Abstract
In large scale mass spectrometry-based phosphoproteomics, a current bottleneck is the unambiguous assignment of the phosphorylation site within the peptide. An additional problem is that it has been reported that under conditions wherein peptide ions are collisionally activated the phosphate group may migrate to a nearby phosphate group acceptor, thus causing ambiguity in site assignment. Here, we generated and analyzed a statistically significant number of phosphopeptides. Starting with a human cell lysate, we obtained via strong cation exchange fractionation nearly pure phosphopeptide pools from trypsin and Lys-N digestions. These pools were subjected to nano-LC-MS using an Orbitrap mass spectrometer that is equipped with both CID and electron transfer dissociation with supplemental activation (ETcaD) functionality. We configured a method to obtain sequentially both ETcaD and CID spectra for each peptide ion. We exploited the resistant nature of ETcaD toward rearrangement of phosphate groups to evaluate whether there is potentially phosphate group relocation occurring during CID. We evaluated a number of peptide and spectral annotation properties and found that for ∼75% of the sequenced phosphopeptides the assigned phosphosite was unmistakably identical for both the ETcaD and CID spectra. For the remaining 25% of the sequenced phosphopeptides, we also did not observe evident signs of relocation, but these peptides exhibited signs of ambiguity in site localization, predominantly induced by factors such as poor fragmentation, sequences causing inefficient fragmentation, and generally poor spectrum quality. Our data let us derive the conclusion that both for trypsin- and Lys-N-generated peptides there is little relocation of phosphate groups occurring during CID.
Signaling events, many of which are regulated by dynamic protein phosphorylation, form the basis of inter- and intracellular communication. Cellular activation, for instance, by a specific activation of a membrane receptor often induces changes in phosphorylation in that receptor but also in hundreds of other proteins connected to that receptor in downstream pathways. Obtaining in-depth phosphopeptide profiles consisting of significant numbers of sites on thousands of proteins has now become accessible to the specialized proteomics community (1). A number of key technologies and strategies have led to this breakthrough. Peptide complexity reduction and/or phosphopeptide enrichment can be considered to be one of the most improved aspects of these phosphoproteomics profiling methodologies. A classic strategy for a phosphoproteome screen is to perform a fractionation of the sample followed by enrichment using a reagent with affinity for the phosphate group (2). Strong cation exchange (SCX)1- (3–5) and SDS-PAGE (“GeLCMS”) (6, 7)-based fractionation are the most common, although hydrophilic interaction chromatography (8, 9) and isoelectric focusing (IEF) (10, 11) provide attractive alternatives. In the case of SCX and hydrophilic interaction chromatography, one can obtain nearly “uncontaminated” phosphopeptide enrichment when appropriate conditions and materials are chosen (5, 12). The classic choice for the second step has been immobilized affinity chromatography (IMAC) (13), which can have its selectivity improved even further through the use of additives (14) or methyl esterification of the peptide population (15). In recent years, one of the most popular technologies used for enrichment of phosphopeptides has been titanium dioxide (TiO2) (16) often with additives (17, 18), and it has been applied either in an off-line form or on line as part of the final nano-LC-MS step (19–21). Alternative metal oxides such as those of zirconium (22) have proven to be quite successful too. In the specific case of phosphotyrosine profiling, antibodies against phosphotyrosine, at the peptide level, have been demonstrated to be very effective (23–25). The increase in throughput of phosphoproteomics data has caused the community to focus on high throughput analysis of the mass spectrometric data to expedite interpretation. One aspect that has received much attention is the dominant neutral loss peak in the fragmentation spectra of phosphopeptides as obtained by traditional CID experiments (26, 27). The neutral loss issue is somewhat exacerbated for ion trap-based collision-induced dissociation primarily because of the energetic regime used and the need to use a discrete limited ion population (28). The neutral loss peak can often suppress sequence-diagnostic ion peaks, causing identification of the peptide to become difficult or even impossible. Because the use of ion traps currently represents the most common way of performing phosphoproteome screens, there have been various attempts to alleviate this specific problem. Modified fragmentation regimes have been introduced such as neutral loss-triggered MS3 (2, 4, 29) and “multistage activation” (30), which alleviate the neutral loss issue. Both of these methods fragment the neutral loss peak of the precursor ion further to generate more backbone cleavages, which then form the more diagnostic source for peptide sequencing. Alternatively, there has also been a re-exploration of tandem in space collision-induced dissociation in the form of the higher-energy collisional activated dissociation cell (31), which exhibits lower levels of neutral loss because the activation step occurs at higher energies and in shorter time frames (28). Electron transfer dissociation (ETD) and electron capture dissociation have also shown great promise because the phosphate group remains attached during and after activation (32–35). Many detected phosphopeptides contain multiple serine/threonine/tyrosine residues, representing the likely possibility that there is more than one possible location for the site of phosphorylation within the peptide. The abundant neutral loss observed in low energy CID can hamper the correct assignment of the phosphosites in such peptides. Therefore, a concerted effort has been made to understand, in detail, the rules of phosphopeptide fragmentation. Currently, there are a number of tools that allow automated site assignment (36, 37), including some that can exploit MSn (38) and electron capture dissociation/ETD data (39). Reid and co-workers (40) recently investigated phosphopeptide fragmentation from a more mechanistic point of view. Interestingly, their results demonstrated that the phosphate group neutral loss pathway observed in low energy CID operates, for the greater part, through an SN2 mechanism and not through the β-elimination pathway. This charge-directed mechanism requires proton abstraction as an initiation step and neighboring group participation. Worryingly, further mechanistic work, using synthetic phosphopeptides to explore the phosphate neutral loss pathway, allowed the identification of rearrangement reactions involving the phosphate group during low energy CID fragmentation. The rearrangement takes the form of a relocation of the phosphate group to an alternative hydroxyl group within the peptide. Thus, when performing a comprehensive annotation of the spectra, one would be able to observe multiple possibilities for the location of the phosphate group, referred to as phosphate group scrambling (41). The authors note that the issue is more apparent when there are no mobile protons (42) and that the scrambling will be more dominant when low energy millisecond time frames are used for CID such as in an ion trap. These observations evidently have raised even more concerns regarding the possible accuracy available for phosphosite localization.
Here, we investigate the extent of rearrangement reactions involving the phosphate group during low energy CID fragmentation by generating a large scale phosphoproteomics data set consisting of phosphopeptides fragmented sequentially and independently by both low energy CID and electron transfer dissociation with supplemental activation (ETcaD) using an ETD-enabled LTQ Orbitrap instrument. We utilize the ETcaD spectrum to allocate the “correct” phosphate group position and compare it with the position allocated by the CID spectrum.
MATERIALS AND METHODS
Digestion of Cell Lysate
1 mg of human cell lysate in 8 m urea was reduced with DTT (45 mm; 30-min incubation at 50 °C) and alkylated with iodoacetamide (100 mm; 30-min incubation). For the trypsin digestion, 1 mg of lysate was digested with Lys-C (1.25 μg; 4-h incubation at 37 °C) in 7 m urea followed by digestion with trypsin (15 μg; incubation overnight at 37 °C) after dilution to 2 m urea. For the Lys-N digestion, 1 mg of lysate was digested with Lys-N (5 μg; 3-h incubation at 37 °C) in 5 m urea followed by digestion with Lys-N (5 μg; incubation overnight at 37 °C) after dilution to 2 m urea.
Peptide Prefractionation by SCX
SCX was performed as described previously (5, 43). In brief, peptides from each digest were loaded onto two C18 cartridges using an Agilent 1100 HPLC system. The flow rate applied was 100 μl/min using water, pH 2.7 as solvent. After that, peptides were eluted from the trapping cartridges with 80% acetonitrile, pH 2.7 onto a PolySULFOETHYL A 200 × 2.1-mm column (PolyLC) for 10 min at the same flow rate. Separation of different peptide populations was performed using a non-linear 65-min gradient as follows: 0–10 min, 100% solvent A (5 mm KH2PO4, 30% acetonitrile, pH 2.7); 10–15 min, up to 26% solvent B (5 mm KH2PO4, 30% acetonitrile, 350 mm KCl, pH 2.7); 15–40 min, to 35% solvent B; and 40–45 min, to 60% solvent B. At 49 min, the concentration of solvent B was 100%. The column was subsequently washed for 6 min with high salt concentration and finally equilibrated with 100% solvent A for 9 min. The flow rate applied during the SCX gradient was 200 μl/min. Fractions were collected in 1-min intervals for 40 min. After evaporation of the solvents, fractionated peptides were resuspended in 10% formic acid.
Mass Spectrometry
SCX fractions were analyzed on a reversed phase nano-LC-coupled LTQ Orbitrap XL ETD instrument (Thermo Fisher Scientific). An Agilent 1200 series HPLC system was equipped with a 20-mm Aqua C18 (Phenomenex) trapping column (packed in house; 100-μm inner diameter, 5-μm particle size) and a 400-mm ReproSil-Pur 120 C18-AQ (Dr. Maisch GmbH) analytical column (packed in house; 50-μm inner diameter, 3-μm particle size). Trapping was performed at 5 μl/min solvent C (0.1 m acetic acid in water) for 10 min, and elution was achieved with a gradient from 10 to 30% (v/v) solvent D (0.1 m acetic acid in 1:4 acetonitrile:water) in solvent C in 110 min followed by a gradient of 30–50% (v/v) solvent D in solvent C in 30 min, a gradient of 50–100% (v/v) solvent D in solvent C in 5 min, and finally 100% solvent D for 2 min. The flow rate was passively split from 0.45 ml/min to 100 nl/min. Nanoelectrospray was achieved using a distally coated fused silica emitter (360-μm outer diameter, 20-μm inner diameter, 10-μm tip inner diameter; New Objective) biased to 1.7 kV. The LTQ Orbitrap ETD instrument was operated in the data-dependent mode to automatically switch between MS and MS/MS. Survey full-scan MS spectra were acquired from m/z 350 to m/z 1500 in the Orbitrap with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000 in the linear ion trap. The two most intense ions at a threshold of above 500 were fragmented in the linear ion trap using CID at a target value of 30,000 and ETcaD at a target value of 50,000. The ETcaD reagent target value was set to 100,000, and the reaction time was set to 50 ms.
Data Processing
From every raw data file recorded by the mass spectrometer, representing a single SCX fraction, two different peak lists containing either CID or ETcaD fragmentation data were generated using Proteome Discoverer (version 1.0, Thermo Fisher Scientific) with a signal-to-noise threshold of 3 and the following settings for the ETcaD non-fragment filter: precursor peak removal with 4 Da, charge-reduced precursor removal with 8 Da, and removal of known neutral losses from charge-reduced precursor with 8 Da within a window of 120 Da. Single fraction peak lists of the major phosphopeptide-containing SCX fractions for trypsin-derived and Lys-N-derived peptides were then merged into four larger peak lists, namely trypsin CID, trypsin ETcaD, Lys-N CID, and Lys-N ETcaD. Mascot (version 2.2.04, Matrix Science) was used to search these peak lists against an in-house built database (150,852 entries) assembled from the International Protein Index human database (version 3.54, http://www.ebi.ac.uk/ipi) plus all sequence-reversed entries of the latter. The following parameters were used for database searching: 5-ppm precursor mass tolerance; 0.6-Da fragment ion tolerance; up to two miscleavages allowed; carbamidomethylcysteine as a fixed modification; and phosphorylated serine, threonine, or tyrosine as a variable modification. The enzyme was specified as either trypsin or Lys-N, and the fragment ion type was specified as either ESI-TRAP or ETD-TRAP. Peptide matches were established at a significance level of <0.05 and above or equal to an ion score of 20. Only first ranking and bold peptides were considered for identification. The false discovery rate for this peptide identification process was estimated as the fraction of all peptides that matches to a reversed database entry and is between 0.6 and 1.0% for all four data sets. PTM scoring was performed only on the trypsin CID and Lys-N CID data sets using MSQuant (version 2.0a81, http://msquant.alwaysdata.net) (44). Further data processing was performed with Excel (supplemental Table 1 and Table 2). Scaffold data files (www.proteomesoftware.com) of fragmentation spectra are publicly available in the repository Tranche (https://proteomecommons.org/) using the following hash code: E64Vo7Il4PbzgMb2yUYidoarn/1yFptVEcStGn4OBDS61nbcM7/9WXcyRgT9mF6ASht9hBItFumDLKW6/FKVgLmCRfEAAAAAAAAGsA==.
RESULTS AND DISCUSSION
To study the potential effect of phosphate group rearrangement on the identification of a phosphorylation site, we performed in a single run sequentially both CID and ETCaD (45) on all peptides. The experiment was carried out using the following proteomics work flow. The phosphopeptide mixture was generated by digestion of a lysate of human cells with Lys-N (46) or trypsin followed by SCX, which was fine tuned for the enrichment of singly net charged phosphorylated peptides as described previously (5). Selected SCX fractions were further separated by on-line reversed phase nano-LC and analyzed on an LTQ Orbitrap ETD instrument that was configured to fragment every peptide precursor with both CID and ETcaD. The time required for the full CID or ETcaD scan, typically a few hundred milliseconds, was short compared with the elution period of a peptide, which is ∼1 min. Thus, the peptide precursor isolated for fragmentation by ETcaD was considered identical to the precursor isolated for fragmentation by CID. Two different peak lists, comprising either CID or ETcaD fragmentation data, were generated for both the Lys-N and trypsin analyses and searched separately with Mascot. After database searching, the peptide identifications obtained from CID and ETcaD peak lists were merged, resulting in a Lys-N data set and a trypsin data set.
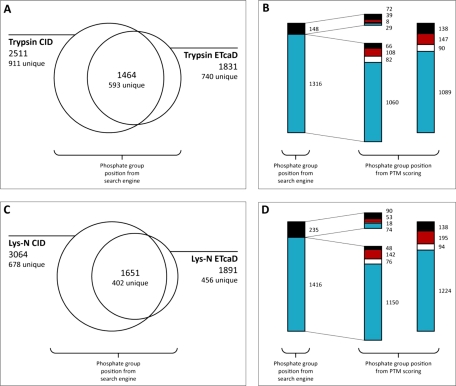
In the trypsin data set (Fig. 1A), 2511 singly phosphorylated peptides (of these, 911 unique) were identified by CID, and 1831 phosphorylated peptides (of these, 740 unique) were identified by ETcaD. From these two groups, 1464 phosphorylated peptides (of these, 593 unique) were identified with CID and ETcaD consecutive fragment ion spectra. For these peptides, for which sequence information derived from complementary fragmentation modes is available, 1316 peptides (of these, 561 unique) or 90% had been assigned identical phosphate group positions by Mascot without any further site validation or inspection of the Mascot delta score (Fig. 1B). The remaining 148 peptides (of these, 93 unique) or 10% had been assigned different phosphate group positions. To obtain a higher level of certainty about the localization of the phosphate group when derived from the CID spectrum, assuming the ETcaD spectrum provided the correct site assignments, we also evaluated the phosphate group position by PTM scoring as implemented in MSQuant (44), which is based on an algorithm that uses the four most intense ions per 100 m/z in an MS2 spectrum. When analyzing the phosphorylated 1464 peptides with PTM scoring, 1089 peptides or 74% had been assigned identical phosphate group positions, and 138 peptides or 9% had been assigned different phosphate group positions. The remaining 237 peptides or 17% could either not be scored by the PTM scoring algorithm or the position of the phosphate group in the CID spectrum could not be unambiguously determined, which was reflected by multiple highest scoring phosphate group positions with nearly equal scores. In this case, no decision was made between agreeing or disagreeing phosphate group positions because the CID spectra alone did not allow distinguishing between two or more possibilities. This group of peptides with ambiguous phosphate group positions might potentially contain spectra with highly abundant fragment ions indicative of phosphate group rearrangement; this is discussed in more detail below.
Fig. 1.
Number of phosphorylated peptides identified in trypsin and Lys-N data sets. A, comparison of the number of singly phosphorylated tryptic peptides identified from CID or ETcaD fragmentation spectra. The overlap represents the number of peptides that were identified from both CID and ETcaD fragmentation spectra in the cases where the ETcaD scan directly followed the CID scan. Peptides with different amino acid sequences were counted as unique irrespective of the position of the phosphate group. B, classification of peptides within the overlap according to the agreement or disagreement of the phosphate group positions. The left bar shows the fraction of peptides for which the position of the phosphate group was in agreement between CID and ETcaD (blue) and the fraction of peptides for which there was disagreement over the position (black) when taking the phosphate group position directly from the Mascot search engine. The right bar shows this classification according to PTM scoring. Peptides for which PTM scoring could not distinguish between two possible phosphate group positions are classified as ambiguous (red), and the groups of peptides that were not scored by PTM scoring are displayed in white. The middle bar depicts the same grouping but for the classes of agreeing and non-agreeing phosphate group positions when taken from the search engine. C and D show the same information as A and B but for the Lys-N data set.
The Lys-N data set (Fig. 1C) gave a picture very similar to that of the trypsin data set. Approximately 3000 singly phosphorylated peptides (of these, 678 unique) were identified by CID, and 1891 peptides (of these, 456 unique) were identified by ETcaD. The overlap is formed by 1651 peptides (of these, 402 unique), which were identified by both CID and subsequent ETcaD. For 1416 of these peptides (of these, 376 unique) or 86%, an identical position of the phosphate group had been derived from both the CID and ETcaD spectra, whereas for 235 peptides (of these, 109 unique) or 14%, the site was different (Fig. 1D). As for the trypsin data set, the CID spectra were processed with PTM scoring to validate the position of the phosphate group, resulting in the following figures. From the total of 1651 peptides, 1224 or 74% had been assigned identical phosphate group positions, and 138 or 8% displayed different phosphate group positions. The remaining 289 peptides or 18% either could not be scored or had multiple highest scoring phosphate group positions in the CID spectrum, classifying these peptides as ambiguous. Similar to the trypsin data set, the group of phosphopeptides containing ambiguous phosphate group positions is potentially most interesting.
In both data sets, the position of the phosphate group was identical for most peptides that had been identified from their CID and subsequent ETcaD spectra. Specifically, this was true for 86% (Lys-N) or 90% (trypsin) of all peptides with complementary sequencing information if the phosphate group position was taken directly from the search engine and for 74% if the position derived from the CID spectrum was validated by PTM scoring. As rearrangement of phosphate groups is suspected to take place only during CID in the ion trap but not during ETcaD, only CID spectra can potentially contain fragment ions indicative of gas phase rearrangement. If these features would have been more abundant, both in terms of number of CID spectra affected and in terms of intensities of the features in the CID spectra, we would have expected to see many more peptides with disagreeing phosphate group positions. In the cases the CID phosphate group position was evaluated by PTM scoring, we would have expected a higher level of peptides with ambiguous phosphate group position in CID. From all our observations, we conclude that if some gas phase rearrangement had taken place it did not affect the identification of the phosphate group position in the majority of cases; i.e. identical phosphate group positions were observed between CID and ETcaD spectra. Irrespective of this conclusion, gas phase rearrangement might still have taken place during CID fragmentation with or without having affected the determination of the correct phosphate group position by the search engine or by PTM scoring. One possible way is that the group of peptides for which identical phosphate group positions were obtained might still contain CID spectra with features indicative of phosphate group rearrangement, but these features might have been of such low abundance that they did not hinder the search engine or PTM scoring reporting the correct phosphate group position. Alternatively, within the groups of peptides for which different or ambiguous phosphate group positions were reported, the indicative fragment ions corresponding to phosphate relocation might have been dominant in the CID spectra such that the correct phosphate group position was not reported by the search engine or PTM scoring. In the case of phosphate group position ambiguity, the PTM scoring algorithm might not have been able to distinguish between the correct and the rearranged phosphate group position, leading to multiple highest scoring positions.
If rearrangement had taken place, in all three peptide groups (identical position, different position, and ambiguous position), it would only have affected the CID spectra but not the ETcaD spectra because ETD has limited intramolecular energy dissipation (47). Therefore, the probability of a less likely peptide match with identical amino acid sequence but different phosphate group position should have increased for a peptide match when derived from the CID spectrum but not when derived from the ETcaD spectrum. This difference in probability is directly reflected in the Mascot delta ion score of a peptide, which is the difference of the ion score of the most likely, i.e. identified, peptide match and the ion score of the next ranking, sequence-identical peptide match. We used this delta ion score as a measure to compare the significance of a phosphate group positioning in CID and ETcaD (28). For calculating the delta ion score, we also used peptide sequence matches with a rank lower than the upper two matches if the matched amino acid sequence was identical to the highest ranking peptide sequence match. In this case, as the two peptides sequences for which the delta ion score is calculated are identical in their primary amino acid sequence and only differ in the position of the phosphate group, the delta ion score is directly related to the presence and intensity of the fragment ions that can distinguish the two possible phosphorylation sites.
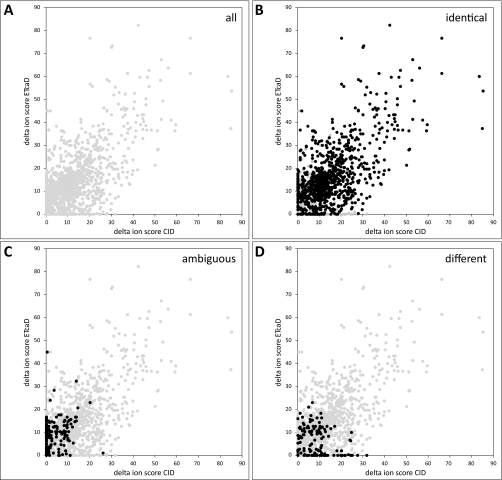
In the trypsin data set, for 1167 (or 90%) of the 1374 phosphorylated peptides for which complementary sequencing information was available and which could be grouped by PTM scoring, a delta ion score could be calculated for both CID and ETcaD. The remaining 207 (or 10%) of the peptides did not have a next lower ranking, sequence-identical peptide match in either CID, ETcaD, or both. In the Lys-N data set, for 1257 (or 78%) of 1618 peptides, a delta ion score could be calculated for both CID and ETcaD, whereas the remaining 361 (or 22%) of the peptides had no delta ion score in either CID, ETcaD, or both. In both data sets, many peptides without a delta ion score contain only a single serine, threonine, or tyrosine residue and hence cannot show rearrangement per se. In the trypsin data set, the comparison of the delta ion score for CID versus the delta ion score for ETcaD, broken down to the three peptide groups that were generated by PTM scoring, showed that all peptides are relatively evenly distributed over the delta ion score space (Fig. 2A). Peptides for which identical phosphate group positions had been determined showed the same distribution (Fig. 2B). Peptides for which non-identical phosphate group positions had been determined showed a mild preference toward a small delta ion score in ETcaD coupled with a more widely distributed delta ion score in CID (Fig. 2D). Logically, peptides with ambiguous phosphate group positions had a mild bias toward a larger delta ion score in ETcaD (Fig. 2C). The Lys-N data set displayed very similar behavior in all groups of peptides (supplemental Fig. 1); that is the same distortions of the delta score distributions were observed for peptides for which ambiguous or non-identical phosphate group positions had been found although slightly less clear cut than in the trypsin data set.
Fig. 2.
Ambiguity of phosphosite assignment evaluated by delta ion scores. All plots show delta ion scores for ETcaD versus delta ion scores for CID for the trypsin data set. A, all peptides (gray). B, peptides with identical phosphate group position (black) versus all peptides (gray). C, peptides for which the phosphate position in CID was ambiguous (black) versus all peptides (gray). D, peptides with non-identical phosphate group position (black) versus all peptides (gray).
Based on the analysis of the delta ion score distributions of peptides with non-identical phosphate group positions, we concluded that these peptides were classified as such because the CID and ETcaD spectra did not provide sufficient certainty to distinguish different potential phosphorylation sites. The delta ion score distributions of the group of peptides with ambiguous phosphate group positions gave a similar impression. In both groups (ambiguous and non-identical), the uncertainty of phosphate group position, as reflected by the delta ion score, was not related to the spectral quality as reflected by the ion score (supplemental Fig. 2). Both the CID and ETcaD ion scores of peptides in these two groups were evenly distributed over the whole score range, and most had a delta score below 15. This observation was also true when the CID and ETcaD ion scores were normalized for the peptide length (supplemental Fig. 3), a normalization method that takes into account that peptide ion scores generally increase with peptide length even though spectral quality does not improve (34). However, when manually inspecting spectra from both peptide groups (ambiguous and non-identical), we found that many peptides had either a low CID or a low ETcaD ion score, reflecting either low quality CID or low quality ETcaD spectra; i.e. one of the pair of spectra was poor. We also observed that many peptides showing ambiguous or non-identical phosphate group positions had other properties that had hindered the clear determination of the phosphate group in either the CID or the ETcaD spectra. One of the most influential properties is the number of serine/threonine/tyrosine residues and their relative locations. As the number of residues increases, the possibility of unambiguously detecting the phosphate group position correctly in CID decreases. In both data sets, we observed that peptides with ambiguous or non-identical phosphate group positions are biased toward containing higher numbers of phosphorylatable residues (supplemental Fig. 4). Another noteworthy property that hinders the assignment of the phosphate group position, particularly in ETcaD spectra, because of the fragmentation regime is the localization of phosphorylatable residues close to the termini of the peptide as small m/z fragment ions are unlikely to be detected abundantly, if at all, in the ion trap (48–50). To conclude, when manually inspecting CID and ETcaD spectra of peptides with ambiguous or non-identical phosphate group position, no fragment ions that could have clearly indicated phosphate group rearrangement could be found. In fact, for every peptide investigated in these two groups, one or more of the factors prevented the unambiguous or even correct identification of the phosphate group in either the CID or ETcaD spectrum or both. Moreover, in cases where a fragment ion indicative of phosphate group rearrangement might have been found, it was not distinguishable from noise.
Although our observations for singly phosphorylated peptides suggested a minimal effect of scrambling, this may not be the case for all types of phosphorylated peptide. We extended our approach for evaluating phosphorylation site scrambling to peptides containing more than one phosphate group. Access to such peptides is made possible by the SCX strategy, which provided the above phosphorylated peptide pools (5). Essentially, multiply phosphorylated peptides elute close to the beginning of the SCX separation. Eight of the early fractions, which should contain multiply phosphorylated peptides, were subjected to the alternating CID/ETcaD analysis regime. In this trypsin data set (supplemental Fig. 5 and supplemental Table 3), 483 doubly phosphorylated peptides (of these, 141 unique) were identified by CID, and 210 (of these, 79 unique) were identified by ETcaD. Please note that all phosphorylated peptide spectra are available in Tranche in the form of a Scaffold file. From these two groups, however, only 29 doubly phosphorylated peptides (of these, 17 unique) were identified with CID and ETcaD from consecutive fragment ion spectra. The spectra of multiply phosphorylated peptides subjected to low energy ion trap CID are dominated by neutral loss peaks, whereas ETcaD struggles because the peptides are often ionized as doubly charged (9), explaining the poor overlap. Nevertheless, 26 peptides of the overlapping 29 had been assigned identical phosphorylation sites by Mascot without any further site validation. However, manual validation showed that most spectra and consequently phosphorylation site assignments were rather poor due to the generally worse fragmentation of multiple phosphorylated peptides. This was also reflected in many peptides having very small Mascot delta ion scores. In agreement with the finding for the singly phosphorylated peptides, no indications for phosphate group rearrangement could be found in this small set of doubly phosphorylated peptides, making our conclusion more general.
Our SCX approach has been refined to isolate phosphopeptides with a solution net charge of 0 (doubly phosphorylated) and 1+ (singly phosphorylated) in which the major population of peptides contains two basic moieties (5). Considering that our peptides in the gas phase will have a minimum of two charges for tandem MS to occur, then the majority of the peptides contained within our data set possessed “mobile” protons (42). Thus, our findings are in agreement with Palumbo and Reid (41) who observed that fragment ions indicating phosphate group rearrangement are much more prominent for peptides in a non-mobile proton situation than for peptides in a partially mobile or even less mobile proton situation.
In summary, we have assessed the effect of gas phase phosphate group rearrangement under LC-MS conditions typically used in a proteomics experiment. By comparing phosphate group positions derived from CID and ETcaD spectra, we observed in both data sets that for 74% of all phosphopeptides for which complementary sequencing information exists assignments of phosphate group positions were identical. Moreover, close inspection of the remaining 26% also revealed no clear evidence for significant phosphate group relocation. The spectrum quality of these 26% was generally lower due to factors such as poor fragmentation and sequences causing inefficient fragmentation. This indicates that, although we cannot exclude that some phosphate group rearrangement might have occurred, it did not affect the identification of the correct phosphosites in large data sets of trypsin- and Lys-N-generated peptides.
Supplementary Material
* This work was supported in part by the Netherlands Proteomics Centre, which is part of the Netherlands Genomics Initiative.
 This article contains supplemental Figs. 1–5 and Tables 1–3.
This article contains supplemental Figs. 1–5 and Tables 1–3.
1 The abbreviations used are:
- SCX
- strong cation exchange
- ETcaD
- electron transfer dissociation with supplemental activation
- ETD
- electron transfer dissociation
- PTM
- post-translational modification.
REFERENCES
- 1.Reinders J., Sickmann A. (2005) State-of-the-art in phosphoproteomics. Proteomics 5, 4052–4061 [DOI] [PubMed] [Google Scholar]
- 2.Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 3.Motoyama A., Xu T., Ruse C. I., Wohlschlegel J. A., Yates J. R., 3rd (2007) Anion and cation mixed-bed ion exchange for enhanced multidimensional separations of peptides and phosphopeptides. Anal. Chem. 79, 3623–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 6.Everley P. A., Bakalarski C. E., Elias J. E., Waghorne C. G., Beausoleil S. A., Gerber S. A., Faherty B. K., Zetter B. R., Gygi S. P. (2006) Enhanced analysis of metastatic prostate cancer using stable isotopes and high mass accuracy instrumentation. J. Proteome Res. 5, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 7.Elortza F., Nühse T. S., Foster L. J., Stensballe A., Peck S. C., Jensen O. N. (2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 8.Gilar M., Olivova P., Daly A. E., Gebler J. C. (2005) Orthogonality of separation in two-dimensional liquid chromatography. Anal. Chem. 77, 6426–6434 [DOI] [PubMed] [Google Scholar]
- 9.Boersema P. J., Taouatas N., Altelaar A. F., Gouw J. W., Ross P. L., Pappin D. J., Heck A. J., Mohammed S. (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell. Proteomics 8, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cargile B. J., Talley D. L., Stephenson J. L., Jr. (2004) Immobilized pH gradients as a first dimension in shotgun proteomics and analysis of the accuracy of pI predictability of peptides. Electrophoresis 25, 936–945 [DOI] [PubMed] [Google Scholar]
- 11.Krijgsveld J., Gauci S., Dormeyer W., Heck A. J. (2006) In-gel isoelectric focusing of peptides as a tool for improved protein identification. J. Proteome Res. 5, 1721–1730 [DOI] [PubMed] [Google Scholar]
- 12.McNulty D. E., Annan R. S. (2008) Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics 7, 971–980 [DOI] [PubMed] [Google Scholar]
- 13.Stensballe A., Andersen S., Jensen O. N. (2001) Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics 1, 207–222 [DOI] [PubMed] [Google Scholar]
- 14.Stensballe A., Jensen O. N. (2004) Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides. Rapid Commun. Mass Spectrom. 18, 1721–1730 [DOI] [PubMed] [Google Scholar]
- 15.Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 16.Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 17.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 18.Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 19.Pinkse M. W., Mohammed S., Gouw J. W., van Breukelen B., Vos H. R., Heck A. J. (2008) Highly robust, automated, and sensitive on line TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 20.Cantin G. T., Shock T. R., Park S. K., Madhani H. D., Yates J. R., 3rd (2007) Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal. Chem. 79, 4666–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammed S., Kraiczek K., Pinkse M. W., Lemeer S., Benschop J. J., Heck A. J. (2008) Chip-based enrichment and nanoLC-MS/MS analysis of phosphopeptides from whole lysates. J. Proteome Res. 7, 1565–1571 [DOI] [PubMed] [Google Scholar]
- 22.Kweon H. K., Håkansson K. (2008) Metal oxide-based enrichment combined with gas-phase ion-electron reactions for improved mass spectrometric characterization of protein phosphorylation. J. Proteome Res. 7, 749–755 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 24.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 25.Boersema P. J., Foong L. Y., Ding V. M., Lemeer S., van Breukelen B., Philp R., Boekhorst J., Snel B., den Hertog J., Choo A. B., Heck A. J. (2010) In-depth qualitative and quantitative profiling of tyrosine phosphorylation using a combination of phosphopeptide immunoaffinity purification and stable isotope dimethyl labeling. Mol. Cell. Proteomics 9, 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huddleston M. J., Annan R. S., Bean M. F., Carr S. A. (1993) Selective detection of phosphopeptides in complex-mixtures by electrospray liquid-chromatography mass-spectrometry. J. Am. Soc. Mass Spectrom. 4, 710–717 [DOI] [PubMed] [Google Scholar]
- 27.DeGnore J. P., Qin J. (1998) Fragmentation of phosphopeptides in an ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 9, 1175–1188 [DOI] [PubMed] [Google Scholar]
- 28.Boersema P. J., Mohammed S., Heck A. J. (2009) Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 44, 861–878 [DOI] [PubMed] [Google Scholar]
- 29.Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 30.Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 31.Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap-Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammed S., Lorenzen K., Kerkhoven R., van Breukelen B., Vannini A., Cramer P., Heck A. J. (2008) Multiplexed proteomics mapping of yeast RNA polymerase II and III allows near-complete sequence coverage and reveals several novel phosphorylation sites. Anal. Chem. 80, 3584–3592 [DOI] [PubMed] [Google Scholar]
- 35.Sweet S. M., Bailey C. M., Cunningham D. L., Heath J. K., Cooper H. J. (2009) Large scale localization of protein phosphorylation by use of electron capture dissociation mass spectrometry. Mol. Cell. Proteomics 8, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 37.Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 38.Ruttenberg B. E., Pisitkun T., Knepper M. A., Hoffert J. D. (2008) PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J. Proteome Res. 7, 3054–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey C. M., Sweet S. M., Cunningham D. L., Zeller M., Heath J. K., Cooper H. J. (2009) SLoMo: automated site localization of modifications from ETD/ECD mass spectra. J. Proteome Res. 8, 1965–1971 [DOI] [PubMed] [Google Scholar]
- 40.Palumbo A. M., Tepe J. J., Reid G. E. (2008) Mechanistic insights into the multistage gas-phase fragmentation behavior of phosphoserine- and phosphothreonine-containing peptides. J. Proteome Res. 7, 771–779 [DOI] [PubMed] [Google Scholar]
- 41.Palumbo A. M., Reid G. E. (2008) Evaluation of gas-phase rearrangement and competing fragmentation reactions on protein phosphorylation site assignment using collision induced dissociation-MS/MS and MS3. Anal. Chem. 80, 9735–9747 [DOI] [PubMed] [Google Scholar]
- 42.Wysocki V. H., Tsaprailis G., Smith L. L., Breci L. A. (2000) Mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 35, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 43.Taouatas N., Altelaar A. F., Drugan M. M., Helbig A. O., Mohammed S., Heck A. J. (2009) Strong cation exchange-based fractionation of Lys-N-generated peptides facilitates the targeted analysis of post-translational modifications. Mol. Cell. Proteomics 8, 190–200 [DOI] [PubMed] [Google Scholar]
- 44.Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 45.Swaney D. L., McAlister G. C., Wirtala M., Schwartz J. C., Syka J. E., Coon J. J. (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taouatas N., Drugan M. M., Heck A. J., Mohammed S. (2008) Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat. Methods 5, 405–407 [DOI] [PubMed] [Google Scholar]
- 47.Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 49.Molina H., Matthiesen R., Kandasamy K., Pandey A. (2008) Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal. Chem. 80, 4825–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hennrich M. L., Boersema P. J., van den Toorn H., Mischerikow N., Heck A. J., Mohammed S. (2009) Effect of chemical modifications on peptide fragmentation behavior upon electron transfer induced dissociation. Anal. Chem. 81, 7814–7822 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.