Abstract
Objective
This paper has two primary goals. First, a brief tutorial on behavioral and molecular genetic methods is provided for readers without extensive training in these areas. To illustrate the application of these approaches to developmental disorders, etiologically-informative studies of reading disability (RD), math disability (MD), and attention-deficit/hyperactivity disorder (ADHD) are then reviewed. Implications of the results for these specific disorders and for developmental disabilities as a whole are discussed, and novel directions for future research are highlighted.
Method
Previous family and twin studies of RD, MD, and ADHD are reviewed systematically, and the extensive molecular genetic literatures on each disorder are summarized. To illustrate four novel extensions of these etiologically-informative approaches, new data are presented from the Colorado Learning Disabilities Research Center, an ongoing twin study of the etiology of RD, ADHD, MD, and related disorders.
Conclusions
RD, MD, and ADHD are familial and heritable, and co-occur more frequently than expected by chance. Molecular genetic studies suggest that all three disorders have complex etiologies, with multiple genetic and environmental risk factors each contributing to overall risk for each disorder. Neuropsychological analyses indicate that the three disorders are each associated with multiple neuropsychological weaknesses, and initial evidence suggests that comorbidity between the three disorders is due to common genetic risk factors that lead to slow processing speed
Keywords: Reading, math, ADHD, genetics, twins
INTRODUCTION
Previous etiological models of complex disorders such as reading disability (RD), math disability (MD), and attention-deficit/hyperactivity disorder (ADHD) often implicated simple linear causal pathways in which a single genetic or environmental risk factor led to a single cognitive deficit that was necessary and sufficient to cause all of the symptoms of the disorder. These models worked well for single-gene disorders such as Huntington’s Disease and phenylketonuria, but a growing literature consistently suggests that single-deficit models do not provide a satisfactory explanation for most developmental disorders1. In this paper we review several lines of etiological research that suggest that developmental disorders may be better conceptualized as heterogeneous conditions that arise from the additive and interactive effects of multiple genetic and environmental risk factors.
This paper is divided into five sections. The first section briefly describes the Colorado Learning Disabilities Research Center (CLDRC), an ongoing twin study funded by a Center grant from the National Institute for Child Health and Human Development2;3. Data from the CLDRC are used to illustrate many of the methodological approaches described in the paper. The second and third sections first describe each behavioral and molecular genetic method for readers without specific training in this area, then systematically review studies of RD, MD, or ADHD that used the approach. In the fourth section we present new data from the CLDRC to illustrate how novel extensions of these behavioral and molecular genetic methods may provide important new information regarding the complex etiologies of learning disorders and ADHD. The final section of the paper discusses the clinical implications of these results and describes several areas in which additional research is needed.
Colorado Learning Disabilities Research Center
Due to the paucity of well designed twin studies of reading disability, a twin study was initiated in 1982 as part of the Colorado Reading Project4. This project was incorporated into the CLDRC when it was initiated in 1991, and twins have been tested continuously since that time. The sample now includes over 1,280 twin pairs selected because at least one of the twins met screening criteria for RD or ADHD, 450 biological siblings of the selected twins, and 790 pairs of control twins in which neither twin met criteria for RD or ADHD. More stringent criteria based on psychometric testing are then applied to identify the final group of probands with RD or ADHD, as described in the subsequent section.
Participants
In collaboration with administrators in 22 Colorado school districts that have agreed to participate in the study, all twin pairs in each district are identified without regard to reading or ADHD status. After initial parental consent is obtained, independent screening procedures are conducted to identify twin pairs in which at least one twin meets criteria for ADHD, RD, or both disorders. If either member of a twin pair has a history of reading difficulties or meets screening criteria for ADHD, the pair and any biological siblings between 8 and 18 years of age are invited to participate in the full study. Each twin that participates in the full study completes an extensive test battery that includes a complete standardized IQ test, psychometric measures of reading, spelling, and mathematics achievement, measures of reading-related language processes, diagnostic measures of ADHD, and measures of key cognitive domains that may be related to one of the disorders3.
A matched comparison group of control twins is selected from the overall sample of pairs who did not meet the screening criteria for RD or ADHD. Because the primary focus of the CLDRC is the etiology of RD and ADHD, pairs at risk for one or both disorders are oversampled to increase statistical power for analyses of these extreme groups.
Definitions of RD, MD, and ADHD
The definitions of RD and MD in the Diagnostic and Statistical Manual for Mental Disorders, Fourth Edition5 specify that an individual's reading or math achievement must be significantly discrepant from their overall intelligence. However, the utility of IQ scores as part of the diagnosis of learning disabilities is a long-standing area of controversy, and most experts argue against the use of an IQ-discrepancy criterion6;7. For the examples in this paper we defined RD by a cutoff score 1.25 SD below the estimated population mean on an age-adjusted composite measure of word reading derived from the Peabody Individual Achievement Test (PIAT) Reading Recognition subtest8 and a time-limited word reading test9. Similarly, MD was defined by a score 1.25 SD below the population mean on a composite measure of math calculations derived from the Math subtests on the PIAT and the Wide Range Achievement Test10.
The DSM-IV definition of ADHD includes three subtypes based on differential elevations of inattention and hyperactivity-impulsivity symptoms. The predominantly inattentive type is characterized by significant elevations of inattention but not hyperactivity-impulsivity, whereas the predominantly hyperactive-impulsive type exhibits significant hyperactivity-impulsivity but not inattention, and individuals with the combined type have clinically significant elevations on both symptom dimensions. Parent and teacher ratings11 of ADHD symptoms and associated impairment were combined based on the algorithm from the DSM-IV field trials for the disruptive behavior disorders12.
BEHAVIORAL GENETIC STUDIES OF RD, MD, AND ADHD
Family Studies
Family studies test whether the rate of a disorder is significantly higher in the biological family members of individuals with the disorder than in the family members of individuals without the diagnosis. If the disorder occurs more frequently in family members of individuals with the disorder, this suggests that familial factors increase susceptibility for the disorder.
Correlations between biological siblings are moderate to high on dimensional measures of reading (r = .40 – .70), math (r = .40 – .80), and ADHD symptoms (r = .20 – .50)13–16. In family studies of categorical diagnoses, the relative risk of RD is 4 – 8 times higher in first-degree relatives of probands with RD than in relatives of individuals without RD17;18, and similar familiality is reported in studies of ADHD19;20. Fewer studies have tested the familiality of MD, but initial results suggest that the relative risk is 5 to 10 times higher in the biological relatives of probands with versus without MD21;22.
Twin studies
Because members of intact biological families share both genetic influences and the home environment, other methods are needed to disentangle the relative contributions of genetic and environmental influences. By comparing the similarity of monozygotic (MZ) twins, who share all of their genes, to dizygotic (DZ) twins, who share half of their segregating genes on average, twin studies provide estimates of the extent to which a disorder is due to genetic or environmental influences23.
Concordance rates
The most straightforward test for genetic influences on a clinical disorder compares the rate of concordance in pairs of MZ versus DZ twins. If a disorder is influenced by genes, the proportion of pairs that are concordant for the disorder will be higher in MZ pairs than DZ pairs. Consistent with this hypothesis, all previous studies of RD, ADHD, and MD (with one exception24) found that the probandwise concordance rate was higher in MZ twin pairs than DZ twin pairs, providing strong evidence that all three disorders are influenced by genes (Table 1).
Table 1.
Concordance rates for RD and ADHD in pairs of MZ and DZ twins
| Number of pairs |
Probandwise Concordance |
|||
|---|---|---|---|---|
| MZ | DZ | MZ | DZ | |
| Reading Disorder | ||||
| Bakwin (1973)126 | 31 | 31 | 91% | 45% |
| Harlaar et al. (2005)32 | 308 | 246 | 72% | 45% |
| Hawke et al. (2006)127 | 306 | 247 | 66% | 35% |
| Stevenson et al. (1987)24 | 14 – 18a | 27 – 38a | 33 – 50%a | 29 – 54%a |
| Zerbin-Rudin (1967)128 | 17 | 34 | 100% | 52% |
| Weighted average | 70% | 41% | ||
| ADHD | ||||
| Goodman & Stevenson (1989)129 | 39 | 54 | 51% | 33% |
| Levy et al. (1997)49 | 57 | 46 | 82% | 38% |
| Levy et al. (2001)130 | 138 | 109 | 67% | 42% |
| Sherman et al. (1997)131 | 69 | 32 | 58% | 31% |
| Thapar et al. (2001)132 | 175b | 410b | 79% | 54% |
| Todd et al. (2001)133 | 72 | 135 | 68% | 22% |
| Willcutt et al. (2000)25 | 88 | 82 | 78% | 38% |
| Willcutt et al. (2007)38 | 83 | 78 | 68% | 24% |
| Weighted average | 71% | 41% | ||
| Math Disorder | ||||
| Alarcón et al. (1997)134 | 63 | 32 | 76% | 56% |
| Kovas et al. (2007)16 | 93 | 83 | 40% | 24% |
| Weighted average | 55% | 33% | ||
concordance rates were provided for several different definitions of RD. The average MZ and DZ concordance was use for the weighted average.
specific Ns were not provided, so Ns are estimated based on the total sample and the 80th percentile threshold used to define the extreme ADHD probands.
Etiology of individual differences
Although the simplicity of a comparison of concordance rates is appealing, increasing evidence suggests that RD, MD, ADHD, and most other complex disorders are defined by a diagnostic threshold imposed upon a quantitative measure that is continuously distributed in the population16;25. Transformation of continuous measures such as reading or math performance or ADHD symptoms into a categorical diagnosis results in the loss of important information pertaining to both severity within the disorder and variability in subthreshold symptomatology. Therefore, several authors have developed more powerful variance components or multiple regression approaches for analyses of continuous data26–28.
Basic twin models estimate three parameters. Heritability is the proportion of the total phenotypic variance in a trait that is attributable to genetic influences. The proportion of variance due to environmental factors is subdivided to distinguish two types of environmental influences. Shared environmental influences are environmental factors that increase the similarity of individuals within a family in comparison to unrelated individuals in the population. These effects may potentially include environmental influences within the home or any other shared experiences such as mutual friends or shared teachers. In contrast, nonshared environmental influences are environmental factors that that are independent or unique for members of twin pairs. These risk factors could include a head injury or other accident, a traumatic event, or exposure to physical or sexual abuse (if the other twin was not similarly exposed).
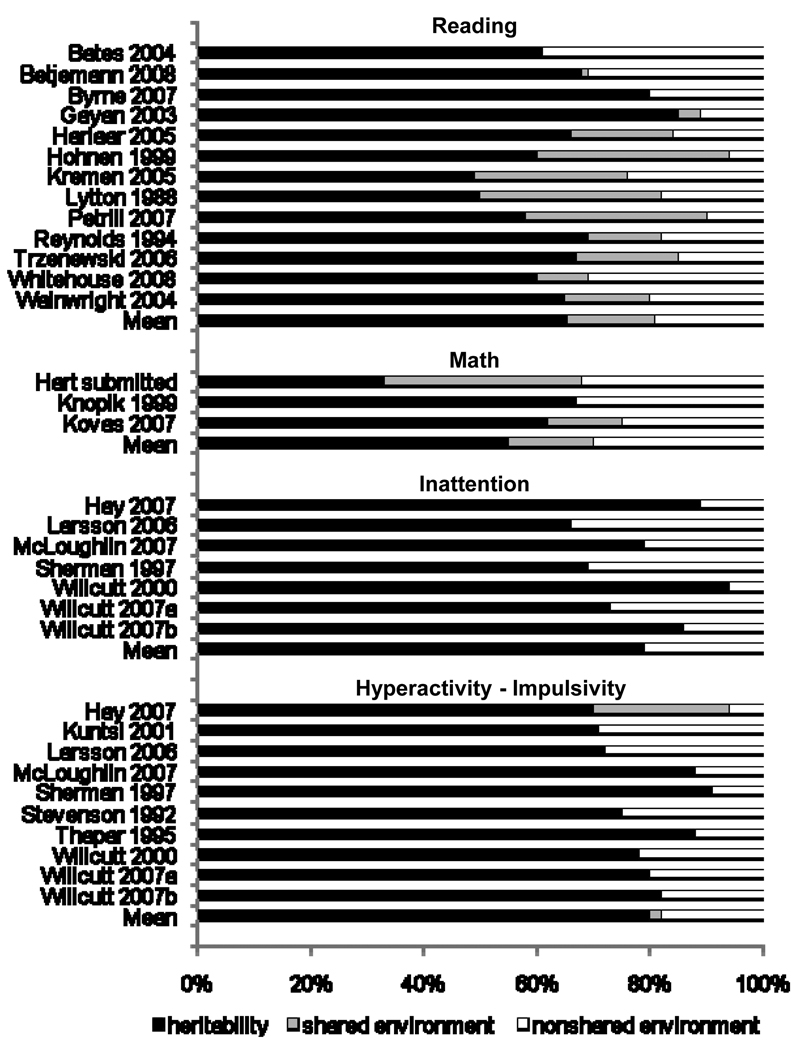
Figure 1 summarizes published twin studies of reading, math, and the two DSM-IV ADHD symptom dimensions13;15;16;25;29–48. Heritability estimates are moderate for individual differences in single-word reading and math, and are consistently high for inattention and hyperactivity-impulsivity. Shared environmental influences account for an additional 10 – 15% of the variance in reading and math, whereas shared environmental influences were not significant for the ADHD symptom dimensions. Nonshared environmental influences and measurement error account for the remaining 20 – 25% of the variance in each of the phenotypes.
Figure 1.
Twin studies of reading, math, and DSM-IV ADHD symptom dimension
Etiology of extreme scores
Although variance components analyses are optimal for analyses of individual differences in unselected or minimally selected samples, this approach is not designed for analyses of extreme groups. Therefore, DeFries and Fulker developed a multiple regression approach to test the etiology of extreme group membership26;27. DeFries-Fulker (DF) analysis is based on the differential regression of MZ and DZ co-twin scores toward the population mean when probands are selected due to an extreme score on a phenotype. Although scores of both MZ and DZ co-twins are expected to regress toward the population mean, scores of DZ co-twins should regress further than scores of MZ co-twins to the extent that the proband deficit is influenced by genes. After appropriate standardization and transformation of scores, the magnitude of differential regression by zygosity provides a direct estimate of the heritability of the extreme group deficit (h2g).
To illustrate the DF approach, univariate models were fitted to composite scores for reading, math, inattention, and hyperactivity-impulsivity in the CLDRC sample (Table 2). The selection criterion for the proband groups for this specific analysis yielded mean MZ and DZ proband scores approximately 2 SD below the population mean. MZ and DZ co-twin means regressed differentially on all four measures, and the multiple regression models revealed significant genetic influences on each group deficit, similar to results that have been reported in other samples for RD32, MD16, and ADHD40;49.
Table 2.
Etiology of group deficits in reading, math, and ADHD symptoms
| MZ pairsa |
DZ pairsa | ||||||
|---|---|---|---|---|---|---|---|
| Proband | Co-twin | Proband | Co-twin | ||||
| Nb | M (SD) | M (SD) | Nb | M (SD) | M (SD) | h2g (SE) | |
| Reading | 106 | −2.00 (0.68) | −1.73 (0.75) | 89 | −1.99 (0.72) | −1.07 (1.23) | 0.65 (0.13)*** |
| Math | 91 | −2.01 (0.56) | −1.38 (0.99) | 84 | −2.04 (0.55) | −0.81 (1.12) | 0.58 (0.15)*** |
| Inattention | 98 | −2.03 (0.58) | −1.46 (1.23) | 83 | −2.07 (0.59) | −0.59 (1.07) | 0.87 (0.14)*** |
| Hyperactivity - impulsivity | 85 | −2.17 (0.57) | −1.47 (0.58) | 65 | −2.09 (0.58) | −0.62 (1.16) | 0.76 (0.15)*** |
Note.
P < .001.
Scores are expressed as standard deviations from the estimated population mean. All scores are scaled so that lower scores indicate greater impairment on all measures.
Total number of pairs in which at least one twin met the criteria for the proband group (score at least 1.25 SD below the population mean).
Conclusions
Both individual differences and extreme scores on measures of math, reading and ADHD are significantly heritable. Although twin analyses cannot test definitively whether the same genetic influences act on extreme scores and individual differences, the similarity of these results is consistent with this hypothesis. In the next section, we summarize results from studies that attempted to localize the specific genes that account for these high heritability estimates.
MOLECULAR GENETIC STUDIES OF RD, MD, AND ADHD
Molecular genetic studies have used three methods to identify genes that increase susceptibility to RD, MD, or ADHD. Space constraints permit only a short description of these methods, but more detailed overviews are provided elsewhere23;50;51. Briefly, the candidate gene approach examines specific genes that are targeted because they play a role in the pathophysiology of the disorder. For example, many candidate gene studies of ADHD have tested for associations with genes in the dopamine system due to the significant effect of stimulant medication on dopamine transmission52. In contrast to the theory-driven candidate gene approach, linkage and association analyses use a dense map of DNA markers to screen the entire genome or targeted chromosomal regions for polymorphisms (differences in the DNA sequence between individuals) that may increase susceptibility to the disorder.
ADHD
Over 200 studies have tested for associations between ADHD and over 100 different genes since the first candidate gene study was completed fifteen years ago53. Although many initial positive results failed to replicate in subsequent studies, a recent meta-analysis implicated seven genes as significant risk factors for ADHD52. The effect size of each of these genes is small (Odds Ratio = 1.1 – 1.3), however, and the combined effects of all seven loci explain only a small amount of the total genetic variance in ADHD. Because these results suggested that additional genes must play a role in ADHD, genome-wide linkage and association analyses were used to screen the entire genome for additional loci54–56. These studies found significant evidence of a susceptibility locus on chromosome 16q, and suggestive evidence for risk loci in nine additional regions of the genome. None of these regions overlapped with the locations of the candidate genes identified by the meta-analysis, and even with the combined effects of the candidate genes and the loci identified by the linkage and association studies the majority of the genetic variance in ADHD symptoms in the population remains unexplained.
RD
In contrast to the dopamine model of ADHD that was derived largely from response to medication, targets for candidate gene studies are not as obvious based on current knowledge about the pathophysiology of RD and MD. Therefore, molecular genetic studies of MD and RD first used linkage and association analyses to identify regions of the genome that may contain a risk locus for the disorder, then targeted fine-mapping procedures were used to test for candidate genes in these regions. Genome-wide and targeted linkage analyses have identified nine locations in the genome that are likely to include risk loci for RD57, and subsequent analyses have identified potential candidate genes in six of these region58–65. Although some of these loci await independent replication, these results suggest that RD is also influenced by multiple genetic risk factors with relatively small effect sizes.
MD
Finally, initial results from the only molecular genetic study of MD are also similar to the results reported for RD and ADHD66. In a genome-wide association study of a population-based sample of 2,449 individuals, a total of 10 single nucleotide polymorphisms were significantly associated with math performance in two separate analyses. In combination, the 10 risk loci accounted for approximately 3% of the phenotypic variance in math performance.
Conclusions
The high heritability of RD, MD, and ADHD led to initial optimism that genes with major effects would be identified for each disorder. Contrary to this prediction, however, results of candidate gene, linkage, and association studies all suggest that the etiologies of RD, MD, and ADHD are complex and polygenic, with multiple genetic and environmental risk factors contributing to the total phenotypic variance in the population. Future molecular genetic studies could still uncover rare polymorphisms with major effects on ADHD, RD, or MD in a subset of the population67, but the current literature argues against single-gene models of each disorder, and is similar to results reported for virtually all other developmental disorders and psychopathology, including pervasive developmental disorders68, bipolar disorder69, major depression70, and schizophrenia71;72. In the next section we discuss four extensions of these basic behavioral and molecular genetic approaches that may help to begin to disentangle the complex etiologies of developmental disorders.
EXTENSIONS OF ETIOLOGICALLY-INFORMATIVE METHODS
Gene × Environment Interactions
Gene × environment (G × E) interactions occur if environmental circumstances modify the expression of an individual's genetic background, either strengthening or weakening genetic influences on a phenotype73. Significant G × E interactions have been reported for several psychopathologies, including conduct disorder74 and depression75. These are both examples of diathesis-stress interactions, which occur when genetic vulnerability (the diathesis) co-occurs with an environmental risk factor, resulting in more severe symptomatology than would be expected based on either risk factor alone or their additive combination.
These exciting initial findings have replicated inconsistently, however, leading others to encourage caution in the interpretation of G × E findings76;77. Among the key concerns that have been raised are statistical issues regarding data transformations and the failure to correct adequately for multiple testing, along with the fact that many putative environmental risk factors are partially heritable, such as parenting behavior, social support, and exposure to stressful life events78. In addition, many studies of humans have reported significant G × E interactions in the absence of a genetic main effect, a phenomenon that is extremely rare in well-controlled studies of nonhuman animals79. Each of these issues is an important caveat for the studies of ADHD and RD that are reviewed in this section, and these points of critique are discussed in detail elsewhere80.
ADHD
Table 3 summarizes results of studies of ADHD that tested for interactions between environmental risk factors and specific candidate genes81–94. Most studies tested for interactions between dopamine genes and prenatal risk factors, and all significant G × E interactions were diathesis-stress interactions. Interactions were significant between several dopamine genes and prenatal smoking, and several of these studies also found a significant interaction between the dopamine transporter gene and prenatal alcohol exposure. In both cases these results appear to be strongest for hyperactivity-impulsivity symptoms and the combined type. The primary postnatal environmental influences that were tested were socioeconomic status and environmental adversity. Although these constructs were measured a variety of different ways, all six studies reported a significant diathesis-stress interaction with a range of different candidate genes, and we recently found a similar diathesis-stress interaction with parental education in the CLDRC sample80.
Table 3.
Studies of candidate gene × environment interactions in ADHD
| Environmental Risk / Study |
Candidate Genesa |
ADHD Phenotype |
Genetic Main Effect |
Sig. G × E Interaction |
|---|---|---|---|---|
| Prenatal Smoking | ||||
| Becker et al. (2008)88 | DAT1 | ADHD | No | Yesb |
| Brookes et al. (2006)93 | DAT1 | ADHD | Yes | No |
| Kahn et al. (2003)89 | DAT1 | Inattention symptoms | No | No |
| Kahn et al. (2003)89 | DAT1 | Hyp-Imp symptoms | No | Yes |
| Langley et al. (2008)90 | DAT1, DRD4, 5HTT, DRD5 | ADHD | No | Noc |
| Neuman et al. (2007)91 | DAT1, DRD4 | Combined Type | No | Yes |
| Neuman et al. (2007)91 | DAT1, DRD4 | Inattentive Type | No | No |
| Todd et al. (2007)92 | CHRNA4 | Combined Type | No | Yes |
| Todd et al. (2007)92 | CHRNA4 | Inattentive Type | No | No |
| Prenatal Alcohol | ||||
| Brookes et al. (2006)93 | DAT1 | ADHD | Yes | Yes |
| Kahn et al. (2003)89 | DAT1 | Inattention symptoms | No | No |
| Kahn et al. (2003)89 | DAT1 | Hyp-Imp symptoms | No | Yes |
| Langley et al. (2008)90 | DAT1, DRD4, 5HTT, DRD5 | ADHD | No | No |
| Neuman et al. (2007)91 | DAT1, DRD4 | Combined Type | No | No |
| Neuman et al. (2007)91 | DAT1, DRD4 | Inattentive Type | No | No |
| Low Birth Weight | ||||
| Langley et al. (2008)90 | 5HTT, DRD4, DAT1, DRD5 | ADHD | No | Nod |
| Season of birth | ||||
| Seeger 200487 | DRD4 | ADHD+CD | No | Yes |
| Brookes 200886 | DRD4 | ADHD | No | No |
| Socioeconomic status / environmental adversity | ||||
| Lasky-Su et al. (2007)81 | BDNF | Inattention symptoms | Yes | Noe |
| Lasky-Su et al. (2007)81 | BDNF | Hyp-Imp symptoms | Yes | Yes |
| Laucht et al. (2007)82 | DAT1 | ADHD | No | Yes |
| Nobile et al. (In press)135 | COMT | ADHD | No | Yes |
| Nigg et al. (2007)94 | DAT1, DRD4, ADRA2Af | ADHD | Yes | Yes |
| Retz et al. (2008)84 | 5HTT | ADHD | Yes | Yes |
| Waldman et al. (2007)85 | DRD2 | ADHD | No | Yes |
ADRA2A = , BDNF = brain-derived neurotrophic factor, CHRNA4 = Nicotinic acetylcholine receptor α-4 , COMT = catechol-O-methyltransferase, DAT1 = dopamine transporter, DRD2 = dopamine D2 receptor, DRD4 = dopamine D4 receptor, DRD5 = dopamine D5 receptor, 5HTT = serotonin transporter,
significant in males only,
the G × E interaction was significant for DRD5 and oppositional defiant disorder,
the interaction was significant interaction for DAT1 and conduct disorder symptoms and for DRD5 and ODD,
marginally significant,
combined genetic risk.
These initial studies of G × E interactions and ADHD are intriguing. However, these results must also be interpreted with caution because most studies did not control for socioeconomic risk factors that may be correlated with prenatal smoking or alcohol use, and many did not test whether the results were explained by comorbid internalizing and externalizing disorders.
RD
Although no studies of RD or MD have tested for interactions between candidate genes and environmental risk factors, several studies have used twin data to test whether the heritability of RD or other cognitive phenotypes vary as a function of specific environmental variables. In contrast to studies of ADHD and other psychopathology, studies of cognitive abilities have typically found bioecological G × E interactions33;95;96. In a bioecological interaction, genetic influences are expressed most strongly in enriched environments due to the lesser impact of environmental risk factors, whereas genetic influences account for less phenotypic variance in high-risk environments due to increased environmental variance97. Recent analyses of the CLDRC and a sample of older adults indicate that the heritability of RD is significantly higher in families with high parental education than families with low parental education, and this result remained significant in the CLDRC even after controlling for potential genetic influences on parental education98.
Conclusions
Few studies have tested G × E interactions for RD or MD, and many G × E interactions for ADHD await independent replication. Nonetheless, initial results for both RD and ADHD suggest that G × E interactions may play an important role in the etiology of developmental disorders. If the effect of genetic risk factors is moderated by environmental influences, this may help to explain the small effect sizes and inconsistent replication in candidate gene studies despite the high heritability estimates for each disorder.
Diagnostic heterogeneity
Etiologically-informative methods also provide a powerful tool for studies that attempt to dissect the marked heterogeneity that characterizes ADHD, learning disabilities, and many other developmental disorders. To illustrate this approach, we examined concordance rates for the DSM-IV ADHD subtypes in MZ and DZ twin pairs. Two key findings would support the validity of the three-subtype model described in DSM-IV. First, if the distinction between the subtypes is valid, the subtypes should “breed true”, such that co-twins tend to meet criteria for the same subtype as the proband. Second, the high heritability of the overall ADHD diagnosis suggests that each subtype should also be strongly influenced by genes. If one of the subtypes is primarily due to environmental influences, it may be better conceptualized as a separate disorder.
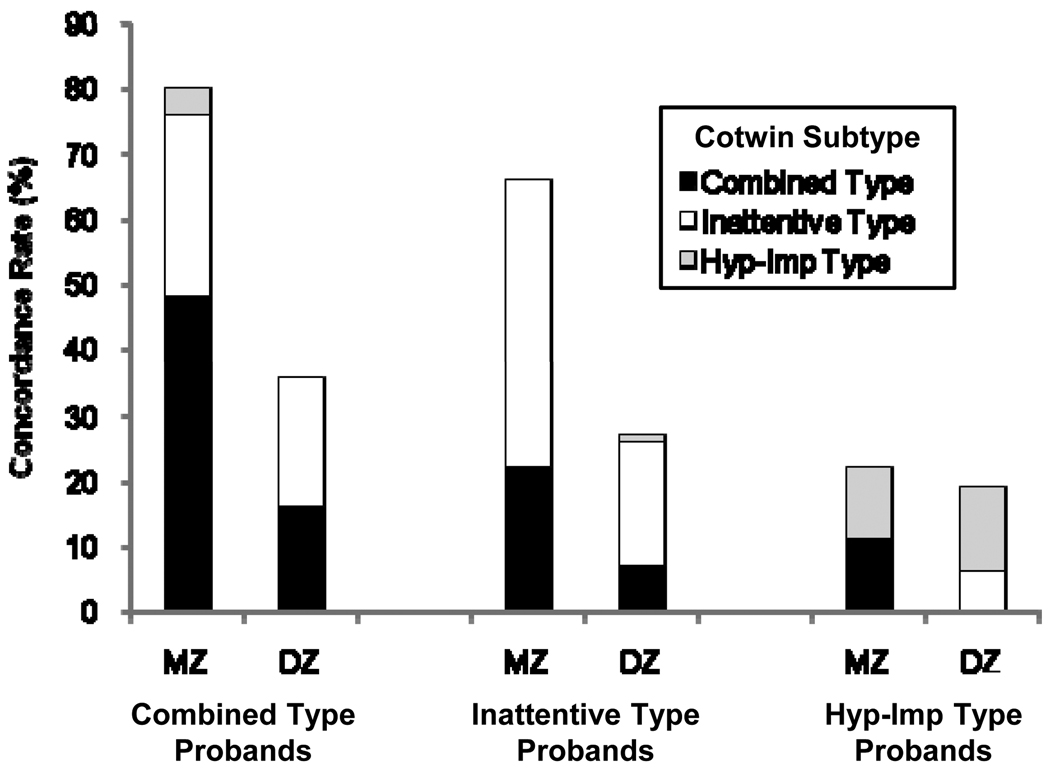
Figure 2 summarizes the ADHD status of co-twins of MZ and DZ probands with each DSM-IV subtype. Whether probands were selected for the combined type or inattentive type, the overall rate of ADHD was significantly higher in MZ co-twins than DZ co-twins. The inattentive and combined subtypes also breed true in twin pairs to some extent. In contrast, probands with the inattentive type also have significantly more co-twins with the combined type than expected by chance, and co-twins of probands with the combined type were equally likely to meet criteria for the combined type or the inattentive type. These results suggest that the subtypes are also influenced by shared genetic influences, a finding that is consistent with the similarity of the academic and neuropsychological profiles of the inattentive and combined types in the CLDRC3;99;100, and with similar results reported for the subtypes in most candidate gene studies and nearly all treatment studies (reviewed by Willcutt, Nigg, et al., under review).
Figure 2.
Rates of DSM-IV ADHD subtypes in the co-twins of MZ and DZ probands who meet criteria for the inattentive type, combined type, or hyperactive-impulsive type.
Results for the hyperactive-impulsive type are quite different. MZ and DZ concordance rates for the hyperactive-impulsive type are nearly identical, suggesting that this subtype is minimally familial and is not significantly heritable. Furthermore, although our results from the CLDRC should be interpreted with caution due to the small sample with the hyperactive-impulsive type, a meta-analysis of 14 studies of the dopamine D5 receptor gene101 and a genome scan of a large sample of affected sibling pairs with DSM-IV ADHD102 both reported that evidence for association and linkage became stronger when probands with the hyperactive-impulsive type were excluded from analyses. Taken together, these results add to a growing literature that challenges the validity of the hyperactive-impulsive type after preschool103.
Etiology of comorbidity
In addition to significant heterogeneity within each disorder, nearly all developmental disorders co-occur with other disorders more frequently than expected by chance. As one of the primary aims of the CLDRC we have used bivariate extensions of DF analysis38 to test the etiology of the significant comorbidity between RD and MD (28–64%)104–107, RD and ADHD (10–40%)44;108;109, and MD and ADHD (12–36%)21;110. Rather than comparing the relative similarity of MZ and DZ twins on the same trait, the bivariate model compares the relation between the proband’s score on the selected trait and the co-twin’s score on a second, unselected trait. If common genetic influences contribute to the association between the two traits, the MZ co-twin score on the unselected trait should regress less than the DZ co-twin score toward the population mean . This differential regression is used to estimate bivariate h2g, an index of the extent to which the proband deficit on the selected measure is due to genetic influences that also contribute to deficits on the unselected measure.
Previous bivariate DF analyses have consistently shown that common genetic influences account for comorbidity between RD and ADHD, with stronger shared genetic influences for inattention symptoms than hyperactivity-impulsivity symptoms15;38;44;111–113. Because few previous studies have examined comorbidity between RD and MD and no studies have tested the etiology of comorbidity between ADHD and MD, we fitted bivariate DF models to test the etiology of these comorbidities (Table 4). These results suggest that comorbidity between RD and MD and between ADHD and MD is also primarily explained by common genetic influences. Similar to our previous results for reading and ADHD, shared genetic influences between ADHD and MD are strongest for inattention symptoms.
Table 4.
Bivariate heritability with math in twin pairs selected for reading or ADHD symptoms
| MZ pairsa |
DZ pairsa |
||||||
|---|---|---|---|---|---|---|---|
| Proband | Co-twin Math |
Proband | Co-twin Math |
Bivariate | |||
| Selected variable | Nb | M (SD) | M (SD) | Nb | M (SD) | M (SD) | h2g (SE) |
| Reading | 106 | −2.00 (0.68) | −1.03 (1.01) | 89 | −1.99 (0.72) | −0.63 (0.99) | 0.40 (0.15)** |
| Inattention | 98 | −2.03 (0.58) | −0.75 (1.13) | 83 | −2.07 (0.59) | −0.28 (1.11) | 0.47 (0.15)** |
| Hyperactivity - impulsivity | 85 | −2.17 (0.57) | −0.47 (1.18) | 65 | −2.09 (0.58) | −0.18 (1.21) | 0.26 (0.16) |
Note.
P < .01.
Scores are expressed as standard deviations from the estimated population mean. All scores are scaled so that lower scores indicate greater impairment on all measures.
Total number of pairs in which at least one twin met the criteria for the proband group (score at least 1.25 SD below the population mean).
Incorporating neuropsychological measures
The inclusion of neuropsychological measures in etiologically-informative analyses provides another useful tool to dissect the complex etiology and neuropsychology of developmental disorders. We recently compared groups with RD, MD, and ADHD in the CLDRC on composite measures of verbal reasoning, naming speed, processing speed, response inhibition, working memory, and phoneme awareness. Consistent with the results of our recent meta-analysis of neuropsychological studies of nine childhood disorders114, groups with RD, MD, and ADHD exhibited significant weaknesses on all six cognitive composites3;99;115;116. These results suggest that rather than unique neuropsychological deficits that are specific to each disorder, RD, MD, and ADHD may be distinguished by more subtle differences in the specific profile or severity of neuropsychological weaknesses across domains that are impaired to some extent in all three disorders.
Phenotypic structural equation models were then used to test which of these neuropsychological weaknesses were independently associated with each disorder when the other cognitive measures were also included in the model (McGrath et al., under review and 116), and multivariate twin analyses were used to test the etiology of any significant associations115;117. Results indicated that RD was independently associated with weaknesses in phonological decoding, verbal reasoning, working memory, naming speed, and processing speed, and MD is independently associated with each of these weaknesses with the exception of naming speed. In contrast, the only neuropsychological measures that independently predicted ADHD were weak response inhibition and slow processing speed. Individual differences on each of the neuropsychological composite scores were significantly heritable, and multivariate twin analyses suggested that comorbidity between ADHD, RD, and MD is due primarily to common genetic influences that lead to slow processing speed13;29;31;38;115–118.
In summary, the neuropsychological correlates of RD, MD, and ADHD are complex and multifactorial, consistent with the findings from molecular genetic studies. The heritability of the neuropsychological measures and their strong relationship with the three disorders suggests that these and other measures of brain functioning may be useful for future studies of the etiology of RD, MD, ADHD, and their comorbidity.
CONCLUSIONS AND FUTURE DIRECTIONS
Family and twin studies clearly show that RD, MD, and ADHD are familial and heritable. Although much more research is needed on the molecular genetic etiology of the three disorders, initial results suggest that each disorder is caused by the additive or interactive effects of multiple genetic and environmental risk factors, each of which may have a relative small effect in isolation. In the remainder of this section we summarize the clinical implications of these results and highlight the need for increased interdisciplinary research in the next generation of studies of the etiology of these disorders.
Clinical implications
There is currently no valid genetic test for RD, MD, or ADHD, and it is unlikely that a definitive diagnostic test will be developed in the near future. Because most developmental disorders have polygenic, multifactorial etiologies in which each risk factor confers only a small increase in susceptibility, it is unlikely that any specific risk factor will have sufficient predictive power to be useful as a diagnostic measure.
Even if behavioral and molecular genetic studies do not identify a definitive genetic test for each disorder, these methods may still have important clinical benefits in the future. It may eventually be possible to develop probabilistic risk profiles based on an individual’s genetic background, family history, environmental circumstances, and other factors. These profiles could be used to identify individuals who are at higher risk for a specific disorder, facilitating primary prevention or early intervention. For example, if a perinatal screening revealed significant susceptibility to RD, early interventions could be provided to reduce the probability that child will develop RD. Similarly, by providing a better understanding of the underlying pathophysiology of ADHD, molecular genetic techniques may inform the development of tertiary pharmacological or psychosocial treatments that directly target the compromised physiological and psychological mechanisms.
Expansion of collaborative research
Multisite molecular genetic networks
One of the most important implications of initial molecular genetic studies of RD, MD, ADHD, and most other complex disorders is that current studies are severely underpowered119. Due to small effect sizes and etiological heterogeneity, procedures for gene localization are likely to require extremely large samples (5,000 – 10,000 individuals or more) that are simply not feasible for a single laboratory to collect in isolation. Fortunately, procedures for DNA collection and genetic analysis continue to become more automated and efficient, and it is now relatively inexpensive for studies without a primary focus on genetics to collect and store DNA for use in future collaborative genetic analyses. A network for collaborative molecular genetic studies already exists for ADHD120, and the initiation of similar networks for RD and MD would provide a useful way to accelerate the progress of the field.
Interdisciplinary research
In conclusion, it is increasingly clear that future progress in understanding the complex etiologies of developmental disorders such as RD, MD, and ADHD is likely to require interdisciplinary research that integrates behavioral and molecular genetic techniques with state-of-the-art clinical, developmental, and cognitive methods. The infrastructure provided by the NICHD Center grant that supports the CLDRC has provided a unique opportunity for innovative interdisciplinary research that would have been difficult or impossible for any of our laboratories to initiate alone. Furthermore, the collaborative synergy within the CLDRC has facilitated the development of important collaborations with a number of groups outside the CLDRC, including molecular genetic studies of RD63;121 and ADHD122, studies of neuropsychological heterogeneity and neurocognitive phenotypes that may be useful for molecular genetic studies of ADHD123;124, and the largest study of the etiology of high intelligence that has been conducted to date125. We hope that these examples and the others described in this paper may stimulate the development of additional innovative collaborations among a larger network of investigators in the field.
Acknowledgments
Sources of support:
Primary funding for this research was provided by NICHD Center grant P50 HD 27802. The authors were also supported by National Institutes of Health grants R01 HD 47264, R01 MH 63207, R01 HD 38526, P50 MH 79485, R01 MH 70037, R01 DA 24002, R01 DC 05190, R0 HD 49027, and T32 HD 07289.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pennington BF. From single to multiple deficit models of developmental disorders. Cognition. 2006;101:385–413. doi: 10.1016/j.cognition.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 2.DeFries JC, Filipek PA, Fulker DW, et al. Colorado Learning Disabilities Research Center. Learning Disabilities: A Multidisciplinary Journal. 1997;8:7–19. [Google Scholar]
- 3.Willcutt EG, Pennington BF, Olson RK, et al. Neuropsychological analyses of comorbidity between reading disability and attention deficit hyperactivity disorder: in search of the common deficit. Dev Neuropsychol. 2005;27:35–78. doi: 10.1207/s15326942dn2701_3. [DOI] [PubMed] [Google Scholar]
- 4.DeFries JC. Colorado reading project. In: Gray DB, Kavanagh JF, editors. Biobehavioral Measures of Dyslexia. Parkton,MD: York Press; 1985. pp. 107–122. [Google Scholar]
- 5.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 6.Fletcher JM, Francis DJ, Rourke BP, et al. The validity of discrepancy-based definitions of reading disabilities. J Learn Disabil. 1992;25:555–561. doi: 10.1177/002221949202500903. 573. [DOI] [PubMed] [Google Scholar]
- 7.Pennington BF, Gilger JW, Olson RK, et al. The external validity of age- versus IQ-discrepancy definitions of reading disability: lessons from a twin study. J Learn Disabil. 1992;25:562–573. doi: 10.1177/002221949202500904. [DOI] [PubMed] [Google Scholar]
- 8.Dunn LM, Markwardt FC. Examiner's Manual: Peabody Individual Achievement Test. Circle Pines, MN: American Guidance Service; 1970. [Google Scholar]
- 9.Olson RK, Forsberg H, Wise B, et al. Measurement of word recognition, orthographic, and phonological skills. In: Lyon GR, editor. Frames of reference for the assessment of learning disabilities: New views on measurement issues. Baltimore, MD: Paul H. Brookes Publishing Company; 1994. pp. 243–277. [Google Scholar]
- 10.Jastak S, Wilkinson GS. Wide Range Achievement Test, Revised: Administration Manual. Wilmington, DE: 1984. [Google Scholar]
- 11.Barkley RA, Murphy K. Attention-deficit hyperactivity disorder: A clinical workbook. New York, NY: Guilford Press; 1998. [Google Scholar]
- 12.Lahey BB, Applegate B, McBurnett K, et al. DSM-IV field trials for attention deficit hyperactivity disorder in children and adolescents. Am J Psychiatry. 1994;151:1673–1685. doi: 10.1176/ajp.151.11.1673. [DOI] [PubMed] [Google Scholar]
- 13.Petrill SA, Deater-Deckard K, Thompson LA, et al. Longitudinal genetic analysis of early reading: The Western Reserve Reading Project. Reading and Writing. 2007;20:127–146. doi: 10.1007/s11145-006-9021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuntsi J, Eley TC, Taylor A, et al. Co-occurrence of ADHD and low IQ has genetic origins. Am J Med Genet B Neuropsychiatr Genet. 2004;124B:41–47. doi: 10.1002/ajmg.b.20076. [DOI] [PubMed] [Google Scholar]
- 15.Willcutt EG, Betjemann RS, Wadsworth SJ, et al. Preschool twin study of the relation between attention-deficit/hyperactivity disorder and prereading skills. Reading and Writing. 2007;20:103–125. [Google Scholar]
- 16.Kovas Y, Haworth CM, Petrill SA, et al. Mathematical ability of 10-year-old boys and girls: genetic and environmental etiology of typical and low performance. J Learn Disabil. 2007;40:554–567. doi: 10.1177/00222194070400060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeFries JC, Singer SM, Foch TT, et al. Familial nature of reading disability. Br J Psychiatry. 1978;132:361–367. doi: 10.1192/bjp.132.4.361. [DOI] [PubMed] [Google Scholar]
- 18.Pennington BF, Lefly DL. Early reading development in children at family risk for dyslexia. Child Dev. 2001;72:816–833. doi: 10.1111/1467-8624.00317. [DOI] [PubMed] [Google Scholar]
- 19.Faraone SV, Biederman J, Mick E, et al. Family study of girls with attention deficit hyperactivity disorder. Am J Psychiatry. 2000;157:1077–1083. doi: 10.1176/appi.ajp.157.7.1077. [DOI] [PubMed] [Google Scholar]
- 20.Friedman MC, Chhabildas N, Budhiraja N, et al. Etiology of the comorbidity between RD and ADHD: exploration of the non-random mating hypothesis. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:109–115. doi: 10.1002/ajmg.b.20029. [DOI] [PubMed] [Google Scholar]
- 21.Monuteaux MC, Faraone SV, Herzig K, et al. ADHD and dyscalculia: Evidence for independent familial transmission. J Learn Disabil. 2005;38:86–93. doi: 10.1177/00222194050380010701. [DOI] [PubMed] [Google Scholar]
- 22.Shalev RS, Manor O, Kerem B, et al. Developmental dyscalculia is a familial learning disability. J Learn Disabil. 2001;34:59–65. doi: 10.1177/002221940103400105. [DOI] [PubMed] [Google Scholar]
- 23.Plomin R, DeFries JC, McClearn GE, et al. Behavioral Genetics. New York: Worth Publishers; 2008. [Google Scholar]
- 24.Stevenson J, Graham P, Fredman G, et al. A twin study of genetic influences on reading and spelling ability and disability. J Child Psychol Psychiatry. 1987;28:229–247. doi: 10.1111/j.1469-7610.1987.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 25.Willcutt EG, Pennington BF, DeFries JC. Etiology of inattention and hyperactivity/impulsivity in a community sample of twins with learning difficulties. J Abnorm Child Psychol. 2000;28:149–159. doi: 10.1023/a:1005170730653. [DOI] [PubMed] [Google Scholar]
- 26.DeFries JC, Fulker DW. Multiple regression analysis of twin data. Behav Genet. 1985;15:467–473. doi: 10.1007/BF01066239. [DOI] [PubMed] [Google Scholar]
- 27.DeFries JC, Fulker DW. Multiple regression analysis of twin data: etiology of deviant scores versus individual differences. Acta Genet Med Gemellol (Roma ) 1988;37:205–216. doi: 10.1017/s0001566000003810. [DOI] [PubMed] [Google Scholar]
- 28.Neale MC, Boker SM, Xie G, et al. Mx: Statistical Modeling. Richmond, VA: Virginia Commonwealth University; 2002. [Google Scholar]
- 29.Betjemann RS, Willcutt EG, Olson RK, et al. Word reading and reading comprehension: stability, overlap and independence. Reading and Writing. 2008;21:539–558. [Google Scholar]
- 30.Byrne B, Samuelsson S, Wadsworth SJ, et al. Longitudinal twin study of early literacy development: preschool through grade 1. Reading and Writing. 2007;20:77–102. [Google Scholar]
- 31.Gayán J, Olson RK. Genetic and environmental influences on individual differences in printed word recognition. J Exp Child Psychol. 2003;84:97–123. doi: 10.1016/s0022-0965(02)00181-9. [DOI] [PubMed] [Google Scholar]
- 32.Harlaar N, Spinath FM, Dale PS, et al. Genetic influences on early word recognition abilities and disabilities: a study of 7-year-old twins. J Child Psychol Psychiatry. 2005;46:373–384. doi: 10.1111/j.1469-7610.2004.00358.x. [DOI] [PubMed] [Google Scholar]
- 33.Kremen WS, Jacobson KC, Xian H, et al. Heritability of word recognition in middle-aged men varies as a function of parental education. Behav Genet. 2005;35:417–433. doi: 10.1007/s10519-004-3876-2. [DOI] [PubMed] [Google Scholar]
- 34.Hay DA, Bennett KS, Levy F, et al. A twin study of attention-deficit/hyperactivity disorder dimensions rated by the strengths and weaknesses of ADHD-symptoms and normal-behavior (SWAN) scale. Biol Psychiatry. 2007;61:700–705. doi: 10.1016/j.biopsych.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 35.Larsson H, Lichtenstein P, Larsson JO. Genetic contributions to the development of ADHD subtypes from childhood to adolescence. J Am Acad Child Adolesc Psychiatry. 2006;45:973–981. doi: 10.1097/01.chi.0000222787.57100.d8. [DOI] [PubMed] [Google Scholar]
- 36.McLoughlin G, Ronald A, Kuntsi J, et al. Genetic support for the dual nature of attention deficit hyperactivity disorder: substantial genetic overlap between the inattentive and hyperactive-impulsive components. J Abnorm Child Psychol. 2007;35:999–1008. doi: 10.1007/s10802-007-9149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman DK, Iacono WG, McGue MK. Attention-deficit hyperactivity disorder dimensions: a twin study of inattention and impulsivity-hyperactivity. J Am Acad Child Adolesc Psychiatry. 1997;36:745–753. doi: 10.1097/00004583-199706000-00010. [DOI] [PubMed] [Google Scholar]
- 38.Willcutt EG, Pennington BF, Olson RK, et al. Understanding comorbidity: a twin study of reading disability and attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:709–714. doi: 10.1002/ajmg.b.30310. [DOI] [PubMed] [Google Scholar]
- 39.Kuntsi J, Stevenson J. Psychological mechanisms in hyperactivity: II. The role of genetic factors. J Child Psychol Psychiatry. 2001;42:211–219. [PubMed] [Google Scholar]
- 40.Stevenson J. Evidence for a genetic etiology in hyperactivity in children. Behav Genet. 1992;22:337–344. doi: 10.1007/BF01066665. [DOI] [PubMed] [Google Scholar]
- 41.Thapar A, Hervas A, McGuffin P. Childhood hyperactivity scores are highly heritable and show sibling competition effects: twin study evidence. Behav Genet. 1995;25:537–544. doi: 10.1007/BF02327577. [DOI] [PubMed] [Google Scholar]
- 42.Wainwright M, Wright MJ, Geffen GM, et al. Genetic and environmental sources of covariance between reading tests used in neuropsychological assessment and IQ subtests. Behav Genet. 2004;34:365–376. doi: 10.1023/B:BEGE.0000023642.34853.cb. [DOI] [PubMed] [Google Scholar]
- 43.Whitehouse AJ, Spector TD, Cherkas LF. No clear genetic influences on the association between dyslexia and anxiety in a population-based sample of female twins. Dyslexia. 2009;15:282–290. doi: 10.1002/dys.378. [DOI] [PubMed] [Google Scholar]
- 44.Trzesniewski KH, Moffitt TE, Caspi A, et al. Revisiting the association between reading achievement and antisocial behavior: new evidence of an environmental explanation from a twin study. Child Dev. 2006;77:72–88. doi: 10.1111/j.1467-8624.2006.00857.x. [DOI] [PubMed] [Google Scholar]
- 45.Bates TC, Castles A, Coltheart M, et al. Behaviour genetic analyses of reading and spelling: A component processes approach. Australian Journal of Psychology. 2004;56:115–126. [Google Scholar]
- 46.Hohnen B, Stevenson J. The structure of genetic influences on general cognitive, language, phonological, and reading abilities. Dev Psychol. 1999;35:590–603. doi: 10.1037//0012-1649.35.2.590. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds CA, Hewitt JK, Erickson MT, et al. The genetics of children's oral reading performance. Journal of Child Psychology and Psychiatry and Allied Disciplines. 1996;37:425–434. doi: 10.1111/j.1469-7610.1996.tb01423.x. [DOI] [PubMed] [Google Scholar]
- 48.Lytton H, Watts D, Dunn BE. Stability of genetic determination from age 2 to 9: a longitudinal twin study. Social Biology. 1988;35:62–73. doi: 10.1080/19485565.1988.9988688. [DOI] [PubMed] [Google Scholar]
- 49.Levy F, Hay DA, McStephen M, et al. Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry. 1997;36:737–744. doi: 10.1097/00004583-199706000-00009. [DOI] [PubMed] [Google Scholar]
- 50.Eley TC, Rijsdijk F. Introductory guide to the statistics of molecular genetics. J Child Psychol Psychiatry. 2005;46:1042–1044. doi: 10.1111/j.1469-7610.2005.01523.x. [DOI] [PubMed] [Google Scholar]
- 51.Eley TC, Craig IW. Introductory guide to the language of molecular genetics. J Child Psychol Psychiatry. 2005;46:1039–1041. doi: 10.1111/j.1469-7610.2005.01522.x. [DOI] [PubMed] [Google Scholar]
- 52.Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009;126:51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- 53.Cook EH, Jr, Stein MA, Krasowski MD, et al. Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet. 1995;56:993–998. [PMC free article] [PubMed] [Google Scholar]
- 54.Lasky-Su J, Neale BM, Franke B, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 55.Neale BM, Lasky-Su J, Anney R, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou K, Dempfle A, Arcos-Burgos M, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith SD, Gilger JW. Dyslexia and Other Specific Learning Disorders. In: Rimoin DL, Conner JM, Pyeritz RE, et al., editors. Emery and Rimoin's Principles and Practice of Medical Genetics. 4th ed. New York, NY: Elsevier; 2007. pp. 2548–2568. [Google Scholar]
- 58.Cope N, Harold D, Hill G, et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anthoni H, Zucchelli M, Matsson H, et al. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet. 2007;16:667–677. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- 61.Couto JM, Gomez L, Wigg K, et al. The KIAA0319-like (KIAA0319L) gene on chromosome 1p34 as a candidate for reading disabilities. J Neurogenet. 2008;22:295–313. doi: 10.1080/01677060802354328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paracchini S, Thomas A, Castro S, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 63.Meng H, Smith SD, Hager K, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taipale M, Kaminen N, Nopola-Hemmi J, et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci U S A. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schumacher J, Anthoni H, Dahdouh F, et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78:52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Docherty SJ, Davis OS, Kovas Y, et al. A genome-wide association study identifies multiple loci associated with mathematics ability and disability. Genes Brain Behav. 2009 doi: 10.1111/j.1601-183X.2009.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Roak BJ, State MW. Autism genetics: strategies, challenges, and opportunities. Autism Res. 2008;1:4–17. doi: 10.1002/aur.3. [DOI] [PubMed] [Google Scholar]
- 69.Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–343. doi: 10.1016/j.neuroscience.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 71.Fanous AH, Kendler KS. Genetics of clinical features and subtypes of schizophrenia: a review of the recent literature. Curr Psychiatry Rep. 2008;10:164–170. doi: 10.1007/s11920-008-0028-z. [DOI] [PubMed] [Google Scholar]
- 72.Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 73.Rutter M, Moffitt TE, Caspi A. Gene-environment interplay and psychopathology: multiple varieties but real effects. J Child Psychol Psychiatry. 2006;47:226–261. doi: 10.1111/j.1469-7610.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- 74.Caspi A, McClay J, Moffitt TE, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 75.Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 76.Eaves LJ. Genotype × environment interaction in psychopathology: Fact or artifact? Twin Res Hum Genet. 2006;9:1–8. doi: 10.1375/183242706776403073. [DOI] [PubMed] [Google Scholar]
- 77.Risch Interaction Between the Serotonin Transporter Gene (5-HTTLPR), Stressful Life Events, and Risk of Depression: A Meta-analysis (vol 301, pg 2462, 2009) Jama-Journal of the American Medical Association. 2009;302:492. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychol Med. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- 79.Valdar W, Solberg LC, Gauguier D, et al. Genetic and environmental effects on complex traits in mice. Genetics. 2006;174:959–984. doi: 10.1534/genetics.106.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pennington BF, McGrath LM, Rosenberg J, et al. Gene × environment interactions in reading disability and attention-deficit/hyperactivity disorder. Dev Psychol. 2009;45:77–89. doi: 10.1037/a0014549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lasky-Su J, Faraone SV, Lange C, et al. A study of how socioeconomic status moderates the relationship between SNPs encompassing BDNF and ADHD symptom counts in ADHD families. Behav Genet. 2007;37:487–497. doi: 10.1007/s10519-006-9136-x. [DOI] [PubMed] [Google Scholar]
- 82.Laucht M, Skowronek MH, Becker K, et al. Interacting effects of the dopamine transporter gene and psychosocial adversity on attention-deficit/hyperactivity disorder symptoms among 15-year-olds from a high-risk community sample. Arch Gen Psychiatry. 2007;64:585–590. doi: 10.1001/archpsyc.64.5.585. [DOI] [PubMed] [Google Scholar]
- 83.Nigg J, Nikolas M, Friderici K, et al. Genotype and neuropsychological response inhibition as resilience promoters for attention-deficit/hyperactivity disorder, oppositional defiant disorder, and conduct disorder under conditions of psychosocial adversity. Dev Psychopathol. 2007;19:767–786. doi: 10.1017/S0954579407000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Retz W, Freitag CM, Retz-Junginger P, et al. A functional serotonin transporter promoter gene polymorphism increases ADHD symptoms in delinquents: interaction with adverse childhood environment. Psychiatry Res. 2008;158:123–131. doi: 10.1016/j.psychres.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 85.Waldman ID. Gene-environment interactions reexamined: does mother's marital stability interact with the dopamine receptor D2 gene in the etiology of childhood attention-deficit/hyperactivity disorder? Dev Psychopathol. 2007;19:1117–1128. doi: 10.1017/S0954579407000570. [DOI] [PubMed] [Google Scholar]
- 86.Brookes KJ, Neale B, Xu X, et al. Differential dopamine receptor D4 allele association with ADHD dependent of proband season of birth. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:94–99. doi: 10.1002/ajmg.b.30562. [DOI] [PubMed] [Google Scholar]
- 87.Seeger G, Schloss P, Schmidt MH, et al. Gene-environment interaction in hyperkinetic conduct disorder (HD + CD) as indicated by season of birth variations in dopamine receptor (DRD4) gene polymorphism. Neurosci Lett. 2004;366:282–286. doi: 10.1016/j.neulet.2004.05.049. [DOI] [PubMed] [Google Scholar]
- 88.Becker K, El-Faddagh M, Schmidt MH, et al. Interaction of dopamine transporter genotype with prenatal smoke exposure on ADHD symptoms. J Pediatr. 2008;152:263–269. doi: 10.1016/j.jpeds.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 89.Kahn RS, Khoury J, Nichols WC, et al. Role of dopamine transporter genotype and maternal prenatal smoking in childhood hyperactive-impulsive, inattentive, and oppositional behaviors. J Pediatr. 2003;143:104–110. doi: 10.1016/S0022-3476(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 90.Langley K, Turic D, Rice F, et al. Testing for gene × environment interaction effects in attention deficit hyperactivity disorder and associated antisocial behavior. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:49–53. doi: 10.1002/ajmg.b.30571. [DOI] [PubMed] [Google Scholar]
- 91.Neuman RJ, Lobos E, Reich W, et al. Prenatal smoking exposure and dopaminergic genotypes interact to cause a severe ADHD subtype. Biol Psychiatry. 2007;61:1320–1328. doi: 10.1016/j.biopsych.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 92.Todd RD, Neuman RJ. Gene-environment interactions in the development of combined type ADHD: evidence for a synapse-based model. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:971–975. doi: 10.1002/ajmg.b.30640. [DOI] [PubMed] [Google Scholar]
- 93.Brookes KJ, Mill J, Guindalini C, et al. A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use of alcohol during pregnancy. Arch Gen Psychiatry. 2006;63:74–81. doi: 10.1001/archpsyc.63.1.74. [DOI] [PubMed] [Google Scholar]
- 94.Nigg JT, Breslau N. Prenatal smoking exposure, low birth weight, and disruptive behavior disorders. J Am Acad Child Adolesc Psychiatry. 2007;46:362–369. doi: 10.1097/01.chi.0000246054.76167.44. [DOI] [PubMed] [Google Scholar]
- 95.Harden KP, Turkheimer E, Loehlin JC. Genotype by environment interaction in adolescents' cognitive aptitude. Behav Genet. 2007;37:273–283. doi: 10.1007/s10519-006-9113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turkheimer E, Haley A, Waldron M, et al. Socioeconomic status modifies heritability of IQ in young children. Psychol Sci. 2003;14:623–628. doi: 10.1046/j.0956-7976.2003.psci_1475.x. [DOI] [PubMed] [Google Scholar]
- 97.Bronfenbrenner U, Ceci SJ. Nature-nurture reconceptualized in developmental perspective: a bioecological model. Psychol Rev. 1994;101:568–586. doi: 10.1037/0033-295x.101.4.568. [DOI] [PubMed] [Google Scholar]
- 98.Friend A, DeFries JC, Olson RK. Parental education moderates genetic influences on reading disability. Psychol Sci. 2008;19:1124–1130. doi: 10.1111/j.1467-9280.2008.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chhabildas N, Pennington BF, Willcutt EG. A comparison of the neuropsychological profiles of the DSM-IV subtypes of ADHD. J Abnorm Child Psychol. 2001;29:529–540. doi: 10.1023/a:1012281226028. [DOI] [PubMed] [Google Scholar]
- 100.Solanto MV, Gilbert SN, Raj A, et al. Neurocognitive functioning in AD/HD, predominantly inattentive and combined subtypes. J Abnorm Child Psychol. 2007;35:729–744. doi: 10.1007/s10802-007-9123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lowe N, Kirley A, Hawi Z, et al. Joint analysis of the DRD5 marker concludes association with attention-deficit/hyperactivity disorder confined to the predominantly inattentive and combined subtypes. Am J Hum Genet. 2004;74:348–356. doi: 10.1086/381561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smalley SL, Kustanovich V, Minassian SL, et al. Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am J Hum Genet. 2002;71:959–963. doi: 10.1086/342732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Willcutt EG, Carlson CL. Diagnostic validity of attention-deficit/hyperactivity disorder. Clinical Neuroscience Review. 2005;5:219–232. [Google Scholar]
- 104.Gross-Tsur V, Manor O, Shalev RS. Developmental dyscalculia: prevalence and demographic features. Dev Med Child Neurol. 1996;38:25–33. doi: 10.1111/j.1469-8749.1996.tb15029.x. [DOI] [PubMed] [Google Scholar]
- 105.Badian NA, Ghublikian M. The personal-social characteristics of children with poor mathematical computation skills. J Learn Disabil. 1983;16:154–157. doi: 10.1177/002221948301600304. [DOI] [PubMed] [Google Scholar]
- 106.Knopik VS, Alarcón M, DeFries JC. Comorbidity of mathematics and reading deficits: evidence for a genetic etiology. Behav Genet. 1997;27:447–453. doi: 10.1023/a:1025622400239. [DOI] [PubMed] [Google Scholar]
- 107.Kovas Y, Haworth CM, Harlaar N, et al. Overlap and specificity of genetic and environmental influences on mathematics and reading disability in 10-year-old twins. J Child Psychol Psychiatry. 2007;48:914–922. doi: 10.1111/j.1469-7610.2007.01748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Willcutt EG, Pennington BF. Comorbidity of reading disability and attention-deficit/hyperactivity disorder: differences by gender and subtype. J Learn Disabil. 2000;33:179–191. doi: 10.1177/002221940003300206. [DOI] [PubMed] [Google Scholar]
- 109.Rucklidge JJ, Tannock R. Neuropsychological profiles of adolescents with ADHD: effects of reading difficulties and gender. J Child Psychol Psychiatry. 2002;43:988–1003. doi: 10.1111/1469-7610.00227. [DOI] [PubMed] [Google Scholar]
- 110.Capano L, Minden D, Chen SX, et al. Mathematical learning disorder in school-age children with attention-deficit hyperactivity disorder. Can J Psychiatry. 2008;53:392–399. doi: 10.1177/070674370805300609. [DOI] [PubMed] [Google Scholar]
- 111.Stevenson J, Pennington BF, Gilger JW, et al. Hyperactivity and spelling disability: testing for shared genetic aetiology. J Child Psychol Psychiatry. 1993;34:1137–1152. doi: 10.1111/j.1469-7610.1993.tb01779.x. [DOI] [PubMed] [Google Scholar]
- 112.Light JG, DeFries JC. Comorbidity of reading and mathematics disabilities: genetic and environmental etiologies. J Learn Disabil. 1995;28:96–106. doi: 10.1177/002221949502800204. [DOI] [PubMed] [Google Scholar]
- 113.Willcutt EG, Pennington BF, DeFries JC. Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. American Journal of Medical Genetics (Neuropsychiatric Genetics) 2000;96:293–301. doi: 10.1002/1096-8628(20000612)96:3<293::aid-ajmg12>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 114.Willcutt EG, Sonuga-Barke EJS, Nigg JT, et al. Recent developments in neuropsychological models of childhood disorders. Advances in Biological Psychiatry. 2008;24:195–226. [Google Scholar]
- 115.Betjemann RS, Willcutt EG, McGrath LM, et al. Etiology of comorbidity between reading disability, math disability, and attention-deficit/hyperactivity disorder. 2009 June [Google Scholar]
- 116.Willcutt EG, Betjemann RS, Bidwell LC, et al. Etiology and neuropsychology of comorbidity between ADHD and Learning Disorders. 2009 June 1; [Google Scholar]
- 117.Willcutt EG, Betjemann RS, McGrath LM, et al. Etiology and neuropsychology of comorbidity between RD and ADHD: The case for multiple-deficit models. 2010 doi: 10.1016/j.cortex.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gayán J, Olson RK. Genetic and environmental influences on orthographic and phonological skills in children with reading disabilities. Dev Neuropsychol. 2001;20:483–507. doi: 10.1207/S15326942DN2002_3. [DOI] [PubMed] [Google Scholar]
- 119.Psychiatric GWAS Consortium Coordinating Committee. Genomewide association studies: History, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Faraone SV. Report from the 4th international meeting of the attention deficit hyperactivity disorder molecular genetics network. Am J Med Genet B Neuropsychiatr Genet. 2003;121B:55–59. doi: 10.1002/ajmg.b.20047. [DOI] [PubMed] [Google Scholar]
- 121.Meng H, Hager K, Held M, et al. TDT-association analysis of EKN1 and dyslexia in a Colorado twin cohort. Hum Genet. 2005;118:87–90. doi: 10.1007/s00439-005-0017-9. [DOI] [PubMed] [Google Scholar]
- 122.Todd RD, Huang H, Smalley SL, et al. Collaborative analysis of DRD4 and DAT genotypes in population-defined ADHD subtypes. J Child Psychol Psychiatry. 2005;46:1067–1073. doi: 10.1111/j.1469-7610.2005.01517.x. [DOI] [PubMed] [Google Scholar]
- 123.Doyle AE, Willcutt EG, Seidman LJ, et al. Attention-deficit/hyperactivity disorder endophenotypes. Biol Psychiatry. 2005;57:1324–1335. doi: 10.1016/j.biopsych.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 124.Nigg JT, Willcutt EG, Doyle AE, et al. Causal heterogeneity in attention-deficit/hyperactivity disorder: do we need neuropsychologically impaired subtypes? Biol Psychiatry. 2005;57:1224–1230. doi: 10.1016/j.biopsych.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 125.Haworth CM, Wright MJ, Martin NW, et al. A twin study of the genetics of high cognitive ability selected from 11,000 twin pairs in six studies from four countries. Behav Genet. 2009;39:359–370. doi: 10.1007/s10519-009-9262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bakwin H. Reading disability in twins. Dev Med Child Neurol. 1973;15:184–187. doi: 10.1111/j.1469-8749.1973.tb15158.x. [DOI] [PubMed] [Google Scholar]
- 127.Hawke JL, Wadsworth SJ, DeFries JC. Genetic influences on reading difficulties in boys and girls: the Colorado twin study. Dyslexia. 2006;12:21–29. doi: 10.1002/dys.301. [DOI] [PubMed] [Google Scholar]
- 128.Zerbin-Rudin E. Kongenitale Worblindheit oder spezifische dyslixie (Congenital Word-Blindness) Bulletin of the Orton Society. 1967;17:47–56. [Google Scholar]
- 129.Goodman R, Stevenson J. A twin study of hyperactivity--II. The aetiological role of genes, family relationships and perinatal adversity. J Child Psychol Psychiatry. 1989;30:691–709. doi: 10.1111/j.1469-7610.1989.tb00782.x. [DOI] [PubMed] [Google Scholar]
- 130.Levy F, McStephen M, Hay DA. The diagnostic genetics of ADHD symptoms and subtypes. In: Levy F, Hay DA, editors. Attention, Genes, and ADHD. Philadelphia, PA: Taylor & Francis; 2001. pp. 35–57. [Google Scholar]
- 131.Sherman DK, McGue MK, Iacono WG. Twin concordance for attention deficit hyperactivity disorder: a comparison of teachers' and mothers' reports. Am J Psychiatry. 1997;154:532–535. doi: 10.1176/ajp.154.4.532. [DOI] [PubMed] [Google Scholar]
- 132.Thapar A, Harrington R, McGuffin P. Examining the comorbidity of ADHD-related behaviours and conduct problems using a twin study design. Br J Psychiatry. 2001;179:224–229. doi: 10.1192/bjp.179.3.224. [DOI] [PubMed] [Google Scholar]
- 133.Todd RD, Rasmussen ER, Neuman RJ, et al. Familiality and heritability of subtypes of attention deficit hyperactivity disorder in a population sample of adolescent female twins. Am J Psychiatry. 2001;158:1891–1898. doi: 10.1176/appi.ajp.158.11.1891. [DOI] [PubMed] [Google Scholar]
- 134.Alarcón M, DeFries JC, Light JG, et al. A twin study of mathematics disability. J Learn Disabil. 1997;30:617–623. doi: 10.1177/002221949703000605. [DOI] [PubMed] [Google Scholar]
- 135.Nobile M, Rusconi M, Bellina M, et al. COMT Val158Met polymorphism and socioeconomic status interact to predict attention deficit/hyperactivity problems in children aged 10–14. Eur Child Adolesc Psychiatry. 2009 doi: 10.1007/s00787-009-0080-1. [DOI] [PubMed] [Google Scholar]