Abstract
Comparative studies of ethnically diverse human populations, particularly in Africa, are important for reconstructing human evolutionary history and for understanding the genetic basis of phenotypic adaptation and complex disease. African populations are characterized by greater levels of genetic diversity, extensive population substructure, and less linkage disequilibrium (LD) among loci compared to non-African populations. Africans also possess a number of genetic adaptations that have evolved in response to diverse climates and diets, as well as exposure to infectious disease. This review summarizes patterns and the evolutionary origins of genetic diversity present in African populations, as well as their implications for the mapping of complex traits, including disease susceptibility.
Keywords: disease susceptibility, African populations, genetic variation, human evolution, linkage disequilibrium
INTRODUCTION
One of the “grand challenges” of the post-genome era is to “develop a detailed understanding of the heritable variation in the human genome” (36). By characterizing genetic variation among individuals and populations, we may gain a better understanding of differential susceptibility to disease, differential response to pharmacological agents, human evolutionary history, and the complex interaction of genetic and environmental factors in producing phenotypes. Africa is an important region to study human genetic diversity because of its complex population history and the dramatic variation in climate, diet, and exposure to infectious disease, which result in high levels of genetic and phenotypic variation in African populations. A better understanding of levels and patterns of variation in African genomes, together with phenotype data on variable traits, including susceptibility to disease and drug response, will be critical for reconstructing modern human origins, the genetic basis of adaptation to diverse environments, and the development of more effective vaccines and other therapeutic treatments for disease. This information will also be important for identifying variants that play a role in susceptibility to a number of complex diseases in people of recent African ancestry (172, 187, 208).
HUMAN EVOLUTIONARY HISTORY IN AFRICA
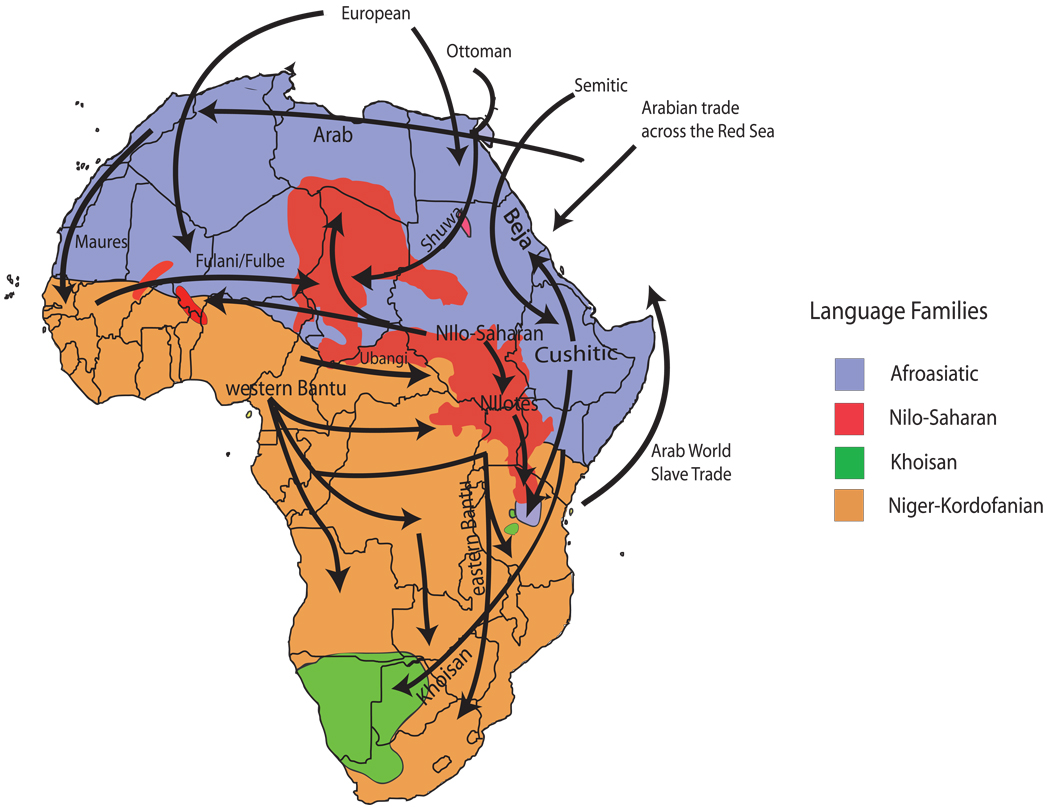
Africa is a region of considerable genetic, linguistic, cultural, and phenotypic diversity. There are more than 2000 distinct ethno-linguistic groups in Africa, speaking languages that constitute nearly a third of the world’s languages (http://www.ethnologue.com/) (Figure 1). These populations practice a wide range of subsistence patterns including various modes of agriculture, pastoralism, and hunting-gathering. Africans also live in climates that range from the world’s largest desert and second largest tropical rainforest to savanna, swamps, and mountain highlands, and these climates have, in some cases, undergone dramatic changes in the recent past (106, 172).
Figure 1.
A map of African language family distributions and hypothesized migration events within and out of Africa. African languages have been classified into four major language families: Niger-Kordofanian (spoken predominantly by agriculturalist populations across a broad geographic distribution in Africa), Afro-Asiatic (spoken predominantly by northern and eastern Africa pastoralists and agropastoralists), Nilo-Saharan (spoken predominantly by eastern and central African pastoralists), and Khoisan (a language containing click-consonants, spoken by southern and eastern African hunter-gatherer populations). Also plotted are the geographic origins of African samples included in the Center d’Etude du Polymorphisme Humain (CEPH) Human Genome Diversity Panel (CEPH-HGDP). Diagram adapted from Reference 170.
According to the Out of Africa (OOA) model of modern human origins, anatomically modern humans originated in Africa and then spread across the rest of the globe within the past ~100,000 years (206). The transition to modern humans within Africa was not sudden; rather, the paleobiological record indicates an irregular mosaic of modern, archaic, and regional morphological and behavioral traits that occurred over a substantial period of time and across a broad geographic range within Africa (127). The earliest known derived suite of morphological traits associated with modern humans appears in fossil remains from Ethiopia, dated to ~150--190 kya (128, 229). However, this finding does not rule out the existence of modern morphological traits in other regions of Africa before 100 kya, particularly where specimens may be less well preserved and/or where extensive archaeological and paleobiological investigations have not been conducted (172). Indeed, a multiregional origin model for modern humans within Africa is not as unlikely as it would be for global populations, considering the greater potential for migration and admixture within a single continental region (172, 241). A more fully modern suite of traits appears in East Africa and Southwest Asia around 90 kya, followed by a rapid spread of modern humans throughout the rest of Africa and Eurasia within the past 40,000--80,000 years (120, 172) (Figure 2).
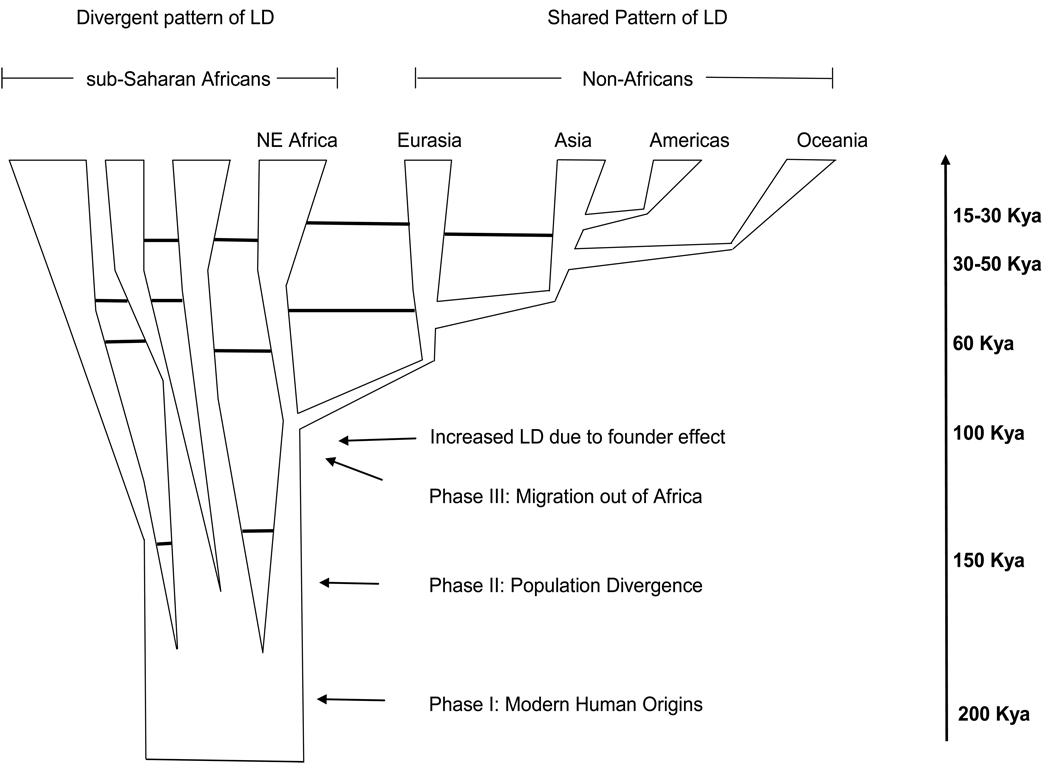
Figure 2.
Ancestral Africans have maintained a large and subdivided population structure and have experienced complex patterns of population expansions, contractions, migration, and admixture during their evolutionary history. The bottleneck associated with the founding of non-African populations (~50–100 kya) resulted in lower levels of genetic diversity, an increase in linkage disequilibrium (LD), and more similar patterns of LD. In addition, several recent studies have suggested that a serial founder model of migration occurred in the history of non-Africans in which the geographic expansion of these populations occurred in many small steps, and each migration involved a sampling of variation from the previous population (36, 90, 109, 167). Solid horizontal lines indicate gene flow between populations and the dashed horizontal line indicates recent gene flow from Asia to Australia/Melanesia.
Two migration routes of modern humans out of Africa have been proposed. The presence of modern humans in Oceania as early as ~50 kya (65, 66), which predates their presence in Europe ~40 kya, has suggested a southern coastal route around the Indian Ocean in which modern humans first left Africa (possibly via Ethiopia) by crossing the Bab-el-Mandeb strait at the mouth of the Red Sea and then rapidly migrated to Southeast Asia and Oceania (62, 172) (Figure 1). This migration model is supported by the presence of very old mtDNA haplotypes in South Asia and their absence in the Levant (120, 168, 197). Other models have traditionally favored a second (or single) northern route via the Sinai Peninsula into the Levant (62, 172) (Figure 1). Regardless of the route of migration of modern humans out of Africa, the shared patterns of genetic diversity among non-African populations [e.g., at the CD4 locus (200)] and the divergent patterns of genetic variation among African populations argue against repeated sampling of African diversity from multiple source populations (172, 200, 206). However, analyses of more independent loci and a larger number of African populations, particularly from East Africa, will be necessary to better estimate the number and source of migration events out of Africa (172). After modern humans migrated from Africa, there could have been some admixture of modern humans with archaic populations in Eurasia, such as Neanderthals. This hypothesis remains a topic of considerable interest and debate and is the subject of a number of recent studies and reviews (46, 59, 71, 73, 77, 144, 158, 172, 184, 185, 224)
The migration of modern humans out of Africa is thought to be accompanied by a population bottleneck. The size of the population(s) migrating out of Africa is estimated to be ~600 effective founding females (i.e., census size of ~1800 females) on the basis of mtDNA evidence (62, 120), to be ~1000 effective founding males and females (i.e., census size of ~3000 individuals) based on the analysis of 783 autosomal microsatellites genotyped in the Center d’Etude du Polymorphisme Humain (CEPH) human genome diversity panel (HGDP) (112), and to be ~1500 (i.e., a census size of ~4500 individuals) based on a combined analysis of mtDNA, Y chromosome, and X chromosome nucleotide diversity data (72). These estimates imply that Eurasians must have rapidly expanded to a larger size to account for estimates of a long-term effective population size (Ne) of ~10,000 individuals (census size of ~30,000 individuals) for global populations (172, 243). Indeed, several recent studies indicate a rapid expansion of Eurasian populations within the past ~50,000 years, whereas Africans have maintained a large effective population size (72, 125, 243).
PATTERNS OF GENETIC VARIATION IN AFRICA
The pattern of genetic variation in modern African populations is influenced by demographic history (e.g., changes in population size, short- and long- range migration events, and admixture) as well as locus-specific forces such as natural selection, recombination, and mutation. For example, the migration of agricultural Bantu speakers from West Africa throughout sub-Saharan Africa within the past ~4000 years and subsequent admixture with indigenous populations has had a major impact on patterns of variation in modern African populations (157, 167, 172, 201, 235a) (Figure 1). Although Africa is critical for understanding modern human origins and genetic risk factors for disease, it has been under-represented in human genetic studies. Much of what we currently know about genetic diversity is from a limited number of the ~2000 ethno-linguistic groups in Africa, and the majority of these data are from mtDNA and Y chromosome studies. Large-scale autosomal studies of African genetic diversity are only now beginning to become available.
Mitochondrial DNA and Y Chromosome Variation
Phylogenetic analyses of both mtDNA and Y chromosome DNA indicate that the oldest lineages are specific to Africa and have a Time to Recent Common Ancestry (TMRCA) of ~200 kya (75, 206). Interestingly, the most ancient mtDNA lineage (L0d) [dated to ~106 kya (75)], which is common in click-speaking southern African Khoisan (SAK) populations, has recently been identified at low frequency (5%) in the click-speaking Sandawe population from Tanzania (75, 201). Maximum likelihood estimates for the time of divergence of these populations based on all mtDNA lineages is ~44 kya, indicating that any common ancestry is quite old. This finding supports studies of classical polymorphisms as well as archeological data that suggest that Khoisan-speaking populations may have originated in eastern Africa and subsequently migrated into southern Africa (26), although a southern African origin of Khoisan-speakers cannot be ruled out.
Phylogenetic analysis indicates that the most recent African specific mtDNA haplogroup lineage, L3, is the likely precursor of modern European and Asian mtDNA haplotypes (226). Indeed a subset of this lineage (M1) is observed at high frequency in Ethiopian populations (101, 168) and may have expanded out of Africa ~60 kya (168). This observation adds strength to the proposal that the dispersal of modern humans out of Africa may have occurred via Ethiopia (117, 200). However, more recent analysis of whole mtDNA genomes suggests that the M1 lineage may have originated in southwestern Asia and then was introduced into East Africa from Asia ~40--45 kya (150), whereas others have argued for a much more recent introduction of the M1 lineage into Africa from the Middle East (63).
Nucleotide and Haplotype Variation
The migration of modern humans out of Africa resulted in a population bottleneck and a concomitant loss of genetic diversity (112, 169). Numerous studies have shown higher levels of nucleotide and haplotype diversity in Africans compared to non-Africans in both nuclear and mitochondrial genomes (40, 72, 93, 111, 200, 202, 206, 208). Non-African populations appear to have a subset of the genetic diversity present in sub-Saharan Africa and more private alleles and haplotypes are observed in Africa relative to other regions (38, 93, 111, 169, 200, 202, 206, 208, 243) as expected under an OOA model. For example, a resequencing study of 3873 genes in 154 chromosomes from European, Latino/Hispanic, Asian, and African American populations observed that African Americans had the highest percentage of rare single nucleotide polymorphisms (SNPs) (64%) and the lowest percentage of common SNPs (36%). Additionally, 44% of all SNPs in this population were private (78). The high level of genetic diversity in African populations is also consistent with a larger long-term effective population size (Ne) compared to non-Africans (72, 195, 196, 202, 206; Ne is estimated to be ~15,000 for Africans and ~7500 for non-Africans based on a resequencing analysis of several 10-kb regions (243) (see Supplemental Material).
Structural Variation
Although most studies of genetic variation in humans have focused on nucleotide and microsatellite diversity, a number of recent studies have demonstrated considerable amounts of structural variation (SV) in the human genome, including both copy number variation (which can include insertions and deletions as well as gene duplications) and inversions (17, 37, 191, 211) (http://projects.tcag.ca/variation/). Some of these structural variants are also associated with phenotypic variability (37, 171, 193). For example, variation in copy number of the amylase gene, which plays a role in digestion of starch, is correlated with enzyme activity level and with diet in ethnically diverse human populations (156). Additionally, SVs may play an important role in susceptibility to common disease (109, 124). A recent study that used high-resolution paired-end mapping to identify SVs in the genomes of a single African (Yoruba from Nigeria) individual and an individual of European descent led to the identification of 1175 insertions/deletions (INDELs) and 122 inversions (103). By extrapolation, these researchers predicted 761 and 887 SVs in the full genomes of these European and African individuals, respectively. Additionally, 45% of the SVs were shared between these samples, suggesting that a large proportion of SV events occurred prior to the divergence of African and non-African populations. The majority of these SVs were less than 10 kb in size, but at least 15% were larger than 100 kb and some SVs were predicted to be several megabases in size in both the European and African sample, indicating that the genomes of healthy individuals may differ by megabases of nucleotide sequence (103). To date, few population genetic studies of SVs across ethnically diverse populations have been performed (37). Instead, most studies have focused on the European, Japanese, Chinese, and African (Yoruba) HapMap populations (37). A study of 67 common copy number variants (CNVs) in these populations indicated that 11% of the variation was due to differences among populations and that many of the variants were shared among populations from different regions, further supporting the argument that these variants existed prior to migration of modern humans out of Africa (171). There are currently no studies of SV variability within and between ethnically diverse African populations. Such knowledge will be informative for reconstructing human evolutionary history and for understanding the role of SVs in normal phenotypic diversity and in susceptibility to disease.
POPULATION STRUCTURE IN AFRICA
Measures of population structure on a global level indicate that only ~10%--16% (Wright’s fixation index, FST = 0.10--0.16) of observed genetic variation is due to differences among populations from Africa, Europe, and Asia (26, 40, 206, 228). Analysis of population structure using the program STRUCTURE, (162,) based on 1048 individuals from the CEPH human diversity panel genotyped for 993 genome-wide microsatellite and insertion/deletion markers, indicates that individuals cluster into five major geographic regions: Africa, Europe/Middle East, East Asia, Oceania, and the New World (175). Two recent studies of >500,000 SNPs genotyped in the CEPH diversity panel support these initial findings (93, 111). Analyses within the African populations indicate that additional substructure exists, particularly between hunter-gatherer and agriculturalist populations (93, 111). However, the CEPH diversity panel includes just eight African populations, four of which are agricultural Bantu-speakers likely to share recent common ancestry (Figure 1). Thus, results from these studies may not reflect the full extent of population structure within Africa.
Several studies of nucleotide and haplotype variation have indicated that ancestral African populations were geographically structured prior to the migration of modern humans out of Africa (72, 73, 82, 158, 200, 241). Additionally, a recent study of 800 short tandem repeat polymorphisms (STRPs) and 400 /INDELs genotyped in more than 3000 geographically and ethnically diverse Africans indicates the presence of at least 13 genetically distinct ancestral populations in Africa and high levels of population admixture in many regions (F.A. Reed and S.A Tishkoff unpublished data). Population clusters are correlated with self-described ethnicity and shared cultural and/or linguistic properties (e.g., Pygmies, Khoisan-speaking hunter-gatherers, Bantu speakers, Cushitic speakers). This study reveals extensive admixture between inferred ancestral populations in most African populations. One exception is among West African Niger-Kordofanian (i.e., Bantu) speakers who are more genetically homogeneous compared with other African populations, likely reflecting the recent and rapid spread of Bantu speakers from a common origin in Cameroon/Nigeria (although fine-scale genetic structure can be detected amongst these populations). Thus, the pattern of genetic diversity in Africa indicates that African populations have maintained a large and subdivided population structure throughout much of their evolutionary history (Figure 2). Historic subdivision among African populations is likely due to ethnic and linguistic barriers, as well as a number of geographic, ecological, and climatic factors (including periods of glaciation and warming) that could have contributed to population expansions, contractions, fragmentations, and extinctions during recent human evolution in Africa (172, 206).
PATTERNS OF LINKAGE DISEQUILIBRIUM
Linkage disequilibrium (LD), the nonrandom association between alleles at different loci, is typically measured using two different estimators: D’ and r2 (161). Levels and patterns of LD depend on a number of demographic factors including population size and structure, as well as locus-specific factors such as selection, mutation, recombination (1, 161, 206, 207), and gene conversion (see Supplemental Material). LD is particularly useful for inferring evolutionary and demographic processes, as well as for mapping disease-susceptibility loci. Therefore, an understanding of levels and patterns of LD has broader implications for studies of human evolutionary history and disease.
Empirical Studies of Linkage Disequilibrium
Several haplotype studies have indicated lower levels of LD in African populations compared to non-Africans (200, 202, 206, 207). Studies of long-range LD between SNP markers at multiple nuclear loci confirmed these initial results and demonstrated that haplotype blocks (where SNPs are in strong LD) extend over greater genomic distances and are more uniform in non-Africans compared to African populations (69, 116, 174, 183)
Given that recombination is an important determinant of the extent of LD, an alternative way to assess LD is to estimate the population recombination rate (ρ = 4Ner, where Ne is effective population size and r is the meiotic recombination rate/kb) (161). Empirical studies have shown that African Americans have higher ρ estimates compared to Europeans and Asians (33, 47, 60), consistent with the results of previous studies that described less LD in Africans relative to non-African populations. The divergent patterns of LD can be explained by the distinct demographic histories of African and non-African populations (206, 208) (Figure 2). Specifically, African populations have shorter blocks of LD because ancestral Africans maintained a larger effective population size (Ne), and because there has been more time for recombination to decrease levels of LD. Greater LD in non-African populations is likely the result of a founding event during expansion of modern humans out of Africa within the past 100,000 years (200, 206, 208).
However, an ongoing challenge has been to characterize patterns of LD among populations within continental regions, especially in Africa. Some evidence has suggested variance in levels and patterns of LD among subpopulations in Africa. Tishkoff and colleagues (200) noted that African populations have divergent patterns of LD; specifically, alleles that were in positive association in one population were in negative association in another. Additionally, a resequencing analysis of the IL-13 gene in 126 geographically diverse Africans identified divergent patterns of LD across West and East African populations (195) These observations suggest that not all African populations are characterized by a single discrete pattern of LD and each may have distinct haplotype block structures (Figure 3). Theoretically, under a model of population subdivision, allelic associations can differ between populations due to the stochastic effects of genetic drift (1).
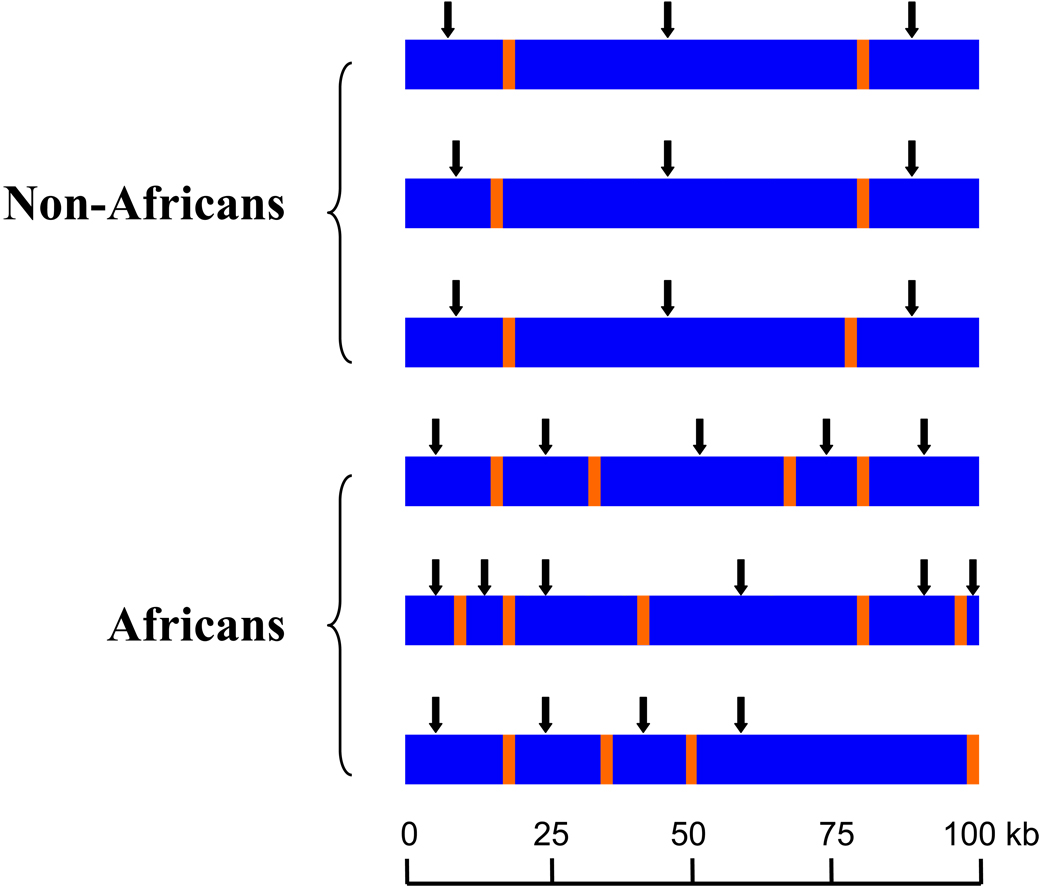
Figure 3.
Several analyses have indicated that haplotype blocks [where single nucleotide polymorphisms (SNPs) are in strong linkage disequilibrium (LD)] extend over greater genomic distances and are more uniform in non-Africans compared to African populations. Additionally, the size and location of haplotype blocks can vary among African samples owing to the distinct demographic histories of populations from different geographic regions in Africa. The blue bars represent haplotype blocks and the thin orange bars denote regions of recombination. Vertical lines indicate SNPs and vertical arrows indicate haplotype tag SNPs (htSNPs). Because haplotype blocks are more variable in Africans compared to non-Africans, identification of htSNPs in diverse African ethnic groups and more dense tag htSNP coverage are needed to detect an association between marker(s) and disease loci in association studies.
Recombination Hotspots
Recombination hotspots, where historical crossing-over events are clustered and separate relatively large haplotype blocks, have been a topic of considerable interest in the scientific literature (12, 42, 43). The occurrence of recombination hotspots in human DNA has been demonstrated empirically from studies of single sperm DNA and from pedigree analyses (12, 43, 94). However, the extent to which this pattern is a general feature of the genome remains unknown, particularly because genetic drift can result in a similar pattern of LD (12, 47).
Several recent studies have observed that the locations of most hotspots tend to be shared between diverse populations (38, 47, 76). However, several datasets have suggested that some hotspots may be population specific (34, 38, 47, 76) and that African and African-American populations have more recombination hotspots relative to non-Africans (34, 47). Given that recombination rates vary between species (165, 223) and even individuals (43, 101a, 102), it is possible that hotspots could also differ among ethnically distinct populations, including Africans. Indeed the identification of haplotypes at the RNF212 gene associated with recombination rate, which occur at different frequencies in the HapMap populations (102), raises the possibility that population-specific genetic variants may influence recombination rates.
Implications of Linkage Disequilibrium for Association Studies
The mapping of complex disease genes relies on the identification of an association of polymorphic markers, either individually or as haplotypes, with disease susceptibility loci (207). The International HapMap Project (http://www.hapmap.org/) has characterized patterns of haplotype structure and LD across the human genome to facilitate mapping of complex disease genes (40, 41, 129). Another goal of this project has been to identify haplotype tag SNPs (htSNPs) that distinguish major haplotypes, thereby reducing the number of SNPs needed for association studies (207).
To date, 3.4 million SNPs have been characterized in 270 individuals from four populations: Yoruba from Nigeria, European-Americans Japanese, and Chinese. Knowledge of the frequency and distribution of these SNPs across ethnically diverse populations is important to assess their usefulness as markers for gene mapping studies in diverse ethnic groups (206, 207). A survey of 3024 SNPs spaced across 36 genomic regions genotyped in 927 unrelated individuals from the CEPH human genetic diversity panel indicates that although haplotype block sharing with the HapMap populations is high in European and East Asian populations, sharing for most other populations is low, particularly for haplotypes in African hunter-gatherer populations (38). These results suggest that development of distinct panels of htSNPs and more dense coverage of SNPs will be needed for African populations (38, 207). Seven additional populations have been added to the HapMap inititative: Luhya (Bantu) and Maasai (NiloSaharan) from Kenya, Tuscans from Italy, Gujarati Indians from Texas, metropolitan Chinese in Denver, people of Mexican ancestry in Los Angeles, and African Americans from the Southwest United States . The characterization of SNP and haplotype diversity in these additional populations will be important for the identification of htSNPs that are more informative across ethnically diverse populations.
Although the SNPs used in the HapMap study have been highly informative for use in association mapping studies, the initial identification of SNPs in one or a few populations can result in an ascertainment bias (AB) toward high-frequency, presumably older, SNPs. Several studies have shown that AB can distort estimates of migration rate (221), mutation rates (140), recombination rates (33, 140), and LD (7, 33). Although the effects of AB can sometimes be corrected (33, 142, 143), these correction methods make a number of assumptions that are not applicable to African populations, including the assumption of no population substructure (142). To more accurately infer human genetic variation it will be necessary to characterize the entire frequency distribution of nucleotide variants in diverse populations. Additionally, because variants associated with disease could be geographically restricted due to new mutation, genetic drift, or regional-specific selection pressure, de novo identification of genetic variation in diverse African populations will be important. The HapMap ENCODE (http://www.hapmap.org) and the proposed “1000 genomes” (http://www.genome.gov) resequencing projects aim to discover novel variation, including rare SNPs and structural variants, in targeted regions of the genome (as well as in whole genomes for a subset of samples) from the extended HapMap populations and other ethnically diverse populations. The extensive levels of substructure identified in Africa will likely require analysis of additional ethnically and geographically diverse African populations.
NATURAL SELECTION
Natural selection, the process by which favorable heritable traits become more common in successive generations, operates to either increase or decrease the frequency of mutations that have an effect on an individual’s fitness. When a mutation is advantageous it can rapidly increase in frequency, together with linked variants (i.e., genetic hitchhiking), due to positive selection and replace pre-existing variation in a given population (i.e., a selective sweep) (83, 141, 180, 206). The strength of selection and local rates of recombination dictate how large of a genomic region is affected by a selective sweep. If selection is recent, there may not be enough time for the selected variant to become fixed in the population, resulting in an incomplete selective sweep. The genetic signatures of a selective sweep include a region of extensive LD [extended haplotype homozygosity (EHH)] and low variation on high-frequency chromosomes with the derived beneficial mutation relative to chromosomes with the ancestral allele (179, 203, 219). After this selective sweep, given enough time, new mutations and recombination will occur, leading to an excess of rare variants and a decrease in the extent of LD. Weak purifying selection is also expected to result in an increase of low frequency variants. Under this scheme of selection, deleterious mutations entering the population generally remain at low frequencies because their adverse effect on fitness makes it unlikely that they will reach high frequencies. In contrast, long-term balancing selection (resulting from greater fitness of heterozygotes or when maintenance of multiple alleles in a population is adaptively advantageous) is expected to result in an excess of alleles at intermediate frequency.
Demographic processes can also cause similar skews in the frequency of polymorphisms. For example, when population size rapidly increases, genetic drift has less effect in a rapidly expanding population, leading to an excess of rare polymorphisms (mimicking the pattern seen under positive or purifying selection). In contrast, a population bottleneck is expected to cause the loss of low-frequency variants, and thus produce an excess of intermediate-frequency variants (mimicking the pattern observed under balancing selection) (141). Although natural selection and demographic history can cause similar departures from a neutral equilibrium model, it is possible to distinguish these forces either by simulating the expected pattern of variation under different demographic scenarios or by using an outlier approach in which targets of selection are identified because they show an unusual pattern of variation or population differentiation compared with an empirical distribution observed at other loci across the genome (98, 141). Given the vast number of studies published on natural selection, the next sections focus on a few case studies of genetic and phenotypic adaptation in sub-Saharan Africa.
Malaria Resistance in Africa
Malaria (caused by infection with the Plasmodium falciparum parasite) is a major cause of mortality in sub-Saharan Africa, resulting in more than 1 million deaths (primarily children) each year (107). Given the enormous impact of malaria in Africa, it is not surprising that this disease has exerted strong selective pressure on African populations during recent human evolutionary history.
A number of genetic variants in African populations have been shown to confer resistance to malaria. One of the best known genetic adaptations is the HbS mutation in the β-globin gene, which causes sickle cell anemia in homozygous individuals. Individuals who are heterozygous for the sickle cell trait are protected against malarial infection and have higher reproductive fitness (107) which results in the maintenance of the HbS allele at high frequency in many malaria endemic regions. A recent genetic study observed long range LD extending over 400 kb at the β-globin locus on haplotypes with the HbS mutation in West African and Caribbean African populations, consistent with the pattern expected under positive selection (81). Other well-known hemoglobin variants associated with malaria resistance in African populations include hemoglobins C (HbC) and E (HbE). Studies have also identified pattens of LD on chromosomes that contain either the HbC variant (236) or the HbE variant (147) that are consistent with recent positive selection.
Glucose-6-phosphate dehydrogenase (G6PD) mutations that result in reduced enzyme activity are also associated with malaria resistance (205). The most common G6PD mutation in Africa, G6PD A-, occurs at a frequency of ~25% in malaria endemic regions (205). Several empirical studies have found evidence for recent selection at the G6PD locus in African populations. For example, a study of SNP and microsatellite haplotype variability demonstrated a signature of strong positive selection of the A- variant (205). On the basis of the breakdown of LD between the microsatellite markers and the G6PD A- variant, the age of this variant was estimated to be between 3840 and 11,760 years (205). Similarly, nucleotide sequence analyses of the G6PD locus in Africa also showed patterns of variation consistent with recent positive selection (181, 217). A signature of a recent partial selective sweep was also supported by two studies that showed extensive LD extending >400 kb on chromosomes with the G6PD A- mutation (179, 182). A comparative analysis of human and nonhuman primates suggested that signatures of selection at the A- allele are unique to humans (218). Overall, these data are consistent with other evidence suggesting that the malaria parasite has had a significant impact on humans only within the past 10,000 years (96, 220), possibly corresponding with the development of agriculture and/or pastoralism in Africa (205).
The Duffy gene on chromosome 1 confers resistance to malaria caused by Plasmodium vivax, which is not prevalent in Africa today but may have been in the past. The Duffy gene encodes a receptor on the surface of erythrocytes and is characterized by three alleles (FY*A, FY*B, and FY*O). The frequency of the FY*O allele is at or close to fixation in most sub-Saharan African populations, but is very rare outside of Africa (79). A resequencing study of nucleotide variation at the FY locus in five sub-Saharan African populations, and in a comparative Italian population (79), reported that variation at this locus is two- to three- fold lower in Africans than in the Italian sample, which is the opposite pattern observed at most loci. A more extensive resequencing analysis of this locus (80) also revealed reduced sequence variation around the FY*O mutation and an excess of high-frequency derived alleles at linked sites, consistent with a selective sweep of this region. Additionally, researchers observed unusually large FST values for the FY*A and FY*O variants at this locus across African, European, and Asian populations, consistent with local adaptation in different geographic regions. These results have led to the conclusion that positive selection has been a dominant force in shaping the distribution of Duffy alleles among human populations.
Dietary Adaptations in African Populations
Lactase persistence (LP), the ability to digest milk and other dairy products into adulthood, is a classic example of a genetic adaptation in humans. LP varies in frequency in different human populations; it is most common in northern Europeans and certain African and Arabian nomadic tribes that practice pastoralism and is at low frequency in East Asians and West sub-Saharan Africans (88). A number of studies have demonstrated a strong association between LP and the presence of the T allele at a C/T SNP located −13910 kb upstream from the lactase gene (LCT) in European populations (58, 159). In vitro studies also showed that the T-13910 variant enhances gene transcription from the LCT promoter (110, 148). In a study of long-range LD in Europeans, Bersaglieri and colleagues (21) found that haplotypes containing the LP-associated T-13910 variant were largely identical over nearly a 1-Mb region, consistent with a strong selective sweep. In genome-wide scans of selection in the HapMap samples, the LCT gene showed the strongest signal of positive selection in Europeans (180, 219).
Although the T-13910 variant is likely the causal mutation of the lactase persistence trait in Europeans, analyses of this SNP in ethnically and geographically diverse African populations indicated that it is present in only a few West African pastoralist populations, such as the Fulani (or Fulbe) and Hausa from Cameroon (35, 135, 136). These results suggested that the T-13910 allele may not be a strong predictor of lactase persistence in most sub-Saharan Africans. A more recent genotype and phenotype association study in a sample of 43 populations from Tanzania, Kenya, and the Sudan identified three novel SNPs located ~14 kb upstream of LCT that are significantly associated with the LP trait in African populations (203). These SNPs are located within 100 bp of the European LP-associated variant (C/T-13910). One LP-associated SNP (G/C-14010) is common in Tanzanian and Kenyan pastoralist populations, whereas the other two (T/G-13915 and C/G-13907) are common in northern Sudanese and Kenyans. The derived alleles at these loci (C-14010, G-13915, and G-13907) were shown to enhance transcription from the LCT promoter in vitro (203). Genotyping of 123 SNPs across a 3-Mb region in these populations demonstrated that these African LP-associated variants exist on haplotype backgrounds that are distinct from the European LP-associated variant and from each other. In addition, haplotype homozyogisty extends >2 Mb on chromosomes with the LP-associated C-14010 variant, consistent with an ongoing selective sweep over the past 3000--7000 years. An independent study of Sudanese populations also identified a significant association of the G-13915 allele with LP in that region (90) and a recent study confirmed the enhancer effect of the G-13915 variant, together with a C-3712 variant, on the same haplotype background (57). These data indicate a striking example of convergent evolution and local adaptation due to strong selective pressure resulting from shared cultural traits (e.g. cattle domestication and adult milk consumption) in Europeans and Africans. These studies also demonstrate the effect of local adaptation on patterns of genetic variation and the importance of resequencing across geographically and ethnically diverse African populations to identify population-specific variants associated with variable traits, including disease susceptibility.
Another important dietary adaptation in human populations is the ability to taste bitter compounds. A hypothesized selective advantage of bitter taste is that it helps individuals avoid ingesting toxic substances in plants. Variation at the TAS2R genes is associated with sensitivity to bitter taste substances (54). An analysis of nucleotide and haplotype variation at 24 TASR2 genes in 55 globally diverse individuals also identified substantial amino acid diversity, an excess of nonsynonymous substitutions, and high levels of population differentiation at variable sites, suggesting that amino acid variability at these loci may be maintained due to natural selection (99).
There have also been a number of in-depth studies at individual TAS2R loci for which there are known associations with bitter taste perception. For example, the ability to taste phenylthiocarbamide (PTC), a synthetic bitter substance, is a highly variable trait in humans (83). Although several TAS2R loci contribute to variability in PTC taste perception (54, 55), 50%--85% of the phenotypic variance in PTC sensitivity is attributed to variation at the TAS2R38 gene (237).. Studies have identified three amino acid substitutions at TAS2R38 that are in nearly complete LD in non-African populations and that form two common amino acid haplotypes (a taster haplotype PAV and a nontaster haplotype AVI; PAV is dominant). Furthermore, considerably more haplotype variability has been observed in Africa (M.C. Campbell and S.A. Tishkoff, unpublished data, 238) and these haplotypes are associated with a broad range of taste perception phenotypes (M.C. Campbell and S.A. Tishkoff, unpublished data).
Genetic analyses of both African and non-African populations have detected signatures of balancing selection at the TAS2R38 locus, including an excess of intermediate-frequency variants, a low amount of genetic differentiation between the continental populations (FST = 0.056), and an ancient divergence between the major taster and nontaster haplotypes (238). Furthermore, Wooding and coworkers (237) showed that PTC taste sensitivity in chimpanzees is associated with different amino acid haplotypes at the TAS2R38 gene compared to humans, implying a unique origin of the taster/nontaster variants in humans and chimpanzees.
It has also been suggested that low sensitivity to bitter taste may provide a selective advantage against malarial infection in some African populations. Recent data have shown that the K172 allele at the TAS2R16 gene (which is associated with low sensitivity to bitter taste substances, including salicin) occurs at moderately high frequencies in malaria endemic regions in central Africa (189). Furthermore, sequence analysis of the entire TAS2R16 coding region, as well as part of the 5' and 3' untranslated regions (UTRs), in 997 individuals from 60 human populations detected a signature of positive selection on chromosomes with the K172N variant (189). Although the variant driving the signal of positive selection was not conclusively determined, the authors speculated that differential selection in malarial (favoring the K variant) and non-malarial (favoring the N variant) environments has maintained both alleles at relatively high frequencies in Africa. An earlier study suggested that the higher dietary intake of naturally occurring bitter substances, such as organic cyanogens, may be protective against malarial infection in populations from Central and Southeast Africa (91]]). Additionally, an inhibitory effect of cyanide on the normal development of the P. falciparum parasite has been observed in vitro (137a]). Thus, individuals with a low sensitivity to the bitter taste of cyanide compounds may have a survival advantage against malarial infection through a higher intake of this bitter compound (189). However, this hypothesis remains to be tested.
Selection in African Versus Non-African Populations
Several studies have reported more evidence for positive selection in populations outside of Africa relative to those in Africa. For example, Akey and colleagues (6) examined 132 genes in 24 African Americans and 23 European Americans and found evidence for selection at eight genes only in the European-derived population. A number of genome scans for selection (which aim to identify de novo targets of selection) have identified differential patterns of selection in Africans and non-Africans (25, 84, 97, 98, 137, 180, 192, 219, 225, 234). Several of these studies have observed more loci under recent selection in non-African relative to African populations (25, 97, 137, 192, 234). Furthermore, it has been hypothesized that non-African populations have experienced more recent strong local adaptation as modern humans migrated out of Africa into novel and diverse environments (6, 84, 192, 234).
Although an increase in positive selection might conceivably occur in populations that have migrated into new environments, it is premature to conclude that the amount of recent positive selection is greater in non-African versus African populations. For example, Voight and colleagues (219) identified widespread evidence of recent selection in each of the HapMap samples in a genome-wide scan. Furthermore, they observed the strongest signals of selection in the HapMap Nigerian population compared to HapMap European and Asian populations (although the power to detect selection may be greater in larger African populations). Additionally, a recent study demonstrated that non-Africans have an excess proportion of nonsynonymous variation, including many variants that are likely to be deleterious (114), which is attributed to population demographic history rather than increased adaptive evolution. Therefore, demographic factors may have influenced the differential pattern of selection observed in African and non-African populations.
However, the primary limitation in comparing the frequency of selective events in African and non-African populations is that African populations have been severely understudied. For instance, many studies have used African Americans as the sole representative of African populations. However, the statistical power to detect selective sweeps is likely to be lower in studies using AfricanAmerican samples because of their recent admixture with Europeans (25, 234). Demographic parameters such as a population bottleneck in the Eurasian populations may also have mimicked patterns of variation caused by selective events in non-African populations (25). Indeed, one might predict that Africans would have relatively high amounts of local adaptation, considering that Africa has the highest levels of genetic diversity and contains populations living in a wide range of environments and with high exposure to infectious disease. However, signatures of selection in African populations may be missed because studies have relied mainly on one or two African populations. To gain a clearer understanding of genetic and phenotypic adaptations in Africa, it is important to scan for genetic signatures of selection in a broad range of ethnically diverse African populations living in distinct environments. Additionally, it will be important to clearly identify functional variants that are likely to be targets of selection and to verify their impact on phenotypic variation (32) before the relative number of selection events in African and non-African populations can be clearly determined .
INFECTIOUS DISEASE
Host genetic variation plays a key role in influencing susceptibility to many infectious diseases in humans. Through recurrent exposure to different pathogens, a number of genetic adaptations have evolved that provide resistance to infection. Although the number of known candidate genes related to infectious disease has expanded, progress in the identification of genes that influence infectious disease susceptibility and/or resistance in diverse African populations has been slow. Understanding the genetic basis of infectious disease in Africans may provide useful insight into devising effective strategies to combat these diseases that have a large impact on African populations. Here we focus on three infectious diseases that cause the highest number of deaths in Africa: acquired immune deficiency syndrome (AIDS) tuberculosis (TB) and malaria.
Acquired Immune Deficiency Syndrome
It is estimated that 42 million people are infected worldwide with HIV, the virus that causes AIDS (146). Moreover, greater than 75% of HIV-1 infections and 84% of all AIDS-related deaths occur in sub-Saharan Africa (235). Although most individuals exposed to HIV rapidly progress to more advanced stages of this disease, researchers have identified a number of Africans who do not progress to AIDS despite exposure to HIV (146). This observation suggests that polymorphisms associated with disease susceptibility and resistance may be present in African populations (Table 1).
Table 1.
Infectious disease loci
| Infectious disease | Known genes associated with disease susceptibility in Africans | Effect of genetic loci on infection or disease progression | References |
|---|---|---|---|
| HIV/AIDS | Chemokine receptor 2 (CCR2) | CCR-2 64I allele associated with ↓ disease progression | (146, 235) |
| Human leukocyte antigen (HLA) Locus | HLA A2/6802 supertype and HLA DRB1*01 associated with ↓ HIV infection in female sex workers from East Africa | (121) | |
| Killer immunoglobin- like receptors (KIR) | KIR3DS1 in combination with HLA-B Bw4--80Ile associated with ↓ disease progression in AfricanAmericans and West African sex workers | (95, 126) | |
| Interferon regulatory factor (IRF-1) | Three polymorphisms (located at 619, microsatellite region, and 6516 of genes) associated with ↓ susceptibility to infection in East African sex workers | (15) | |
| TRIM5α | 136Q and 43Y alleles protect against HIV infection in African Americans | (108) | |
| APOBEC3G | 186R allele associated with ↑ disease progression in African Americans | (8) | |
| CUL5 | Haplotype10 associated with ↑ disease progression of HIV-1 in African Americans | (9) | |
| Tuberculosis | Human homolog of natural-resistance--- associated macrophage protein 1 (NRAMP1) | Several polymorphisms (5’ microsatellite repeat, INT4,, and a deletion in 3’ UTR) associated with ↑ risk for tuberculosis in East and West Africans | ( 19, 20a, 188) |
| UBE3A | 7-bp deletion at 5’ end of UBE3A gene associated with ↑ susceptibility in West African families | (27) | |
| Chromosome 15 and X chromosome | Unknown loci on these chromosomes associated with ↑ tuberculosis susceptibility | (20) | |
| Human leukocyte antigen (HLA) locus | DRB1*1302 allele and DQB1*0301— 0304 alleles associated with ↑ tuberculosis susceptibility in Venda population from South Africa | (115) | |
| Vitamin D receptor (VDR) locus | F-b-A-T haplotype associated with ↓ susceptibility in a South African population | (115) | |
| Vitamin D receptor VDR) locus | Polymorphism at codon 352 of VDR gene associated with ↓susceptibility in Gambian population | (20) | |
| Vitamin D receptor (VDR) locus | Polymorphisms (FokI and ApaI) associated with ↑ susceptibility in West African families. | (149) | |
| CD209 | Promoter −336G allele associated with ↓ susceptibility in West Africa and Malawi | (215) | |
| Promoter −336A and −871G alleles associated with ↓ susceptibility in admixed colored population from South Africa | (18) | ||
| Intron 6 variant associated with ↑ susceptibility in West African population | (149) | ||
| Pentraxin 3 (PTX3) | G-A-G haplotype associated with ↓ susceptibility in West Africa | (149) | |
| Malaria | β-globin gene | HbS, HbC, and HbE alleles associated with ↓ malaria susceptibility in East and West Africans | (13a, 216a) |
| α-globin gene | α+-thalassemias associated with ↓ malaria susceptibility in East Africans | (58a) | |
| β-globin/α-globin genes | HbS and α+-thalassemia variants inherited together associated with ↑ susceptibility in East Africans | ( 232) | |
| Duffy gene | FY*O allele associated with ↓ malaria susceptibility in Africans | (83, 86) | |
| Glucose 6-phosphate dehydrogenase (G6PD) | G6PD A- allele associated with ↓ malaria susceptibility in West Africans | (67) | |
| Human leukocyte antigen (HLA) locus | HLA-B53 and HLA-DR.B1_1302 alleles associated with ↓ malaria susceptibility in West Africans | (67) | |
| Chromosome 10 | Chromosome 10p15.3--10p14 associated with ↑ malaria susceptibility in Africans | (199) | |
| Interleukin-4 (IL-4) | IL-4 −590 T allele associated with ↓ malaria susceptibility in the Fulani of Mali and Burkino Faso | (213) |
Chemokine receptors, which aid in the entry of HIV into the host cell, play a role in AIDS susceptibility (146). The protective effect of the Δ32 mutation at the chemokine receptor 5 (CCR5) gene has been well established in populations of Northern European ancestry (108). Because this mutation occurs at a relatively low frequency in sub-Saharan Africa, it is unlikely to play a major role in disease resistance in that region. However, several studies have demonstrated that mutations at the CCR2 gene play a role in resistance to HIV in Africans; for example, the CCR2–64I allele is associated with delayed HIV disease progression in African and AfricanAmerican populations (118, 233), although this effect was not observed in a large number of individuals from Uganda (170), indicating that underlying genetic and/or environmental factors that affect resistance may vary across African populations. See Table 1 for more details about other genes identified in African popluations.
Tuberculosis
Mycobacterium tuberculosis infection leading to TB causes significant mortality throughout the world, particularly in resource-poor countries. Furthermore, HIV infection is strongly associated with an increased risk for TB in sub-Saharan Africa (45, 119). In Africa, the rates of tuberculosis range from 50 to greater than 300 cases per 100,000 individuals (215). Genetic variation has been shown to influence susceptibility to TB in African populations. For example, polymorphisms in the NRAMP1 gene have been associated with increased TB susceptibility in ethnically diverse populations from the Gambia (20a ) and a single population in northern Tanzania (188). Other genes that are thought to play a role in TB susceptibility are shown in Table 1.
Malaria
As previously mentioned, malaria infection has been a strong selective force in recent human evolution. Approximately 40% of the world is at risk for malaria infection, and approximately 90% of all malaria deaths occur in sub-Saharan Africa (67). It is estimated that 500 million new cases of malarial illness caused by the Plasmodium falciparum parasite occur every year in Africa (http://www.rbm.who.int/amd2003/amr2003/ch1.htm).
Most of the common variants associated with resistance to malarial infection in Africans are expressed in red blood cells or play a role in immune response. These variants include hemoglobin HbS, HbC, HbE and α+-thalassemia, the G6PD A- allele, the FY*O Duffy allele (which prevents P. vivax from invading erythrocytes), and a number of HLA alleles (67, 83, 86).
Interestingly, several studies have shown that ethnically diverse African populations may differ in regard to genetic susceptibility to malarial infection. A variant in the promoter region of the IL4 gene, for example, is associated with a decrease in P. falciparum infection in the pastoralist Fulani from Mali, as evidenced by lower parasite load, but no such genetic association is observed in the neighboring agriculturalist Dogon population (52, 213). Other studies also reported lower prevalence of malaria parasites and fewer clinical attacks of malaria among the Fulani compared to other ethnically distinct populations in neighboring villages (107, 133, 152a). Differences in the expression profile of genes involved in immune response in the Fulani have been suggested to explain the distinct resistance to malaria in this population, but not in neighboring African populations (210). Therefore, novel genetic adaptations to malaria have evolved in genetically distinct populations in response to differential exposure to pathogens. These studies demonstrate that ethnically diverse African populations may have different resistance or susceptibility alleles (Table 1), motivating the need to include a large number of genetically distinct populations in studies of infectious disease susceptibility in Africa.
OTHER COMPLEX DISEASES IN AFRICAN POPULATIONS
There are a number of useful approaches for mapping complex disease traits that involve analyses of the association of markers and disease traits in pedigrees or parent/offspring trios [i.e., linkage and transmission disequilibrium test (TDT) analyses], and/or in populations (i.e., case/control association studies) (87). Several genome-wide SNP panels have been developed for use in genome-wide association studies (GWAS) in populations. Association mapping for complex disease relies on having some a priori knowledge of genetic diversity, population structure, and LD in both case and control populations to identify polymorphisms associated with disease genes and to avoid erroneous associations (122). Given the high levels of substructure and admixture between genetically distinct populations in Africa, even within small geographic regions, it is particularly important to control for population heterogeneity and substructure in GWAS of African populations [using, for example, programs such as PLINK (166)]. Another useful method for mapping complex traits in highly admixed populations (e.g., African Americans who have African and European ancestry) is mapping by admixture linkage disequilibrium (MALD) (see Supplemental Material). The MALD approach assumes that the genomic region that contains the susceptibility allele for a given disease will have enriched ancestry from populations in which the disease phenotype is more prevalent. Thus, detailed characterization of allelic variation across ancestral West African populations, which will enable accurate inference of African ancestry, will be important for the success of this approach.
Overall, the genetic factors underlying complex diseases are still poorly understood. To date, two models of complex disease have been proposed. The common disease/common variant (CD/CV) hypothesis posits that alleles influencing complex diseases are relatively common and are therefore found in multiple populations (51, 206). In contrast, some data have suggested that complex disease is caused by rare susceptibility alleles at many loci with small effect (50, 160). Additionally, gene × environment interactions as well as epistatic interactions among loci likely influence complex disease susceptibility (including infectious disease susceptibility) (44, 230, 232). Due to local adaptation, there may be population or regional specific susceptibility alleles underlying some complex diseases in Africa. Here we discuss three complex diseases that are common in populations of recent African descent (see Supplemental Material for a review of genetic susceptibility to prostate cancer in African descent populations and Reference 187 for a detailed review of additional genetic disease susceptibility studies in Africa).
Obesity
Obesity is a multifactorial disease that disproportionately affects African Americans and Afro-Caribbeans living in the United States (151). Moreover, this disease is increasing in prevalence in sub-Saharan Africa (13), particularly among urban residents . A recent study reported that the incidence of obesity in urban West Africa has more than doubled (114%) over the past 15 years (2). Obesity is a serious health concern because it is closely associated with other common disorders, such as type 2 diabetes and hypertension (208).
Although environmental factors are important determinants of obesity (92, 132), studies have also identified candidate loci that contribute to the onset of this disease. For example, the human uncoupling gene UCP3 has been correlated with obesity and lower resting energy expenditure in African Americans and the Mende tribe of Sierra Leone (10, 100); in contrast, risk for obesity associated with this gene varied among non-African populations (48, 134, 152). Another closely linked human uncoupling gene, UCP2, was also correlated with obesity in AfricanAmerican children (240). However, whether or not these tightly linked genes exert an independent effect on obesity in populations of African ancestry has not been firmly established.
Other candidate gene studies have reported associations between polymorphisms in promoter regions and weight-related phenotypes in populations of African descent. For example, polymorphisms in the promoter of the angiotensin-converting enzyme (ACE) gene are correlated with obesity in Nigerians and African Americans (105). Also, allelic variation in the promoter of the agouti-related protein (AGRP) gene was found to affect gene expression and was implicated in the regulation of body weight in people of African origin (11, 22).
Genome-wide linkage analyses of obesity-related phentoypes have indicated strong linkage to chromosomes 1, 2, 5, 7, 8, and 11 in West Africans (4, 29). These results demonstrate that multiple loci, together with environmental factors, likely contribute to the phenotypic variance of obesity-related traits.
Type 2 Diabetes Mellitus
Type 2 diabetes mellitus (T2DM) is a late-onset metabolic disorder characterized by reduced insulin secretion and insulin action (89). African Americans have a twofold increase in risk for T2DM compared to other populations in the United States. Furthermore, the prevalence of T2D is lower in Africa (~1%--2%) than among people of African descent in industrialized nations (~11%--13%)(177).
One of the earliest T2DM susceptibility loci implicated in this disease is the calpain 10 (CAPN10) gene on chromosome 2 in Mexican Americans (89). Subsequent association studies, however, have yielded inconsistent results among geographically distinct populations (70). In addition, the risk associated with this gene also differs between ethnically diverse populations from West Africa. Specifically, a CAPN10 haplotype defined by known risk polymorphisms was associated with T2DM in populations from Nigeria, but not in distinct populations from Ghana (31). Further study in a larger set of Africans will be needed to resolve this discrepancy.
Other recently identified candidate loci associated with diabetes or diabetes-related traits in Africans and African Americans include the AGRP gene (22) the transcription factor 7-like 2 (TCF7L2) gene (85), and the proprotein convertase subtilisin/kexin-type 2 (PCSK2) gene (108a). Genome-wide linkage analyses in West African families have also indicated suggestive linkage to diabetes on chromosomes 12 and 19 as well as stronger evidence of linkage on chromosome 20 (177). Other studies have reported strong linkage for several quantitative traits that contribute to diabetes on chromosomes 4, 6, 8, 10, 15, 16, 17, and 18 (28, 30, 178).
Increased risk for type 2 diabetes in Africans and other indigenous populations is also suggested to be due to changes in selective pressure (i.e., the thrifty gene hypothesis) (139). Prior to 10,000 years ago, all modern humans subsisted as hunter-gatherers who likely experienced frequent cycles of feast and famine. According to the thrifty gene hypothesis, ancestral genetic variants that once promoted the efficient absorption, storage, or utilization of nutrients in this ancestral environment are now maladaptive in more modern environments, increasing risk for disease (51, 153). Although genetic evidence for this hypothesis has been inconclusive (34, 68, 214), recent data from the Macaque Genome Project indicated that a number of polymorphisms in the macaque correspond to known disease-predisposing alleles in humans (39). These results suggest that ancestral variants may influence disease susceptibility in humans .
Hypertension
Hypertension (HT) disproportionately affects people of African descent living in Western environments. For example, African Americans have a 1.6-fold higher prevalence of HT than European Americans (49). However, HT was not considered to be a major disease in sub-Saharan Africa until recently. Studies have now reported a growing incidence of HT, especially in urban settings in Nigeria, Cameroon, and Tanzania (56, 212).
To date, candidate gene studies have shown inconsistent associations with HT. For example, increased risk for disease was correlated with the G protein β3-subunit (GNB3) gene, angiotensin (AGT), and angiotensin II receptor (AGTR1) in some populations of African ancestry (53, 123, 186, 245), but these results were not replicated in other studies (61, 216, 231). A study of the heritability of AGT and ACE levels in Nigerians and African Americans indicated that heritability is high in Nigerians (77% for AGT and 67% for ACE) but low in African Americans (18% for AGT and ACE), suggesting a strong environmental component to variability in African American populations (44).
Other evidence has suggested that interactions between susceptibility loci may play an important role in HT susceptibility. For example, polymorphisms in the ACE, ACE4, and ACE8 genes were jointly associated with blood pressure through epistatic interaction in Nigerian families (244). Moreover, interaction between genes (ACE and GRK4) on different chromosomes was also found to influence blood pressure in a cohort of West African families (231). These studies suggest a complex model of disease susceptibility that involves epistatic interactions and may account for inconsistent associations in previous studies.
The results of genome-wide linkage studies have also suggested that chromosome 6q24 and 21q21 likely contain genes that influence risk for hypertension in African Americans. In another study, fine mapping of HT susceptibility loci on chromosome 6 uncovered polymorphisms in the vanin 1 (VNN1) gene that were correlated with HT in African Americans and Mexican Americans (246,).
From an evolutionary perspective, salt retention (a characteristic of HT) has been proposed to represent a phenotypic adaptation to heat. Specifically, ancient human populations living in hot, humid areas, who consumed low levels of dietary salt, were theorized to have adapted to their environment by retaining salt, while populations in cooler, temperate climates adapted to conditions of higher sodium levels (14). Under this scenario, polymorphisms that promote salt retention will increase in frequency in hot and humid environments due to selective pressure. This hypothesis was recently supported by several studies that observed the highest frequency of variants associated with salt retention and/or high blood pressure in Africans and decreasing allele frequency outside of Africa (138, 198, 242).
Pharmacogenetics
Individuals with distinct ancestry are known to vary in drug response. For example, drugs commonly used to treat heart disease are known to be less effective in individuals of African descent relative to individuals of European descent (see Supplemental Material). To better understand variation in drug response, it is important to characterize levels and patterns of genetic diversity at genes that may influence drug response, drug metabolism, and/or transport in ethnically diverse populations (23). To date, some of the most intensively studied drug metabolism enzyme (DME) loci include cytochrome P450s (CYPs), N-acetyl transferases (NATs), and multidrug transporters (MDR). These genes are highly polymorphic and their variation results in proteins with increased, normal, or decreased activity (187).
Studies have shown differences in the distribution of variation at these DME loci in African and non-African populations. For example, the CYP2B6 gene is involved in the metabolism of several clinically important drugs, including artemisinin and efavirenz used to treat malaria and HIV infection, respectively. Common CYP2B6 polymorphisms vary in frequency between ethnically diverse populations from West Africa, as well as between African and non-African populations. The functional significance of most of these variants is not yet known (131). However, recent studies have shown that two polymorphisms (983T>C and 516G>T), found at a higher frequency in African populations relative to non-Africans, are associated with a reduction in CYP2B6 protein expression as well as a large increase in levels of the antiretroviral drug efavirenz in the plasma of African HIV patients (130, 145, 239).
Additionally, the frequency of functional variants at the NAT2 gene, known to play a role in the metabolism of the drug isoniazid (used to treat TB), was found to vary among ethnically diverse African populations. In particular, haplotypes associated with the fast-acetylation and the slow-acetylation phenotypes at the NAT2 gene differ in frequency among African populations, particularly between hunter-gatherer and agriculturalist populations (H.M. Mortensen and S.A.Tishkoff, unpublished data, 154, 155). However, Africans have high levels of haplotype diversity and the effect of many of these haplotypes remains unknown. These studies imply that ethnically diverse Africans may differ in response to drugs used to treat infectious disease due to variation at genes involved in drug metabolism or transport. Given the high numbers of deaths due to infectious disease in Africa, the study of variation at genes that play a role in response to these drugs across ethnically diverse Africans is of critical importance.
FUTURE DIRECTIONS
To date, only a fraction of the ~2000 linguistically distinct ethnic groups in Africa has been extensively studied for genome-wide variation. Extensive sampling of East African populations will be informative for testing models of the origin and dispersal of modern humans out of Africa, whereas in depth sampling of West African populations will be informative for determining African American ancestry and for the identification of markers and populations useful in MALD mapping. It is important to use an ethical approach when collecting samples to be used for genetic diversity studies, particularly if those samples and genetic data will be made publicly available. In addition to obtaining research permits from the local African governments and informed consent from individual participants (including benefits and risks involved in use of samples in current and future studies), results should ideally be made available to participants after translation into the local language. Additionally, efforts should be made to train local African scientists and to build resources across Africa for independent human genetics research. The African Society of Human Genetics was recently formed in 2004 to help achieve many of these goals (176) (http://www.afshg.org).
As we begin to build diverse sets of samples, a shift toward genome-wide studies of genetic diversity will be particularly informative for making inferences about African demographic history and the genetic basis of complex traits. A better understanding of the distribution and frequency of structural variation and its role in regard to phenotypic diversity will be of particular interest. The development of high-throughput SNP genotyping methodology (i.e., the Affymetrix 6.0 and Illumina 1M SNP chips), which is rapidly becoming less expensive, will facilitate the possibility of GWAS in a large number of Africans. Indeed, several recent GWAS in Europeans that have identified genetic variants associated with traits such as height, skin pigmentation, and eye color indicate that very large sample sizes (>3000--5000) are required to identify variants associated with complex traits influenced by multiple loci and the environment (194, 227). However, to date no GWAS have been perfomed on a comparable scale in Africa. Such studies will be highly informative for identifying genes that play a role in a number of common traits (e.g., height), as well as for identifying genes that play a role in susceptibility to infectious and other complex diseases. The high levels of genetic substructure in Africa, even within small geographic regions ( F.A. Reed and S.A. Tishkoff, unpublished data), require determination of individual ancestry and proper correction for substructure in association studies.
Genotyping ethnically diverse African populations living in distinct climates and with distinct subsistence patterns for these high density SNP panels will also be useful for conducting wholegenome scans of selection to identify genes that have played a role in local adaptation and disease. The continued development of statistical and computational methods for inferring demographic history and natural selection will shed light on human evolutionary history in Africa. Approaches that incorporate detailed geographic information such as natural boundaries (i.e., mountain ranges, rivers, deserts, etc.) will be particularly useful for inferring African demographic history (163, 172).
Given that African populations possess a large fraction of population-specific alleles and may have experienced local adaptation, resequencing across diverse African populations will be important for identifying population-specific functional variants. Targeted resequencing of genes that play a key role in disease susceptibility and drug response will be particularly important for the design of more effective treatments in individuals of recent African descent. Additionally, whole genome resequencing (a goal of the 1000 genomes project) will be informative for identifying large-scale structural variants and rare variants that may play an important role in disease and for reconstructing human evolutionary history.
Supplementary Material
ACKNOWLEDGMENTS
We thank S.M. Williams, J.M. Akey, J.S. Friedlaender, and N.A. Rosenberg for critical review of sections of the manuscript and/or figures and for helpful suggestions. The authors are funded by the U.S. National Science Foundation (NSF) grant BSC-0552486, U.S. National Institutes of Health (NIH) grant R01GM076637, and a David and Lucile Packard Career Award to S.A.T.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abecasis GR, Ghosh D, Nichols TE. Linkage disequilibrium: ancient history drives the new genetics. Hum. Hered. 2005;59:118–124. doi: 10.1159/000085226. [DOI] [PubMed] [Google Scholar]
- 2.Abubakari AR, Lauder W, Agyemang C, Jones M, Kirk A, Bhopal RS. Prevalence and time trends in obesity among adult West African populations: a meta-analysis. Obes. Rev. 2008 doi: 10.1111/j.1467-789X.2007.00462.x. doi:10.1111/j.1467-789X.2007.00462.x. [DOI] [PubMed] [Google Scholar]
- 3.Abu-Raddad LJ, Patnaik P, Kublin JG. Dual infection with HIV and malaria fuels the spread of both diseases in sub-Saharan Africa. Science. 2006;314:1603–1606. doi: 10.1126/science.1132338. [DOI] [PubMed] [Google Scholar]
- 4.Adeyemo A, Luke A, Cooper R, Wu X, Tayo B, et al. A genome-wide scan for body mass index among Nigerian families. Obes. Res. 2003;11:266–273. doi: 10.1038/oby.2003.40. [DOI] [PubMed] [Google Scholar]
- 5.Deleted in proof.
- 6.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akey JM, Zhang K, Xiong M, Jin L. The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequilibrium. Mol. Biol. Evol. 2003;20:232–242. doi: 10.1093/molbev/msg032. [DOI] [PubMed] [Google Scholar]
- 8.An P, Bleiber G, Duggal P, Nelson G, May M, et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An P, Duggal P, Wang LH, O’Brien SJ, Donfield S, et al. Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argyropoulos G, Brown AM, Willi SM, Zhu J, He Y, et al. Effects of mutations in the human uncoupling protein 3 gene on the respiratory quotient and fat oxidation in severe obesity and type 2 diabetes. J. Clin. Invest. 1998;102:1345–1351. doi: 10.1172/JCI4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argyropoulos G, Rankinen T, Bai F, Rice T, Province MA, et al. The agouti-related protein and body fatness in humans. Int. J. Obes. Relat. Metab. Disord. 2003;27:276–280. doi: 10.1038/sj.ijo.802201. [DOI] [PubMed] [Google Scholar]
- 12.Arnheim N, Calabrese P, Nordborg M. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 2003;73:5–16. doi: 10.1086/376419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aspray TJ, Mugusi F, Rashid S, Whiting D, Edwards R, et al. Rural and urban differences in diabetes prevalence in Tanzania: the role of obesity, physical inactivity and urban living. Trans. R. Soc. Trop. Med. Hyg. 2000;94:637–644. doi: 10.1016/s0035-9203(00)90216-5. [DOI] [PubMed] [Google Scholar]
- 13a.Ayodo G, Price AL, Keinan A, Ajwang A, Otieno MF, et al. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am. J. Hum. Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balaresque PL, Ballereau SJ, Jobling MA. Challenges in human genetic diversity: demographic history and adaptation. Hum. Mol. Genet. 2007;16(Spec. No. 2):R134–R139. doi: 10.1093/hmg/ddm242. [DOI] [PubMed] [Google Scholar]
- 15.Ball TB, Ji H, Kimani J, McLaren P, Marlin C, et al. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS. 2007;21:1091–1101. doi: 10.1097/QAD.0b013e3280ef6ae1. [DOI] [PubMed] [Google Scholar]
- 16.Deleted in proof.
- 17.Bansal V, Bashir A, Bafna V. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 2007;17:219–230. doi: 10.1101/gr.5774507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barreiro LB, Neyrolles O, Babb CL, Tailleux L, Quach H, et al. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006;3:e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. 2003;4:4–11. doi: 10.1038/sj.gene.6363915. [DOI] [PubMed] [Google Scholar]
- 20.Bellamy R, Beyers N, McAdam KP, Ruwende C, Gie R, et al. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc. Natl. Acad. Sci. USA. 2000;97:8005–8009. doi: 10.1073/pnas.140201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N. Engl. J. Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 21.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonilla C, Panguluri RK, Taliaferro-Smith L, Argyropoulos G, Chen G, et al. Agouti-related protein promoter variant associated with leanness and decreased risk for diabetes in West Africans. Int. J. Obes. 2006;30:715–721. doi: 10.1038/sj.ijo.0803047. [DOI] [PubMed] [Google Scholar]
- 23.Brockmoller J, Tzvetkov MV. Pharmacogenetics: data, concepts and tools to improve drug discovery and drug treatment. Eur. J. Clin. Pharmacol. 2008;64:133–157. doi: 10.1007/s00228-007-0424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deleted in proof.
- 25.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavalli-Sforza LL, Piazza A, Menozzi P. History and Geography of Human Genes. Princeton: Princeton Univ. Press; 1994. [Google Scholar]
- 27.Cervino AC, Lakiss S, Sow O, Bellamy R, Beyers N, et al. Fine mapping of a putative tuberculosis-susceptibility locus on chromosome 15q11-13 in African families. Hum. Mol. Genet. 2002;11:1599–1603. doi: 10.1093/hmg/11.14.1599. [DOI] [PubMed] [Google Scholar]
- 28.Chen G, Adeyemo A, Zhou J, Chen Y, Huang H, et al. Genome-wide search for susceptibility genes to type 2 diabetes in West Africans: potential role of C-peptide. Diabetes Res. Clin. Pract. 2007;78:e1–e6. doi: 10.1016/j.diabres.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Adeyemo AA, Johnson T, Zhou J, Amoah A, et al. A genome-wide scan for quantitative trait loci linked to obesity phenotypes among West Africans. Int. J. Obes. 2005;29:255–259. doi: 10.1038/sj.ijo.0802873. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, Adeyemo AA, Zhou J, Chen Y, Doumatey A, et al. A genome-wide search for linkage to renal function phenotypes in West Africans with type 2 diabetes. Am. J. Kidney Dis. 2007;49:394–400. doi: 10.1053/j.ajkd.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Kittles R, Zhou J, Chen G, Adeyemo A, et al. Calpain-10 gene polymorphisms and type 2 diabetes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Ann. Epidemiol. 2005;15:153–159. doi: 10.1016/j.annepidem.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Cho MK, Sabeti PC, Tishkoff SA. Natural selection in humans: ethical issues and methodological challenges. Trends Genet. 2008 In review. [Google Scholar]
- 33.Clark AG, Nielsen R, Signorovitch J, Matise TC, Glanowski S, et al. Linkage disequilibrium and inference of ancestral recombination in 538 single-nucleotide polymorphism clusters across the human genome. Am. J. Hum. Genet. 2003;73:285–300. doi: 10.1086/377138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark VJ, Ptak SE, Tiemann I, Qian Y, Coop G, et al. Combining sperm typing and linkage disequilibrium analyses reveals differences in selective pressures or recombination rates across human populations. Genetics. 2007;175:795–804. doi: 10.1534/genetics.106.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, et al. Microsatellite variation and evolution of human lactase persistence. Hum. Genet. 2005;117:329–339. doi: 10.1007/s00439-005-1322-z. [DOI] [PubMed] [Google Scholar]
- 36.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 37.Conrad DF, Hurles ME. The population genetics of structural variation. Nat. Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 39.Consort. RMGSAA. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 40.Consort. Int. HapMap. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consort. Int. HapMap. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–862. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coop G, Przeworski M. An evolutionary view of human recombination. Nat. Rev. Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 43.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 44.Cooper RS, Guo X, Rotimi CN, Luke A, Ward R, et al. Heritability of angiotensin-converting enzyme and angiotensinogen: A comparison of US blacks and Nigerians. Hypertension. 2000;35:1141–1147. doi: 10.1161/01.hyp.35.5.1141. [DOI] [PubMed] [Google Scholar]
- 45.Corbett EL, Marston B, Churchyard GJ, De Cock KM. Tuberculosis in sub-Saharan Africa: opportunities, challenges, and change in the era of antiretroviral treatment. Lancet. 2006;367:926–937. doi: 10.1016/S0140-6736(06)68383-9. [DOI] [PubMed] [Google Scholar]
- 46.Cox MP, Mendez FL, Karafet TM, Pilkington MM, Kingan SB, et al. Testing for archaic hominin admixture on the X chromosome: model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent. Genetics. 2008;178:427–437. doi: 10.1534/genetics.107.080432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 48.de Luis DA, Aller R, Izaola O, González Sagrado M, Conde R, Pérez Castrillón JL. Lack of association of −55CT polymorphism of UCP3 gene with fat distribution in obese patients. Ann. Nutr. Metab. 2006;51:374–378. doi: 10.1159/000107685. [DOI] [PubMed] [Google Scholar]
- 49.Deo RC, Patterson N, Tandon A, McDonald GJ, Haiman CA, et al. A high-density admixture scan in 1,670 African Americans with hypertension. PLoS Genet. 2007;3:e196. doi: 10.1371/journal.pgen.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Rienzo A. Population genetics models of common diseases. Curr. Opin. Genet. Dev. 2006;16:630–636. doi: 10.1016/j.gde.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Dolo A, Modiano D, Maiga B, Daou M, Dolo G, et al. Difference in susceptibility to malaria between two sympatric ethnic groups in Mali. Am. J. Trop. Med. Hyg. 2005;72:243–248. [PubMed] [Google Scholar]
- 53.Dong Y, Zhu H, Sagnella GA, Carter ND, Cook DG, Cappuccio FP. Association between the C825T polymorphism of the G protein β3-subunit gene and hypertension in blacks. Hypertension. 1999;34:1193–1196. doi: 10.1161/01.hyp.34.6.1193. [DOI] [PubMed] [Google Scholar]
- 54.Drayna D. Human taste genetics. Annu. Rev. Genomics Hum. Genet. 2005;6:217–235. doi: 10.1146/annurev.genom.6.080604.162340. [DOI] [PubMed] [Google Scholar]
- 55.Drayna D, Coon H, Kim UK, Elsner T, Cromer K, et al. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum. Genet. 2003;112:567–572. doi: 10.1007/s00439-003-0911-y. [DOI] [PubMed] [Google Scholar]
- 56.Edwards R, Unwin N, Mugusi F, Whiting D, Rashid S, et al. Hypertension prevalence and care in an urban and rural area of Tanzania. J. Hypertens. 2000;18:145–152. doi: 10.1097/00004872-200018020-00003. [DOI] [PubMed] [Google Scholar]
- 57.Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 58a.Enevold A, Alifrangis M, Sanchez JJ, Carneiro I, Roper C, et al. Associations between α+-thalassemia and Plasmodium falciparum malarial infection in northeastern Tanzania. J. Infect. Dis. 2007;196:451–459. doi: 10.1086/519390. [DOI] [PubMed] [Google Scholar]
- 59.Eswaran V, Harpending H, Rogers AR. Genomics refutes an exclusively African origin of humans. J. Hum. Evol. 2005;49:1–18. doi: 10.1016/j.jhevol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 60.Evans DM, Cardon LR. A comparison of linkage disequilibrium patterns and estimated population recombination rates across multiple populations. Am. J. Hum. Genet. 2005;76:681–687. doi: 10.1086/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fejerman L, Wu X, Adeyemo A, Luke A, Zhu X, et al. The effect of genetic variation in angiotensinogen on serum levels and blood pressure: a comparison of Nigerians and US blacks. J. Hum. Hypertens. 2006;20:882–887. doi: 10.1038/sj.jhh.1002077. [DOI] [PubMed] [Google Scholar]
- 62.Forster P, Matsumura S. Evolution. Did early humans go north or south? Science. 2005;308:965–966. doi: 10.1126/science.1113261. [DOI] [PubMed] [Google Scholar]
- 63.Forster P, Romano V. Timing of a back-migration into Africa. Science. 2007;316:50–53. doi: 10.1126/science.316.5821.50. [DOI] [PubMed] [Google Scholar]
- 64.Deleted in proof.
- 65.Friedlaender JS, Friedlaender FR, Hodgson JA, Stoltz M, Koki G, et al. Melanesian mtDNA complexity. PLoS ONE. 2007;2:e248. doi: 10.1371/journal.pone.0000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friedlaender JS, Friedlaender FR, Reed FA, Kidd KK, Kidd JR, et al. The genetic structure of Pacific islanders. PLoS Genetics. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frodsham AJ, Hill AV. Genetics of infectious diseases. Hum. Mol. Genet. 2004;13(Spec No 2):R187–R194. doi: 10.1093/hmg/ddh225. [DOI] [PubMed] [Google Scholar]
- 68.Fullerton SM, Bartoszewicz A, Ybazeta G, Horikawa Y, Bell GI, et al. Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am. J. Hum. Genet. 2002;70:1096–1106. doi: 10.1086/339930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 70.Garant MJ, Kao WH, Brancati F, Coresh J, Rami TM, et al. SNP43 of CAPN10 and the risk of type 2 diabetes in African-Americans: the Atherosclerosis Risk in Communities Study. Diabetes. 2002;51:231–237. doi: 10.2337/diabetes.51.1.231. [DOI] [PubMed] [Google Scholar]
- 71.Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat. Rev. Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- 72.Garrigan D, Kingan SB, Pilkington MM, Wilder JA, Cox MP, et al. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garrigan D, Mobasher Z, Severson T, Wilder JA, Hammer MF. Evidence for archaic Asian ancestry on the human X chromosome. Mol. Biol. Evol. 2004;22:189–192. doi: 10.1093/molbev/msi013. [DOI] [PubMed] [Google Scholar]
- 74.Deleted in proof.
- 75.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 76.Graffelman J, Balding DJ, Gonzalez-Neira A, Bertranpetit J. Variation in estimated recombination rates across human populations. Hum. Genet. 2007;122:301–310. doi: 10.1007/s00439-007-0391-6. [DOI] [PubMed] [Google Scholar]
- 77.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 78.Guthery SL, Salisbury BA, Pungliya MS, Stephens JC, Bamshad M. The structure of common genetic variation in United States populations. Am. J. Hum. Genet. 2007;81:1221–1231. doi: 10.1086/522239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamblin MT, Thompson EE, Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hanchard N, Elzein A, Trafford C, Rockett K, Pinder M, et al. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52. doi: 10.1186/1471-2156-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harding RM, McVean G. A structured ancestral population for the evolution of modern humans. Curr. Opin. Genet. Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 83.Harris EE, Meyer D. The molecular signature of selection underlying human adaptations. Am. J. Phys. Anthropol Suppl. 2006;43:89–130. doi: 10.1002/ajpa.20518. [DOI] [PubMed] [Google Scholar]
- 84.Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc. Natl. Acad. Sci. USA. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 86.Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 2006;40:469–486. doi: 10.1146/annurev.genet.40.110405.090546. [DOI] [PubMed] [Google Scholar]
- 87.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 88.Hollox EJ, Poulter M, Zvarik M, Ferak V, Krause A, et al. Lactase haplotype diversity in the Old World. Am. J. Hum. Genet. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 90.Ingram CJ, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum. Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 91.Jackson F. Two evolutionary models for the interactions of dietary organic cyanogens, hemoglobins, and falciparum malaria. Am. J. Hum. Biol. 1990;2:521–532. doi: 10.1002/ajhb.1310020508. [DOI] [PubMed] [Google Scholar]
- 92.Jackson M, Walker S, Cruickshank JK, Sharma S, Cade J, et al. Diet and overweight and obesity in populations of African origin: Cameroon, Jamaica and the UK. Public Health Nutr. 2007;10:122–130. doi: 10.1017/S1368980007246762. [DOI] [PubMed] [Google Scholar]
- 93.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 94.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 95.Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 96.Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 97.Kayser M, Brauer S, Stoneking M. A genome scan to detect candidate regions influenced by local natural selection in human populations. Mol. Biol. Evol. 2003;20:893–900. doi: 10.1093/molbev/msg092. [DOI] [PubMed] [Google Scholar]
- 98.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum. Mutat. 2005;26:199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- 100.Kimm SY, Glynn NW, Aston CE, Damcott CM, Poehlman ET, et al. Racial differences in the relation between uncoupling protein genes and resting energy expenditure. Am. J. Clin. Nutr. 2002;75:714–719. doi: 10.1093/ajcn/75.4.714. [DOI] [PubMed] [Google Scholar]
- 101.Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, et al. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears. Am. J. Hum. Genet. 2004;75:752–770. doi: 10.1086/425161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101a.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 102.Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, et al. Sequence variants in the RNF212 gene associate with genomewide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 103.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korenromp EL, Williams BG, de Vlas SJ, Gouws E, Gilks CF, et al. Malaria attributable to the HIV-1 epidemic, sub-Saharan Africa. Emerg. Infect. Dis. 2005;11:1410–1419. doi: 10.3201/eid1109.050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kramer H, Wu X, Kan D, Luke A, Zhu X, et al. Angiotensin-converting enzyme gene polymorphisms and obesity: an examination of three black populations. Obes. Res. 2005;13:823–828. doi: 10.1038/oby.2005.94. [DOI] [PubMed] [Google Scholar]
- 106.Kuper R, Kropelin S. Climate-controlled Holocene occupation in the Sahara: motor of Africa’s evolution. Science. 2006;313:803–807. doi: 10.1126/science.1130989. [DOI] [PubMed] [Google Scholar]
- 107.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lama J, Planelles V. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology. 2007;4:52. doi: 10.1186/1742-4690-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108a.Leak TS, Keene KL, Langefeld CD, Gallagher CJ, Mychaleckyj JC, et al. Association of the proprotein convertase subtilisin/kexin-type 2 (PCSK2) gene with type 2 diabetes in an African American population. Mol. Genet. Metab. 2007;92:145–150. doi: 10.1016/j.ymgme.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 110.Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT. T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum. Mol. Genet. 2005;14:3945–3953. doi: 10.1093/hmg/ddi418. [DOI] [PubMed] [Google Scholar]
- 111.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 112.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deleted in proof.
- 114.Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lombard Z, Brune AE, Hoal EG, Babb C, Van Helden PD, et al. HLA class II disease associations in southern Africa. Tissue Antigens. 2006;67:97–110. doi: 10.1111/j.1399-0039.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 116.Lonjou C, Zhang W, Collins A, Tapper WJ, Elahi E, et al. Linkage disequilibrium in human populations. Proc. Natl. Acad. Sci. USA. 2003;100:6069–6074. doi: 10.1073/pnas.1031521100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lovell A, Moreau C, Yotova V, Xiao F, Bourgeois S, et al. Ethiopia: between sub-Saharan Africa and western Eurasia. Ann. Hum. Genet. 2005;69:275–287. doi: 10.1046/j.1529-8817.2005.00152.x. [DOI] [PubMed] [Google Scholar]
- 118.Ma L, Marmor M, Zhong P, Ewane L, Su B, Nyambi P. Distribution of CCR2-64I and SDF1-3’A alleles and HIV status in 7 ethnic populations of Cameroon. J. Acquired Immune Defic. Syndr. 2005;40:89–95. doi: 10.1097/01.qai.0000157008.66584.d6. [DOI] [PubMed] [Google Scholar]
- 119.Maartens G, Wilkinson RJ. Tuberculosis. Lancet. 2007;370:2030–2043. doi: 10.1016/S0140-6736(07)61262-8. [DOI] [PubMed] [Google Scholar]
- 120.Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 121.MacDonald KS, Fowke KR, Kimani J, Dunand VA, Nagelkerke NJ, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 2000;181:1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 122.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 123.Markovic D, Tang X, Guruju M, Levenstien MA, Hoh J, et al. Association of angiotensinogen gene polymorphisms with essential hypertension in African-Americans and Caucasians. Hum. Hered. 2005;60:89–96. doi: 10.1159/000088657. [DOI] [PubMed] [Google Scholar]
- 124.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 127.McBrearty S, Brooks A. The revolution that wasn’t: a new interpretation of the origin of modern human behavior. J. Hum. Evol. 2000;39:453–563. doi: 10.1006/jhev.2000.0435. [DOI] [PubMed] [Google Scholar]
- 128.McDougall IBF, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 129.McVean G, Spencer CC, Chaix R. Perspectives on human genetic variation from the HapMap Project. PLoS Genet. 2005;1:e54. doi: 10.1371/journal.pgen.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mehlotra RK, Bockarie MJ, Zimmerman PA. CYP2B6 983T>C polymorphism is prevalent in West Africa but absent in Papua New Guinea: implications for HIV/AIDS treatment. Br. J. Clin. Pharmacol. 2007;64:391–395. doi: 10.1111/j.1365-2125.2007.02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mehlotra RK, Ziats MN, Bockarie MJ, Zimmerman PA. Prevalence of CYP2B6 alleles in malaria-endemic populations of West Africa and Papua New Guinea. Eur. J. Clin. Pharmacol. 2006;62:267–275. doi: 10.1007/s00228-005-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mendez MA, Wynter S, Wilks R, Forrester T. Under- and overreporting of energy is related to obesity, lifestyle factors and food group intakes in Jamaican adults. Public Health Nutr. 2003;7:9–19. doi: 10.1079/phn2003508. [DOI] [PubMed] [Google Scholar]
- 133.Modiano D, Luoni G, Sirima BS, Lanfrancotti A, Petrarca V, et al. The lower susceptibility to Plasmodium falciparum malaria of Fulani of Burkina Faso (west Africa) is associated with low frequencies of classic malaria-resistance genes. Trans. R. Soc. Trop. Med. Hyg. 2001;95:149–152. doi: 10.1016/s0035-9203(01)90141-5. [DOI] [PubMed] [Google Scholar]
- 134.Mottagui-Tabar S, Hoffstedt J, Brookes AJ, Jiao H, Arner P, Dahlman I. Association of ADRB1 and UCP3 gene polymorphisms with insulin sensitivity but not obesity. Horm. Res. 2008;69:31–36. doi: 10.1159/000111793. [DOI] [PubMed] [Google Scholar]
- 135.Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am. J. Hum. Genet. 2004;74:1102–1110. doi: 10.1086/421050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Myles S, Bouzekri N, Haverfield E, Cherkaoui M, Dugoujon JM, Ward R. Genetic evidence in support of a shared Eurasian-North African dairying origin. Hum. Genet. 2005;117:34–42. doi: 10.1007/s00439-005-1266-3. [DOI] [PubMed] [Google Scholar]
- 137.Myles S, Tang K, Somel M, Green RE, Kelso J, Stoneking M. Identification and analysis of genomic regions with large between-population differentiation in humans. Ann. Hum. Genet. 2008;72:99–110. doi: 10.1111/j.1469-1809.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 137a.Nagel RL, Raventos C, Tanowitz HB, Wittner M. Effect of sodium cyanate on Plasmodium falciparum in vitro. J. Parasitol. 1980;66:483–487. [PubMed] [Google Scholar]
- 138.Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, et al. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am. J. Hum. Genet. 2004;74:898–916. doi: 10.1086/420793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 140.Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nielsen R, Hubisz MJ, Clark AG. Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics. 2004;168:2373–2382. doi: 10.1534/genetics.104.031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nielsen R, Signorovitch J. Correcting for ascertainment biases when analyzing SNP data: applications to the estimation of linkage disequilibrium. Theor. Popul. Biol. 2003;63:245–255. doi: 10.1016/s0040-5809(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 144.Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nyakutira C, Röshammar D, Chigutsa E, Chonzi P, Ashton M, et al. High prevalence of the CYP2B6 516G-->T(*6) variant and effect on the population pharmacokinetics of efavirenz in HIV/AIDS outpatients in Zimbabwe. Eur. J. Clin. Pharmacol. 2007;64:357–365. doi: 10.1007/s00228-007-0412-3. [DOI] [PubMed] [Google Scholar]
- 146.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat. Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 147.Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Brittenham G, et al. Extended linkage disequilibrium surrounding the hemoglobin E variant due to malarial selection. Am. J. Hum. Genet. 2004;74:1198–1208. doi: 10.1086/421330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum. Mol. Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 149.Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes. Immun. 2007;8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 150.Olivieri A, Achilli A, Pala M, Battaglia V, Fornarino S, et al. The mtDNA legacy of the Levantine early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. [DOI] [PubMed] [Google Scholar]
- 151.Osei K. Metabolic consequences of the West African diaspora: lessons from the thrifty gene. J. Lab. Clin. Med. 1999;133:98–111. doi: 10.1016/s0022-2143(99)90002-1. [DOI] [PubMed] [Google Scholar]
- 152.Otabe S, Clement K, Dina C, Pelloux V, Guy-Grand B, et al. A genetic variation in the 5' flanking region of the UCP3 gene is associated with body mass index in humans in interaction with physical activity. Diabetologia. 2000;43:245–249. doi: 10.1007/s001250050037. [DOI] [PubMed] [Google Scholar]
- 152a.Paganotti GM, Babiker HA, Modiano D, Sirima BS, Verra F, et al. Genetic complexity of Plasmodium falciparum in two ethnic groups of Burkina Faso with marked differences in susceptibility to malaria. Am. J. Trop. Med. Hyg. 2004;71:173–178. [PubMed] [Google Scholar]
- 153.Paradies YC, Montoya MJ, Fullerton SM. Racialized genetics and the study of complex diseases: the thrifty genotype revisited. Perspect. Biol. Med. 2007;50:203–227. doi: 10.1353/pbm.2007.0020. [DOI] [PubMed] [Google Scholar]
- 154.Patin E, Barreiro LB, Sabeti PC, Austerlitz F, Luca F, et al. Deciphering the ancient and complex evolutionary history of human arylamine N-acetyltransferase genes. Am. J. Hum. Genet. 2006;78:423–436. doi: 10.1086/500614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Patin E, Harmant C, Kidd KK, Kidd J, Froment A, et al. Sub-Saharan African coding sequence variation and haplotype diversity at the NAT2 gene. Hum. Mutat. 2006;27:720. doi: 10.1002/humu.9438. [DOI] [PubMed] [Google Scholar]
- 156.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pilkington MM, Wilder JA, Mendez FL, Cox MP, Woerner A, et al. Contrasting signatures of population growth for mitochondrial DNA and Y chromosomes among human populations in Africa. Mol. Biol. Evol. 2008;25:517–525. doi: 10.1093/molbev/msm279. [DOI] [PubMed] [Google Scholar]
- 158.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, et al. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann. Hum. Genet. 2003;67:298–311. doi: 10.1046/j.1469-1809.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 160.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Deleted in proof.
- 165.Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 166.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Quintana-Murci L, Quach H, Harmant C, Luca F, Massonnet B, et al. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc. Natl. Acad. Sci. USA. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 169.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ramaley PA, French N, Kaleebu P, Gilks C, Whitworth J, Hill AV. HIV in Africa (Communication arising): chemokine-receptor genes and AIDS risk. Nature. 2002;417:140. doi: 10.1038/417140a. [DOI] [PubMed] [Google Scholar]
- 171.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reed FA, Tishkoff SA. African human diversity, origins and migrations. Curr. Opin. Genet. Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 173.Deleted in proof.
- 174.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 175.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 176.Rotimi CN. Inauguration of the African Society of Human Genetics. Nat. Genet. 2004;36:544. doi: 10.1038/ng0604-544. [DOI] [PubMed] [Google Scholar]
- 177.Rotimi CN, Chen G, Adeyemo AA, Furbert-Harris P, Parish-Gause D, et al. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Diabetes. 2004;53:838–841. doi: 10.2337/diabetes.53.3.838. [DOI] [PubMed] [Google Scholar]
- 178.Rotimi CN, Chen G, Adeyemo AA, Jones LS, Agyenim-Boateng K, et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest. Ophthalmol. Vis. Sci. 2006;47:3262–3267. doi: 10.1167/iovs.05-1537. [DOI] [PubMed] [Google Scholar]
- 179.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 180.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saunders MA, Hammer MF, Nachman MW. Nucleotide variability at G6pd and the signature of malarial selection in humans. Genetics. 2002;162:1849–1861. doi: 10.1093/genetics/162.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Saunders MA, Slatkin M, Garner C, Hammer MF, Nachman MW. The extent of linkage disequilibrium caused by selection on G6PD in humans. Genetics. 2005;171:1219–1229. doi: 10.1534/genetics.105.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, et al. Linkage disequilibrium patterns vary substantially among populations. Eur. J. Hum. Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- 184.Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:E57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shimada MK, Panchapakesan K, Tishkoff SA, Nato AQ, Hey J., Jr Divergent haplotypes and human history as revealed in a worldwide survey of X-linked DNA sequence variation. Mol. Biol. Evol. 2007;24:687–698. doi: 10.1093/molbev/msl196. [DOI] [PubMed] [Google Scholar]
- 186.Siffert W, Forster P, Jockel KH, Mvere DA, Brinkmann B, et al. Worldwide ethnic distribution of the G protein β3 subunit 825T allele and its association with obesity in Caucasian, Chinese, and black African individuals. J. Am. Soc. Nephrol. 1999;10:1921–1930. doi: 10.1681/ASN.V1091921. [DOI] [PubMed] [Google Scholar]
- 187.Sirugo G, Hennig BJ, Adeyemo A, Matimba A, Newport MJ, et al. Genetic studies of African populations: an overview. 2008 doi: 10.1007/s00439-008-0511-y. In review. [DOI] [PubMed] [Google Scholar]
- 188.Soborg C, Andersen AB, Range N, Malenganisho W, Friis H, et al. Influence of candidate susceptibility genes on tuberculosis in a high endemic region. Mol. Immunol. 2007;44:2213–2220. doi: 10.1016/j.molimm.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 189.Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, et al. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 190.Deleted in proof.
- 191.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 192.Storz JF, Payseur BA, Nachman MW. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol. Biol. Evol. 2004;21:1800–1811. doi: 10.1093/molbev/msh192. [DOI] [PubMed] [Google Scholar]
- 193.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 195.Tarazona-Santos E, Tishkoff SA. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes. Immun. 2005;6:53–65. doi: 10.1038/sj.gene.6364149. [DOI] [PubMed] [Google Scholar]
- 196.Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, et al. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- 198.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am. J. Hum. Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Timmann C, Evans JA, Konig IR, Kleensang A, Ruschendorf F, et al. Genome-wide linkage analysis of malaria infection intensity and mild disease. PLoS Genet. 2007;3:e48. doi: 10.1371/journal.pgen.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 201.Tishkoff SA, Gonder MK, Henn BM, Mortensen H, Knight A, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 202.Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat. Genet. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- 203.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Deleted in proof.
- 205.Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 206.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 207.Tishkoff SA, Verrelli BC. Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr. Opin. Genet. Dev. 2003;13:569–575. doi: 10.1016/j.gde.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 208.Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat. Rev. Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 209.Deleted in proof.
- 210.Torcia MG, Santarlasci V, Cosmi L, Clemente A, Maggi L, et al. Functional deficit of T regulatory cells in Fulani, an ethnic group with low susceptibility to Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. USA. 2008;105:646–651. doi: 10.1073/pnas.0709969105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 212.Unwin N, Setel P, Rashid S, Mugusi F, Mbanya JC, et al. Noncommunicable diseases in sub-Saharan Africa: where do they feature in the health research agenda? Bull. WHO. 2001;79:947–953. [PMC free article] [PubMed] [Google Scholar]
- 213.Vafa M, Maiga B, Berzins K, Hayano M, Bereczky S, et al. Associations between the IL-4 −590 T allele and Plasmodium falciparum infection prevalence in asymptomatic Fulani of Mali. Microbes Infect. 2007;9:1043–1048. doi: 10.1016/j.micinf.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 214.Vander Molen J, Frisse LM, Fullerton SM, Qian Y, Del Bosque-Plata L, et al. Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am. J. Hum. Genet. 2005;76:548–560. doi: 10.1086/428784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vannberg FO, Chapman SJ, Khor CC, Tosh K, Floyd S, et al. CD209 genetic polymorphism and tuberculosis disease. PLoS ONE. 2008;3:e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Velez DR, Guruju M, Vinukonda G, Prater A, Kumar A, Williams SM. Angiotensinogen promoter sequence variants in essential hypertension. Am. J. Hypertens. 2006;19:1278–1285. doi: 10.1016/j.amjhyper.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 216a.Verra F, Simpore J, Warimwe GM, Tetteh KK, Howard T, et al. Haemoglobin C and S role in acquired immunity against Plasmodium falciparum malaria. PLoS ONE. 2007;2:e978. doi: 10.1371/journal.pone.0000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A, et al. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am. J. Hum. Genet. 2002;71:1112–1128. doi: 10.1086/344345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Verrelli BC, Tishkoff SA, Stone AC, Touchman JW. Contrasting histories of G6PD molecular evolution and malarial resistance in humans and chimpanzees. Mol. Biol. Evol. 2006;23:1592–1601. doi: 10.1093/molbev/msl024. [DOI] [PubMed] [Google Scholar]
- 219.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Volkman SK, Barry AE, Lyons EJ, Nielsen KM, Thomas SM, et al. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- 221.Wakeley J, Nielsen R, Liu-Cordero SN, Ardlie K. The discovery of single-nucleotide polymorphisms-and inferences about human demographic history. Am. J. Hum. Genet. 2001;69:1332–1347. doi: 10.1086/324521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Deleted in proof.
- 223.Wall JD, Frisse LA, Hudson RR, Di Rienzo A. Comparative linkage-disequilibrium analysis of the β-globin hotspot in primates. Am. J. Hum. Genet. 2003;73:1330–1340. doi: 10.1086/380311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wall JD, Hammer MF. Archaic admixture in the human genome. Curr. Opin. Genet. Dev. 2006;16:606–610. doi: 10.1016/j.gde.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 225.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Watson E, Forster P, Richards M, Bandelt HJ. Mitochondrial footprints of human expansions in Africa. Am. J. Hum. Genet. 1997;61:691–704. doi: 10.1086/515503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.White T, Asfaw B, DeGusta D, Gilbert H, Richards G, et al. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
- 230.Williams SM, Haines JL, Moore JH. The use of animal models in the study of complex disease: all else is never equal or why do so many human studies fail to replicate animal findings? BioEssays. 2004;26:170–179. doi: 10.1002/bies.10401. [DOI] [PubMed] [Google Scholar]
- 231.Williams SM, Ritchie MD, Phillips JA, 3rd, Dawson E, Prince M, et al. Multilocus analysis of hypertension: a hierarchical approach. Hum. Hered. 2004;57:28–38. doi: 10.1159/000077387. [DOI] [PubMed] [Google Scholar]
- 232.Williams TN, Mwangi TW, Wambua S, Peto TE, Weatherall DJ, et al. Negative epistasis between the malaria-protective effects of α+-thalassemia and the sickle cell trait. Nat. Genet. 2005;37:1253–1257. doi: 10.1038/ng1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Williamson C, Loubser SA, Brice B, Joubert G, Smit T, et al. Allelic frequencies of host genetic variants influencing susceptibility to HIV-1 infection and disease in South African populations. AIDS. 2000;14:449–451. doi: 10.1097/00002030-200003100-00020. [DOI] [PubMed] [Google Scholar]
- 234.Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Winkler C, An P, O’Brien SJ. Patterns of ethnic diversity among the genes that influence AIDS. Hum. Mol. Genet. 2004;13(Spec No. 1):R9–R19. doi: 10.1093/hmg/ddh075. [DOI] [PubMed] [Google Scholar]
- 235a.Wood ET, Stover DA, Ehret C, Destro-Bisol G, Spedini G, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 236.Wood ET, Stover DA, Slatkin M, Nachman MW, Hammer MF. The β-globin recombinational hotspot reduces the effects of strong selection around HbC, a recently arisen mutation providing resistance to malaria. Am. J. Hum. Genet. 2005;77:637–642. doi: 10.1086/491748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- 238.Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am. J. Hum. Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J. Antimicrob. Chemother. 2008;61:914–918. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yanovski JA, Diament AL, Sovik KN, Nguyen TT, Li H, et al. Associations between uncoupling protein 2, body composition, and resting energy expenditure in lean and obese African American, white, and Asian children. Am. J. Clin. Nutr. 2000;71:1405–1420. doi: 10.1093/ajcn/71.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yotova V, Lefebvre JF, Kohany O, Jurka J, Michalski R, et al. Tracing genetic history of modern humans using X-chromosome lineages. Hum. Genet. 2007;122:431–443. doi: 10.1007/s00439-007-0413-4. [DOI] [PubMed] [Google Scholar]
- 242.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, et al. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhao Z, Yu N, Fu YX, Li WH. Nucleotide variation and haplotype diversity in a 10-kb noncoding region in three continental human populations. Genetics. 2006;174:399–409. doi: 10.1534/genetics.106.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhu X, Bouzekri N, Southam L, Cooper RS, Adeyemo A, et al. Linkage and association analysis of angiotensin I-converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am. J. Hum. Genet. 2001;68:1139–1148. doi: 10.1086/320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zhu X, Chang YP, Yan D, Weder A, Cooper R, et al. Associations between hypertension and genes in the renin-angiotensin system. Hypertension. 2003;41:1027–1034. doi: 10.1161/01.HYP.0000068681.69874.CB. [DOI] [PubMed] [Google Scholar]
- 246.Zhu X, Cooper RS. Admixture mapping provides evidence of association of the VNN1 gene with hypertension. PLoS ONE. 2007;2:e1244. doi: 10.1371/journal.pone.0001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.