions in polygenic obesity susceptibility in French Europeans.
Strong evidence for a genetic contribution to monogenic early-onset human obesity has been brought by the identification of rare mutations with major functional defects in genes involved in the melanocortinergic pathways (1). Furthermore, it has been shown that frequent single-nucleotide polymorphism (SNP) variation in or near some of these genes can increase the risk or in the contrary protect from more common forms of obesity (2–6).
We previously performed a genome-wide scan in 115 multiplex French obese white families having at least two obese sibs and reported the most significant evidence of linkage for chromosome 6q13–q24 locus (7). SIM1 lies in this locus. This gene encodes a human homolog of Drosophila sim (single-minded), a transcription factor that belongs to the bHLH (basic helix-loop-helix) family of protein (8). This gene, consisting of 11 exons and spanning 75 kilobases of genomic DNA, is highly expressed in the paraventricular nucleus of the hypothalamus known for its pivotal role in food intake (8).
In rodents, SIM1 gene plays an important role in the downstream activity of the leptin/melanocortin pathway (9–13). Heterozygous sim1(+/−) knockout mice develop hyperphagic obesity, enhanced sensitivity to diet-induced obesity but have normal metabolism and energy expenditure (11–13). Expression of shRNA directed against SIM1 into the paraventricular nucleus by stereotaxic adenovirus delivery resulted in a significant increase in food intake, whereas adenovirus-mediated overexpression of SIM1 induced a decrease in food intake (14). In comparison to wild-type mice, transgenic mice overexpressing human SIM1 gene are resistant to diet-induced obesity on a high-fat diet (15). The SIM1 transgene also completely rescued the hyperphagia and partially rescued the obesity of agouti yellow (A(y)) mice in which melanocortin signaling is abrogated (15). In humans, a balanced translocation disrupting the SIM1 gene on the 6q locus has been shown to induce profound obesity (16), and we recently found that rare coding deleterious mutations were associated with inherited Prader–Willi-like syndrome or monogenic obesity (F. Stutzmann et al., unpublished data).
So far, genome-wide association studies excluded a major role of SIM1 common SNP variation in BMI variation or obesity risk in European populations (6, 17, 18). However, a recent study of the SIM1 locus performed in 6,000 Pima Indians provided strong evidence of association of a linkage disequilibrium (LD) block spanning from 5′UTR to intron 8 with obesity risk (P < 10−7) (19). This prompted us to analyze the contribution of SIM1 exonic, 5′ and 3′ UTR variation in 1,275 obese children and severely obese adults, in 1,395 control subjects and in 578 obesity-selected pedigrees of French European origin.
METHODS
Population used for the sequencing of the gene
We sequenced all exons, exon/intron boundaries, 870 base pairs (bp) of the putative promoter, and 1,095 bp of the 3′UTR in 143 unrelated obese children and 24 unrelated nonobese French white adults. The obese subset of children included 47 French children from families with evidence for linkage of childhood obesity to 6q (maximum likelihood score >1) and 96 additional obese children (z score of BMI >97th percentile).
Population used for association studies and TDT
All subjects were French whites. Association studies with childhood and adulthood obesity were performed for the eight frequent variants (minor allele frequency ≥5%) using a set of 602 obese children chosen from the cohort of 849 obese children available (male/female = 402/447, age = 10.7 ± 3.60 years, BMI = 28.84 ± 6.56 kg/m2, and z score of BMI = 4.16 ± 1.32), 673 severely obese adults (male/female = 171/502, mean age = 45.95 ± 12.06, BMI = 47.69 ± 7.22 kg/m2), and 1,395 nonobese normoglycemic adults (male/female = 558/837, age = 41.37 ± 15.93, and BMI = 22.38 ± 2.35 kg/m2). All case and control subjects were unrelated.
Obese children cohort
The pool of obese children used for case/control analysis was constituted for a first set of 420 unrelated obese children collected from 420 pedigrees with at least one obese child and an obese first-degree relative at the CNRS-UMR8090 Unit in Lille; a second set of 92 unrelated obese children recruited at the Children’s Hospital, Toulouse; and a third set of 90 children collected from the Trousseau Hospital. Children with BMI greater than the 97th percentile for age and sex reported on the tables of Rolland-Cachera et al. (French general population) were defined as obese as recommended by the European Childhood Obesity Group (20,21).
Obese adults cohort
The obese adult subgroup was constituted by 673 severely obese (BMI ≥40 kg/m2) adults collected at the Department of Nutrition of the Hotel Dieu Hospital in Paris or at the CNRS-Institut Pasteur Unit in Lille.
Families used for TDT
We used 420 pedigrees with childhood obesity (645 childhood obesity trios—two parents and one obese child) and 158 pedigrees with adulthood obesity including 514 individuals (303 obese (BMI ≥30 kg/m2) subjects) for transmission disequilibrium test (TDT) analysis for obesity status. There was an overlap of 158 obese adults and of 420 obese children between the case–control and the TDT studies.
Control adults cohort
The same adult control group was used for both association studies in obese children and adults as this group had a longer environmental exposure and still remains nonobese. This group consisted of 1,395 nonobese (BMI <27 kg/m2) normoglycemic (fasting glycemia <5.56 mmol/l) French white adults pooled from four separate studies; 394 unrelated nonobese and nondiabetic subjects were recruited at the CNRS-UMR8090 Unit in Lille, 265 were recruited by the “Fleurbaix-Laventie Ville Santé” study (22), 365 from the Haguenau study (23), and 371 from the SUVIMAX study (24). Absence of stratification among the different case and control cohorts was verified using 47 neutral polymorphic markers disseminated across the genome (data not shown). The genetic study was approved by the ethical committee’s of Hôtel Dieu Hospital in Paris and the CHRU (Centre Hospitalier Regional Universitaire of Lille).
Sequencing and genotyping
The screening of the SIM1 gene was performed using overlapping PCR fragments that cover all SIM1 exons, exon/intron junctions, 870 bp of the putative promoter, and 1,095 bp in the 3′UTR (untranslated region). Primer details and PCR optimization conditions are available from the authors. PCR amplifications were inspected for single bands of expected sizes on agarose gels before purification with Montage PCR384 Multiscreen S384PCR (Millipore, Billerica, MA). Sequencing was performed using the automated ABI Prism 3730 DNA sequencer in combination with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA) and purification sequencing reaction with MultiScreen SEQ384 filter plates (Millipore).
Eight SNPs with a minor allele frequency >5% were genotyped in all case and control groups using direct sequencing for the SNPs +2,004 TGA −/insT, +2,215 TGA A/G, and +2,222 TGA G/A and using Light-Cycler/Typer technology (Roche, Basel, Switzerland) for the SNPs −127 ATG T/C, IVS5 −15 A/C, IVS7 −53 A/G, P352T C/A, and +113 TGA A/T. Genotyping error rates calculated from duplicate genotypes of 250 individuals were 0% for P352T C/A, +2,004 TGA −/insT, +2,215 TGA A/G, +2,222 TGA G/A, −127 ATG T/C SNPs; 0.4% for IVS5 −15 A/C and IVS7 −53 A/G SNPs; and 0.7% for the +113 TGA A/T SNP. No recurrent Mendelian inconsistencies were detected in the pedigrees for the three analyzed SNPs using the PedCheck version 1.1 software (http://watson.hgen.pitt.edu/register/docs/pedcheck.html, developed by Jeff O’Connell, University of Pittsburgh, Pittsburgh, PA).
Plasmid constructions
Double mutant P352T/A371V was generated by site-directed mutagenesis from pcDNA3.1-SIM1-wt vector, using the “Quick change site-directed Mutagenesis Kit” (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. All plasmids were sequence-verified using a 3730 sequencer (Applied Biosystems).
Cell culture and transfection
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol) fetal calf serum, 4.5 g/l glucose, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 1 mmol/l glutamine (Life Technologies, Carlsbad, CA). Cells were incubated at 37 °C in humidified air containing 5% CO2, and transfections were carried out using the FuGene6 reagent (Roche) as described by the manufacturer.
SIM1 transcriptional activity
Reporter gene assay
HeLa cells were first transfected in suspension with 75 ng of the pHRE-LUC firefly reporter construct, 450 ng of the ARNT2 expressing vector (hARNT2 (TC114814) - origene), and 0.1 ng of the pRL-SV40 plasmid and seeded in 12-well plates. Twenty-four hours after transfection, cells were dien transfected widi 500 ng of WT or P352T/A371T mutant SIM1 expressing vectors.
After 48 hours, cells were lysed, and the luciferase activities were measured using the Dual Luciferase Assay System (Promega, Madison, WI) with a Berthold Luminometer (Lumat LB 9507; Berthold Detection Systems, Huntsville, AL). Firefly luciferase activity was normalized to Renilla luciferase activity, and results were expressed relative to the wild-type. SIM1 protein transcriptional activity compared to basal mock transfected cells measured activity.
Statistical analysis
Tests for deviation from Hardy–Weinberg equilibrium and for association were performed with the De Finetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). All SNPs were in Hardy–Weinberg equilibrium (0.02< P value <0.7). We compared all cases against all control individuals as well as obese adults and obese children separately against the control group. These analyses were done by comparing allelic frequencies of the SNPs between cases and controls. Familial analysis on binary traits was performed by the TDTPHASE method implemented in the UNPHASED software (http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/, developed by Frank Dudbridge). In order to evaluate the effect of the three associated frequent variants on linkage, we used the genotype IBD sharing test procedure. SPSS 10.1 software (SPSS, Chicago, IL) was used for general statistical analysis. Differences in transcriptional activities were compared with the Wilcoxon test (paired test on ranks). All P values are two-sided, P values <0.05 were considered to indicate statistical significance.
In order to deal with the multiple testing problem (case/control analysis, quantitative trait analysis, and linkage and TDT analysis), we performed a Bonferroni correction test. The conventional P value of 0.05 was divided by the total number of tests performed in the present study (87 tests) accounting for a new P value threshold of 0.00057. The 87 tests were calculated as follows: eight SNPs were tested for association with obesity in each of the three affected traits (childhood obesity, adulthood obesity, and pooled data) accounting for 24 independent tests. These analyses were performed in both sexes raising the amount of tests to 48. The contribution of the eight SNPs to linkage on the 6q locus was assessed in the additive, dominant, and recessive models (24 tests). Finally, the three associated SNPs were tested for association with BMI in the three subgroups of populations (9 tests) and in transmission disequilibrium in pedigrees with childhood and adulthood obesity (6 tests).
RESULTS
Screening of the gene
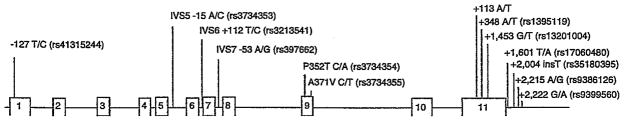
Thirteen frequent variants were identified in total (minor allele frequency >5%). The location and frequency of these SNPs are displayed in Table 1 and Figure 1.
Table 1.
Positions and frequencies of the 13 frequent variants identified within the SIM1 gene in the initial screened set
| SNPs (corresponding rs) | Frequency (%) |
|---|---|
| −127 T/C(rs41315244) | 5.3 |
| IVS5 −15 A/C (rs3734353) | 28.4 |
| IVS6 +112 T/C (rs3213541) | 24.2 |
| IVS7 −53 A/G (rs397662) | 21.6 |
| P352T C/A (rs3734354) | 13.3 |
| A371V C/T (rs3734355) | 13.3 |
| +113 A/T (rs41318039) | 7.0 |
| +348 A/T (rs1395119) | 34.5 |
| +1,453 G/T (rs13201004) | 13.1 |
| +1,601 T/A (rs17060480) | 21.1 |
| +2,004 insT (rs35180395) | 14.1 |
| +2,215 A/G (rs9386126) | 20.7 |
| +2,222 G/A (rs9399560) | 34.3 |
SNP positions in the first column are indicated in relation to the initiation codon ATG or the stop codon TGA (in the UCSC genome browser, NCBI Build 36.1).
SNP, single-nucleotide polymorphism.
Figure 1.
Schematic representation of SIM1 gene. The 13 SNPs shown above the figure are those identified after the sequencing of SIM1 gene. These variants had an MAF >5% and were studied in case/control analysis. The size of the genomic region represented was of 76.77 kilobases (870 bp of the putative promoter, exons, introns, and 1,095 bp in the 3′UTR). bp, base pair; SNP, single-nucleotide polymorphism.
LD analysis of the 13 common variants revealed that five pairs of SNPs were in high LD (r2 >0.8): IVS5 −15 A/C and IVS6 +112T/C, P352T C/A and A371V C/T, +348A/T and +2,222 G/A, +1,601 T/A and +2,215 A/G, and +1,453 G/T and +2,004 −/insT. Therefore, genotyping of eight SNPs was sufficient for further analyses.
Association studies of frequent SNPs with obesity
The genotyping was performed for the eight SNPs in 2,670 French whites (602 unrelated obese children, 673 unrelated severely obese adults, and 1,395 normoglycemic nonobese control adults). No heterogeneity was found between obese children and obese adults for all SNPs (0.32 < P < 0.95, data not shown). The C allele of the P352T C/A SNP showed nominal evidence of association with adulthood obesity (P = 0.02) with a similar trend in children (P = 0.07). The combined analysis of childhood/adult obesity showed a putative protective effect of the A allele (P = 0.01, OR = 0.81, 95% CI = 0.70–0.95; Table 2). The “ins T” allele of the +2,004 −/insT SNP only showed nominal evidence for association with childhood/adult obesity after the comparison of the pooled case set to controls (P = 0.02, OR = 1.21, 95% CI = 1.02–1.43; Table 2). Finally, the A allele of the +2,215 A/G SNP was associated with childhood (P = 0.02) and adult severely obesity (P = 0.009), and the association was stronger when comparing the combined case group to control subjects (P = 0.002, OR = 0.81, 95% CI = 0.71–0.93; Table 2). There was no significant association (P > 0.05) between P352T C/A, +2,004 −/insT, +2,215 A/G SNPs and quantitative BMI in the obese children, severely obese adults or nonobese control adult subgroups (data not shown). Subgroup analyses of the eight SNPs did not reveal gender-specific associations with obesity (data not shown). The eight SNPs did not provide any evidence for participation to the linkage observed on the 6q locus with childhood obesity risk (P > 0.1, data not shown).
Table 2.
Association (P < 0.05) of genotypes and alleles of SIM1 gene SNPs with obesity
| Variants | Genotypes | Allelic frequency | OR (P value) (IC) | ||||
|---|---|---|---|---|---|---|---|
| Promoter | −127 ATG T/C | TT | TC | CC | T | C | |
| Set 1 | Obese children (N = 596) | 536 | 59 | 1 | 0.95 | 0.05 | 0.84 (0.26) |
| Control (N = 1,334) | 1,177 | 153 | 4 | 0.94 | 0.06 | (0.62–1.13) | |
| Set 2 | Severely obese (N = 673) | 594 | 77 | 2 | 0.94 | 0.06 | 1.0 (0.98) |
| Control (N = 1,334) | 1,177 | 153 | 4 | 0.94 | 0.06 | (0.76–1.31) | |
| Set 1 + 2 | Obese (N = 1,269) | 1,130 | 136 | 3 | 0.94 | 0.06 | 0.923 (0.50) |
| Control (N = 1,334) | 1,177 | 153 | 4 | 0.94 | 0.06 | (0.73–1.16) | |
| Intron 5 | IVS5-15A/C | AA | AC | CC | A | C | |
| Set 1 | Obese children (N = 562) | 294 | 206 | 62 | 0.71 | 0.29 | 0.97 (0.70) |
| Control (N = 1,175) | 587 | 471 | 117 | 0.70 | 0.30 | (0.83–1.13) | |
| Set 2 | Severely obese (N = 631) | 323 | 255 | 53 | 0.71 | 0.29 | 0.93 (0.38) |
| Control (N = 1,175) | 587 | 471 | 117 | 0.70 | 0.30 | (0.80–1.9) | |
| Set 1 + 2 | Obese (N = 1,193) | 617 | 461 | 115 | 0.7.1 | 0.29 | 0.95 (0.43) |
| Control (N = 1,175) | 587 | 471 | 117 | 0.70 | 0.30 | (0.84–1.10) | |
| Intron 7 | IVS7-53A/G | AA | AG | GG | A | G | |
| Set 1 | Obese children (N = 586) | 413 | 151 | 22 | 0.83 | 0.17 | 1.13 (0.19) |
| Control (N = 1,367) | 994 | 336 | 37 | 0.85 | 0.15 | (0.94–1.36) | |
| Set 2 | Severely obese (N = 672) | 475 | 181 | 16 | 0.84 | 0.16 | 1.07 (0.48) |
| Control (N = 1,367) | 994 | 336 | 37 | 0.85 | 0.15 | (0.89–1.28) | |
| Set 1 + 2 | Obese (N = 1,258) | 619 | 462 | 115 | 0.68 | 0.32 | 1.10 (0.22) |
| Control (N = 1,367) | 994 | 336 | 37 | 0.85 | 0.15 | (0.94–1.27) | |
| Exon 9 | P352T C/A | CC | CA | AA | C | A | |
| Set 1 | Obese children (N = 597) | 450 | 138 | 9 | 0.87 | 0.13 | 0.83 (0.07) |
| Control (N = 1,339) | 955 | 358 | 26 | 0.85 | 0.15 | (0.68–1.01) | |
| Set 2 | Severely obese (N = 668) | 513 | 141 | 14 | 0.87 | 0.13 | 0.80 (0.02) |
| Control (N = 1,339) | 955 | 358 | 26 | 0.85 | 0.15 | (0.66–0.97) | |
| Set 1 + 2 | Obese (N = 1,265) | 963 | 279 | 23 | 0.87 | 0.13 | 0.81 (0.01) |
| Control (N = 1,339) | 955 | 358 | 26 | 0.85 | 0.15 | (0.70–0.95) | |
| Exon 11 | +113 A/T | AA | AT | TT | A | T | |
| Set 1 | Obese children (N = 594) | 539 | 54 | 1 | 0.95 | 0.05 | 1.09 (0.60) |
| Control (N = 1,395) | 1,276 | 117 | 2 | 0.96 | 0.04 | (0.80–1.51) | |
| Set 2 | Severely obese (N = 669) | 604 | 63 | 2 | 0.95 | 0.05 | 1.63 (0.33) |
| Control (N = 1,395) | 1,276 | 117 | 2 | 0.96 | 0.04 | (0.86–1.58) | |
| Set 1 + 2 | Obese (N = 1,263) | 1,143 | 117 | 3 | 0.95 | 0.05 | 1.13 (0.35) |
| Control (N = 1,395) | 1,276 | 117 | 2 | 0.96 | 0.04 | (0.87–1.46) | |
| Exon 11 | +2004 −/insT | −/− | −/insT | insT/insT | - | insT | |
| Set 1 | Obese children (N = 555) | 409 | 132 | 14 | 0.86 | 0.14 | 1.23 (0.05) |
| Control (N = 1,243) | 960 | 266 | 17 | 0.88 | 0.12 | (1.0–1.5) | |
| Set 2 | Severely obese (N = 613) | 458 | 137 | 18 | 0.86 | 0.14 | 1.20 (0.08) |
| Control (N = 1,243) | 960 | 266 | 17 | 0.88 | 0.12 | (0.98–1.46) | |
| Set 1 + 2 | Obese (N = 1,168) | 867 | 269 | 32 | 0.86 | 0.14 | 1.21 (0.02) |
| Control (N = 1,243) | 960 | 266 | 17 | 0.88 | 0.12 | (1.02–1.43) | |
| Exon 11 | +2,215 A/G | AA | AG | GG | A | G | |
| Set 1 | Obese children (N = 567) | 363 | 180 | 24 | 0.80 | 0.20 | 0.82 (0.02) |
| Control (N = 1,274) | 749 | 451 | 74 | 0.76 | 0.24 | (0.69–0.97) | |
| Set 2 | Severely obese (N = 632) | 407 | 200 | 25 | 0.80 | 0.20 | 0.80 (0.009) |
| Control (N = 1,274) | 749 | 451 | 74 | 0.76 | 0.24 | (0.68–0.95) | |
| Set 1 + 2 | Obese (N = 1,199) | 770 | 380 | 49 | 0.80 | 0.20 | 0.81 (0.002) |
| Control (N = 1,274) | 749 | 451 | 74 | 0.76 | 0.24 | (0.71–0.93) | |
| Exon 11 | +2,222 G/A | GG | GA | AA | G | A | |
| Set 1 | Obese children (N = 572) | 250 | 255 | 67 | 0.66 | 0.34 | 0.93 (0.39) |
| Control (N = 1,312) | 551 | 592 | 169 | 0.65 | 0.35 | (0.81–1.09) | |
| Set 2 | Severely obese (N = 633) | 278 | 281 | 74 | 0.66 | 0.34 | 0.93 (0.34) |
| Control (N = 1,312) | 551 | 592 | 169 | 0.65 | 0.35 | (0.81–1.07) | |
| Set 1 + 2 | Obese (N = 1,205) | 528 | 536 | 141 | 0.66 | 0.34 | 0.94 (0.26) |
| Control (N = 1,312) | 551 | 592 | 169 | 0.65 | 0.35 | (0.83–1.05) | |
Cases: Set 1 = 602 French white children with BMI higher than the obesity threshold of the 97th percentile. Cases: Set 2 = 673 French white adults with BMI ≥40. Control set = 1,395 nonobese and normoglycemic French white adults.
OR, odds ratio; SNP, single-nucleotide polymorphism.
TDT analysis in trios with childhood and adulthood obesity
TDT analysis of 420 pedigrees with childhood obesity and 158 pedigrees with adulthood obesity were then performed for the three associated variants. We found evidence for an over-transmission of the at-risk C allele of the P352T C/A SNP to obese children (64.6% vs. 35.4%, P = 0.04) in 48 informative families and to severely obese adults (83% vs. 17%, P = 0.02) in 15 informative families. The risk allele +2,004 insT of the variant +2,004 −/insT was overtransmitted to severely obese adults (100% vs. 0%, P = 0.003) in 16 informative families. However, the associated SNP +2,215 A/G did show significant allelic transmission distortion neither in obese children nor in adults.
We then performed a Bonferroni correction test taking into account the 87 tests performed in this study (see Methods section). None of the three variants remained significantly associated with obesity (P corrected = 0.00057) implying that SIM1 is not a major gene for polygenic obesity.
Functional characterization of the SIM1 P352T/A371V haplotype
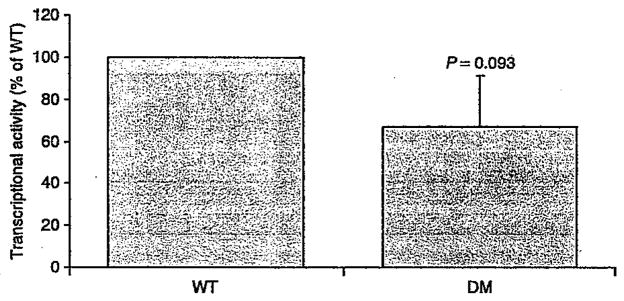
Association studies between the P352T/A371V haplotype and obesity-related traits provided inconsistent results in literature (19,25) and in the current study. To further investigate the putative implication of the haplotype in obesity susceptibility, we characterized the transcriptional activity of the SIM1 double-mutant protein. The aryl hydrocarbon receptor nuclear translocator 2 (ARNT2) is a required dimerization partner for SIM1 signaling (26). The functional effect of P352T/A371V haplotype on the transcriptional properties of this complex was then assessed using a pHRE-Luc gene reporter assay (27). Compared to mock, the SIM1 wt expressing vector was able to promote a HRE-mediated firefly expression when co-transfected with ARNT2. In this experimental setting, the protein carrying the P352T/A371V haplotype did not show any significant decrease in transcriptional activity compared to that of the wild-type protein (66.9%, P = 0.093; Figure 2).
Figure 2.
Effect of the P352T/A371T double mutation (DM) on the transcriptional activity of SIM1. Relative luciferase activities are expressed as the percentage of SIM1 Wild-type (WT) transcriptional activity measured in each experiment. Results represent the mean ± standard deviation of eight independent experiments, each performed in quadruplicate.
DISCUSSION
In the French population, we found a nominal association of three common variants (P352T, +2,004 −/insT TGA, and +2,215 TGA A/G) with polygenic forms of obesity. Although two of these associations were supported by TDT tests, none of the associations described above remained significant after multiple testing Bonferroni correction suggesting that SIM1 common variants are unlikely to play a major role in polygenic obesity susceptibility.
The current study therefore excludes a major role of SIM1 exons, exon/intron junctions, promoter, and 3′UTR SNP variation in polygenic obesity risk in the French population. Extension of the screening of SIM1 to intronic and intergenic regions followed by large-scale association would be helpful to complete this study.
The P352T polymorphism is in complete LD with the non-synonymous variant A371V (r2 = 1). In comparison to P352T C/A, die C allele of the variant A371V C/T and its corresponding alanine amino acid are highly evolutionary conserved across human, mice, and rats. However, it is noteworthy that a previous study in the British population reported controversial results with our findings. Indeed, male subjects carrying the alternative A-T allele of the P352T C/A/A371V C/T haplotype were found to have a slightly higher BMI (25). Moreover, female subjects homozygous for this mutated haplotype gained more weight over a period of 4.5 and 10 years in comparison to noncarriers (25). More recently, a large-scale association study in Pima Indians identified conclusive evidence of association with BMI for noncoding SNPs, but no significant association for the P352T and A371V variants (19). Altogether, these controversial data suggest that the association of P352T and A371V SNPs with obesity-related traits is likely to be false positive. Functional data strengthened this conclusion because the P352T C/A/A371V C/T haplotype did not show any significant effect on transcriptional activity compared to that of the wild-type protein (Figure 2).
In summary, we have excluded a major contribution of SIM1 common gene variation in exons, exon/intron junctions, promoter, and 3′UTR in polygenic obesity risk in the French population.
Acknowledgments
We are indebted to all families who participated in this study. We also thank the ALFEDIAM (Association de Langue Française pour l’Etude du Diabète et des Maladies Métaboliques), “200 Families pour Vaincre le Diabète et l’Obésite,” and “Association Française des Diabétiques” charities; and “le Conseil National de la Recherche Scientifique Libanais (CNRS-L)” and “le Conseil Régional Nord Pas de Calais/FEDER” institutions for their financial support.
Footnotes
DISCLOSURE
The authors declared no conflict of interest.
References
- 1.Farooqi IS, O’Rahiliy S. Mutations in ligands and receptors of the leptin-melanocortin pathway that lead to obesity. Nat Clin Pract Endocrinol Metab. 2008;4:569–577. doi: 10.1038/ncpendmet0966. [DOI] [PubMed] [Google Scholar]
- 2.Challis BG, Pritchard LE, Creemers JW, et al. A missense mutation disrupting a dibasic prohormone processing site in pro-opiomelanocortin (POMC) increases susceptibility to early-onset obesity through a novel molecular mechanism. Hum Mol Genet. 2002;11:1997–2004. doi: 10.1093/hmg/11.17.1997. [DOI] [PubMed] [Google Scholar]
- 3.Geller F, Relchwald K, Dempfle A, et al. Melanocortin-4 receptor gene variant 1103 is negatively associated with obesity. Am J Hum Genet. 2004;74:572–581. doi: 10.1086/382490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stutzmann F, Vatin V, Cauchi S, et al. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum Mol Genet. 2007;16:1837–1844. doi: 10.1093/hmg/ddm132. [DOI] [PubMed] [Google Scholar]
- 5.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 7.Meyre D, Lecoeur C, Delplanque J, et al. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31–q23.2. Diabetes. 2004;53:803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 8.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balthasar N, Dalgaard LT, Lee CE, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 10.Baskin DG. Single-minded view of melanocortin signaling In energy homeostasis. Endocrinology. 2006;147:4539–4541. doi: 10.1210/en.2006-0807. [DOI] [PubMed] [Google Scholar]
- 11.Holder JL, Jr, Zhang L, Kublaoui BM, et al. Sim1 gene dosage modulates the homeostatic feeding response to increased dietary fat in mice. Am J Physiol Endocrinol Metab. 2004;287:E105–E113. doi: 10.1152/ajpendo.00446.2003. [DOI] [PubMed] [Google Scholar]
- 12.Kublaoui BM, Holder JL, Jr, Gemelli T, Zinn AR. Sim1 haploinsufciency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons. Mol Endocrinol. 2006;20:2483–2492. doi: 10.1210/me.2005-0483. [DOI] [PubMed] [Google Scholar]
- 13.Michaud JL, Boucher F, Melnyk A, et al. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum Mol Genet. 2001;10:1465–1473. doi: 10.1093/hmg/10.14.1465. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Gagnon D, Vachon P, et al. Adenoviral-mediated modulation of Sim1 expression In the paraventricular nucleus affects food intake. J Neurosci. 2006;26:7116–7120. doi: 10.1523/JNEUROSCI.0672-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kublaoui BM, Holder JL, Jr, Tolson KP, Gemelli T, Zinn AR. SIM1 overexpression partially rescues agouti yellow and diet-induced obesity by normalizing food intake. Endocrinology. 2006;147:4542–4549. doi: 10.1210/en.2006-0453. [DOI] [PubMed] [Google Scholar]
- 16.Holder JL, Jr, Butte NF, Zinn AR. Profound obesity associated with a balanced translocation that disrupts the SIM1 gene. Hum Mol Genet. 2000;9:101–108. doi: 10.1093/hmg/9.1.101. [DOI] [PubMed] [Google Scholar]
- 17.Wilier CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyre D, Delplanque J, Chèvre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 19.Traurig M, Mack J, Hanson RL, et al. Common variation in SIM1 is reproducibly associated with BMI in Pima Indians. Diabetes. 2009;58:1682–1689. doi: 10.2337/db09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolland-Cachera MF, Cole TJ, Sempé M, et al. Body mass index variations: centiles from birth to 87 years. Eur J Clin Nutr. 1991;45:13–21. [PubMed] [Google Scholar]
- 21.Poskitt EM. Defining childhood obesity: the relative body mass index (BMI). European Childhood Obesity group. Acta Paediatr. 1995;84:961–963. doi: 10.1111/j.1651-2227.1995.tb13806.x. [DOI] [PubMed] [Google Scholar]
- 22.Maillard G, Charles MA, Lafay L, et al. Macronutrient energy intake and adiposity in non obese prepubertal children aged 5–11 y (the Fleurbaix Laventie Ville Santé Study) Int J Obes Relat Metab Disord. 2000;24:1608–1617. doi: 10.1038/sj.ijo.0801446. [DOI] [PubMed] [Google Scholar]
- 23.Vu-Hong TA, Durand E, Deghmoun S, et al. The INS VNTR locus does not associate with smallness for gestational age (SGA) but interacts with SGA to increase insulin resistance in young adults. J Clin Endocrinol Metab. 2006;91:2437–2440. doi: 10.1210/jc.2005-2245. [DOI] [PubMed] [Google Scholar]
- 24.Hercberg S, Preziosi P, Briançon S, et al. A primary prevention trial using nutritional doses of antioxidant vitamins and minerals in cardiovascular diseases and cancers in a general population: the SU.VI.MAX study—design, methods, and participant characteristics. Supplementation en Vltamines et Minéraux AntioXydants. Control Clin Trials. 1998;19:336–351. doi: 10.1016/s0197-2456(98)00015-4. [DOI] [PubMed] [Google Scholar]
- 25.Hung CC, Luan J, Sims M, et al. Studies of the SIM1 gene in relation to human obesity and obesity-related traits. Int J Obes (Lond) 2007;31:429–434. doi: 10.1038/sj.ijo.0803443. [DOI] [PubMed] [Google Scholar]
- 26.Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech Dev. 2000;90:253–261. doi: 10.1016/s0925-4773(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 27.Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J Biol Chem. 2002;277:10236–10243. doi: 10.1074/jbc.M110752200. [DOI] [PubMed] [Google Scholar]