Abstract
Granulocyte colony-stimulating factor (G-CSF) mediates “emergency” granulopoiesis during infection, a process that is mimicked by clinical G-CSF use, yet we understand little about the intracellular signaling cascades that control demand-driven neutrophil production. Using a murine model with conditional deletion of signal transducer and activator of transcription 3 (STAT3) in bone marrow, we investigated the cellular and molecular mechanisms of STAT3 function in the emergency granulopoiesis response to G-CSF administration or infection with Listeria monocytogenes, a pathogen that is restrained by G-CSF signaling in vivo. Our results show that STAT3 deficiency renders hematopoietic progenitor cells and myeloid precursors refractory to the growth-promoting functions of G-CSF or L monocytogenes infection. STAT3 is necessary for accelerating granulocyte cell-cycle progression and maturation in response to G-CSF. STAT3 directly controls G-CSF–dependent expression of CCAAT-enhancer-binding protein β (C/EBPβ), a crucial factor in the emergency granulopoiesis response. Moreover, STAT3 and C/EBPβ coregulate c-Myc through interactions with the c-myc promoter that control the duration of C/EBPα occupancy during demand-driven granulopoiesis. These results place STAT3 as an essential mediator of emergency granulopoiesis by its regulation of transcription factors that direct G-CSF–responsive myeloid progenitor expansion.
Introduction
Neutrophils serve a critical function in the innate immune system by maintaining a frontline defense against bacterial and fungal pathogens. However, precise control of circulating neutrophil amounts is important in preserving a balance between effective immunity versus immunosuppression or inflammation. For example, congenital or therapy-induced neutropenias render persons sensitive to life-threatening infections, whereas neutrophil overproduction or hyperactivation contributes to inflammation and, potentially, autoimmunity.1,2 To maintain appropriate neutrophil numbers, the process of granulopoiesis is tightly regulated. In steady state conditions, the bone marrow continuously produces granulocytes and stores a reserve of mature neutrophils, whereas circulating neutrophil numbers are maintained within a narrow range (3.5-4.5 × 109/L for humans and approximately 0.488 × 109/L for mice).3 The neutrophil production pathway is responsive to environmental stresses such as infection, which stimulate a rapid increase in granulopoiesis, enhanced neutrophil migration from bone marrow, and a subsequent rise in peripheral neutrophils (eg, circulating neutrophils > 8 × 109/L in human infection).4
Neutrophil development from hematopoietic stem cells (HSCs) is guided by granulocyte colony-stimulating factor (G-CSF), which activates a high-affinity type I cytokine receptor, G-CSF receptor (G-CSFR), expressed on HSCs, myeloid progenitor cells, granulocyte precursors, and mature neutrophils.5 In homeostatic conditions, G-CSF–G-CSFR signals maintain mature neutrophil numbers in circulation by regulating the proliferation and differentiation of myeloid progenitors as well as survival of postmitotic bone marrow neutrophils.6,7 During infection, G-CSF serum levels can increase 10- to 20-fold, stimulating enhanced neutrophil production in the bone marrow and neutrophil mobilization to the peripheral blood.8 This response, termed “emergency” or demand-driven granulopoiesis, is characterized by enhanced cell-cycle progression of granulocyte precursors and efflux of mature neutrophils from the bone marrow, relative to steady state conditions.9 The signal transduction pathways that regulate emergency granulopoiesis are of significant interest because G-CSF is used therapeutically to increase circulating neutrophil counts; however, the underlying mechanisms directing G-CSF–responsive myeloid progenitor expansion are poorly understood.
Janus kinase and signal transducer and activator of transcription (STAT) pathway components are principal intermediates in the G-CSFR signaling cascade. STAT3 activation predominates; thus, attention has focused on its role in granulopoiesis in vivo. In steady state, STAT3 serves a negative regulatory function by suppressing peripheral neutrophilia.10 This activity is attributed to STAT3-dependent regulation of suppressor of cytokine signaling-3 (SOCS3), a crucial feedback inhibitor of G-CSFR signaling.10,11 By contrast, during G-CSF–driven emergency granulopoiesis STAT3 is required to boost immature neutrophil numbers in bone marrow and to regulate acute neutrophil mobilization.12,13 Although we have shown that these functions are independent of SOCS3,13 the underlying cellular and molecular mechanisms were not resolved. It is important to understand the full action of STAT3 in directing emergency granulopoiesis because this pathway is critical for effective responses to infection.
Transcription factors in the CCAAT-enhancer–binding protein (C/EBP) family, including C/EBPα, C/EBPβ, and C/EBPϵ, regulate steady state granulopoiesis as well as demand-driven neutrophil production.14,15 In homeostatic conditions, C/EBPα is essential for transition from the common myeloid progenitor to the granulocyte-macrophage progenitor (GMP),16 whereas terminal neutrophil differentiation critically depends on C/EBPϵ.17 C/EBPα and C/EBPϵ control genes that are necessary for myeloid progenitor growth and neutrophil function, including cytokine receptors, proteases, and granule proteins.18–21 In addition, C/EBPα restrains proliferation of developing granulocytes by inhibiting cell-cycle regulators such as c-Myc.22 Conversely, C/EBPβ is dispensable for steady state neutrophil production, yet it has a crucial role in emergency granulopoiesis in response to G-CSF administration or Candida albicans infection.23 The discrete function of C/EBPβ in demand-driven granulopoiesis is related to its ability to support sustained G-CSF–responsive proliferation, including prolonged c-Myc expression, compared with C/EBPα.22,23
Previous studies have shown C/EBPα and C/EBPβ as downstream targets of the Janus kinase–STAT3 signaling pathway in response to G-CSF or interleukin-6 (IL-6), respectively.24,25 Moreover, G-CSF stimulates Cebpb mRNA in GMPs, suggesting G-CSF–dependent signaling pathways control inducible Cebpb transcription.23 Aspects of the phenotype of bone marrow conditional STAT3 knockout mice resemble C/EBPβ deficiency, eg, impaired emergency granulopoiesis.13,23 These data suggested to us that STAT3 might regulate G-CSF–driven granulopoiesis by its induction of C/EBPβ. To examine this and to explore mechanisms by which STAT3 regulates emergency granulopoiesis, we used mice with conditional STAT3 deletion in hematopoietic cells. Our results show that STAT3 promotes G-CSF–responsive proliferation of immature granulocytes by stimulating G1-S phase transition and enhances terminal granulocyte maturation. The growth-stimulatory function of G-CSF–STAT3 signaling is also observed in pluripotent lin− Sca-1+ c-kit+ (LSK) bone marrow progenitors, which amplify in a STAT3-dependent manner during G-CSF administration in vivo. STAT3 directly controls G-CSF–responsive induction of C/EPBβ expression in myeloid progenitor cells. Furthermore, STAT3 regulates G-CSF–dependent c-myc transcription by direct promoter binding, and by facilitating interaction of C/EBPβ and displacement of C/EBPα at proximal promoter elements. These results place STAT3 as an essential mediator of G-CSF–driven granulopoiesis through its control of growth-enhancing transcription factors in developing granulocytes.
Methods
Mouse strains
Hematopoietic STAT3-deficient mice (Tg(Tek-cre)12Flv, Stat3f/Δ) were obtained by breeding Tg(Tek-cre)12Flv transgenic mice26 and Stat3wt/f mice,27 as described.13 Wild-type littermates containing 2 intact Stat3 alleles were used as controls.13 Animals were housed in a specific pathogen-free barrier facility. Experimental procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas M. D. Anderson Cancer Center (MDACC).
Plasmids
Murine C/EBPβ cDNA was generously provided by Dr Jessica Schwartz (University of Michigan) and was used to generate pRV-C/EBPβ. pRV-C/EBPα was kindly provided by Dr Chen Dong (MDACC). The Cebpb and c-myc promoter fragments encompassing respective STATx sites were amplified with AccuPrime Pfx DNA Polymerase (Invitrogen) and cloned into pGL3-Basic (Promega). Murine STAT3 or a DNA binding mutant28 (STAT3-DN) in the pMX-IRES-GFP vector were used. Plasmid inserts were verified by DNA sequencing.
G-CSF administration in vivo, bone marrow collection, and immunoblotting
Mice were injected subcutaneously with 250 μg/kg recombinant human G-CSF (Amgen) in phosphate-buffered saline (PBS) containing 1% endotoxin-free bovine serum albumin (BSA) as described.13 Control mice received an equivalent volume of 1% BSA/PBS. Bone marrow cells were obtained as described.13 Whole cell lysates were subjected to immunoblotting with antibodies for pSTAT3 (Cell Signaling Technology), STAT3 (C-20), or C/EBPβ (H-7; Santa Cruz Biotechnology) as described.13,23
Infection with Listeria monocytogenes
Experiments were performed with L monocytogenes strain 1043S, generously provided by Drs Chen Dong (MDACC) and Hao Shen (University of Pennsylvania). L monocytogenes was grown to an optical density of 0.1 at 600 nm in brain heart infusion broth. Bacteria were suspended in sterile PBS at 1 × 105 cells/mL; 200 μL was injected intravenously as described.29 Animals were killed 12 to 24 hours after infection.
Antibody staining, flow cytometry, and fluorescence-activated cell sorting
Bone marrow cells were stained with fluorescently labeled antibodies as described13 and analyzed on an LSRII machine (BD Bioscience). Immature (Gr-1lo) and mature (Gr-1hi) granulocytes were purified from bone marrow with the use of a FACSAria (BD Biosciences).13 LSKs and GMPs were analyzed as described.30 Briefly, lin− bone marrow progenitors were stained with biotinylated rat anti–mouse CD3 (CT-CD3), CD11b (M1/70), Gr-1 (RB6-8C5), CD45R/B220 (RA36B2), and Ter119 and negatively selected with anti–rat microbeads (Miltenyi Biotec). The selected population was then stained with fluorescein isothiocyanate–CD34+ (RAM34), phycoerythrin–FcγRII/III (2.4G2), peridin chlorophyll proteincyanine 5.5–Sca-1 (E13-161.7), allophycocyanin cyanine 7–c-kit (2B8), and pacific blue–IL-7 receptor (A7R34; eBioscience) to label hematopoietic progenitors, and streptavidin-allophycocyanin to label lin+ cells. LSKs and GMPs (lin− Sca-1− c-kit+ CD34+ FcγRII/III+ cells) were analyzed on an LSRII machine or isolated by fluorescence-activated cell sorting.
5,6-Carboxyfluorescein diacetate, succinimidyl ester dilution experiments, cell-cycle analysis
Purified immature granulocytes and GMPs were labeled with 2.5mM 5,6-carboxyfluorescein diacetate, succinimidyl ester (CFSE; Molecular Probes) in RPMI 1640, cultured 3 days in RPMI/fetal calf serum containing 2.5 ng/mL G-CSF and analyzed by flow cytometry. For cell-cycle analysis, purified immature granulocytes were stimulated with G-CSF, fixed with 70% ethanol at −20°C overnight, and stained with propidium iodide (20 μg/mL; Sigma-Aldrich) in 1% RNase buffer (Sigma) for 30 minutes at room temperature. Flow cytometry and analysis by ModFit LT v3.0 (BD Biosciences) were used to determine the proportion of cells in S phase.
RNA isolation and quantitative polymerase chain reaction
Total RNA was extracted by Trizol and reverse-transcribed with iScript (Bio-Rad). Quantitative polymerase chain reaction (PCR) was performed with SYBR Green PCR Mix (Bio-Rad) and a sequence detector (Bio-Rad 5000). Primer sequences are available on request. Threshold cycle values were normalized to 18s RNA; relative mRNA abundance was determined by the equation 1.8(18sRNA − GENE) × 10 000 000.31
Retroviral transduction, electrophoretic mobility shift assays, chromatin immunoprecipitations, and reporter gene assays
The murine myeloblast cell line 32D was cultured as described.28 To generate G-CSFR–expressing cells (32D.G-CSFR), 32D cells were infected with retrovirus generated from the bicistronic vector pMX-G-CSFR-GFP; GFP+ cells were isolated by fluorescence-activated cell sorting at 48 hours after infection. Chromatin immunoprecipitations (ChIPs) were performed with antibodies to STAT3 (c-20 and H119), C/EBPβ (H7), or C/EBPα (14AA; Santa Cruz Biotechnology) with the use of a ChIP assay kit and the manufacturer's instructions (Millipore). Nuclear extracts were used in electrophoretic mobility shift assays (EMSAs) with 32P-labeled oligonucleotides corresponding to STATx binding sites in the Cebpb or c-myc promoter regions (Cebpb distal element, 5′-CAGAGGCTGCTGGGAAGGCCA-3′; Cebpb proximal element, 5′-GTGGAGGGCTGGGATCCAGG-3′; c-myc, 5′-GCATCTTCCCAGAACCTGGA-3′). Competitor oligonucleotides or STAT3 antibodies were included as indicated in the figure legends. For reporter assays, 32D.G-CSFR cells were electroporated with the Cell Line Nucleofector Kit V (Lonza) and the Nucleofector Device (Lonza), with indicated plasmids (see figure legends). Cells were cultured in IL-3–containing medium for 24 hours and treated with or without 25 ng/mL G-CSF for 2 hours; luciferase activity was measured as described.32
C/EBPβ short hairpin RNA lentiviral vector production
C/EBPβ short hairpin RNA (shRNA) lentiviral constructs were purchased from Sigma. 293T cells were transfected with packaging plasmids and lentiviral constructs (Sigma), and viral supernatants were collected after 48 hours. Purified immature granulocytes were infected and analyzed as indicated in Figure 4.
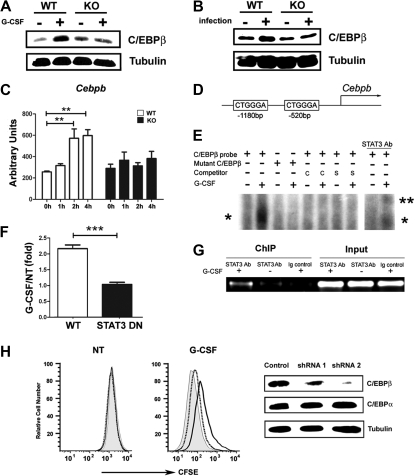
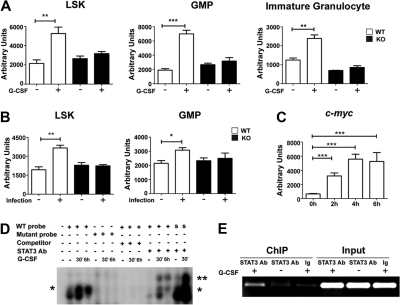
Figure 4.
STAT3 directly regulates C/EBPβ expression during emergency granulopoiesis. (A-B) C/EBPβ expression was measured in Gr-1+ granulocytes isolated from wild-type (WT) and STAT3-deficient (KO) mice 6 hours after treatment with G-CSF (+) or BSA buffer (−) in vivo (A) or 24 hours after infection with L monocytogenes (+) or in untreated controls (−) (B). The dominant LAP (liver-enriched transcriptional activator protein) isoform of C/EBPβ was detected by immunoblotting. Tubulin was used as a loading control. Data are representative of 3 independent experiments. (C) Cebpb mRNA expression was measured in immature Gr-1lo granulocytes from WT or STAT3-deficient mice, after treatment with G-CSF ex vivo for 0 to 4 hours, using quantitative PCR (n = 3 for WT and KO for each condition). Average values from 3 independent experiments are shown. **P < .01 compared with control. (D) Schematic diagram of the murine Cebpb promoter, showing the location of 2 IL-6 RE II sites. (E) 32D.G-CSFR cells were stimulated with G-CSF (25 ng/mL) for 1 hour or left untreated, as indicated. Nuclear extracts were used in EMSAs with a radiolabeled oligonucleotide corresponding to the IL-6 RE II consensus sequence at position −1180 in the Cebpb promoter (C/EBPβ probe) or a mutant radiolabeled C/EBPβ oligonucleotide (mutant C/EBPβ) in the presence or absence of unlabeled competitor C/EBPβ probe (C), an unlabeled competitor oligonucleotide corresponding to the STAT3 consensus site in the murine Socs3 promoter (S), or STAT3 antibodies, as indicated. The migration positions of STAT3 dimers (*) and supershifted STAT3 complexes (**) are shown. Data are representative of 3 independent experiments. (F) 32D.G-CSFR cells were electroporated with pGL3-C/EBPβ, pTK-Renilla and pMX-STAT3 or pMX-STAT3 DN (DNA-binding mutant). After 24 hours, cells were treated with or without G-CSF for 2 hours and assayed for luciferase activity. The ratio of firefly:renilla relative light units (RLU) from G-CSF–treated and unstimulated cells (NT) was averaged from 3 independent experiments. Error bars represent SEM. (G) 32D.G-CSFR cells were treated as indicated in panel E. ChIPs were performed with STAT3 antibodies (STAT3 Ab) or an irrelevant IgG (Ig). PCR reactions were performed with primers specific for the murine Cebpb promoter on total cell lysates (input) or immunoprecipitated samples, as indicated. Data are representative of 3 independent experiments. (H) Wild-type Gr-1lo cells were infected with lentiviral shRNA vectors (control vector, gray shading; shRNA 1, dotted line; shRNA 2, black line), stained with CFSE, analyzed immediately (NT) by flow cytometry or cultured in G-CSF for 4 days (G-CSF), and evaluated by flow cytometry. Expression of C/EBPβ, C/EBPα, and tubulin was determined by immunoblotting. Data are representative of 3 independent experiments.
Statistical analysis
Data are presented as mean ± SEM. The statistical significance between groups was calculated by a 2-tailed t test with the use of GraphPad Prism 4 software (www.graphpad.com).
Results
STAT3 controls G-CSF–responsive cell-cycle progression in immature granulocytes
We previously showed that STAT3 is necessary for the accumulation of immature Gr-1lo granulocytes in the bone marrow during G-CSF administration.13 These data could be explained by ineffective G-CSF–responsive proliferation or an enhanced rate of differentiation; thus, the underlying cellular response remained unclear. To assess STAT3 function in G-CSF–dependent growth, immature granulocytes were used in CFSE dilution experiments ex vivo. Gr-1lo or Gr-1hi expression has been used to distinguish immature and mature granulocytes in the marrow, respectively.13,33 We confirmed that greater than 80% of bone marrow Gr-1lo cells from wild-type or STAT3-deficient mice show a granulocyte precursor phenotype (ie, Gr-1lo CD11b+ CD115−) and immature morphology.13,32 Most STAT3-deficient immature granulocytes showed suppressed G-CSF–responsive growth relative to wild-type cells, as judged by CFSE dilution (Figure 1A). To independently assess proliferation, we examined cell-cycle distribution during a time course of G-CSF treatment ex vivo. Wild-type immature granulocytes entered S phase semisynchronously 2 to 4 hours after G-CSF addition, subsequently exited S phase, and accumulated in G0/G1 by 12 to 24 hours (Figure 1B; data not shown). By contrast, STAT3-deficient immature granulocytes showed a suppressed response to G-CSF (Figure 1B). These results indicate that STAT3 has a cell autonomous role in controlling G-CSF–dependent proliferation.
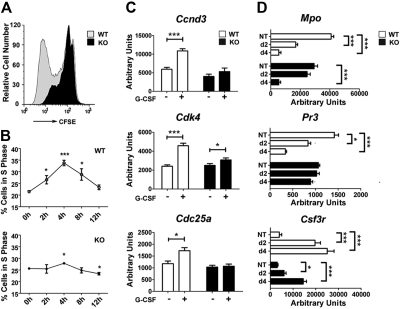
Figure 1.
STAT3 regulates G-CSF–responsive proliferation of immature granulocytes and the kinetics of granulocyte maturation. (A) Immature Gr-1lo granulocytes were isolated from the bone marrow of wild-type (WT) and STAT3-deficient (KO) mice by fluorescence-activated cell sorting. Cells were labeled with CFSE, cultured in G-CSF (2.5 ng/mL) for 4 days, and analyzed by flow cytometry. Results of a representative experiment (1 of 3) are shown. (B) Gr-1lo cells were cultured in G-CSF (25 ng/mL) for the indicated times or analyzed directly after isolation (0 hour). Cells were stained with propidium iodide and analyzed by flow cytometry. The percentage of cells in S phase was determined by ModFit LT v3.0 (n = 3 for WT and KO for each condition). (C) Bone marrow Gr-1lo cells were isolated 4 hours after mice received a single dose of G-CSF (250 μg/kg; +) or BSA carrier alone (−). Gene expression was measured by quantitative PCR, with normalization to 18s RNA (n = 3 for WT and KO for each condition). (D) Gr-1lo cells were cultured in G-CSF for 2 or 4 days or analyzed directly after isolation (NT). The expression of neutrophil differentiation markers was measured at the indicated times by quantitative PCR (n = 3 for WT and KO for each condition). (B-D) Average values from 3 independent experiments are shown. Error bars represent SEM. *P < .05 compared with untreated or BSA-treated controls. **P < .01, ***P < .001 compared with control.
To investigate the molecular basis of this proliferative defect, we examined the expression of candidate cell-cycle regulators in wild-type and STAT3-deficient immature granulocytes in vivo, after administering G-CSF or control BSA-containing buffer. These experiments showed G-CSF stimulated Ccnd3, Cdk4, and Cdc25, which control G1/S-phase transition,34,35 in wild-type immature granulocytes but had little or no effect on STAT3-deficient cells (Figure 1C). STAT3-dependent induction of these genes in vivo coincided with the peak time point of S-phase accumulation observed in wild-type granulocytes stimulated with G-CSF ex vivo.
To assess the kinetics of immature granulocyte maturation, we analyzed the expression of neutrophil differentiation markers, including Csf3r, Mpo, and Pr3, in G-CSF culture conditions that promote terminal neutrophil development.13 Freshly isolated immature granulocytes from both genotypes did not show appreciable differences in marker protein expression (Figure 1D), as reported previously.13 By contrast, we found disparate regulation in wild-type and STAT3-deficient granulocytes on G-CSF–induced maturation. In wild-type granulocytes, G-CSF stimulated Csf3r and suppressed Mpo and Pr3 expression by 2 days, similar to their regulation during human granulopoiesis.36 In comparison, STAT3-deficient granulocytes failed to induce Csf3r or suppress Mpo and Pr3 as efficiently as wild-type cells by 2 days of G-CSF treatment. By 4 days, Mpo expression appeared similar in granulocytes from both genotypes, whereas Csf3r was induced in STAT3-deficient neutrophils, although at lower levels relative to wild-type cells. Pr3 expression was unchanged in STAT3-deficient cells yet declined significantly in wild-type cells by 4 days (Figure 1D). These results indicate STAT3-deficient granulocytes have delayed maturation, consistent with their aberrant G-CSF responsiveness.
STAT3 controls G-CSF–driven expansion of hematopoietic progenitor cells in bone marrow
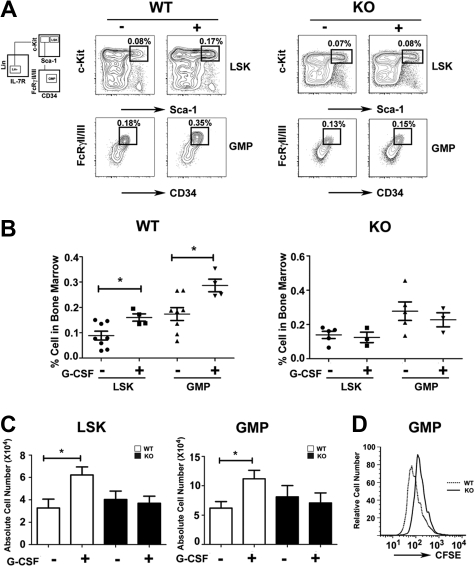
The impaired response of STAT3-deficient immature granulocytes to G-CSF suggested myeloid progenitor cells might require STAT3 for G-CSF–dependent proliferation in vivo. We found that the frequency and absolute number of LSKs and GMPs increased approximately 2-fold in G-CSF–treated wild-type mice by 24 hours (Figure 2A-C). By contrast, STAT3-deficient mice failed to induce LSK and GMP amounts on G-CSF treatment (Figure 2A-C). In resting conditions, LSK and GMP proportions were increased slightly in STAT3-deficient mice relative to wild-type animals, although absolute numbers were similar in both genotypes (Figure 2A-C). CFSE dilution experiments showed that purified STAT3-deficient GMPs have suppressed G-CSF–responsive growth ex vivo, compared with wild-type GMPs (Figure 2D). Thus, our results suggest that STAT3 regulates G-CSF–dependent expansion of the LSK and GMP subsets in the bone marrow by enhancing progenitor proliferation.
Figure 2.
Role of STAT3 in hematopoietic progenitor responses to G-CSF. Wild-type (WT) and STAT3-deficient (KO) mice were injected with G-CSF (250 μg/kg; +) or BSA carrier alone (−), and bone marrow cells were collected 24 hours after treatment. (A) The proportion of LSKs and GMPs was determined by flow cytometry, as indicated. Data are representative of 3 independent experiments. (B) The frequency of LSKs (BSA, ●; G-CSF, ■) and GMPs (BSA, ▲; G-CSF, ▼) within individual mice is indicated for wild-type or STAT3-deficient animals. (C) Absolute numbers of bone marrow LSKs and GMPs in 2 femurs and 2 tibiae were determined by enumeration. (B-C) Average values from 3 independent experiments are shown. Error bars represent SEM (n = 8, WT BSA; n = 4, WT G-CSF; n = 5, KO BSA; n = 3, KO G-CSF). *P < .05 compared with BSA-treated controls. (D) GMPs from WT and STAT3-deficient mice were stained with CFSE, cultured in G-CSF for 3 days, and analyzed by flow cytometry. Data are representative of 3 independent experiments.
Expansion of hematopoietic progenitor subsets during L monocytogenes infection requires STAT3
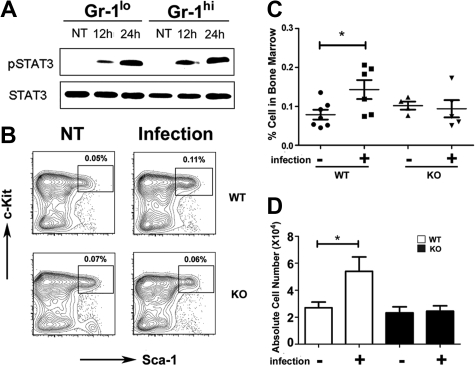
To determine whether STAT3 participates in the host response to bacterial infection we used L monocytogenes, which is controlled by G-CSF signaling in vivo.8 STAT3 was activated by L monocytogenes infection, as judged by inducible STAT3 tyrosine phosphorylation in immature Gr-1lo and mature Gr-1hi granulocytes from infected mice versus uninfected controls (Figure 3A). L monocytogenes infection induced the frequency and absolute number of LSKs 1.5- to 2.0-fold in wild-type mice by 12 hours and 24 hours after infection, whereas no increase was observed in LSKs in STAT3-deficient mice (Figure 3B-D; data not shown). Lin− progenitors were present at similar proportions in the peripheral blood of wild-type and STAT3-deficient mice at 24 hours after infection (data not shown), indicating that differences in progenitor mobilization did not contribute to impaired LSK induction in the bone marrow of L monocytogenes–infected STAT3-deficient mice. These data collectively suggest that STAT3 mediates accumulation of LSKs during early phases of infection, which may contribute to the diminished response of STAT3-deficient mice to L monocytogenes.32
Figure 3.
STAT3 controls hematopoietic progenitor responses during L monocytogenes infection. Wild-type (WT) or STAT3-deficient (KO) mice were infected with 20 000 colony-forming units of L monocytogenes by intravenous injection. (A) Immature Gr-1lo (Gr-1lo) and mature Gr-1hi (Gr-1hi) granulocytes were isolated from untreated controls (NT) or L monocytogenes–infected mice at 12 or 24 hours after infection, as indicated. Tyrosine-phosphorylated and total STAT3 was detected by immunoblotting, as shown. Data are representative of 3 independent experiments. (B) The frequency of LSKs was determined 12 hours after infection or in untreated controls (NT), as indicated. Results from a representative experiment (1 of 4) are shown. (C) LSK proportions within individual mice is indicated for untreated (−) or infected (+) WT or STAT3-deficient animals. The horizontal bar indicates average values, determined from 4 independent experiments. (D) Absolute numbers of LSKs in 2 femurs and 2 tibiae from untreated (−) or infected (+) mice were determined by enumeration (n = 7 for untreated WT; n = 6 for infected WT; n = 4 for untreated or infected KO). Average values are shown, determined from 4 independent experiments. (C-D) Error bars represent SEM. *P < .05 compared with untreated controls.
STAT3 directly controls C/EBPβ expression during emergency granulopoiesis
C/EBPβ is required for emergency granulopoiesis; moreover, Cebpb mRNA levels are selectively up-regulated in GMPs in response to G-CSF treatment in vivo.23 These results suggested Cebpb as a target gene of the G-CSF–STAT3 signaling cascade during emergency granulopoiesis. We examined C/EBPβ protein and mRNA expression in wild-type and STAT3-deficient granulocytes in response to G-CSF treatment or L monocytogenes infection. G-CSF administration in vivo stimulated C/EBPβ expression in wild-type bone marrow granulocytes within 6 hours, yet it did not induce C/EBPβ amounts appreciably in STAT3-deficient cells (Figure 4A). L monocytogenes infection also up-regulated C/EBPβ in wild-type granulocytes but not in STAT3-deficient cells (Figure 4B). Furthermore, we found that STAT3 was required for the rapid induction of Cebpb mRNA in immature granulocytes after G-CSF stimulation ex vivo or G-CSF administration in vivo (Figure 4C; data not shown). These data collectively show that STAT3 has a cell autonomous function in controlling inducible Cebpb expression in neutrophil precursors during G-CSF– or L monocytogenes–driven granulopoiesis.
The rapid induction of Cebpb mRNA (Figure 4C) indicated the potential for Cebpb to be a direct STAT3 target gene. The murine Cebpb promoter lacks a classical STAT consensus site within its proximal 10 kilobase region as judged by nucleotide-sequence analysis (data not shown); however, we found 2 atypical IL-6 response elements II (IL-6 RE II), CTGGGA (Figure 4D), which serve as STAT3 interaction sites in the fibrinogen promoter.37 EMSAs showed G-CSF–stimulated STAT3 binding to the distal IL-6 RE II (−1180 base pair) but not the proximal element (−520 base pair) in the Cebpb promoter (Figure 4E; data not shown). Mutation of the distal IL-6 RE II abrogated G-CSF–responsive STAT3 interaction, whereas binding to the intact distal IL-6 RE II was competed with excess unlabeled oligonucleotide or an oligonucleotide containing the STAT3 binding site from the Socs3 promoter (Figure 4E). STAT3 antibody supershifts confirmed G-CSF–responsive STAT3 binding to the distal IL-6 RE II (Figure 4E). Reporter gene assays indicated G-CSF–responsive transcription from the proximal Cebpb promoter required functional STAT3, as judged by attenuated reporter activity in the presence of a STAT3 DNA binding mutant (Figure 4F). ChIP experiments detected G-CSF–responsive STAT3 accumulation at the region encompassing the distal IL-6 RE II in vivo (Figure 4G). These data indicate that STAT3 directly stimulates Cebpb transcription in response to G-CSF by interaction with the distal IL-6 RE II in the Cebpb promoter.
To examine whether C/EBPβ was required for G-CSF–driven proliferation of immature granulocytes, we used shRNA-mediated C/EBPβ depletion. Efficient C/EBPβ knockdown correlated with impaired G-CSF–responsive growth of immature granulocytes, whereas shRNA targeting that failed to suppress C/EBPβ did not affect G-CSF–driven proliferation (Figure 4H). These data support a fundamental role for C/EBPβ in G-CSF–responsive myeloid progenitor proliferation,23 and thus suggest that STAT3-mediated transcriptional control of C/EBPβ is important for G-CSF–driven granulopoiesis.
STAT3 controls c-myc expression in hematopoietic progenitors during emergency granulopoiesis
c-Myc has been implicated as an important regulator of emergency granulopoiesis and a potential target of C/EBPβ during the G-CSF response.23 STAT3-deficient hematopoietic progenitors show defects consistent with impaired c-Myc function, including the inability to induce the c-Myc target genes Cdc25a and Cdk438 and suppressed progression of G1/S phase on G-CSF stimulation (Figure 1B-C). Thus, in light of the role for STAT3 in C/EBPβ induction (Figure 4), we hypothesized that c-Myc may be controlled by a G-CSF–STAT3–C/EBPβ signaling cascade in myeloid progenitors. Consistent with this, we found that G-CSF stimulated c-myc mRNA expression in vivo in wild-type LSKs, GMPs, and immature granulocytes, whereas STAT3-deficient hematopoietic progenitors and immature granulocytes were impaired in their ability to up-regulate c-myc in response to G-CSF (Figure 5A). Similarly, L monocytogenes infection stimulated c-myc expression in wild-type hematopoietic progenitors but failed to induce c-myc in STAT3-deficient cells (Figure 5B).
Figure 5.
STAT3 controls c-myc expression during emergency granulopoiesis. (A) Wild-type (WT) or STAT3-deficient (KO) mice were treated with G-CSF or BSA as indicated in Figure 1. c-myc mRNA expression was determined in bone marrow LSK and GMP subsets isolated 24 hours after treatment or in immature Gr-1lo granulocytes at 4 hours after treatment, by quantitative PCR (n = 3 for WT and KO for each condition). (B) WT or STAT3-deficient mice were infected with L monocytogenes or left untreated. c-myc expression was analyzed in LSKs and GMPs purified 12 hours after infection, using quantitative PCR (n = 3 for WT and KO for each condition). (A-B) Average values from 3 independent experiments are shown. *P < .05, **P < .01, ***P < .001 compared with control. (C) 32D.G-CSFR cells were treated 0 to 6 hours with G-CSF; c-myc RNA was measured by quantitative PCR. Average values from 3 independent experiments are shown. ***P < .001 compared with control. (D) Nuclear extracts were generated from untreated (−) or G-CSF–treated (+) 32D.G-CSFR cells at the indicated times and used in EMSAs with a radiolabeled oligonucleotide corresponding to a putative STAT3 consensus site in the c-myc promoter (WT), a mutated radiolabeled c-myc promoter oligonucleotide (mutant), or a radiolabeled oligonucleotide corresponding to the STAT3 binding site in the murine Socs3 promoter (S). EMSAs were performed in the presence or absence of STAT3 antibodies or competitor WT oligonucleotides, as indicated. The migration positions of STAT3 dimers (*) and supershifted STAT3 complexes (**) are shown. Data are representative of 3 independent experiments. (E) 32D.G-CSFR cells were treated as indicated in Figure 4E. ChIPs were performed with STAT3 antibodies (STAT3) or an irrelevant IgG (Ig), as indicated, followed by PCR with primers specific for the murine c-myc promoter. Control PCR reactions were performed on total lysate (input). Data are representative of 3 independent experiments.
Using 32D.G-CSFR cells, we found rapid induction of c-myc mRNA on G-CSF stimulation (Figure 5C), suggesting c-myc might be directly regulated by STAT3. Analysis of the c-myc promoter identified a conserved STATx consensus sequence TTCxxxGAA39 approximately 2 kilobase upstream of the transcriptional start site (Figure 6A-B). EMSAs showed that STAT3 bound this consensus sequence in response to G-CSF stimulation (Figure 5D). STAT3 binding was competed with an excess of unlabeled oligonucleotide and was not observed with an oligonucleotide containing a mutated STATx element (Figure 5D). Supershift experiments confirmed G-CSF–responsive STAT3 binding to the STATx site in the c-myc promoter, as well as the positive control Socs3 promoter STATx element (Figure 5D). ChIP experiments showed that STAT3 bound the c-myc promoter in vivo in proximity to the STATx element on G-CSF stimulation (Figure 5E). Reporter gene assays indicted STAT3 regulated G-CSF–responsive c-myc transcription, as judged by repression with a STAT3 DNA binding mutant (Figure 6E). Thus, collectively, our results indicate that STAT3 directly regulates c-myc, in addition to Cebpb, in myeloid progenitor cells in response to G-CSF stimulation.
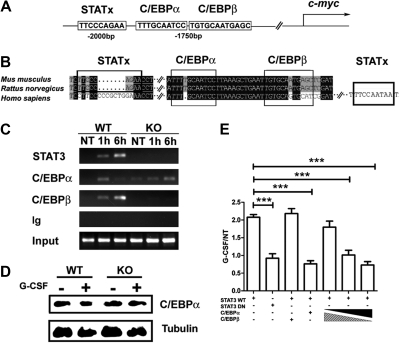
Figure 6.
STAT3 modulates C/EBPα and C/EBPβ recruitment to the c-myc promoter in response to G-CSF. (A) A schematic diagram shows the location of putative STAT3, C/EBPα, and C/EBPβ binding sites in the murine c-myc promoter. (B) Sequence alignment of the murine, rat, and human c-myc promoter regions encompassing the putative STAT3, C/EBPα, and C/EBPβ binding sites. (C) Bone marrow cells from wild-type (WT) or STAT3-deficient (KO) mice were treated with G-CSF (25 ng/mL) for 1 hour or 6 hours, as indicated. ChIPs were performed with antibodies to STAT3, C/EBPβ, C/EBPα, or an irrelevant IgG (Ig). PCR reactions were performed with primers specific for the murine c-myc promoter region encompassing the putative STAT3, C/EBPα, and C/EBPβ binding sites, using immunoprecipitation or input samples, as indicated. Data are representative of 2 independent experiments. (D) Wild-type (WT) or STAT3-deficient (KO) mice were treated with G-CSF or BSA buffer in vivo. C/EBPα expression was determined in bone marrow Gr-1+ granulocytes at 6 hours after treatment by immunoblotting (n = 3 for WT and KO for each condition). Data are representative of 3 independent experiments. (E) 32D.G-CSFR cells were electroporated with pGL3-myc, pTK-Renilla, pMX-STAT3, and pMX-STAT3 DN as indicated. pRV-C/EBPβ or pRV-C/EBPα were included individually in some samples or in a 9:1, 1:1, or 1:9 ratio, as illustrated. Cells were treated with or without G-CSF for 2 hours and assayed for luciferase activity. Relative luciferase unit induction in G-CSF–treated vs unstimulated (NT) cells was averaged from 3 independent experiments. Error bars represent SEM.
G-CSF signaling by STAT3 controls c-myc transcription by regulating the ratio of C/EBPα to C/EBPβ at the c-myc promoter
C/EBPβ and C/EBPα have been previously reported to have opposing effects on c-myc regulation, maintaining or suppressing c-myc expression, respectively.22,23 These results suggested STAT3-dependent induction of C/EBPβ might contribute to c-myc regulation in granulocytes. Sequence analysis of the c-myc promoter showed conserved C/EBPβ and C/EBPα consensus sites in proximity to the STAT3 binding site (Figure 6A-B). To determine whether G-CSF regulates C/EBPα or C/EBPβ binding to the c-myc promoter and to assess the function of STAT3, we analyzed in vivo interactions with the use of ChIP assays with wild-type or STAT3-deficient bone marrow cells. These experiments showed that STAT3, C/EBPα, and C/EBPβ were rapidly recruited to the c-myc promoter in wild-type cells by 1 hour of G-CSF treatment. STAT3 and C/EBPβ further accumulated by 6 hours, whereas C/EBPα was largely disassociated by this time (Figure 6C). ChIP assays with 32D.G-CSFR cells showed a similar pattern of sustained STAT3 and C/EBPβ recruitment and transient C/EBPα binding at the c-myc promoter on G-CSF stimulation (data not shown). By contrast, G-CSF–responsive C/EBPβ recruitment to the c-myc promoter was suppressed considerably in STAT3-deficient cells, consistent with their inability to induce C/EBPβ (Figure 6C and Figure 4A,C). Moreover, C/EBPα association was prolonged in G-CSF–stimulated STAT3-deficient cells, whereas STAT3 recruitment was abrogated as expected (Figure 6C). C/EBPα protein amounts were similar in wild-type and STAT3-deficient cells (Figure 6D), indicating that C/EBPα stability was not regulated by G-CSF. Thus, the data suggest that STAT3 activation and induction of C/EBPβ leads to C/EBPα dissociation from the c-myc promoter.
To test whether C/EBPβ and C/EBPα differentially regulate G-CSF–dependent transcription from the proximal c-myc promoter, we used reporter gene assays. These experiments showed that overexpression of C/EBPα relative to C/EBPβ (9:1 ratio) abrogated G-CSF–dependent c-myc reporter activity (Figure 6E), confirming the suppressive action of C/EBPα.22 By contrast, overexpression of C/EBPβ versus C/EBPα (9:1 ratio) enabled G-CSF–responsive c-myc promoter activity, indicating C/EBPβ can override the inhibitory function of C/EBPα. These results suggest a model in which C/EBPα and C/EBPβ have opposing inhibitory or stimulatory effects, respectively, on c-myc proximal promoter activity. Our data therefore suggest that c-myc transcription is activated on G-CSF stimulation by direct STAT3 interaction with the c-myc proximal promoter and by STAT3-dependent induction of C/EBPβ; induction of C/EBPβ or recruitment of STAT3 or both STAT3 and C/EBPβ is required to stimulate dissociation of the repressive factor C/EBPα from the c-myc proximal promoter. This mechanism may contribute to enhanced myeloid progenitor proliferation during emergency granulopoiesis.
Discussion
The appropriate control of granulopoiesis is crucial for maintaining a functional innate immune response. G-CSF signals are necessary for basal and emergency granulopoiesis,6,8,40 although little has been known about the intracellular signaling pathways downstream of the G-CSFR that control neutrophil production in vivo. We clarify the role of STAT3 in emergency granulopoiesis by demonstrating its function in myeloid progenitor proliferation and by revealing a unique signaling pathway, by C/EBPβ and c-Myc, used in demand-driven conditions. Moreover, we identify a novel mechanism of c-Myc regulation through STAT3-dependent control of the ratio of C/EBPα to C/EBPβ at the c-myc promoter.
The neutrophilia of STAT3-deficient mice in homeostatic conditions, attributed to STAT3-dependent control of SOCS3, initially implicated STAT3 as a negative regulator of granulopoiesis.10 However, the amount of circulating neutrophils is the net result of production from myeloid progenitors, survival of developing and mature neutrophils, and neutrophil lifetime in the circulation. Our results show that STAT3 has a more complex role in granulopoiesis than previously recognized, with an obligate function in regulating granulocyte progenitor proliferation during emergency granulopoiesis. By contrast, basal neutrophilia in STAT3-deficient mice is not likely to result from increased progenitor numbers. We found that LSK and GMP numbers are similar in wild-type and STAT3-deficient mice under homeostatic conditions. Similarly, Lee et al10 reported that STAT3-deficiency did not affect resting granulocyte precursor amounts. Moreover, additional factors beyond the negative STAT3-SOCS3 feedback loop may contribute to basal neutrophilia in STAT3-deficient mice. STAT3 has a fundamental role in controlling neutrophil mobilization and chemotaxis, which appears to affect the ability of neutrophils to marginate or enter peripheral tissues.13,32 Defects in neutrophil migratory capabilities thus may contribute to neutrophil accumulation in the blood of STAT3-deficient animals. Therefore, the results to date show multiple roles for STAT3 affecting different stages of granulopoiesis, including a cell autonomous role in G-CSF–responsive progenitor proliferation, control of neutrophil migration,13,32 and regulation of the negative feedback inhibitor SOCS3.10
STAT3-dependent induction of LSKs during G-CSF– or L monocytogenes-driven granulopoiesis suggests the potential for STAT3 to control HSC proliferation in response to environmental stimuli,41 which may contribute to the enhanced production of myeloid cells during G-CSF therapy or infection.8 STAT3-deficient mice fail to accumulate immature granulocytes during G-CSF treatment,13 supporting the idea that STAT3-dependent regulation of progenitor growth responses is necessary for G-CSF–driven production of nascent granulocytes. Conversely, we considered whether HSCs and progenitor cell exhaustion because of excessive or sustained proliferation could underlie impaired emergency granulopoiesis in STAT3-deficient mice. We think this is unlikely because growth-promoting genes (eg, c-myc, Ccnd3, Cdk4, Cdc25a) are not constitutively active in STAT3-deficient hematopoietic progenitors and, alternatively, show dependence on STAT3 for G-CSF–responsive expression. In addition, we ruled out a role for suppressed G-CSFR expression in the refractory response of STAT3-deficient mice because myeloid progenitors show similar amounts of Csf3r mRNA and G-CSF–dependent STAT5 activation under resting conditions (data not shown). Moreover, SOCS3-deficient myeloid progenitors have increased STAT3 activation and undergo an extra round of cell division on G-CSF stimulation.42 These results provide additional evidence for STAT3-dependent growth-promoting signals in myelopoiesis and furthermore indicate that loss of the negative regulator SOCS3 is accompanied by STAT3 overactivation. Thus, the neutrophilia of SOCS3-deficient mice11 could result in part from excessive STAT3 growth-promoting signals. In terms of STAT3 function, it will be important to determine whether STAT3 is critical for G-CSF–mediated HSC self-renewal or exit from dormancy or both, which are poorly understood processes. This can be addressed by analysis of STAT3 function and STAT3 target gene activation in the HSC compartment in vivo.
Because C/EBPβ is an essential regulator of neutrophil production under demand conditions23 and both G-CSF and L monocytogenes infection require STAT3 to stimulate C/EBPβ expression, we propose that STAT3-dependent C/EBPβ induction may be a common response pathway used to induce myeloid output in stress conditions. Consistent with this, we found that suppression of C/EBPβ inhibited G-CSF–dependent proliferation of immature granulocytes. However, the mechanism of inducible Cebpb regulation by myeloid cytokines has been unclear. We demonstrated that STAT3 mediates G-CSF–responsive Cebpb induction in myeloid progenitor cells by interacting with the Cebpb proximal promoter. STAT3 associates with an atypical sequence, in agreement with studies that have shown plasticity in STAT3 interaction sites in vivo.43 Thus, our results suggest a direct molecular pathway initiated by extracellular G-CSF and executed by STAT3 to induce Cebpb transcription during emergency granulopoiesis.
One consequence of C/EBPβ up-regulation in myeloid cells is sustained c-myc expression, as indicated by C/EBPβ overexpression in K562 cells.23 By contrast, the related transcription factor C/EBPα abrogates c-myc transcription, has antiproliferative function and induces granulocyte differentiation.22,44 C/EBPα was shown to block c-myc transcription indirectly by inhibiting E2F function.22 We found C/EBPα was recruited to the endogenous c-myc proximal promoter. Moreover, C/EBPα overexpression abrogated G-CSF– and STAT3-responsive c-myc transcription from the c-myc proximal promoter. Thus, our results indicate C/EBPα inhibits c-myc transcription by a direct mechanism in addition to indirect regulation by E2F.
G-CSF stimulates STAT3 and C/EBPβ binding to the c-myc proximal promoter, while concomitantly shortening the duration of C/EBPα interaction. These results suggest that the G-CSF–STAT3–C/EBPβ signaling cascade attenuates the suppressive effect of C/EBPα on c-myc transcription, allowing for sustained c-myc expression during emergency granulopoiesis. Gene reporter assays confirmed opposing effects of C/EBPβ and C/EBPα on proximal c-myc promoter activity, with C/EBPβ overexpression overriding C/EBPα-mediated suppression to drive G-CSF–responsive c-myc transcription. Collectively, our results suggest STAT3 sustains c-myc transcription and thus myeloid progenitor cell-cycle progression by direct binding to the c-myc promoter, as well as attenuating C/EBPα and enhancing C/EBPβ interaction at c-myc promoter elements through control of C/EBPβ protein levels.
Regulation of the ratio of C/EBPα to C/EBPβ may control the timing of expression of neutrophil differentiation genes, such as the C/EBPα targets Csf3r and Pr3,22,45 which showed aberrant expression in STAT3-deficient immature granulocytes. Thus, proliferation and differentiation control mechanisms may not be distinctly separable or mutually exclusive. Rather, myeloid progenitor proliferative capacity and maturation status may be on a continuum, resulting from gradual changes in gene expression because of cell intrinsic events or microenvironmental cues or both. It is necessary to gain further insight into mechanisms controlling C/EBPα and C/EBPβ, as well as other myeloid transcriptional regulators, to understand the molecular control of granulopoiesis. G-CSF was recently shown to have instructive function on isolated primary hematopoietic progenitors,46 suggesting a potential for G-CSF–dependent signals such as STAT3 in granulocytic lineage specification. However, it remains unclear whether G-CSF instructive function is accompanied by induction of C/EBPα, C/EBPβ, or other myeloid transcriptional regulators. Furthermore, it is well established that granulopoiesis in vivo can be supported by G-CSF–independent pathways6,40; similarly, neutrophil development is STAT3 independent under steady state conditions,10,13 indicating that redundant pathways exist. The identity of granulopoiesis-inducing or -sustaining signals and their role in regulating critical myeloid transcription factors requires further investigation and may best be served by unbiased approaches involving genetic screens.47,48
In summary, our data indicate that STAT3 has a distinct role in G-CSF– or infection-driven granulopoiesis by activating the expression of the C/EBPβ and c-Myc transcriptional regulators and by promoting hematopoietic progenitor growth responses in vivo. Numerous studies have illustrated the critical role of STAT3 in immune regulation. The importance of STAT3 is further underscored by the identification of dominant-negative mutations in the human STAT3 gene in patients with hyperimmunoglobulin E syndrome (HIES), a primary immunodeficiency characterized by recurrent skin abscesses and lung infections.49 Innate immune responses of patients with HIES such as emergency granulopoiesis or neutrophil function have not been investigated extensively, to our knowledge. Deciphering the function of STAT3 during emergency granulopoiesis may lead to greater understanding of host defense mechanisms in patients with HIES, as well as persons with severe congenital neutropenia undergoing G-CSF therapy.
Acknowledgments
We thank Dr Shao-Cong Sun and Dr Nahum Puebla-Osorio for critical advice; Dr Jessica Schwartz, Dr Chen Dong, and Dr Hao Shen for providing reagents; L. Zhang for technical assistance; and Karen Ramirez and David He in the flow core facility at MDACC (supported by NCI P30CA16672) for assistance.
This work was supported by a predoctoral training grant (T32-CA-09598-16; H.N.-J. and A.D.P), a National Institutes of Health grant (AI073587), and a seed grant from the Center for Stem Cell and Developmental Biology at MDACC (S.S.W.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.Z. designed and performed experiments, analyzed data, and wrote the paper; P.J.M. designed and performed experiments and analyzed data; H.N.-J., A.D.P., and H.S.L. performed experiments and provided STAT3-deficient mice; S.S.W. designed experiments, supervised the project, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: This study was funded through an investigator-initiated Preclinical Research Agreement with Amgen Inc.
The current affiliation for A.D.P. is Gene Expression Laboratory, The Salk Institute for Biological Studies, La Jolla, CA.
Correspondence: Stephanie S. Watowich, Department of Immunology, The University of Texas M. D. Anderson Cancer Center, PO Box 301402, Unit 902, Houston, TX 77030-1903; e-mail: swatowic@mdanderson.org.
References
- 1.Boxer L, Dale DC. Neutropenia: causes and consequences. Semin Hematol. 2002;39(2):75–81. doi: 10.1053/shem.2002.31911. [DOI] [PubMed] [Google Scholar]
- 2.Eyles JL, Hickey MJ, Norman MU, et al. A key role for G-CSF-induced neutrophil production and trafficking during inflammatory arthritis. Blood. 2008;112(13):5193–5201. doi: 10.1182/blood-2008-02-139535. [DOI] [PubMed] [Google Scholar]
- 3.von Vietinghoff S, Ley K. Homeostatic regulation of blood neutrophil counts. J Immunol. 2008;181(8):5183–5188. doi: 10.4049/jimmunol.181.8.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Gwaiz LA, Babay HH. The diagnostic value of absolute neutrophil count, band count and morphologic changes of neutrophils in predicting bacterial infections. Med Princ Pract. 2007;16(5):344–347. doi: 10.1159/000104806. [DOI] [PubMed] [Google Scholar]
- 5.McKinstry WJ, Li CL, Rasko JE, Nicola NA, Johnson GR, Metcalf D. Cytokine receptor expression on hematopoietic stem and progenitor cells. Blood. 1997;89(1):65–71. [PubMed] [Google Scholar]
- 6.Lieschke GJ, Grail D, Hodgson G, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84(6):1737–1746. [PubMed] [Google Scholar]
- 7.Richards MK, Liu F, Iwasaki H, Akashi K, Link DC. Pivotal role of granulocyte colony-stimulating factor in the development of progenitors in the common myeloid pathway. Blood. 2003;102(10):3562–3568. doi: 10.1182/blood-2003-02-0593. [DOI] [PubMed] [Google Scholar]
- 8.Zhan Y, Lieschke GJ, Grail D, Dunn AR, Cheers C. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood. 1998;91(3):863–869. [PubMed] [Google Scholar]
- 9.Lord BI, Bronchud MH, Owens S, et al. The kinetics of human granulopoiesis following treatment with granulocyte colony-stimulating factor in vivo. Proc Natl Acad Sci U S A. 1989;86(23):9499–9503. doi: 10.1073/pnas.86.23.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17(1):63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 11.Croker BA, Metcalf D, Robb L, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20(2):153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 12.McLemore ML, Grewal S, Liu F, et al. Stat-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 2001;14(2):193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- 13.Panopoulos AD, Zhang L, Snow JW, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108(12):3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 15.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Iwasaki-Arai J, Iwasaki H, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6):853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Yamanaka R, Barlow C, Lekstrom-Himes J, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc Natl Acad Sci U S A. 1997;94(24):13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gombart AF, Shiohara M, Kwok SH, Agematsu K, Komiyama A, Koeffler HP. Neutrophil-specific granule deficiency: homozygous recessive inheritance of a frameshift mutation in the gene encoding transcription factor CCAAT/enhancer binding protein-epsilon. Blood. 2001;97(9):2561–2567. doi: 10.1182/blood.v97.9.2561. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P, Iwama A, Datta MW, Darlington GJ, Link DC, Tenen DG. Upregulation of interleukin 6 and granulocyte colony-stimulating factor receptors by transcription factor CCAAT enhancer binding protein alpha (C/EBP alpha) is critical for granulopoiesis. J Exp Med. 1998;188(6):1173–1184. doi: 10.1084/jem.188.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996;16(9):4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Wang X, Ward AC, Touw IP, Friedman AD. C/EBPalpha and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15(5):779–786. doi: 10.1038/sj.leu.2402094. [DOI] [PubMed] [Google Scholar]
- 22.Johansen LM, Iwama A, Lodie TA, et al. c-Myc is a critical target for C/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21(11):3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirai H, Zhang P, Dayaram T, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7(7):732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 24.Numata A, Shimoda K, Kamezaki K, et al. Signal transducers and activators of transcription 3 augments the transcriptional activity of CCAAT/enhancer-binding protein alpha in granulocyte colony-stimulating factor signaling pathway. J Biol Chem. 2005;280(13):12621–12629. doi: 10.1074/jbc.M408442200. [DOI] [PubMed] [Google Scholar]
- 25.Niehof M, Streetz K, Rakemann T, et al. Interleukin-6-induced tethering of STAT3 to the LAP/C/EBPbeta promoter suggests a new mechanism of transcriptional regulation by STAT3. J Biol Chem. 2001;276(12):9016–9027. doi: 10.1074/jbc.M009284200. [DOI] [PubMed] [Google Scholar]
- 26.Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193(6):741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161(9):4652–4660. [PubMed] [Google Scholar]
- 28.Panopoulos AD, Bartos D, Zhang L, Watowich SS. Control of myeloid-specific integrin alpha Mbeta 2 (CD11b/CD18) expression by cytokines is regulated by Stat3-dependent activation of PU.1. J Biol Chem. 2002;277(21):19001–19007. doi: 10.1074/jbc.M112271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busch DH, Vijh S, Pamer EG. Animal model for infection with Listeria monocytogenes. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1909s36. Chapter 19:Unit 19. [DOI] [PubMed] [Google Scholar]
- 30.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 31.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28(4):509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen-Jackson H, Panopoulos AD, Zhang H, Li HS, Watowich SS. STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood. 2010;115(16):3354–3363. doi: 10.1182/blood-2009-08-240317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hestdal K, Ruscetti FW, Ihle JN, et al. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J Immunol. 1991;147(1):22–28. [PubMed] [Google Scholar]
- 34.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13(6):738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 35.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 36.Theilgaard-Monch K, Jacobsen LC, Borup R, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105(4):1785–1796. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Fuentes NL, Fuller GM. Characterization of the IL-6 responsive elements in the gamma fibrinogen gene promoter. J Biol Chem. 1995;270(41):24287–24291. doi: 10.1074/jbc.270.41.24287. [DOI] [PubMed] [Google Scholar]
- 38.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 39.Hattori M, Abraham LJ, Northemann W, Fey GH. Acute-phase reaction induces a specific complex between hepatic nuclear proteins and the interleukin 6 response element of the rat alpha 2-macroglobulin gene. Proc Natl Acad Sci U S A. 1990;87(6):2364–2368. doi: 10.1073/pnas.87.6.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5(5):491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 41.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 42.Croker BA, Mielke LA, Wormald S, et al. Socs3 maintains the specificity of biological responses to cytokine signals during granulocyte and macrophage differentiation. Exp Hematol. 2008;36(7):786–798. doi: 10.1016/j.exphem.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallania F, Schiavone D, Dewilde S, et al. Genome-wide discovery of functional transcription factor binding sites by comparative genomics: the case of Stat3. Proc Natl Acad Sci U S A. 2009;106(13):5117–5122. doi: 10.1073/pnas.0900473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27(4):619–628. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88(4):1234–1247. [PubMed] [Google Scholar]
- 46.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325(5937):217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- 47.Kile BT, Hilton DJ. The art and design of genetic screens: mouse. Nat Rev Genet. 2005;6(7):557–567. doi: 10.1038/nrg1636. [DOI] [PubMed] [Google Scholar]
- 48.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9(7):554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 49.Minegishi Y, Saito M, Tsuchiya S, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]