Abstract
1. The objective of this study was to characterize Huh7 cells' baseline capacity to metabolize drugs and to investigate whether the drug metabolism was enhanced upon treatment with dimethyl sulfoxide (DMSO).
2. The messenger RNA (mRNA) levels of major Phase I and Phase II enzymes were determined by quantitative real-time-polymerase chain reaction (RT-PCR), and activities of major drug-metabolizing enzymes were examined using probe drugs by analysing relevant metabolite production rates.
3. The expression levels of drug-metabolizing enzymes in control Huh7 cells were generally very low, but DMSO treatment dramatically increased the mRNA levels of most drug-metabolizing enzymes as well as other liver-specific proteins. Importantly, functionality assays confirmed concomitant increases in drug-metabolizing enzyme activity. Additionally, treatment of the Huh7 cells with 3-methylcholanthrene induced cytochrome P450 (CYP) 1A1 expression.
4. The results indicate that DMSO treatment of Huh7 cells profoundly enhances their differentiation state, thus improving the usefulness of this common cell line as an in vitro hepatocyte model.
Keywords: Drug metabolism, Huh7 cells, dimethyl sulfoxide (DMSO), differentiation
Introduction
Hepatocytes, the main cell type of the liver, carry out vital liver functions through the synthesis, degradation and storage of a wide variety of substances. Thus, the inability of the liver to function normally has significant adverse effects on health, and hepatic diseases such as hepatitis and cirrhosis can be fatal. For this reason, experimental in vitro cell culture systems for understanding liver functions and pathogenesis are critically needed and the use of human liver-derived cells has become an integral part of hepatology research.
Primary human hepatocytes are the ‘gold standard’ system for examining hepatic metabolism of drugs, drug toxicity, and the potential for drug–drug interactions (Gomez-Lechon et al. 2004), as well as to investigate progression of liver diseases such as hepatitis C (Duverlie & Wychowski 2007). However, healthy primary hepatocytes are scarce and not readily available to all investigators. Moreover, among different hepatocyte preparations, huge variations in the state of differentiation maintained, specifically the expression and function of drug-metabolizing enzymes as well as the varying donors genetics and limited longevity of the culture, have been problematic (Gomez-Lechon et al. 2007).
In contrast, hepatoma cell lines have indefinite proliferative capacity and thus could potentially serve as the basis of more practical alternative experimental systems. However, while different hepatoma cell lines do retain certain liver-specific functions, these cells typically fail to exhibit sufficient hepatocyte function. For example, HepG2 cells, a hepatocellular carcinoma cell line, have been widely used in the study of liver physiology, but expression levels of various liver-specific drug-metabolizing enzymes and transcription factors are low in these cells, making them a less than ideal substitute for primary hepatocytes (Alexandre et al. 1999; Barbier et al. 2003; Gardner-Stephen et al. 2004; Hewitt & Hewitt 2004). Accordingly, drug toxicity that is mediated by its reactive metabolites may not be as evident in HepG2 cells as it is in human hepatocytes due to low amount of toxic metabolites generated by the HepG2 cells (Castell et al. 1997). Another hepatocyte cell line that has been recently characterized as a potential in vitro hepatocyte model is HepaRG. HepaRG cells have been reported to express Phase I and II enzymes at levels comparable to those in human hepatocytes (Aninat et al. 2006; Guillouzo et al. 2007; Kanebratt and Andersson 2008). While the expression of drug-metabolizing enzymes in these cells were also shown to be readily inducible by typical enzyme inducers (Aninat et al. 2006), rendering HepaRG as a possible alternative hepatocyte model for drug metabolism studies, their use thus far has been limited.
Huh7 is another continuous hepatocyte cell line which was established in 1982 from a 57-year-old male with a well-differentiated hepatocellular carcinoma (Nakabayashi et al. 1982). Unlike HepaRG cells, Huh7 cells are widely distributed, commercially available (Japan Health Science Research Resources Bank, catalogue number JCRB0403), and cheaper to culture. Huh7 cells have been used to investigate liver toxicity of drugs (Kogure et al. 2004), the molecular mechanisms of hepatic gene regulation and drug efficacy (Gunton et al. 2003; Goldring et al. 2006; Huang et al. 2007), as well as the pathogenesis of hepatitis virus infection (Zhong et al. 2005; Gottwein et al. 2007). These cells retain the functional activities of various carbohydrate-metabolizing enzymes and are capable of secreting plasma proteins such as albumin (Nakabayashi et al. 1982); however, despite their widespread use in hepatology research, the expression and function of drug-metabolizing enzymes in Huh7 cells has not yet been extensively characterized. Interestingly, it was recently shown that Huh7 cells, when incubated in the presence of dimethyl sulfoxide (DMSO), stopped dividing, entered a G0 state, and remained viable in culture without splitting for over 60 days. Under these conditions the Huh7 cells obtained a more ‘differentiated’ hepatocyte state characterized by up-regulation of liver-specific genes such as albumin, transthyretin, HNF4α, and α1-antitrypsin (Sainz & Chisari 2006). The potential implication of liver-specific genes being induced by DMSO treatment is high, particularly if this includes drug-metabolizing enzymes, as the metabolizing activities they exhibit not only may play an important role in modulating the drug efficacy or toxicity studies performed in these commonly used cells, but also because DMSO treated Huh7 cells might then represent an alternative model system specifically for drug metabolism studies.
Hence, in the present study we examined the expression and function of major Phase I and II drug-metabolizing enzymes in actively growing Huh7 cells, as well as in non-growing DMSO-treated Huh7 cells. The results demonstrate that basal expression levels and activity of drug-metabolizing enzymes in growing Huh7 cells are low, but that DMSO treatment induced both dramatically. While the DMSO Huh7 cells failed to reach the absolute levels of drug metabolism observed in primary human hepatocytes, the data demonstrate that Huh7 cells did achieve a high state of differentiation when incubated in the presence of DMSO, and thus highlight the potential utility of the Huh7-DMSO cell culture system as an improved in vitro hepatocyte model for a variety of different studies.
Materials and methods
Chemicals and reagents
Diclofenac, indomethacin, S-mephenytoin, 4-hydroxy mephenytoin, S-nirvanol, dextrorphan, levallorphan, phenytoin, 3-methylcholanthrene (3-MC), caffeine, paraxanthine, and rifampin were obtained from Sigma (St. Louis, MO, USA). Midazolam, prazepam, morphine, morphine 6-glucuronide, and [2H3] morphine 3-glucuronide were purchased from Cerilliant (Round Rock, TX, USA). 4′-Hydroxy diclofenac and dextromethorphan were purchased from Axxora (San Diego, CA, USA). CITCO, i.e. 6-(4-chlorophenyl) imidazo[2,1-b] [1,3]thiazole-5-carbaldehyde O-3,4-dichlorobenzyl) oxime, was purchased from Biomol (Plymouth Meeting, PA, USA). Bufuralol and 1-hydroxybufuralol were purchased from BD Biosciences (Franklin Lakes, NJ, USA). P450-Glo™ CYP1A2 assay system was purchased from Promega (Madison, WI, USA). Lamotrigine and lamotrigine-N2-glucuronide were generous gifts from GlaxoSmithKline (Research Triangle Park, NC, USA). Formic acid (ACS grade) and methanol (Optima grade) were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Cells and treatments
Huh7 cells were cultured in complete DMEM which consists of 10% fetal bovine serum, 100 units ml−1 penicillin, 100 mg ml−1 streptomycin, 10 mM Hepes, 2 mM L-glutamine, and nonessential amino acid. For all DMSO experiments, Huh7 cells were seeded in twelve-well collagen-coated BioCoat dishes (BD Biosciences) at a cell density of 8 × 104 cells per well. At 95% confluence, culture medium was replaced with 3 ml complete DMEM containing 1% DMSO (Sigma-Aldrich). Cultures were incubated for 20 days, as previously described (Sainz & Chisari 2006), during which time complete DMEM containing 1% DMSO was replenished every 3 days. Non-DMSO treated Huh7 cells were seeded 24 h before use at a cell density of 2 × 104 cells per well in twelve-well BioCoat dishes in 2 ml complete DMEM. For comparison of gene expression in Huh7 cells with primary human hepatocytes, pooled hepatocytes were purchased from Celsis (Baltimore, MD, USA; catalogue number X008052; five donors of mixed gender).
Isolation of RNA and quantitative reverse transcription real-time (RT) PCR
Total cellular RNA was isolated from Huh7 cells as well as from pooled human hepatocytes by the guanidine thiocyanate method using standard protocols (Chomczynski & Sacchi 1987). A total of 1 μg of RNA was used for cDNA synthesis using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA, USA), followed by real-time PCR quantification using an Applied Biosystems 7300 real-time thermocycler (Applied Biosystems). Thermal cycling consisted of an initial denaturation step for 10 min at 95°C followed by 40 cycles of denaturation (15 s at 95°C) and annealing/extension (1 min at 60°C). Expression levels were estimated using the ΔΔCt method and normalized to β-actin. ΔΔCt was transformed into fold induction (compared with non-DMSO treated, growing Huh7 cells) with the following formula: fold change = 2−ΔΔCt. The PCR primers used are listed in Table 1.
Table 1.
Primer sequences for quantitative reverse transcriptase-polymerase chain reaction (RT-PCR).
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| CYP1A1 | TCC-AGA-GAC-AAC-AGG-TAA-AAC-A | AGG-AAG-GGC-AGA-GGA-ATG-TGA-T |
| CYP1A2 | GCT-TCT-ACA-TCC-CCA-AGA-AAT | TCC-CAC-TTG-GCC-AGG-ACT |
| CYP2B6 | ATG-GGG-CAC-TGA-AAA-AGA-CTG-A | AGA-GGC-GGG-GAC-ACT-GAA-TGA-C |
| CYP2C8 | AGA-TCA-GAA-TTT-TCT-CAC-CC | AAC-TTC-GTG-TAA-GAG-CAA-CA |
| CYP2C9 | CAG-ATC-TGC-AAT-AAT-TTT-TCT-C | CTT-TCA-ATA-GTA-AAT-TCA-GAT-G |
| CYP2C19 | TTG-AAT-GAA-AAC-ATC-AGG-ATT-G | GAG-GGT-TGT-TGA-TGT-CCA-TC |
| CYP2D6 | GGT-GTG-ACC-CAT-ATG-ACA-TC | CTC-CCC-GAG-GCA-TGC-ACG |
| CYP3A4 | CCA-AGC-TAT-GCT-CTT-CAC-CG | TCA-GGC-TCC-ACT-TAC-GGT-GC |
| CYP3A5 | AGT-GTT-CTT-TCC-TTC-ACT-TTA | TTC-AAC-ATC-TTT-CTT-GCA-AGT |
| UGT1A1 | GGC-CTC-TCT-CCT-CTC-ATT-CA | GGA-ATT-CTG-AGG-CAA-GGG-TT |
| UGT1A4 | TAC-CCT-CTG-GCC-CTG-TCC-TA | GAA-CAG-CCA-CAC-GGA-TGC |
| UGT1A6 | GCA-GAA-GCC-CAG-ACC-CT | GGG-CTC-CAA-CAA-ATT-AAC-AA |
| UGT1A9 | GGT-TGT-AGT-CAT-GCC-AGA-GG | ACT-CCC-GGT-CCA-GAT-CC |
| UGT2B7 | CTG-GGG-TCA-ATG-GTC-AGT-AA | GAA-CCT-TTT-GTG-GGA-TCT-GG |
| CAR | AGT-GCT-TAG-ATG-CTG-GCA-TGA-GGA | TGC-TCC-TTA-CTC-AGT-TGC-ACA-GGT |
| PXR | CAA-GCG-GAA-GAA-AAG-TGA-ACG | CTG-GTC-CTC-GAT-GGG-CAA-GTC |
| AhR | GGC-CGT-GTC-GAT-GTA-TCA-GTG | GTA-CTG-GAT-TGT-ACT-GCA-TCT-GAC |
| RXRα | AAG-GAC-CGG-AAC-GAG-AAT-GA | ATC-CTC-TCC-ACC-GGC-ATG-T |
| β-Actin | ATC-CTG-GCC-TCG-CTG-TCC | CTC-CTG-CTT-GCT-GAT-CCA-CAT |
| Albumin | CGC-CTG-AGC-CAG-AGA-TTT-C | GCC-CTG-TCA-TCA-GCA-CAT-TC |
| Transthyretin (TTR) | CCG-GTG-AAT-CCA-AGT-GTC-CT | GCA-CGG-CCA-CAT-TGA-TG |
| HNF4α | ACA-TTC-GGG-CGA-AGA-AGA-TT | ACT-TGG-CCC-ACT-CAA-CGA-G |
| α1-Antitrypsin (A1AT) | TGC-TGC-CCA-GAA-GAC-AGA-TA | GGC-GGT-ATA-GGC-TGA-AGG |
Determination of drug-metabolizing enzyme activities
Various probe drugs (Table 2) were added to growing or DMSO-treated Huh7 cells and culture media were sampled at various time points for up to 48 h post-treatment. For all metabolic pathways, the concentration of metabolites was determined by LC/MS/MS (Agilent 1200 HPLC interfaced with Applied Biosystems Qtrap 3200) using an electrospray ion source. The mobile phase consisted of water [0.1% (v/v) formic acid] and methanol. For most analytes, separation was performed with a Zorbax Eclipse XDB-C8 column (4.6 × 50 mm, 3.5 μm; Agilent Technologies) at a flow rate of 0.4 ml min−1. For morphine, Eclipse XDB-18 (4.6 × 50 mm, 3.5 μm; Agilent Technologies) was used at a flow rate of 0.6 ml min−1. MS detection of the parent drugs and metabolites was followed by examining multiple MRM pairs in the positive ion mode (Table 2). At the end of the 48-h experiment, the cells were trypsinized and cell numbers were counted to normalize the rate of metabolite formation. The limit of quantification for most analytes was 1 ng ml−1. The metabolic activity of CYP1A2 was further determined using a Promega P450-Glo Assay for CYP1A2 according to the manufacturer's protocol (Promega) using a luminometer (Synergy 4, Biotek).
Table 2.
Probe drugs used to characterize the activities of Phase I and Phase II drug-metabolizing enzymes.
| CYP1A1/2 | CYP1A2 | CYP2B6 | CYP2C9 | CYP2C19 | CYP2D6 | CYP2D6 | CYP3A4 | UGT1A4 | UGT2B7 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Parent drug | Caffeine | Luciferin-ME | S-mephenytoin | Diclofenac | S-mephenytoin | Dextromethorphan | Bufuralol | Midazolam | Lamotrigine | Morphine |
| Concentration (μM) | 75 | 100 | 60 | 100 | 60 | 80 | 20 | 9.2 | 40 | 10.5 |
| Metabolites (analyte) | Paraxanthine | Luciferin | S-nirvanol | 4-Hydroxy diclofenac | 4-Hydroxy mephenytoin | Dextrorphan | 1-Hydroxy bufuralol | 1-Hydroxy midazolam | Lamotrigine-N-glucuronide | Morphine-6-glucuronide |
| Internal Standard | 7-(β-Hydroxyethyl) theophylline | n.a. | Phenytoin | Indomethacin | Phenytoin | Levallophan | 1-Hydroxy midazolam | Prazepam | 1-Hydroxy midazolam | [2H3]morphine-3-glucuronide |
| Mobile phase %B (min) | 5(0)→95 (5) | n.a. | 30 (0)→95 (5) | 30 (0)→90 (6) | 25 (0)→85 (7) | 10 (0)→90 (7) | 25 (0)→85 (5) | 30 (0)→90 (6) | 15 (0)→90 (7) | 5 (0)→90 (5) |
| Analyte MRM | 181.1/124.1 | n.a. | 205.1/134.2 | 311.9/230.1 | 235.2/150.1 | 258.4/157.0 | 278.4/186.3 | 341.9/324.0 | 432.2/256.0 | 462.3/286.3 |
| Internal Standard MRM | 225.4/181.2 | n.a. | 253.2/182.2 | 357.9/139.1 | 253.2/182.2 | 284.3/157.2 | 341.9/324.0 | 325.0/271.1 | 341.9/324.0 | 465.3/289.3 |
For enzyme induction studies, growing or DMSO-treated Huh7 cells were incubated with enzyme inducers — rifampin (10 μM), 3-MC (1 μM), or CITCO (100 nM) — or vehicle control (DMSO, in a final concentration of 0.1%) for 48 h. The media were then changed to contain caffeine, S-mephenytoin, or midazolam. The rate of metabolite formation was determined by measuring concentrations of relevant metabolites in the media by using LC/MS/MS as described above.
Results
Baseline expression of drug-metabolizing enzyme genes
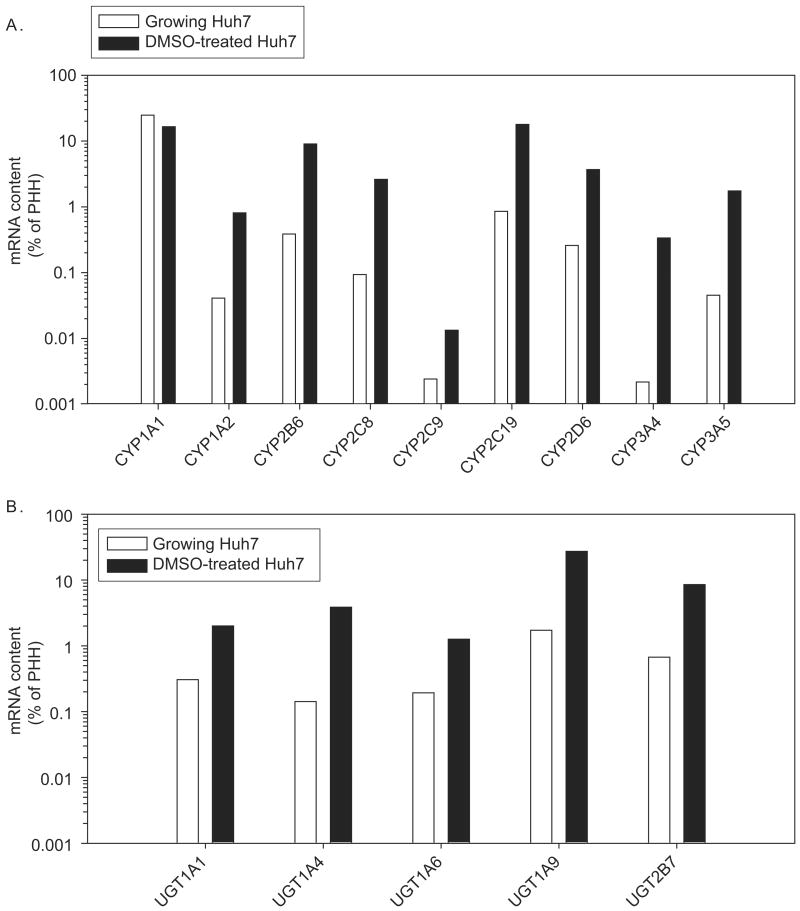
To determine baseline expression of major drug-metabolizing enzymes in Huh7 cells, transcripts of nine P450s (CYP1A1, CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5) and five UDP glucuronosyltransferases (UGT1A1, UGT1A4, UGT1A6, UGT1A9, and UGT2B7) were analysed by reverse transcription followed by quantitative RT-PCR, in Huh7 cells as well as in pooled human hepatocytes (Figure 1). In actively growing Huh7 cells, mRNA expression levels of the Phase I enzymes were less than 1% of the level of mRNA in primary human hepatocytes, except for CYP1A1 (25%). Likewise, the mRNA levels of Phase II enzymes were low, ranging from 0.74% to 1.7% of the mRNA content in hepatocytes.
Figure 1.
Expression of CYP (A) and UGT (B) in growing and DMSO-treated Huh7 cells. Total RNA was isolated from Huh7 cells or pooled human hepatocytes (PHH, purchased from Celsis). Specific mRNA levels were determined by quantitative RT-PCR. The mRNA levels were normalized to β-actin levels.
DMSO-mediated induction of drug-metabolizing enzyme expression
To examine whether the baseline expression of drug-metabolizing enzymes in Huh7 cells is inducible by DMSO treatment, Huh7 cells were also grown in 1% DMSO over 20 days (i.e. the culture condition previously shown to induce Huh7 cells into a more differentiated, growth arrested state (Sainz & Chisari 2006)), and transcripts for Phase I and II enzymes were compared in parallel with the growing Huh7 cultures above (Figure 1). DMSO treatment increased the mRNA levels of most CYP and UGT enzymes by 5.5- (CYP2C9) to 155-fold (CYP3A4) as compared with the levels in actively growing Huh7 cells, except no enhancement of the already relatively highly expressed CYP1A1 gene was detected after DMSO treatment (Figure 1).
DMSO-mediated induction of drug-metabolizing enzyme activity
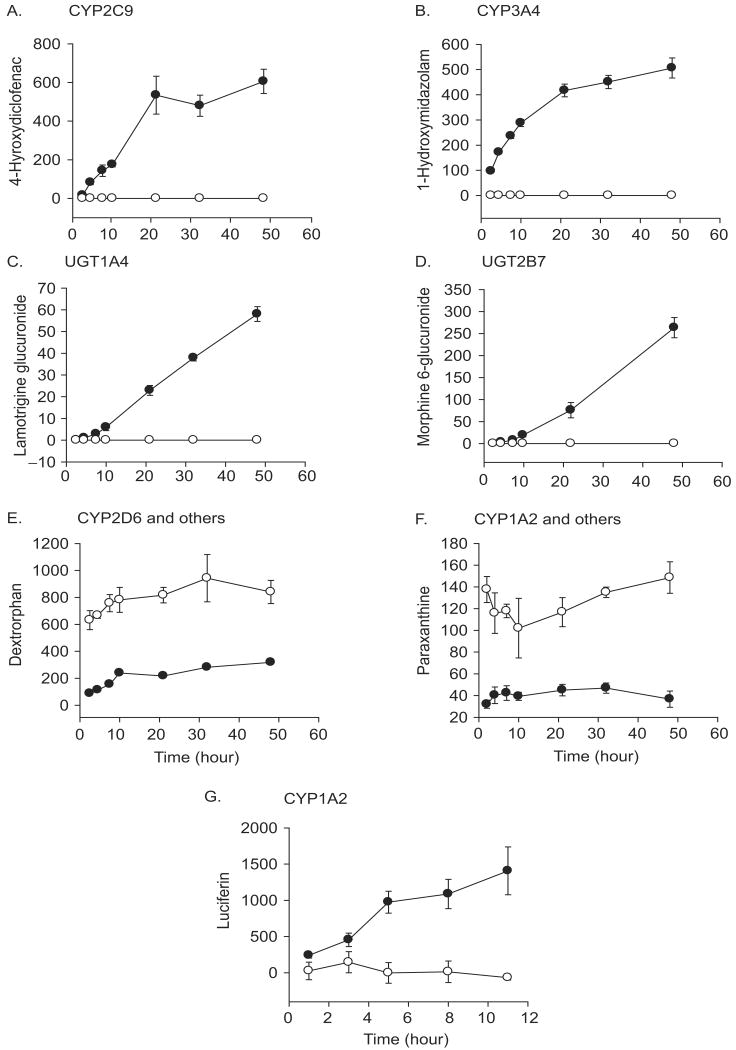
To examine whether the increased mRNA levels observed for these known drug-metabolizing genes in the DMSO-Huh7 cells lead to enhanced function of the various associated metabolic pathways, metabolite production rates from probe drugs were determined (Figure 2). To this end, we chose a list of drugs that are commonly used to characterize known metabolic pathways for xenobiotics (Table 2). Specifically, we characterized metabolic pathways of Phase I (CYP1A1/2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4) and Phase II (UGT1A4 and UGT2B7) reactions in both growing and DMSO-treated Huh7 cultures. In actively growing Huh7 cells, the amount of metabolites produced from the majority of drugs, diclofenac (CYP2C9), S-mephenytoin (CYP2C19), midazolam (CYP3A4), lamotrigine (UGT1A4), and morphine (UGT2B7), were below quantitation limits. DMSO treatment of the Huh7 cells, however, led to a dramatic increase in the metabolic activities of CYP2C9, CYP3A4, and UGT1A4, UGT2B7 (Figures 2A–D). Interestingly, hydroxy metabolites of S-mephenytoin (CYP2C19) were not detected in DMSO-treated Huh7 cells, potentially due to the inhibitory effects of DMSO on the reaction (Yuan et al. 2002).
Figure 2.
Effects of DMSO treatment on functional activities of drug-metabolizing enzymes in Huh7 cells. Growing (open circle) or DMSO-treated (closed circle) Huh7 cells were incubated with various probe drugs (Table 2). The media were sampled at the indicated time points and metabolic activities of CYP1A2, CYP2C9, CYP2D6, and CYP3A4 were determined by measuring concentrations of relative metabolites, i.e. paraxanthine or luciferin, 4-hydroxydiclofenac, dextrorphan, and 1-hydroxymidazolam, respectively. In paraxanthine (F) or dextrorphan (E) production, additional CYP enzymes other than CYP1A2 or CYP2D6 may be involved (see text for more detail). Activities of UGT1A4 and UGT2B7 were also determined by measuring concentrations of lamotrigine N2-glucuronide and morphine 6-glucuronide, respectively, in the media. Data presented show the amount of metabolite produced, expressed in pmol per 106 cells except for luciferin (G) which shows luminescence unit (the mean of triplicate experiment ± standard deviation).
Unexpectedly, metabolite production rates of CYP1A1/2 and CYP2D6 initially appeared to be decreased in DMSO-treated Huh7 cells as compared with the growing Huh7 cells when caffeine and dextromethorphan, respectively, were used as probe drugs (Figures 2E and 2F). However, further characterization of CYP1A2 activities in these cells using luciferin-ME indicated an enhanced CYP1A2-mediated metabolite production in the DMSO-treated Huh7 cells (Figure 2G). In addition, characterization of the CYP2D6 pathway using a different probe drug, bufuralol, exhibited a little, but similar amount of metabolite (hydroxybufuralol) production in both the growing and DMSO-treated Huh7 cells; 4.5 ± 0.5 versus 4.1 ± 0.3 ng per 106 cells/48 h, respectively (data not shown). The slow metabolic rates of bufuralol appear to indicate that Huh7 cells were derived from an individual who may be a poor metabolizer of the CYP2D6 pathway. In total, however, these data suggest that DMSO treatment of Huh7 cells not only up-regulates mRNA levels of drug-metabolizing genes, but also exhibits a corresponding increase in drug-metabolizing enzyme activity.
Baseline expression of liver-specific receptors and other differentiation-associated genes
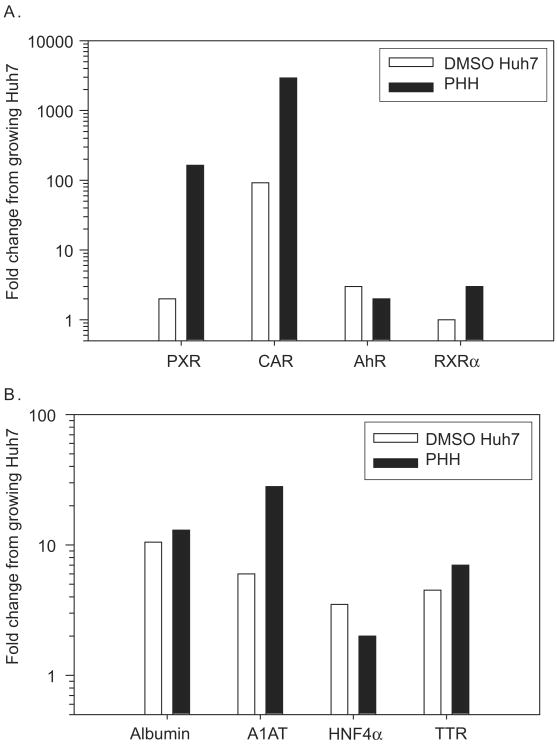
Expression levels of four orphan receptors involved in regulation of drug-metabolizing enzymes — aromatic hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), pregnane X receptor (PXR), and retinoid X receptor α (RXRα) — and four common markers of liver differentiation — albumin, transthyretin, hepatocyte nuclear factor 4α (HNF4α), and α1-antitrypsin — were also analysed in growing and DMSO-treated Huh7 cells and compared with levels in primary human hepatocytes (Figure 3). Generally, expression levels of these genes in growing Huh7 cells were much lower as compared with primary hepatocytes. Notably, however, DMSO treatments led to an increase in the mRNA levels of all the nuclear receptors, except RXRα the levels of which were only marginally higher in primary human hepatocytes relative to the growing Huh7 cells (Figure 3A). The increase was most dramatic in CAR which showed 92-fold enhancement in its expression in DMSO-treated cells; however, the transcript level was still significantly lower compared with that detected in primary human hepatocytes. On the other hand, expression levels of AhR in DMSO-treated Huh7 cells were comparable with those in primary human hepatocytes. Likewise, as seen previously (Sainz & Chisari 2006), the levels of the hepatocyte differentiation markers were comparable to those detected in the primary human hepatocytes confirming the effectiveness of our DMSO treatment (Figure 3B).
Figure 3.
Expression levels of hepatic transcription factors in Huh7 cells. Expression levels of transcription factors (A) and liver-specific genes (B) were compared among Huh7 cells and pooled human hepatocytes (PHH), by using RT-PCR. The data are fold difference of the gene expression relative to growing Huh7 cells. PXR: pregnane X receptor, CAR: constitutive androstane receptor, AhR: aromatic hydrocarbon receptor, RXRα: retinoid X receptor α, A1AT: α1-antitrypsin, HNF4α: hepatocyte nuclear factor 4α, TTR: transthyretin.
Effects of enzyme inducers on expression of drug-metabolizing enzymes
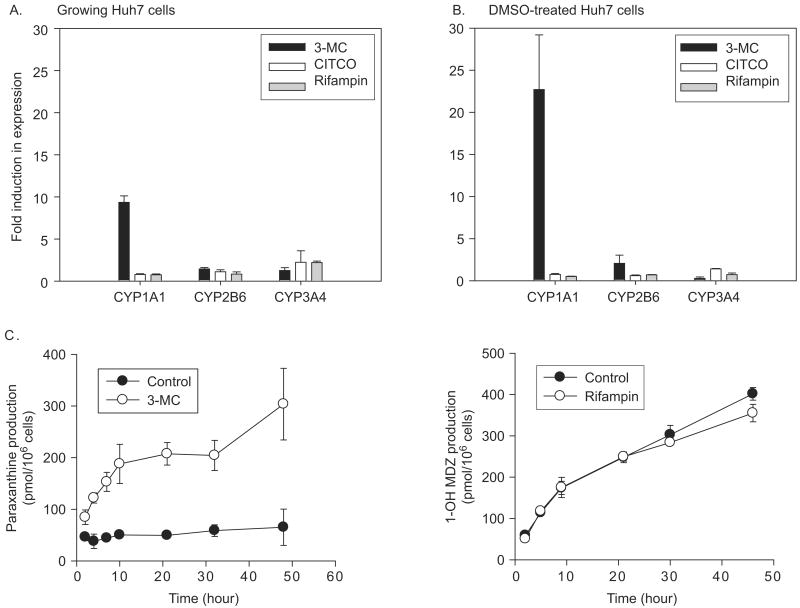
Because the receptors AhR, CAR, and PXR, which are known to target CYP1A1, CYP2B6, and CYP3A4, were up-regulated in DMSO-treated Huh7 cells in the experiment above, we proceeded to determine the inducibility of these Phase I enzymes in growing versus DMSO-treated Huh7 cells. The expression and function of CYP1A1, CYP2B6, and CYP3A4 were determined after treatment of the cells with standard enzyme inducers, i.e. 3-MC, CITCO, or rifampin, which induce expression of their target genes by binding to AhR, CAR, and PXR, respectively. 3-MC exhibited the greatest effect among the three inducers (Figures 4A and 4B), as expected from the relatively high AhR expression levels in the Huh7 cells compared with primary human hepatocytes (Figure 3A). Specifically, 3-MC induced CYP1A1 mRNA levels by 9.4- and 22.7-fold in growing and differentiated Huh7 cells, respectively; these magnitudes of induction are comparable with the level of induction typically observed in primary human hepatocytes (six- to 20-fold) (Donato et al. 1995; Gomez-Lechon et al. 2007). Rifampin led to a marginal induction in the expression of CYP3A4 in growing Huh7 cells (by 2.2-fold), although the induced CYP3A4 expression level was still much lower compared with the level in DMSO-treated Huh7 cells (Figure 4A). Surprisingly, despite the fact that CAR was induced 92-fold in DMSO-treated Huh7 cells (Figure 3A), treatment with its ligand CITCO had insignificant effects on CYP2B6 expression in both growing and differentiated Huh7 cells (Figure 4A). The lack of significant effect of CITCO and rifampin may be due to the lower absolute expression of relevant nuclear receptors (CAR and PXR, respectively) in Huh7 cells as compared with the primary human hepatocytes.
Figure 4.
Effects of typical enzyme inducers on mRNA and activity levels of CYPs in growing and DMSO-treated Huh7 cells. Growing (A) and DMSO-treated (B) Huh7 cells were treated with 3-MC (AhR ligand and CYP1A1 inducer, 1 μM), CITCO (CAR ligand and CYP2B6 inducer, 100 nM), rifampin (PXR ligand and CYP3A4 inducer, 10 μM), or vehicle (DMSO in final concentration of 0.1%) for 48 hours. mRNA levels of CYP1A1, CYP2B6, and CYP3A4 (target genes of AhR, CAR, and PXR, respectively) were determined by real-time PCR. Fold induction was calculated by comparing β-actin-normalized mRNA levels of the relevant genes between the vehicle-treated and inducer-treated groups. (C) DMSO-treated Huh7 cells were incubated with 3-MC or rifampin, and the differences in metabolite production were determined. Production of paraxanthine from caffeine (CYP1A1/2) and that of 1-hydroxymidazolam (1-OH MDZ) from midazolam (CYP3A4) are shown. Data presented are the mean of triplicate experiment ± standard deviations.
To confirm the above findings, a functional study was conducted using probe drugs for CYP1A1, CYP2B6, and CYP3A4, i.e. caffeine, S-mephenytoin, and midazolam, respectively. The results of caffeine and midazolam were consistent with the findings of mRNA levels in that expression of CYP1A1, but not of CYP3A4, was enhanced in DMSO-treated Huh7 cells by treatment with inducers (Figure 4C). Finally, S-nirvanol, a CYP2B6-mediated metabolite of S-mephenytoin, was undetectable in the media before and after CITCO treatment of either growing or differentiated Huh7 cells (data not shown).
Intrinsic metabolic activities of DMSO-treated Huh7 cells
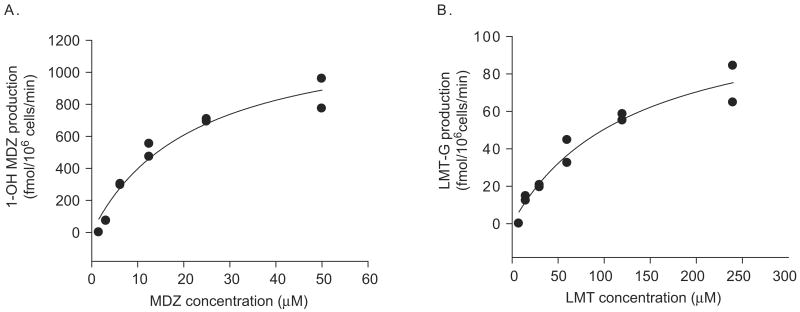
To compare the enhanced intrinsic metabolic activities of the enzymes in the DMSO-treated Huh7 cells with those in primary human hepatocytes, we obtained intrinsic clearance parameters of two representative metabolic pathways, i.e. CYP3A4 and UGT1A4, by examining concentration dependent metabolite production rates. Concentrations of 1-hydroxymidazolam and lamotrigine glucuronide were measured in the culture media, and approximate Michaelis–Menten constants were estimated from the metabolite production profile (Figure 5). Relevant Km and Vmax for CYP3A4 and UGT1A4 reactions were 26.4 μM and 1560 fmol min−1 per 106 cells for CYP3A4; 109 μM and 108 fmol min−1 per 106 cells for UGT1A4 reactions. Intrinsic clearance values (= Vmax/Km) were then calculated (Table 3). Also included in Table 3 are metabolite production rates reported for primary human hepatocytes expressed in pmol min−1 per million cells, for midazolam and diclofenac. In general, the intrinsic metabolic activities of the enzymes were much lower in DMSO-treated Huh7 cells when compared with those in primary human hepatocytes.
Figure 5.
Concentration dependent metabolite production in DMSO-treated Huh7 cells. DMSO-treated Huh7 cells were incubated with various concentrations of midazolam (MDZ; A) or lamotrigine (LMT; B) for 4 and 18 hours, respectively. The media were sampled at the end of incubation, and concentrations of 1-hydroxymidazolam (1-OH MDZ) and lamotrigine N2-glucuronide (LMT-G) in the media were determined by LC/MS/MS. Data presented were obtained from a duplicate experiment. Best-fit curves for Michaelis–Menten equations are also shown.
Table 3.
Intrinsic metabolic activities of DMSO-treated Huh7 cells.
| Differentiated Huh7 | Human hepatocytes | References | |
|---|---|---|---|
| Midazolam | |||
| 1-Hydroxymidazolam production rate (pmol min−1 per 106 cells) | 0.87 | 10–28 | Kanebratt and Andersson (2008), Ledirac et al. (2000), |
| Intrinsic clearance (μl min−1 per 106 cells) | 0.00059 | 0.0015–0.04 | Hallifax et al. (2005), Li et al. (1997) |
| Lamotrigine | |||
| Intrinsic clearance (μl min−1 per 106 cells) | 0.00099 | 0.11 (liver microsome)a | Rowland et al. (2006) |
| Diclofenac | |||
| 4-Hydroxydiclofenac production rate (pmol min−1 per 106 cells) | 0.189 | 300 | Hewes et al. (2006) |
Based on previous reports indicating that 1 g of liver tissue is equivalent to 40 mg microsomal proteins (Hakooz et al. 2006) and 108 hepatocytes (Wilson et al. 2003).
Discussion
The use of primary human hepatocyte, although the closest to the liver physiologically, is limited by their scarce availability, inherent donor-based variability, and inconsistent differentiation state. On the other hand, low expression levels of drug-metabolizing enzymes in the more accessible, stable hepatoma cell lines limit their use in drug metabolism studies. However, we recently reported that DMSO treatment of the widely used Huh7 hepatoma cells induces expression of four differentiation markers (Sainz & Chisari 2006). Therefore, we characterized the baseline functions of drug-metabolizing enzymes in the widely used Huh7 hepatoma cell line, as well as parallel DMSO-treated Huh7 cells. Consistent with previous reports, we found that the baseline mRNA expression levels of all CYP and UGT genes investigated (except for CYP1A1) were extremely low in growing Huh7 cells (Phillips et al. 2005; Olsavsky et al. 2007). Importantly, however, we show that DMSO treatment of the cells significantly enhances xenobiotic metabolism and the inducibility of some major drug-metabolizing enzymes by known enzyme inducers. Not only does this validate that DMSO significantly increases the state of Huh7 cell differentiation both at the level of gene expression and function, but also it indicates that DMSO-treated Huh7 cells might serve as an improved in vitro hepatocyte model for a wide range of studies in which hepatocyte physiology is of relevance (for example, hepatitis C virus–host cell interactions, hepatocyte differentiationspecific gene expression and drug–drug interaction studies). Notably, however, it was clear that CYP and UGT expression was still higher (approximately tenfold or more) in the primary human hepatocytes leaving primary human hepatocytes as the ‘gold standard’ for drug metabolism studies.
The one exception to the general low baseline gene expression pattern observed for the CYP and UGT genes in growing Huh7 cells was CYP1A1, for which we found relatively high expression level in the absence of DMSO treatment. Not only was basal expression of CYP1A1 relatively high in growing Huh7 cells as compared with the low expression of other CYP enzymes, but also CYP1A1 expression was not influenced by DMSO treatment in Huh7 cells, as were the other CYP enzymes which showed significant increases in their expression in DMSO-treated Huh7 cell cultures (Figure 1). These findings suggest that expression of CYP1A1 is not governed by the differentiation state of the hepatocytes. Consistent with this idea, a previous report did note that after isolation hepatocytes maintained CYP1A1 expression in contrast to expression of other major CYP enzyme, such as CYP3A4, which were rapidly lost (Bowen et al. 2000).
Of interest, although CYP1A1 and CYP1A2 are both on chromosome 15, have overlapping substrate specificity over various carcinogens and environmental toxicants (Ma & Lu 2007), and are regulated similarly by AhR ligands and inflammatory mediators (Morgan 2001; Ma & Lu 2007), expression of CYP1A2 was enhanced by 20-fold by DMSO treatment of Huh7 cells (Figure 1). Hence, despite the high similarity of these two genes, expression of CYP1A1 and CYP1A2 is clearly differentially regulated, with CYP1A2 expression being governed by liver differentiation similar to most CYP genes whereas CYP1A1 is not. In agreement with this hypothesis, CYP1A2 is primarily expressed in hepatic tissues whereas CYP1A1 expression is found in extra-hepatic tissues (Martignoni et al. 2006).
Just as basal expression levels of drug-metabolizing enzymes are important in determining a baseline potential to metabolize xenobiotics, expression of ligand-activated transcription factors, such as PXR, CAR, and AhR (Pascussi et al. 2008), is important in mediating up-regulation of drug-metabolizing enzymes. These transcription factors, upon binding of their ligands (for example, drugs), regulate distinct but overlapping sets of target genes, including the CYPs and UGTs, which may lead to unwarranted drug–drug interaction. Although DMSO tended to up-regulate expression of PXR and CAR, the mRNA levels detected in DMSO-treated Huh7 cells remained lower than those detected in the primary human hepatocyte controls (except for AhR). Accordingly, rifampin and CITCO (ligands of PXR and CAR, respectively) caused only a marginal or no induction in target gene expression (Figure 4). However, in agreement with our findings that the expression level of AhR in Huh7 cells was comparable with the level in primary hepatocytes, 3-MC, a ligand of AhR, led to a significant induction of its target gene (that is, CYP1A1) expression (Figure 4). This result suggests the potential use of Huh7 cells as a model for AhR-mediated drug metabolism induction studies.
Interestingly, comparing the magnitude of changes in mRNA levels of specific drug-metabolizing enzymes (Figure 1) with the increase in activity of the relevant pathways (Figure 2), we found that the increases in enzyme activity detected in DMSO-treated Huh7 cells far exceeded the magnitude of mRNA increase of specific enzymes. For example, a 155-fold increase in CYP3A4 mRNA level led to a greater than 500-fold increase in hydroxymidazolam production in DMSO-treated Huh7 cells when compared with the growing Huh7 control cells (Figures 1 and 2). This finding appears inconsistent with previous reports that showed when human hepatocytes were treated with an enzyme inducer, e.g. rifampin, the increase in CYP3A4 mRNA levels exceeded the increase in CYP3A4 activity (for example, 25-fold increase in mRNA level versus a four-fold increase in enzyme activity) (Gomez-Lechon et al. 2007). This may be due to the relatively short incubation time used for the enzyme inducer as compared with DMSO in this study, i.e. 48–72 h versus over 20 days. In other words, considering that the half-lives of human CYP enzymes range from 39 to 51 h (Venkatakrishnan et al. 2007), the typical incubation time of 72 h for human hepatocytes may be too short for the proteins to reach steady-state expression levels. Alternatively, DMSO treatment may modulate expression levels of other hepatocyte-specific factors, such as microRNAs, which act in the CYP3A4 pathway and potentiate the activity of the enzyme (Takagi et al. 2008).
Of note, there was a prominent lag time in the glucuronide metabolite production in DMSO-treated Huh7 cells (Figures 2C and 2D). This unusual metabolism kinetics may be due to low expression levels of glucuronide transporters on the endoplasmic reticulum or plasma membrane of the cells. UGT enzymes, located on the membranes of endoplasmic reticulum, have their active sites exposed to the lumen of the endoplasmic reticulum (Radominska-Pandya et al. 2005). Thus, it has often been speculated that glucuronide conjugate metabolites, once produced by UGT, are transported to cytosol by yet unidentified active transporters for excretion (Csala et al. 2004; Radominska-Pandya et al. 2005). On the other hand, the transport of glucuronide conjugates from cytosol to the bile or blood is mediated by transporters on hepatocyte cell membrane, e.g. multidrug resistance associated proteins (Deeley et al. 2006). In DMSO-treated Huh7 cells, it appears plausible to speculate that the expression levels of the glucuronide transporters on endoplasmic reticulum or plasma membranes are lower as compared with the rate of metabolite production, leading to the lag time in metabolite appearance in the media.
Consistent with previous claims that choice of probe drug used can be problematic in the interpretation of drug metabolism studies (Yuan et al. 2002), our results indicated that caffeine and dextrormethorphan were not ideal probe compounds to characterize activities of CYP1A2 and CYP2D6 enzymes, respectively, in Huh7 cells (Figures 2E and 2F). First, CYP1A2-mediated production of paraxanthine from caffeine was lower in DMSO-treated Huh7 cells as compared with the growing cells despite the fact that mRNA levels of CYP1A2 were increased by 20-fold by DMSO treatment. Importantly, however, use of a specific CYP1A2 probe compound (i.e. luciferin-ME) led to a dramatic increase in the CYP1A2-mediated metabolite formation rate in differentiated Huh7 cells (Figure 2G). The seemingly opposite effect initially observed in the caffeine metabolism assay could be because, although CYP1A2 is the major CYP enzyme metabolizing caffeine in human livers, multiple CYP enzymes including CYP3A4 can catalyse caffeine elimination to produce minor metabolites (Tassaneeyakul et al. 1994). Hence, potentially, the DMSO-mediated increase in CYP3A4 expression (155-fold; Figure 1) may have contributed to caffeine metabolism more significantly than usually occurs in human liver tissues. Analogously, the disproportionally high CYP1A1 expression in control and DMSO-treated Huh7 cells may have masked the contribution of increased CYP1A2 expression to caffeine metabolism. Similar speculations can be made regarding the use of dextrormethorphan as a probe for CYP2D6 because, in addition to CYP2D6, dextromethorphan also can be metabolized by other CYP enzymes (for example, CYP3A4) to produce different metabolites (Von Moltke et al. 1998; Yu & Haining 2001). In this case, however, when a different CYP2D6 probe drug bufuralol was used, the metabolite production rates were similar between the growing and differentiated Huh7 cells, a finding that still appears inconsistent with the 14-fold increase in CYP2D6 mRNA levels in the differentiated Huh7 cells (Figure 1). This is potentially due to (1) other CYP enzymes involved in bufuralol metabolism (Mankowski 1999), (2) inhibitory effects of DMSO on bufuralol metabolism (Yuan et al. 2002), (3) unknown regulatory mechanisms governing CYP2D6 activity that may be absent in the DMSO-Huh7 cells, or (4) the possibility that Huh7 cells were derived from a poor metabolizer of the CYP2D6 activity. CYP2D6 activities in poor metabolizers have been shown to be less influenced by regulatory stimulus for the enzyme (Wadelius et al. 1997).
In summary, we characterized baseline expression and function of drug-metabolizing enzymes in Huh7 cells and demonstrated that DMSO treatment of Huh7 cells induces their capability to metabolize drugs. The results indicate that a highly differentiated state can be achieved in Huh7 cells by DMSO treatment with the expression of liver-specific genes, such as drug-metabolizing enzymes and liver-enriched transcription factors. Unfortunately, when compared with primary hepatocytes the activities and expression levels of the most drug-metabolizing enzymes in DMSO-treated Huh7 cells were still significantly lower, limiting the utility of Huh7 cells for drug metabolism studies per se. Yet, the enhanced expression of drug metabolism enzymes as well as differential regulation of major drug-metabolizing enzymes and key transcription factors in DMSO-treated Huh7 cell culture system potentially makes them an improved in vitro hepatocyte model for a variety of other studies such as: (1) metabolite profiling of new drug candidates, (2) characterization of potential drug–drug interactions, especially the ones involving AhR, (3) cytotoxicity testing of bioactivated compounds for both short term and long term effects (the plated DMSO-treated Huh7 cells are viable for over a few months), (4) investigation of hepatocyte differentiation process and molecular details of certain hepatocyte-specific functions, as well as (5) the elucidation of important interaction between host hepatocyte pathways and hepatotropic pathogens such as hepatitis B or C viruses.
Footnotes
Declaration of interest: The authors report no conflicts of interest.
References
- Alexandre E, Davida P, Viollonb C, Wolfa P, Jaecka D, Azimzadeha A, Nicodb L, Boudjemaa K, Richertb L. Expression of cytochromes P-450 2E1, 3A4 and 1A1/1A2 in growing and confluent human HepG2 hepatoma cells — effect of ethanol. Toxicol In Vitro. 1999;13:427–435. doi: 10.1016/s0887-2333(99)00007-7. [DOI] [PubMed] [Google Scholar]
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouet S, Morel F, Guguen-Guillouzo C, Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos. 2006;34:75–83. doi: 10.1124/dmd.105.006759. [DOI] [PubMed] [Google Scholar]
- Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem. 2003;278:32852–32860. doi: 10.1074/jbc.M305361200. [DOI] [PubMed] [Google Scholar]
- Bowen WP, Carey JE, Miah A, McMurray HF, Munday PW, James RS, Coleman RA, Brown AM. Measurement of cytochrome P450 gene induction in human hepatocytes using quantitative real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2000;28:781–788. [PubMed] [Google Scholar]
- Castell JV, Gomez-Lechon MJ, Ponsoda X, Bort R. The use of cultured hepatocytes to investigate the mechanisms of drug hepatotoxicity. Cell Biol Toxicol. 1997;13:331–338. doi: 10.1023/a:1007443610025. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Csala M, Staines AG, Banhegyi G, Mandl J, Coughtrie MW, Burchell B. Evidence for multiple glucuronide transporters in rat liver microsomes. Biochem Pharmacol. 2004;68:1353–1362. doi: 10.1016/j.bcp.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- Donato MT, Castell JV, Gomez-Lechon MJ. Effect of model inducers on cytochrome P450 activities of human hepatocytes in primary culture. Drug Metab Dispos. 1995;23:553–558. [PubMed] [Google Scholar]
- Duverlie G, Wychowski C. Cell culture systems for the hepatitis C virus. World J Gastroenterol. 2007;13:2442–2445. doi: 10.3748/wjg.v13.i17.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner-Stephen D, Heydel JM, Goyal A, Lu Y, Xie W, Lindblom T, Mackenzie P, Radominska-Pandya A. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32:340–347. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring CE, Kitteringham NR, Jenkins R, Lovatt CA, Randle LE, Abdullah A, Owen A, Liu X, Butler PJ, Williams DP, et al. Development of a transactivator in hepatoma cells that allows expression of Phase I, Phase II, and chemical defense genes. Am J Physiol Cell Physiol. 2006;290:C104–C115. doi: 10.1152/ajpcell.00133.2005. [DOI] [PubMed] [Google Scholar]
- Gomez-Lechon MJ, Castell JV, Donato MT. Hepatocytes — the choice to investigate drug metabolism and toxicity in man: in vitro variability as a reflection of in vivo. Chem Biol Interact. 2007;168:30–50. doi: 10.1016/j.cbi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Gomez-Lechon MJ, Donato MT, Castell JV, Jover R. Human hepatocytes in primary culture: the choice to investigate drug metabolism in man. Curr Drug Metab. 2004;5:443–462. doi: 10.2174/1389200043335414. [DOI] [PubMed] [Google Scholar]
- Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, Bukh J. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133:1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gunton JE, Delhanty PJ, Takahashi S, Baxter RC. Metformin rapidly increases insulin receptor activation in human liver and signals preferentially through insulin-receptor substrate-2. J Clin Endocrinol Metab. 2003;88:1323–1332. doi: 10.1210/jc.2002-021394. [DOI] [PubMed] [Google Scholar]
- Hakooz N, Ito K, Rawden H, Gill H, Lemmers L, Boobis AR, Edwards RJ, Carlile DJ, Lake BG, Houston JB. Determination of a human hepatic microsomal scaling factor for predicting in vivo drug clearance. Pharm Res. 2006;23:533–539. doi: 10.1007/s11095-006-9531-2. [DOI] [PubMed] [Google Scholar]
- Hallifax D, Rawden HC, Hakooz N, Houston JB. Prediction of metabolic clearance using cryopreserved human hepatocytes: kinetic characteristics for five benzodiazepines. Drug Metab Dispos. 2005;33:1852–1858. doi: 10.1124/dmd.105.005389. [DOI] [PubMed] [Google Scholar]
- Hewes JC, Riddy D, Morris RW, Woodrooffe AJ, Davidson BR, Fuller B. A prospective study of isolated human hepatocyte function following liver resection for colorectal liver metastases: the effects of prior exposure to chemotherapy. J Hepatol. 2006;45:263–270. doi: 10.1016/j.jhep.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Hewitt NJ, Hewitt P. Phase I and II enzyme characterization of two sources of HepG2 cell lines. Xenobiotica. 2004;34:243–256. doi: 10.1080/00498250310001657568. [DOI] [PubMed] [Google Scholar]
- Huang H, Chen Y, Ye J. Inhibition of hepatitis C virus replication by peroxidation of arachidonate and restoration by vitamin E. Proc Natl Acad Sci USA. 2007;104:18666–18670. doi: 10.1073/pnas.0708423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanebratt KP, Andersson TB. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos. 2008;36:1444–1452. doi: 10.1124/dmd.107.020016. [DOI] [PubMed] [Google Scholar]
- Kogure T, Ueno Y, Iwasaki T, Shimosegawa T. The efficacy of the combination therapy of 5-fluorouracil, cisplatin and leucovorin for hepatocellular carcinoma and its predictable factors. Cancer Chemother Pharmacol. 2004;53:296–304. doi: 10.1007/s00280-003-0725-6. [DOI] [PubMed] [Google Scholar]
- Ledirac N, de Sousa G, Fontaine F, Agouridas C, Gugenheim J, Lorenzon G, Rahmani R. Effects of macrolide antibiotics on CYP3A expression in human and rat hepatocytes: interspecies differences in response to troleandomycin. Drug Metab Dispos. 2000;28:1391–1393. [PubMed] [Google Scholar]
- Li AP, Reith MK, Rasmussen A, Gorski JC, Hall SD, Xu L, Kaminski DL, Cheng LK. Primary human hepatocytes as a tool for the evaluation of structure–activity relationship in cytochrome P450 induction potential of xenobiotics: evaluation of rifampin, rifapentine and rifabutin. Chem Biol Interact. 1997;107:17–30. doi: 10.1016/s0009-2797(97)00071-9. [DOI] [PubMed] [Google Scholar]
- Ma Q, Lu AY. CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos. 2007;35:1009–1016. doi: 10.1124/dmd.107.015826. [DOI] [PubMed] [Google Scholar]
- Mankowski DC. The role of CYP2C19 in the metabolism of (+/–) bufuralol, the prototypic substrate of CYP2D6. Drug Metab Dispos. 1999;27:1024–1028. [PubMed] [Google Scholar]
- Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- Morgan ET. Regulation of cytochrome p450 by inflammatory mediators: why and how? Drug Metab Dispos. 2001;29:207–212. [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Olsavsky KM, Page JL, Johnson MC, Zarbl H, Strom SC, Omiecinski CJ. Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol Appl Pharmacol. 2007;222:42–56. doi: 10.1016/j.taap.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Duret C, Daujat-Chavanieu M, Vilarem MJ, Maurel P. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu Rev Pharmacol Toxicol. 2008;48:1–32. doi: 10.1146/annurev.pharmtox.47.120505.105349. [DOI] [PubMed] [Google Scholar]
- Phillips A, Hood SR, Gibson GG, Plant NJ. Impact of transcription factor profile and chromatin conformation on human hepatocyte CYP3A gene expression. Drug Metab Dispos. 2005;33:233–242. doi: 10.1124/dmd.104.001461. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Bratton S, Little JM. A historical overview of the heterologous expression of mammalian UDP-glucuronosyltransferase isoforms over the past twenty years. Curr Drug Metab. 2005;6:141–160. doi: 10.2174/1389200053586127. [DOI] [PubMed] [Google Scholar]
- Rowland A, Elliot DJ, Williams JA, Mackenzie PI, Dickinson RG, Miners JO. In vitro characterization of lamotrigine N2-glucuronidation and the lamotrigine–valproic acid interaction. Drug Metab Dispos. 2006;34:1055–1062. doi: 10.1124/dmd.106.009340. [DOI] [PubMed] [Google Scholar]
- Sainz B, Jr, Chisari FV. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J Virol. 2006;80:10253–10257. doi: 10.1128/JVI.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- Tassaneeyakul W, Birkett DJ, McManus ME, Veronese ME, Andersson T, Tukey RH, Miners JO. Caffeine metabolism by human hepatic cytochromes P450: contributions of 1A2, 2E1 and 3A isoforms. Biochem Pharmacol. 1994;47:1767–1776. doi: 10.1016/0006-2952(94)90304-2. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan K, Obach RS, Rostami-Hodjegan A. Mechanism-based inactivation of human cytochrome P450 enzymes: strategies for diagnosis and drug–drug interaction risk assessment. Xenobiotica. 2007;37:1225–1256. doi: 10.1080/00498250701670945. [DOI] [PubMed] [Google Scholar]
- Von Moltke LL, Greenblatt DJ, Grassi JM, Granda BW, Venkatakrishnan K, Schmider J, Harmatz JS, Shader RI. Multiple human cytochromes contribute to biotransformation of dextromethorphan in-vitro: role of CYP2C9, CYP2C19, CYP2D6, and CYP3A. J Pharm Pharmacol. 1998;50:997–1004. doi: 10.1111/j.2042-7158.1998.tb06914.x. [DOI] [PubMed] [Google Scholar]
- Wadelius M, Darj E, Frenne G, Rane A. Induction of CYP2D6 in pregnancy. Clin Pharmacol Ther. 1997;62:400–407. doi: 10.1016/S0009-9236(97)90118-1. [DOI] [PubMed] [Google Scholar]
- Wilson ZE, Rostami-Hodjegan A, Burn JL, Tooley A, Boyle J, Ellis SW, Tucker GT. Inter-individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. Br J Clin Pharmacol. 2003;56:433–440. doi: 10.1046/j.1365-2125.2003.01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Haining RL. Comparative contribution to dextromethorphan metabolism by cytochrome P450 isoforms in vitro: can dextromethorphan be used as a dual probe for both CTP2D6 and CYP3A activities? Drug Metab Dispos. 2001;29:1514–1520. [PubMed] [Google Scholar]
- Yuan R, Madani S, Wei XX, Reynolds K, Huang SM. Evaluation of cytochrome P450 probe substrates commonly used by the pharmaceutical industry to study in vitro drug interactions. Drug Metab Dispos. 2002;30:1311–1319. doi: 10.1124/dmd.30.12.1311. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]