Abstract
Foodstuffs have traditionally been challenging matrices for conducting immunoassays. Proteins, carbohydrates, and other macromolecules present in food matrices may interfere with both immunoassays and PCR-based tests, and removal of particulate matter may also prove challenging prior to analyses. This has been found true when testing for bacterial contamination of foods using the standard polystyrene microspheres utilized with Luminex flow cytometers. Luminex MagPlex microspheres are encoded with the same dyes as standard xMAP microspheres, but have superparamagnetic properties to aid in preparation of samples in complex matrices. In this work, we present results demonstrating use of MagPlex for sample preparation and identification of bacteria and a toxin spiked into a variety of food samples. Fluorescence-coded MagPlex microsphere sets coated with antibodies for Salmonella, Campylobacter, Escherichia coli, Listeria, and staphylococcal enterotoxin B (SEB) were used to capture these bacteria and toxin from spiked foodstuffs and then evaluated by the Luminex system in a multiplex format; spiked foods included apple juice, green pepper, tomato, ground beef, alfalfa sprouts, milk, lettuce, spinach, and chicken washes. Although MagPlex microspheres facilitated recovery of the microspheres and targets from the complex matrices, assay sensitivity was sometimes inhibited by up to one to three orders of magnitude; for example the detection limits E. coli spiked into apple juice or milk increased 100-fold, from 1000 to 100,000 cfu/mL. Thus, while the magnetic and fluorescent properties of the Luminex MagPlex microspheres allow for rapid, multiplexed testing for bacterial contamination in typically problematic food matrices, our data demonstrate that achieving desired limits of detection is still a challenge.
Keywords: Magnetic microspheres, Luminex, Multiplex, Immunoassay, Complex matrices, Foodborne pathogens
Introduction
Almost 80 million foodborne illnesses occur each year in the United States, and Salmonella, Campylobacter, and Escherichia coli are the major causes of these threats to public health (www.who.int/mediacentre/factsheets/fs237/en/index.html). These pathogens, commonly found in the intestines of birds, reptiles, and mammals, can contaminate other food products during processing (www.cdc.gov/ncidod/dbmd/diseaseinfo/foodborneinfections_g.htm). Large outbreaks of infection by these bacteria in produce, fruit juices, meats, and dairy products receive significant worldwide publicity, while many more seemingly isolated cases go undocumented.
Culture of suspect samples is the primary method for determining bacterial contamination of foods using guidelines found in the FDA Bacteriological Analytical Manual (www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm). Sample preparation and testing is time consuming, requiring hours of pre-enrichment before culturing 1–2 days on agar plates or in broths. The materials and reagent requirements for testing are broad and include a wide array of growth media with complex mixtures of constituents optimized for the variety of bacterial stains. Identifying the pathogenic species necessitates specialized technical expertise of microbiology for classification by morphology and biochemistry for assays.
Microsphere-based flow cytometric assays are sensitive, specific, and can be multiplexed to a high level for the detection of analytes [1]. Compared to bacterial culture methods, microsphere array assays such as the Luminex system are faster (a few hours), may require fewer reagents, and demand less technical expertise. Other types of assays, such as enzyme-linked immunosorbent assays (ELISA) and polymerase chain reaction (PCR), have limitations in time and, in many cases, are designed to detect only a single toxin (ELISA) or microbial analyte (ELISA, PCR). While FDA guidelines maintain that culture methods of identification will remain the gold standard for some time, they recognize that faster alternative methods will be important for the development of efficient new pathogen identification technologies and the modernization of protocols (www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM).
A number of researchers have performed microsphere-based assays to detect potential contaminants of foods: Dunbar et al. [2] detected several bacterial pathogens in phosphate-buffered saline (PBS), Ikeda et al. [3] detected pathogenic bacteria using extracted RNA, Fantozzi et al. [4] detected a genetically modified maize protein, while Haasnoot and du Pre [5] demonstrated a triplex immunoassay for vegetable proteins in a milk powder matrix. Although these studies demonstrated detection of food-borne pathogens, with the exception of Haasnoot and du Pre, application of these assays to a wide range of real-world food matrices was not demonstrated.
In general, particulates found in complex matrices such as foods can make microsphere-based immunoassays impossible because most assays utilizing fluorescently coded microspheres rely on size exclusion filters or centrifugation to collect microspheres while removing excess reagents; under these conditions, food components may not separate from the microspheres. Malkova et al. [6] recognized this issue and developed an immunomagnetic separation technique to enrich samples before detection by ELISA. MagPlex microspheres have recently been developed to assist in and streamline sample preparation; these spheres are fluorescently coded for use in Luminex assays, but also contain superparamagnetic material for collection by a permanent magnet. The concept is to use magnetic force to selectively separate microspheres, leaving the particulate matter and interfering macromolecules generated from the food sample fluids. Bergervoet et al. [7] performed a detection of potato viruses from infected potato leaves using MagPlex microspheres for this purpose. In this work, we apply MagPlex-based assays to a new category of complex samples and demonstrate their use to detect and identify multiple pathogens in food matrices in a single, multiplex assay.
Materials and methods
Instruments and reagents
Carboxy-functionalized standard xMAP and MagPlex microspheres, obtained from Luminex Corporation (Austin, TX), were analyzed on a Luminex 100 flow analyzer system. Phosphate buffered saline (PBS), bovine serum albumin (BSA) and Tween-20 were purchased from Sigma-Aldrich (St. Louis, MO). 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (sulfo-NHS), and NHS-LC-biotin were purchased from Pierce (Rockford, IL). Streptavidin-conjugated phycoerythrin (SA-PE) was supplied by Prozyme (San Leandro, CA). Heat-deactivated bacteria Escherichia coli O157: H7, Salmonella typhimurium, Campylobacter jejuni, and Listeria monocytogenes as well as goat anti-E. coli, anti-Campylobacter, anti-Salmonella, and anti-Listeria IgGs were kindly provided by Kirkegaard & Perry Laboratories, Inc. (Gaithersburg, MD). Staphylococcal enterotoxin B (SEB) was purchased from Toxin Technology. Rabbit anti-SEB was kindly provided by the Naval Medical Research Center (Silver Spring, MD).
Preparation of microspheres and biotinylated antibodies
The carboxy-functionalized xMAP or MagPlex microspheres were coated with antibodies using the manufacturer’s suggested sulfo-NHS/EDC mediated coupling. All steps requiring collection of microspheres and washing were performed by centrifugation for xMAP microspheres, whereas a permanent magnet was used with the MagPlex spheres. Briefly, 0.1 mL of the carboxylated microsphere stock suspension was diluted with 0.1 mL of 0.1 M sodium phosphate buffer (PB), pH 6.0 and washed by centrifugation (xMAP) or using a permanent magnet (MagPlex). The carboxy-coated microspheres were resuspended in 0.1 mL of PB, to which 0.01 mL of EDC (50 g/L in DMSO) and 0.01 mL of sulfo-NHS (50 g/L in water) were added. Following 15 min of incubation, the activated microspheres were washed twice in 0.2 mL PB and resuspended in 0.05 mL PBS containing the desired antibody at 1 mg/mL. After overnight coupling at 4 °C in the dark, the microspheres were washed twice with 0.5 mL PBS and resuspended in 1.0 mL PBS containing 0.05% Tween-20 (PBST) and 0.1% BSA (PBSTB). Microsphere sets 34, 38, 73, 77, and 81 were conjugated to goat anti-Salmonella, goat anti-Campylobacter, goat anti-E. coli, goat anti-Listeria, and rabbit anti-SEB, respectively.
Tracer antibodies were identical to those coated on the beads and were biotinylated using a tenfold molar excess of NHS-LC-biotin in PBS for 1 h. The free biotin was removed by gel filtration using a BioRad Bio-gel P10 column (Hercules, CA) with the final concentrations measured by UV absorption at 280 nm.
Preparation of food matrices
Pepper, tomato, ground beef, alfalfa sprouts, lettuce, and spinach were homogenized by placing 50 g of the food with 50 mL PBS in a blender. The resulting fluid was separated from the debris and placed into a centrifuge at 3000 RPM for 5 min to remove large particulate matter. Apple juice and milk (2% fat content) were diluted to 25% in PBS pH 7.4. Chicken carcass wash was prepared in a manner similar to that described in Taitt et al. [8], except that the liquid contained in the plastic-wrapped chicken was drawn off and used for the testing. For beef samples (30% fat content), 50 g of the product was homogenized in a blender with 50 mL PBS. The debris and fluid were separated by centrifugation at 3000 rpm. The fluids separated from the foodstuffs were stored at 4 °C for short-term use or frozen for long-term use. Aliquots of the fluids were spiked with antigen for analysis without any further treatment. Pathogen-positive controls were spiked into the prepared food matrices 1 h prior to beginning the assay protocol to allow the target antigen time to interact with components of the food matrix before magnetic separation.
Data processing
Mean fluorescence intensity values and associated standard deviations were determined by the Luminex 100 flow cytometer’s data output program. Plotted against concentrations of spiked bacteria and toxins, dose–response curves were generated for the multiplexed assays. The background fluorescence of an assay was the value generated from the assay by a zero concentration of antigen (bacteria or toxins). Background fluorescence intensities between microsphere sets can vary, thus the background values for the limit-of-detection were measured for the specific capture microsphere set used for the assay.
MagPlex sandwich immunoassays
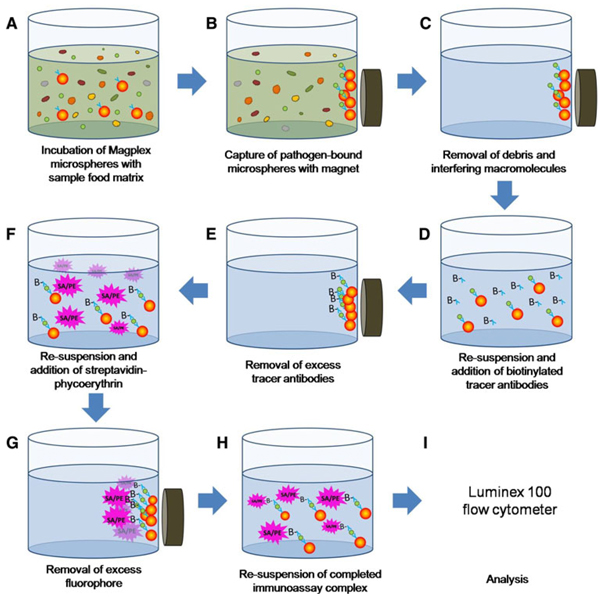
As depicted in Fig. 1, the assay was performed by preparing dilutions of the target in the matrix being tested (90 µL/well) in wells of a flat-bottom 96-microwell plate suitable for use with a plate magnet (Qiagen, Valencia, CA). To each well, the desired mixture of antibody-coated microspheres was added (5 µL/well, ≥200 microspheres of each set) and then incubated for 30 min at room temperature in the dark. Unbound antigen and matrix were removed by placing the plate on the magnet for at least 2 min and then withdrawing the solution from the center of the well using a multipipettor. The MagPlex microspheres were washed once using 100 µL of PBSTB, then the appropriate mixture of biotinylated tracer antibodies was added to each well (50 µL at 10 mg/L). After 30 min of incubation (room temperature), the excess antibody was removed, then SA-PE (5 mg/L) was added and the plate incubated again at room temperature in the dark for 30 min. The excess SA-PE was removed and the Magplex microspheres re-suspended in 85 µL of PBST. MagPlex sandwich immunoassays were measured on a Luminex 100 instrument with the PMT on the standard setting. At least 100 microspheres of each set were measured per data point of the dose–response experiment in each sample matrix. Data were collected using the xPONENT software version 3.0.380.0.
Fig. 1.
A schematic representation of the protocol used in preparing sandwich assays in MagPlex beads for detecting pathogens in food matrices
Determining limits-of-detection
Following analysis of samples spiked with bacteria or toxins, limits-of-detection (LODs) were determined as the lowest concentration tested with a signal greater the mean background fluorescence plus three times the standard deviation. Assays run in PBST were considered true measures of performance. Assays run in spiked food matrices demonstrated the inhibitory effect of interfering debris and soluble macromolecules.
Results and discussion
Luminex 100 utilizes polystyrene microspheres encoded with two internal fluorophores to create a two-dimensional array for a high degree of multiplexing; if each encoded bead set is decorated with an antibody directed against a different target, the presence of up to 100 different targets can be determined by Luminex’s ability to discriminate between these beads sets. MagPlex microspheres are analogous to xMAP beads, but possess superparametric properties that enable easy separation from liquids and complex mixtures. Multiplexed assays were performed using a mixture of MagPlex beads coated with different antibodies; after the beads were incubated with samples, target antigens bound to the beads were detected using a mixture of biotinylated tracer antibodies, followed by a streptavidin–phycoerthrin fluorescent tag. The microspheres were then interrogated for phycoerythrin fluorescence using the Luminex 100 flow cytometer.
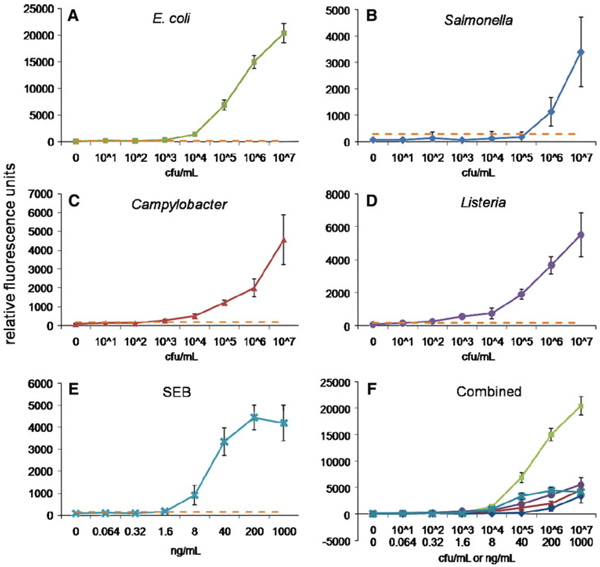
Spiked PBST samples were analyzed as controls of our multiplexed magnetic microsphere-based immunoassay. Figure 2 shows the dose–response curves of the fully multiplexed assays for E. coli, Salmonella, Campylobacter, Listeria, and SEB performed in buffer; analogous results were obtained for individual assays using only a single target-specific tracer antibody (data not shown). The LODs were determined as the concentration measured with a mean fluorescence signal greater the mean background fluorescence signal plus three times the standard deviation. Table 1 shows the MagPlex LODs in buffer for E. coli, Salmonella, Campylobacter, Listeria, and SEB compared with data measured previously using standard, non-magnetic microspheres [9]. Our results with MagPlex microspheres and the standard xMAP microspheres provided similar results in buffer for SEB and E. coli. An improvement was observed with detection of Listeria, whereas sensitivity for Salmonella decreased; as the MagPlex beads were coated with antibodies different from those used previously, these effects are likely antibody-specific.
Fig. 2.
Dose–response curves in buffer using MagPlex microspheres for a E. coli, b Salmonella, c Campylobacter, d Listeria, e SEB, and f all bacteria and SEB. Bacteria concentrations are reported in colony forming units (CFU) per milliliter and SEB in nanograms per milliliters. The dashed lines show the fluorescence threshold used to calculate the LODs for each target (lowest tested concentration that has a mean fluorescence signal greater the mean background fluorescence signal plus three times the standard deviation). Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Table 1.
Detection limits for bacterial and toxin antigens detected in the MagPlex and standard non-magnetic (xMAP) microspheres in a PBST matrix
| Antigen | LOD (MagPlex) | LOD (xMAP) |
|---|---|---|
| Escherichia coli | 103 cfu/mL | 103 cfu/mL [9] |
| Salmonella | 106 cfu/mL | 105 cfu/mL [9] |
| Campylobacter | 103 cfu/mL | |
| Listeria | 102 cfu/mL | 106 cfu/mL [9] |
| SEB | 0.064 ng/mL | 0.064 ng/mL [9] |
MagPlex microsphere LODs were determined as the concentration measured that had a mean relative fluorescence signal greater the mean background fluorescence signal plus three times the standard deviation. LODs using xMAP microspheres were previously reported
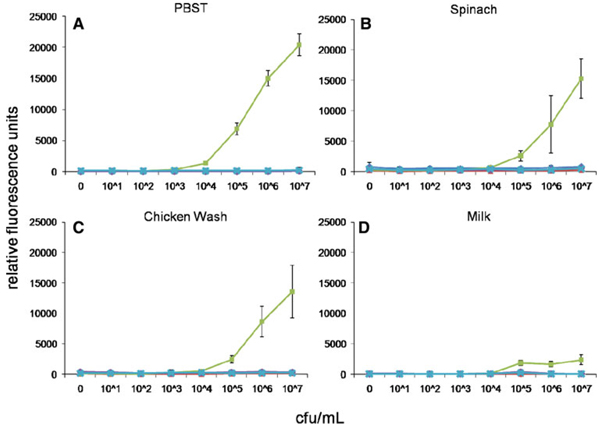
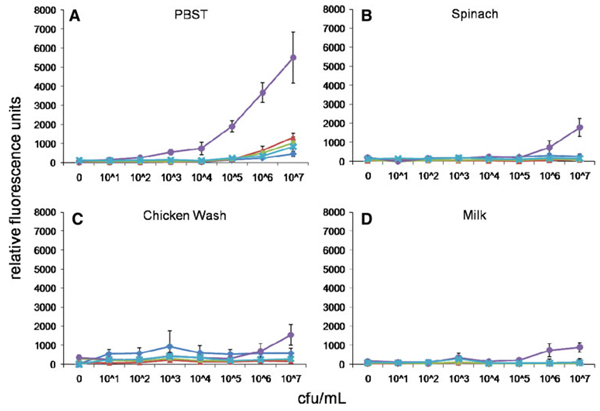
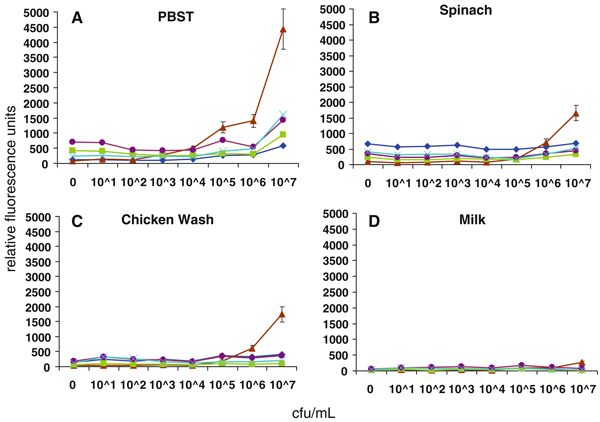
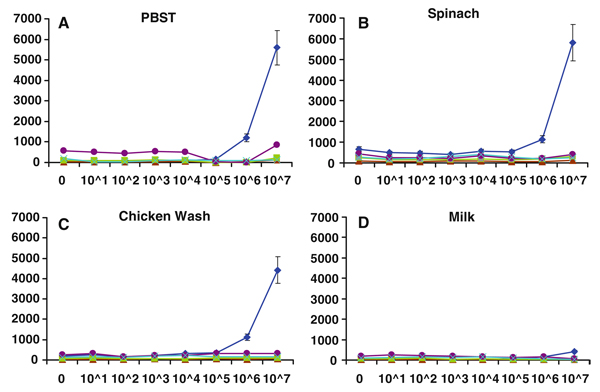
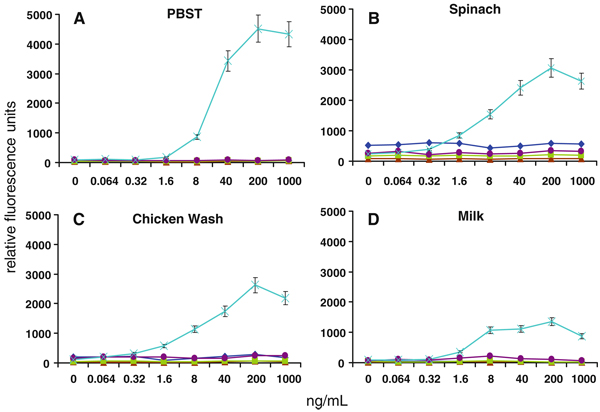
We next assessed the ability of MagPlex spheres to detect the same analytes in the presence of food matrices. Common food matrices of both animal and plant origin were prepared by homogenization or washing, then spiked with the target pathogens or toxin and analyzed using the MagPlex protocol. Shown in Figs. 3, 4, 5, 6, and 7 are dose–response curves for the five targets spiked into three food matrices representative of plant-based, animal-based, and high-fat foods in which contamination has been documented [10–13]. PBST is shown as a control. In general, assays were specific for the targets tested at low-to-intermediate concentrations, as indicated by the low levels of fluorescence on the microspheres directed against the other targets. Although not observed with SEB, E. coli, and Salmonella, high concentrations of Listeria and Campylobacter resulted in dose-dependent signal increases on all bead sets (specific and nonspecific), presumably due to adherence to all surfaces (Figs. 4a, 5a). This nonspecific binding was not as evident in the spiked foods, but was still occasionally noted.
Fig. 3.
Dose–response curves of MagPlex microspheres detecting E. coli in multiplexed assays in matrices: a PBST, b spinach, c chicken wash, and d milk. Concentrations are reported in colony forming units per milliliter. Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Fig. 4.
Dose–response curves of MagPlex microspheres detecting Listeria in multiplexed assays in matrices: a PBST, b spinach, c chicken wash, and d milk. Concentrations are reported in colony forming units per milliliter. Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Fig. 5.
Dose–response curves of MagPlex microspheres detecting Campylobacter in multiplexed assays in matrices: a PBST, b spinach, c chicken wash, and d milk. Concentrations are reported in colony forming units per milliliter. Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Fig. 6.
Dose–response curves of MagPlex microspheres detecting Salmonella in multiplexed assays in matrices: a PBST, b spinach, c chicken wash, and d milk. Concentrations are reported in colony forming units per milliliter. Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Fig. 7.
Dose–response curves of MagPlex microspheres detecting SEB in multiplexed assays in matrices: a PBST, b spinach, c chicken wash, and d milk. Concentrations are reported in colony forming units per milliliter. Microsphere sets are coated with antibodies to the antigens indicated using the following code: E. coli (square), Salmonella (diamond), Campylobacter (triangle), Listeria (circle), SEB (cross)
Some generalized increases in background signals on both target-specific and non-target-specific microspheres were also noted—most often on Salmonella-specific microspheres in the presence of spinach (cf. Figs. 5b, 6b, 7b). Since the presence of these food matrices decreased target-specific signal intensities and increased background signals for all assays, poorer limits of detection were often observed when compared to buffer controls (Table 2). The E. coli assay suffered the greatest decrease in signal-to-noise ratio when performed in food matrices, but since signals on E. coli-specific microspheres were high (Fig. 2), the decrease in overall assay performance was not as significant as those suffered by the other assays. SEB assays, on the other hand, had comparable signals to those levels found in assays for Salmonella, Campylobacter, and Listeria, but demonstrated less overall inhibition by the food matrices. While detection limits for several of the bacterial targets increased by up to 105-fold, sensitivity for SEB was decreased 125-fold (lettuce) or less. In fact, detection limits of less than 2 ng/mL for SEB were retained in eight of the nine food matrices tested. It is likely that SEB, a small water soluble 28 kDa protein, has less tendency to adsorb to the food matrices and remains available for the detection assay, while the killed cells are more sticky.
Table 2.
Limits-of-detection (LODs) for bacterial and toxin antigens in PBST and complex food matrices using MagPlex microspheres
| PBST | Alfalfa | Lettuce | Spinach | Tomato | Pepper | Apple juice | Milk | Chicken wash | Ground beef | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (cfu/mL) | 103 | 105 | 105 | 105 | 104 | 103 | 105 | 105 | 104 | 105 |
| Salmonella (cfu/mL) | 106 | 106 | 104 | 106 | 107 | 107 | 106 | 107 | 106 | 106 |
| Campylobacter (cfu/mL) | 103 | >107a | >107 | 106 | >107 | 107 | 107 | 107 | 106 | 107 |
| Listeria (cfu/mL) | 102 | 107 | >107 | 106 | 104 | 105 | 104 | 106 | 106 | 104 |
| SEB (ng/mL) | 0.064 | 0.32 | 8.0 | 0.32 | 0.064 | 1.6 | 0.32 | 1.6 | 1.6 | 0.32 |
Bacteria LODs are measured in colony forming units per milliliter and toxin LODs in nanograms in milliliters. LODs were determined as the concentration actually measured that has a mean fluorescence signal greater the mean background fluorescence signal plus three times the standard deviation
Not detectable within the concentration range of 101–107 cfu/mL
Inhibition in many cases can be attributed to the release of interfering proteins, carbohydrates, and small molecules from the food matrix during sample preparation. For example, high levels of polyphenols found in many fruits and vegetables, have been shown to interfere with immunoassays [11, 13]. This effect tracks well with the extent of inhibition in the plant-based matrices we tested. Milk matrices have also been found to decrease biosensor performance in some, but not all, assays. [12, 14, 15] The effect of pH in canned foods was previously shown to have a significant detrimental effect on the testing of botulinum toxoid A, SEB, and ricin [16, 17]; thus we maintained pH of our samples at 7.2 to maximize assay sensitivity. Additionally, the presence of other contaminating microbes (or naturally occurring microflora) may have been responsible for some decrease in assay performance; however, our results showing minimal cross-reactivity between the different bead sets in the presence of our targets, suggest that the presence of additional microbial species probably did not play a significant role.
MagPlex microspheres have recently become available as materials with tremendous potential for rapid preparation of complex samples. The experiments described here are the first to apply these microspheres for multiplexed analysis of a range of foodstuffs. These experiments demonstrate that MagPlex microspheres can be used in immunoassays in complex food matrices, but attaining the limits of detection achieved in standard buffers is not trivial. There is considerable interference when assays are conducted in homogenized foods; thus the impact on the limit for each assay needs to be ascertained prior to implementation of a testing protocol. Decreased levels of performance may range from 10- to 20-fold, such as in the case to E. coli and SEB, to significantly higher extents depending on the matrix and analyte tested (e.g., Salmonella, Campylobacter, and Listeria). Examination of loss of signal and increases in background show minimal correlations. For many foodstuffs, performing a simple wash as opposed to homogenization would provide a useful sample; assay inhibition would be minimized and sample processing simplified, leading to an overall more rapid assay.
Alternatively, one could employ an enrichment step; many “rapid” detection systems require a 24- to 36-h enrichment step to increase the number of target cells present. Depending on the target, the growth time, and the medium used, post-enrichment microbial targets can reach 106–109 cfu/mL [18–20]. Inclusion of even a shortened enrichment step prior to analysis using MagPlex would increase cell numbers to within our range of detectable concentrations. Moreover, the ability of the MagPlex-based assays to detect contaminants in the presence of the foodstuffs (without significant dilution) may allow growth media to be added directly to the food sample, with direct analysis of cultures immediately after enrichment and without removal of the food components; although not tested here, such a streamlined processing protocol would significantly decrease the logistical burden for on-site testing. As for toxic contaminants not affected by an enrichment step, the low- to sub-ng/mL detection limits here are in the same range as other rapid detection systems [12, 14, 16, 21], demonstrating the applicability of this methodology for rapid detection of toxins. However, when attempting to multiplex assays for microbial and toxic targets, each assay—as well as each method for sample preparation—must be validated, and preferably optimized, to establish appropriate limits of detection.
Acknowledgments
JSK is an American Society for Engineering Education Postdoctoral Fellow. We would like to thank KPL for generously providing reagents. The work presented here was performed under NIH Grant UO1 A1075489 and ONR/NRL 6.2 work unit 6336. The views presented here are those of the authors and do not represent the opinion or policy of the National Institutes of Health, the US Navy or the Department of Defense.
References
- 1.Vignali DA. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods. 2000;243(1–2):243–255. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 2.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex Lab-MAP system. J. Microbiol. Methods. 2003;53(2):245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda M, Yamaguchi N, Tani K, Nasu M. Rapid and simple detection of food poisoning bacteria by bead assay with a microfluidic chip-based system. J. Microbiol. Methods. 2006;67(2):241–247. doi: 10.1016/j.mimet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Fantozzi A, Ermolli M, Marini M, Scotti D, Balla B, Querci M, Langrell SR, Van den Eede G. First application of a microsphere-based immunoassay to the detection of genetically modified organisms (GMOs): quantification of Cry1Ab protein in genetically modified maize. J. Agric. Food Chem. 2007;55(4):1071–1076. doi: 10.1021/jf061506p. [DOI] [PubMed] [Google Scholar]
- 5.Haasnoot W, du Pre JG. Luminex-based triplex immunoassay for the simultaneous detection of soy, pea, and soluble wheat proteins in milk powder. J. Agric. Food Chem. 2007;55(10):3771–3777. doi: 10.1021/jf063281o. [DOI] [PubMed] [Google Scholar]
- 6.Malkova K, Rauch P, Wyatt GM, Morgan MRA. Combined immunomagnetic separation and detection of Salmonella enteritidis in food samples. Food Agric. Immunol. 1998;10(3):271–280. [Google Scholar]
- 7.Bergervoet JH, Peters J, van Beckhoven JR, van den Bovenkamp GW, Jacobson JW, van der Wolf JM. Multiplex microsphere immuno-detection of potato virus Y, X and PLRV. J. Virol. Methods. 2008;149(1):63–68. doi: 10.1016/j.jviromet.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Taitt CR, Shubin YS, Angel R, Ligler FS. Detection of Salmonella enterica serovar typhimurium by using a rapid, array-based immunosensor. Appl. Environ. Microbiol. 2004;70(1):152–158. doi: 10.1128/AEM.70.1.152-158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JS, Anderson GP, Erickson JS, Golden JP, Nasir M, Ligler FS. Multiplexed detection of bacteria and toxins using a microflow cytometer. Anal. Chem. 2009;81(13):5426–5432. doi: 10.1021/ac9005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waxman HA, DeLauro R. FDA, Fresh Spinach Safety, Report to US House of Representatives Committee on Oversight and Government Reform 2008, 111th Congress; pp. 1–10. [Google Scholar]
- 11.Ogunjimi AA, Choudary PV. Adsorption of endogenous polyphenols relieves the inhibition by fruit juices and fresh produce of immuno-PCR detection of Escherichia coli O157:H7. FEMS Immunol. Med. Microbiol. 1999;23(3):213–220. doi: 10.1111/j.1574-695X.1999.tb01241.x. [DOI] [PubMed] [Google Scholar]
- 12.Shriver-Lake LC, Shubin YS, Ligler FS. Detection of staphylococcal enterotoxin B in spiked food samples. J. Food Prot. 2003;66(10):1851–1856. doi: 10.4315/0362-028x-66.10.1851. [DOI] [PubMed] [Google Scholar]
- 13.Henning SM, Aronson W, Niu Y, Conde F, Lee NH, Seeram NP, Lee RP, Lu J, Harris DM, Moro A, Hong J, Pak-Shan L, Barnard RJ, Ziaee HG, Csathy G, Go VL, Wang H, Heber D. Tea polyphenols and theaflavins are present in prostate tissue of humans and mice after green and black tea consumption. J. Nutr. 2006;136(7):1839–1843. doi: 10.1093/jn/136.7.1839. [DOI] [PubMed] [Google Scholar]
- 14.Homola J, Dostalek J, Chen S, Rasooly A, Jiang S, Yee SS. Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int. J. Food Microbiol. 2002;75(1–2):61–69. doi: 10.1016/s0168-1605(02)00010-7. [DOI] [PubMed] [Google Scholar]
- 15.Son JR, Kim G, Kothapalli A, Morgan MT, Ess D. Detection of Salmonella enteritidis using a miniature optical surface plasmon resonance biosensor. J. Phys. 2007;61:1086–1090. [Google Scholar]
- 16.Sapsford KE, Taitt CR, Loo N, Ligler FS. Biosensor detection of botulinum toxoid A and staphylococcal enterotoxin B in food. Appl. Environ. Microbiol. 2005;71(9):5590–5592. doi: 10.1128/AEM.71.9.5590-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson GP, Liu JL, Hale ML, Bernstein RD, Moore M, Swain MD, Goldman ER. Development of anti-ricin single domain antibodies toward detection and therapeutic reagents. Anal. Chem. 2008;80(24):9604–9611. doi: 10.1021/ac8019398. [DOI] [PubMed] [Google Scholar]
- 18.Bhaduri S, Cottrell B. Sample preparation methods for PCR detection of Escherichia coli O157:H7, Salmonella typhimurium, and Listeria monocytogenes on beef chuck shoulder using a single enrichment medium. Mol. Cell. Probes. 2001;15(5):267–274. doi: 10.1006/mcpr.2001.0370. [DOI] [PubMed] [Google Scholar]
- 19.Bhagwat AA. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 2003;84(2):217–224. doi: 10.1016/s0168-1605(02)00481-6. [DOI] [PubMed] [Google Scholar]
- 20.Valadez AM, Lana CA, Tu SI, Morgan MT, Bhunia AK. Evanescent wave fiber optic biosensor for Salmonella detection in food. Sensors. 2009;9(7):5810–5824. doi: 10.3390/s90705810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim DV. Detection of microorganisms and toxins with evanescent wave fiber-optic biosensors. Proc. IEEE. 2003;91(6):902–907. [Google Scholar]