Abstract
Blue native electrophoresis is one of the most popular techniques for mass estimation of native membrane proteins, but the use of non-optimal mass markers and acrylamide gels can compromise accuracy and reliability of the results. We present short protocols taking 10–30 min to prepare optimal sets of membrane protein markers from chicken, rat, mouse, and bovine heart. Especially heart materials from local supermarkets or butcher's shops, e.g. chicken or bovine heart, are ideal sources of high mass membrane protein standards. Considerable discrepancies between the migration behavior of membrane and soluble markers suggest using membrane protein markers for mass estimation of membrane proteins. Soluble standard proteins can be used, with some limitations, when soluble proteins are the focus. Principles and general rules for the determination of mass and oligomeric state of native membrane and soluble proteins are elaborated, and potential pitfalls are discussed.
Blue native electrophoresis (BNE),1 also known as blue native polyacrylamide gel electrophoresis (BN-PAGE; Refs. 1–3), separates native proteins and protein complexes in the mass range of 10 kDa to 10 MDa (2, 3). Clear native electrophoresis (CNE; Refs. 2, 4) and high resolution clear native electrophoresis (hrCNE; Refs. 5, 6) are non-colored variants of the same electrophoresis system that differ essentially by the composition of the cathode buffer, the upper buffer in the vertical electrophoresis apparatus. A seemingly minor variation of the cathode buffer has a large impact on separation principles, resolution, and general usefulness. BNE is the most robust and preferred variant except for the analysis of fluorescently labeled proteins or in-gel catalytic activity assays for which hrCNE seems optimal. CNE is rarely applied because of its limitation to the separation of acidic soluble proteins and considerable aggregation of membrane proteins. Features and applications of blue native and clear native electrophoresis have been reviewed recently (7). Here we focus on the use of BNE and hrCNE to estimate native protein masses and oligomeric states using microgram amounts of partially purified protein.
Proteins migrate according to their intrinsic isoelectric point (pI) in CNE because CNE does not involve charged compounds that could bind to proteins and impose a charge shift. Proteins with pI <7.5 show anodic migration in the native gels with pH 7.5. Proteins with pI >7.5 migrate toward the cathode and are lost. The cathode buffer for BNE is the same as for CNE except that Coomassie Blue G-250 (abbreviated here as Coomassie dye) is added. The anionic dye binds to hydrophobic protein surfaces and induces a charge shift. Therefore, all membrane proteins migrate to the anode, even the basic membrane proteins. Coomassie dye also binds to basic amino acid residues. This is why some but not all soluble proteins bind the dye, and even strongly basic soluble proteins can show anodic migration. hrCNE is a charge shift technique like BNE. The cathode buffer for hrCNE contains mixed anionic micelles formed from a neutral and an anionic detergent that can bind to all hydrophobic and to some soluble proteins similar to the Coomassie dye in BNE.
BNE was originally used to study the five major complexes from solubilized bovine heart mitochondria (BHM) because the mitochondrial complexes were highly abundant and could be detected as strong bands in the gels after Coomassie staining (1). Later on, research groups having access to fresh slaughterhouse material used solubilized BHM also for mass calibration in BNE. Others preferred to use a set of commercially available water-soluble proteins for mass calibration because some selected soluble proteins had been shown to fit the calibration curve defined by membrane protein complexes from BHM under certain conditions (2). This coincidence of the mass/migration plots of soluble and DDM-solubilized membrane proteins was surprising because soluble proteins, in contrast to membrane proteins, mostly do not contain bound lipid, detergent, and Coomassie dye that can equal or even exceed the mass of the membrane protein moiety (7, 8). One would therefore expect considerable error in mass estimation of membrane proteins whenever soluble proteins are used for calibration and vice versa. Remarkably, such error has been observed only for the mass estimation of small membrane proteins so far (8). Here we show that depending on the gel type used and the timing of electrophoresis the calibration curves of membrane and soluble proteins can almost coincide or strongly diverge. This raises some concerns about the accuracy of the vast number of published mass estimations of membrane proteins that used soluble marker proteins for mass calibration in BNE.
Considering these limitations, soluble marker proteins should not be used for mass estimation of membrane proteins except with special gel types and electrophoresis conditions. Membrane protein markers, e.g. from isolated bovine heart mitochondria, seem more reliable. Here we demonstrate that isolation of mitochondria is not required when mitochondrion-rich heart tissue from different animals is homogenized and used directly for the extraction of membrane protein complexes by detergents. As convenient sources of high molecular mass protein standards we identified heart from laboratory mouse and rat and chicken and bovine heart that are available from local supermarkets and butcher's shops.
EXPERIMENTAL PROCEDURES
Electrophoretic Techniques
BNE, CNE, hrCNE, and associated techniques like two-dimensional Tricine-SDS-PAGE and staining were performed as described (3–5).
Casting Acrylamide Gradient Gels for Native Electrophoresis
Linear acrylamide gradient gels were cast in the cold (4–7 °C) using a gradient mixer and solutions as described recently (3). Long cannulas were used to cast the gels from the bottom; i.e. the light solution was pumped in first and then underlayed with acrylamide solution of increasing density. Some further experimental details were added in the following to minimize unintended experimental variation between different laboratories. A larger volume of the low acrylamide solution was added to the exit arm of the gradient mixer (e.g. 18 ml of a 3.5% acrylamide solution), whereas a smaller volume of the high acrylamide solution was added to the other arm (e.g. 15 ml of a 13% acrylamide solution containing glycerol to increase density). Pumping 3 ml of the low acrylamide solution into the preassembled gel casting device guaranteed that all gels started reproducibly with a 3.5% acrylamide layer and that equal fluid levels were reached in both arms. At that time point, the tube connecting both arms was opened, and formation of the linear gradient could start. It is important to note that, at the end of gradient mixing, a certain volume of the acrylamide solution remains in the gradient mixer and in the tubing. This dead volume must be measured at least once to find the actual final acrylamide concentration in the gradient gels. In our hands, this residual volume was around 4 ml. This means that we obtained a 3.5–12% acrylamide gel when we used 13% acrylamide for the high percent acrylamide solution. Similarly, an 18% acrylamide solution was required to cast a 3.5–16% acrylamide gradient gel. Supplemental Fig. S6 schematically depicts the relative portions of the 3.5% acrylamide layer, the linear gradient, and the discarded residual volume and helps to set the required acrylamide concentration in the high percent acrylamide solution.
Homogenization of Chicken and Mammalian Heart for BNE
Chicken heart from the local supermarket is commonly obtained in the non-frozen state. It can be frozen for later use or processed immediately. Mitochondrial membrane protein complexes and supercomplexes were equally well preserved using frozen and non-frozen heart as analyzed by two-dimensional BN/SDS-PAGE under various detergent conditions (not shown).
Fat is removed from one non-frozen or frozen/thawed chicken heart (around 5 g). Around 1–2 g of the heart muscle is cut into thin slices. Similarly, rat heart (around 1 g) or mouse heart (around 50–100 mg each; sufficient for a maximum of 40 mass calibrations) and bovine heart can be processed by the same protocol or on a proportionally reduced or extended scale. The slices are then frayed out using tweezers. It is essential that this tissue disintegration is done very carefully to facilitate further homogenization. Minced heart tissue (1 g) is then suspended using 9 ml of homogenization buffer (250 mm sucrose, 20 mm sodium phosphate, 1 mm EDTA, 2 mm 6-aminohexanoic acid, pH 7.0) and homogenized by a 10-ml motor-driven glass/Teflon Potter-Elvehjem homogenizer (500 cycles/min) to obtain a 10% homogenate. No sediments of unminced tissue should be left on the bottom of the homogenizer. The homogenate is then portioned into 0.05- and 0.1-ml aliquots corresponding to 5 and 10 mg of heart tissue or two and four mass calibrations, respectively. Following centrifugation (5 min, 10,000 × g) the supernatants are discarded. The pellets are rapidly frozen in liquid nitrogen and stored at −80 °C.
Solubilization of Membrane Protein Complexes from Chicken and Mammalian Heart for Mass Estimation by Native Electrophoresis
It is important to use the same detergent that was used for solubilization of the protein of interest also to solubilize the mitochondrial mass markers. Protocols to solubilize individual mitochondrial protein complexes I–V with masses up to 1 MDa or to also solubilize supercomplexes with masses larger than 2 MDa essentially differ only by the detergent used. Dodecylmaltoside and Triton X-100 solubilize the individual mitochondrial complexes from heart homogenates, whereas digitonin also can preserve supramolecular structures as ideal markers for the higher mass range.
Pelleted heart homogenate (pellet from 5 mg of heart) is suspended in 40 μl of low salt buffer (50 mm NaCl, 1 mm EDTA, 2 mm 6-aminohexanoic acid, 50 mm imidazole/HCl, pH 7.0) and solubilized with 4 μl of dodecylmaltoside (10%), 4–8 μl of Triton (10%), or 4 μl of digitonin (20%). Following centrifugation (5 min, 100,000 × g or 10 min, 20,000 × g) the supernatant is collected, and 5 μl of Ponceau S stock (0.1% Ponceau S dye in 70% glycerol) is added to facilitate sample application to BN, clear native, and high resolution clear native gels. Specifically for BNE, Coomassie dye is finally added to the samples to set a 1:8 Coomassie dye/detergent ratio (1–2 μl from a 5% Coomassie Blue G-250 stock suspended in 500 mm 6-aminohexanoic acid). 20 μl of each sample, corresponding to 2 mg of total chicken or mammalian heart, is finally applied to the gel wells (0.15 × 0.5 cm). Native electrophoresis in a cold room or a refrigerated box (4–7 °C) is then started under conditions that do not significantly warm up the gel, e.g. with a constant power of 5 watts using 14 × 14 × 0.15-cm gels.
Mass Spectrometry
Sample preparation for mass spectrometry, MALDI-TOF, and MALDI-TOF/TOF analyses are described in the supplemental methods. Mass spectrometric data are detailed in supplemental Figs. S7–S23 and supplemental Tables S6–S9.
RESULTS
Exploring Principles of Mass Estimation by BNE
Migration velocity of proteins decelerates during BNE because the pore size of the acrylamide gradient gels used decreases gradually with increasing migration distance of the proteins. Depending on their size, proteins are finally stopped at specific acrylamide concentrations or pore size limits in the gel provided that the final acrylamide concentration in the acrylamide gradient gel is sufficiently high.
Determining End Points of Migration of Proteins in Acrylamide Gradient Gels
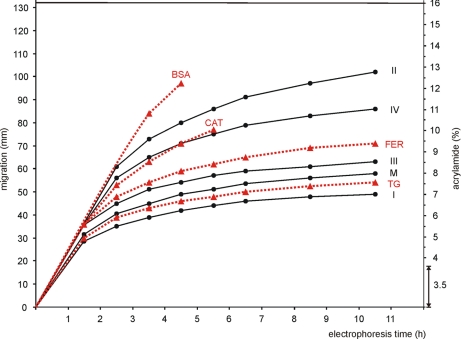
We were interested to measure the acrylamide concentrations in gradient gels that can stop proteins of a given size or mass. For this purpose we applied membrane protein markers (DDM-solubilized complexes from BHM) and a high molecular weight kit of soluble proteins (GE Healthcare) to a linear 3.5–16% acrylamide gradient gel for BNE and followed the migration of protein bands during electrophoresis by their blue color. Native electrophoresis at a constant power of 5 watts was paused repeatedly in 1–2-h intervals, and the actual positions of the visible blue bands were marked on the covering glass plate. After completion of the electrophoresis (10.5 h) the migration distances at the different time points were measured and plotted versus the time of electrophoresis (Table I and Fig. 1). The BN gel was finally fixed and Coomassie-stained (Fig. 2E).
Table I. Mass calibration in BNE using unfixed 3.5–16% acrylamide gels.
Dodecylmaltoside-solubilized complexes from bovine heart mitochondria and soluble proteins of the high molecular weight kit were used as mass markers. Proteins with assigned migration distances were visible as blue bands during BNE. The mass/migration data were used for Fig. 1 and supplemental Fig. S1. Coom., Coomassie binding capability of proteins. Dashes (—) indicate migration that could not be estimated at certain time points.
| Marker proteins | Source | pI | Coom. | Mass | Migration after various electrophoresis times |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.5 h | 2.5 h | 3.5 h | 4.5 h | 5.5 h | 6.5 h | 8.5 h | 10.5 h | |||||
| kDa | mm | |||||||||||
| Membrane protein complexes | ||||||||||||
| Monomeric RC I | BHM | <7 | Yes | ∼1000a | 28.5 | 35.0 | 39.0 | 42.0 | 44 | 46.0 | 48.0 | 49.0 |
| Monomeric complex V | BHM | <7 | Yes | 597a | 31.5 | 40.5 | 45.0 | 49.0 | 51 | 53.5 | 56.0 | 58.0 |
| Dimeric RC III | BHM | <7 | Yes | 482a | 35.5 | 45.0 | 51.0 | 54.0 | 57 | 59.0 | 61.0 | 63.0 |
| Monomeric RC IV | BHM | <7 | Yes | 205a | — | 56.0 | 65.0 | 71.0 | 75 | 79.0 | 83.0 | 86.0 |
| Monomeric RC II | BHM | <7 | Yes | 123a | — | 61.0 | 73.0 | 80.0 | 86 | 91.0 | 97.0 | 102.0 |
| High molecular weight kitb | ||||||||||||
| Thyroglobulin dimer | Porcine thyroid | — | Yes | 669c | 30.0 | 39.0 | 43.0 | 46.0 | 48 | 50.0 | 52.5 | 54.0 |
| Ferritin | Equine spleen | 5.4a; 4.2–4.5d | No | 440c; 476a | 36.0 | 48.0 | 54.0 | 59.0 | 62 | 65.0 | 69.0 | 71.0 |
| Catalase | Bovine liver | 6.8a; 5.4e | Yes | 232c; 239a | — | 53.0 | 63.0 | 71.0 | 77 | — | — | — |
| Lactate dehydrogenase | Bovine heart | 6.0a; ∼5.0f | No | 140c; 146a | — | — | — | — | — | — | — | — |
| BSA | Bovine serum | 5.6a; 4.9d | Yes | 66c; 66a | — | — | 84.0 | 97.0 | — | — | — | — |
a Calculated for the mature protein using the Swiss-Prot database and the pI/Mw tool from the ExPASy proteomics server (www.expasy.ch).
b From GE Healthcare.
c Supplier's data (GE Healthcare).
d Experimental data from Ref. 26.
e Experimental data from Ref. 27.
f Estimated here by IEF using the high molecular weight kit.
Fig. 1.
Protein migration velocity on acrylamide gradient gels decreases during blue native electrophoresis, but no protein is completely immobilized after commonly used electrophoresis times (<5 h). Black dots on solid black lines mark dodecylmaltoside-solubilized membrane protein complexes: respiratory complex I (I; 1 MDa), monomeric ATP synthase (M; 597 kDa), respiratory complex III (III; 482 kDa), respiratory complex IV (IV; 205 kDa), and respiratory complex II (II; 123 kDa). Red triangles on dashed red lines mark soluble proteins from the high molecular weight kit: thyroglobulin (TG; 669 kDa), ferritin (FER; 476 kDa), catalase (CAT; 239 kDa), and BSA (66 kDa). BSA and catalase that were visible as blue bands during BNE for some hours were no longer detected at later stages. A double headed arrow marks the uniform 3.5% acrylamide layer on top of the 3.5–16% acrylamide gradient gel. Mass/migration data were taken from Table I.
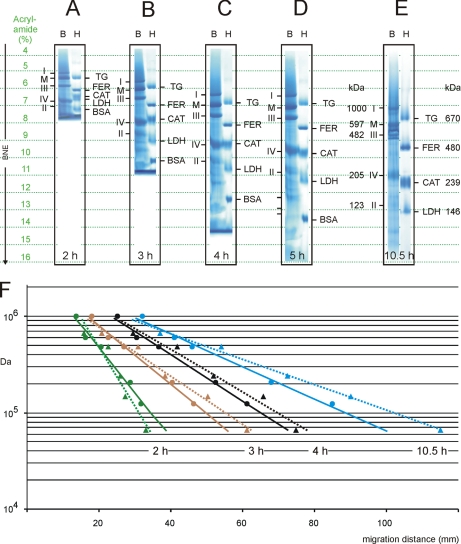
Fig. 2.
Mass calibrations on BN gels by membrane and soluble proteins differ at all electrophoresis times. Dodecylmaltoside-solubilized bovine mitochondrial complexes (lanes B) and soluble proteins of the high molecular weight kit (lanes H) were applied to 3.5–16% acrylamide gradient gels. Assignment of protein bands is as in Fig. 1 and Table I. A–E, following BNE for 2, 3, 4, 5, and 10.5 h, respectively, the gels were fixed and Coomassie-stained. Dashed green lines mark specific acrylamide concentrations in the gradient gels and relate to the actual position of the analyzed proteins. F, regression lines for the calibrations by membrane proteins (dots on solid lines) and soluble proteins (triangles on dashed lines) are indicated for various electrophoresis times (green for gel A (2 h), brown for gel B (3 h), black for gel C (4 h), and blue for gel E (10.5 h). Note the inversion of the slopes for membrane and soluble proteins upon transition from 2 to 3 h of electrophoresis (see “Discussion”). Mass/migration data were taken from supplemental Table S1. CAT, catalase; FER, ferritin; LDH, lactate dehydrogenase; TG, thyroglobulin.
From the plot of migration distance versus time of electrophoresis in the native non-fixed gel (Fig. 1) it was apparent that the migration velocity of all proteins decreased with electrophoresis time and migration distance. However, not even the largest protein complexes like respiratory complex I (∼1 MDa) and ATP synthase (∼600 kDa) were completely immobilized after extended (10.5-h) electrophoresis. The theoretical end points of protein migration, expressed as the limiting acrylamide concentration in the gel, were then identified by a modified plot using a reciprocal time scale (supplemental Fig. S1). The limiting acrylamide concentrations were between 7.5% acrylamide for complex I (∼1 MDa) and around 14% acrylamide for complex II (123 kDa). Catalase and bovine serum albumin (BSA) were detected only at early time points in the unfixed BN gel, and lactate dehydrogenase, which did not bind Coomassie dye, was not detected at all (see Table I for Coomassie binding capabilities of specific proteins). Because of this limited detection of soluble proteins in the unfixed BN gel, protein migration was also measured in fixed and stained BN gels (Fig. 2, A–E, and supplemental Table S1). The estimated theoretical end points of migration of soluble proteins after extended electrophoresis are shown in supplemental Fig. S2. Thyroglobulin (669 kDa; Ref. 9), ferritin (476 kDa), catalase (239 kDa), and lactate dehydrogenase (146 kDa) were found immobilized at 8.2, 10.3, 12.8, and 15% acrylamide, respectively. BSA (66 kDa) was clearly too small to be stopped by the 16% acrylamide gel.
Errors in Mass Estimation Using Membrane or Soluble Proteins for Calibration
After very short electrophoresis on a linear 3.5–16% acrylamide gradient gel (2 h at a constant power of 5 watts; Fig. 2A) the mass markers were already separated from each other. Complex I (∼1 MDa) was just entering the 5% acrylamide area, and the smallest protein, BSA (66 kDa), approached the 7.5% acrylamide range. All proteins were far from being immobilized. Mass versus migration data points were then used for linear regression analysis, resulting in two different regression lines, one for the membrane protein markers (Fig. 2F, solid green line) and the other one for the soluble markers (Fig. 2F, broken green line). Although the migration behavior of membrane and soluble proteins larger than 200 kDa seemed largely comparable, considerable differences were noticed at smaller masses. The 123-kDa membrane protein complex II, for example, showed the migration behavior and apparent mass of 80-kDa soluble proteins. Because the two regression lines diverged toward the low mass range, the relative error increased with decreasing protein mass.
After longer electrophoresis (3 h; Fig. 2B) the migration behavior of membrane and soluble proteins larger than 200 kDa again was comparable, but the migration velocity of smaller membrane and soluble proteins of comparable mass was inverted; i.e. the regression line for the membrane proteins (Fig. 2F, solid brown line) became the steeper line. Theoretically, 80-kDa membrane proteins now migrated as far as 120-kDa soluble proteins. The feature that soluble proteins migrated further than membrane proteins of comparable protein mass was observed up to extremely long (10.5-h) electrophoresis times (Fig. 2, E and F, blue lines) when most proteins approached their theoretical end point of migration. This suggested that membrane and soluble proteins of comparable protein mass differ in their real size due to binding or not binding lipids, dye, and detergent molecules.
The calibration curves of membrane and soluble proteins did not coincide at any point in time using 3.5–16% acrylamide gradient gels. This is in agreement with the findings by Poolman and co-workers (8) who studied proteins after extended electrophoresis on 5–17% acrylamide gels and in contrast to the coincident calibration curves that we found previously for a 5–18% acrylamide gel type (2). It should be noted that we could not confirm this earlier finding in the present study. Instead, we now achieved nearly superimposable calibration curves using 3.5–13% acrylamide gels. We conclude that in general soluble proteins do not fit to the calibration by DDM-solubilized membrane proteins except if special gel types and electrophoresis conditions are used as addressed in more detail below.
Concerning mass calibrations by soluble and membrane protein markers as separate sets, membrane protein markers seemed suitable for mass calibration of membrane proteins even after relatively short and convenient electrophoresis times. At first the same also seemed to hold for mass calibration of soluble proteins using the soluble proteins of the high molecular weight kit. However, fundamental problems with the mass estimation of soluble proteins by BNE became apparent in the course of the following experiments.
Mass Estimation of Small Membrane Proteins
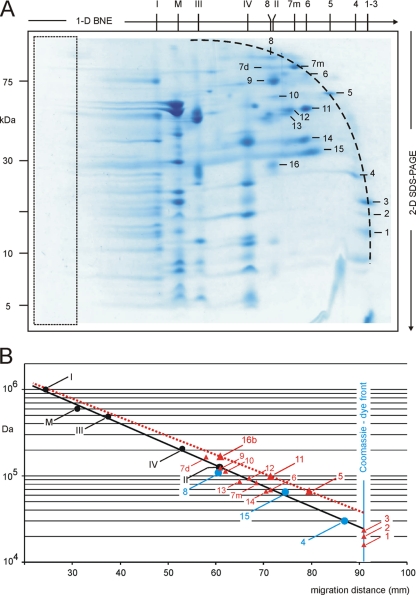
Small membrane protein markers are not commercially available for mass estimation by BNE. Therefore, calibration by soluble proteins and correction for protein-bound components like Coomassie dye were necessary (8). We asked whether it would be possible to simplify things by extending the calibration line of membrane proteins to the small mass range. To test this we excised a BN gel strip from a 3.5–16% acrylamide gel similar to that in Fig. 2C for resolution by second dimensional SDS-PAGE (Fig. 3A) and used this two-dimensional BN/SDS gel for mass estimation of the separated proteins. The area on the left side (Fig. 3A, boxed) corresponding to the 3.5% acrylamide layer on top of one-dimensional BN gels can be cut off. This changes the measured migration distances of proteins and the slope of the calibration lines but does not affect the estimated masses. Two-dimensional BN/SDS gels are preferentially used for mass estimation whenever protein bands cannot be unambiguously assigned in one-dimensional BN gels, and two-dimensional resolution is needed for verification. Large mitochondrial complexes, for example, were identified by their characteristic subunit patterns in two-dimensional BN/SDS gels (Fig. 3A). Smaller membrane and soluble proteins could be identified by mass spectrometry as summarized in Table II and in the supplemental material. Some proteins that were monomeric in the one-dimensional native gel and in the two-dimensional SDS gel as well (protein numbers 1–6, 7m, and 8 in Fig. 3A and Table II) were found in an arclike arrangement (Fig. 3A, dashed line). Other proteins that migrated as homomeric or heteromeric protein complexes in the native gel were dissociated by SDS during two-dimensional SDS-PAGE, and the subunits (protein numbers 7d and 9–16) were therefore found below the dashed arc.
Fig. 3.
Mass estimation of small proteins by BNE. Dodecylmaltoside-solubilized mitochondrial proteins and protein complexes were separated by one-dimensional BNE (A) using a 3.5–16% acrylamide gradient gel with a uniform 3.5% acrylamide layer on top (boxed area). Two-dimensional separation by Tricine-SDS-PAGE revealed the characteristic polypeptide patterns of respiratory complexes I–IV (I, II, III, and IV) and monomeric ATP synthase (M). Several further proteins in the low mass range (protein numbers 1–16) were identified by mass spectrometry (Table II, supplemental Figs. S7–S22, and supplemental Tables S6–S9). Proteins that were monomeric in one-dimensional BNE (protein numbers 1–8) were found in an arclike arrangement. Protein subunits from homo- and heteromeric complexes (protein numbers 7d and 9–16) were dissociated by SDS and found below the arclike structure. B, small membrane proteins (blue dots; protein numbers 4, 8, and 15) and most soluble proteins (small red triangles) fitted the extended regression line for mitochondrial membrane protein complexes I–IV and monomeric ATP synthase (M) in contrast to a few further soluble proteins (large red triangles; protein numbers 5, 11, and 16b). This non-uniform behavior of soluble proteins may relate to a non-uniform binding of Coomassie dye (see “Discussion”). Protein numbers 1–3 (in the 15–20-kDa range) were too small to be separated by the 3.5–16% acrylamide gel used for BNE and comigrated with the Coomassie dye front.
Table II. Use of dodecylmaltoside-solubilized bovine heart mitochondria for mass calibration on two-dimensional BN/SDS gel.
Dodecylmaltoside-solubilized RCs from bovine heart mitochondria were separated by BNE on a linear 3.5–16% acrylamide gradient gel followed by two-dimensional Tricine-SDS-PAGE using a 10% acrylamide gel. Mass/migration data refer to Fig. 3. Protein numbers 1–16 were identified by mass spectrometry as detailed in the supplemental material. Molecular weight search (MOWSE) scores larger than 65 were considered significant (p < 0.05) for protein identification.
| Spot | Identified protein | UniProt accession no. | Massa | pIa | Migration | TMH per monomer |
GRAVY score | MOWSE score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| b | c | d | e | f | ||||||||
| kDa | mm | |||||||||||
| I | RC I | ∼1000 (mon) | <7 | 24.5 | ||||||||
| M | Monomeric complex V | 597 (mon) | <7 | 31.0 | ||||||||
| III | RC III | 482 (dim) | <7 | 37.5 | ||||||||
| IV | RC IV | 205 (mon) | <7 | 53.0 | ||||||||
| II | RC II | 123 (mon) | <7 | 61.0 | ||||||||
| 1a | Hemoglobin α chain | P01966 | 15.1 (mon) | 8.2 | 91.0 | 0 | 0 | 0 | 0 | +0.014 | 97 | |
| 1b | Hemoglobin β chain | P02070 | 16.0 (mon) | 7.0 | 91.0 | 0 | 0 | 0 | 0 | −0.004 | 104 | |
| 2 | Myoglobin | P02192 | 16.9 (mon) | 7.0 | 91.0 | 0 | 0 | 0 | 0 | −0.378 | 83 | |
| 3 | Not identified | 91.0 | ||||||||||
| 4 | Rieske iron-sulfur protein of RC III | P13272 | 29.5 (mon) | 9.0 | 87.0 | 2 | 1 | 0 | 0 | 1 | −0.125 | 208 |
| 5 | Albumin | P02769 | 66.4 (mon) | 5.6 | 79.5 | 0 | 0 | 0 | 0 | −0.475 | 117 | |
| 6 | 75-kDa glucose-regulated protein | Q3ZCH0 | 68.7 (mon) | 5.5 | 71.5 | 1 | 1 | 0 | 0 | −0.398 | 183 | |
| 7m | Aconitase 2, mitochondrial, monomer | P20004 | 82.4 (mon) | 7.2 | 68.5 | 1–2 | 3 | 0 | 0 | −0.353 | 330 | |
| 7d | Aconitase 2, mitochondrial, dimer | P20004 | 164.8 (dim) | 7.2 | 58.0 | 1–2 | 3 | 0 | 0 | −0.353 | 137 | |
| 8 | Nicotinamide nucleotide transhydrogenase | P11024 | 109.0 (mon) | 7.3 | 60.5 | 14 | 17 | 12 | 12 | 14 | +0.307 | 413 |
| 9 | Succinate dehydrogenase, subunit A | P31039 | 123 (in RC II) | 6.4 | 61.0 | 1–2 | 0 | 0 | 0 | −0.283 | 242 | |
| 10 | 3-Oxoacid CoA-transferase 1 | Q24JZ7 | 113 (dim) | 8.6 | 62.0 | 3–4 | 3 | 0 | 0 | −0.099 | 128 | |
| 11 | Citrate synthase, mitochondrial | Q29RK1 | 98.0 (dim) | 7.0 | 71.5 | 2 | 0 | 0 | 0 | −0.228 | 156 | |
| 12 | Isocitrate dehydrogenase 2, mitochondrial | Q04467 | 93.0 (dim) | 8.5 | 67.0 | 1 | 0 | 0 | 0 | −0.382 | 148 | |
| 13 | Creatine kinase M chain | Q9XSC6 | 86.0 (dim) | 6.6 | 65.0 | 0 | 0 | 0 | 0 | −0.573 | 110 | |
| 14 | Malate dehydrogenase 2, mitochondrial | Q32LG3 | 66.4 (dim) | 8.4 | 70.5 | 2–3 | 0 | 0 | 0 | +0.126 | 187 | |
| 15 | Adenine nucleotide translocator 2 | Q8SQH5 | 65.6 (dim) | 9.8 | 74.5 | 3–4 | 4 | 0 | 2 | 6 | +0.032 | 119 |
| 16a | Succinate dehydrogenase, subunit B | Q3T189 | 123 (in RC II) | 8.5 | 61.0 | 1 | 0 | 0 | 0 | −0.491 | 96 | |
| 16b | Enoyl-CoA hydratase, short chain 1, mitochondrial | Q58DM8 | 168.6 (hex) | 6.9 | 61.0 | 1 | 1 | 0 | 0 | +0.027 | 358 | |
a Calculated for the mature protein using the Swiss-Prot/UniProt database and the pI/Mw tool from the ExPASy proteomics server (www.expasy.ch). Information on the oligomeric state of the identified proteins was also taken from the Swiss-Prot/UniProt database. mon, monomer; dim, dimer; hex, hexamer.
b Content of TMHs analyzed by TMpred prediction program.
c Content of TMHs analyzed by HMMTOP prediction program.
d Content of TMHs analyzed by SOSUI prediction program.
e Content of TMHs analyzed by TMHMM prediction program.
Next we plotted the migration distances of the five prominent mitochondrial complexes versus their masses on a logarithmic scale (Fig. 3B, black dots), extended the obtained regression line to the low mass range, and plotted the mass/migration data for three membrane proteins that were identified by mass spectrometry (Fig. 3B, blue dots). These three proteins, the “Rieske” iron-sulfur protein of respiratory complex III (protein number 4; Ref. 10), the adenine nucleotide translocator (protein number 15; Ref. 11), and the nicotinamide nucleotide transhydrogenase (protein number 8; Ref. 12), are true membrane proteins as indicated by a vast body of biochemical and structural data and comprise one, 6, and 14 transmembrane helices (TMHs), respectively. Predictions of TMHs by the TMpred and HMMTOP programs (Table II) were close to the experimental evidence, whereas TMHMM and SOSUI failed to identify one or two proteins, respectively, as membrane proteins. All three membrane proteins fitted the extended regression line fairly well. Three other proteins, bovine serum albumin (protein number 5), mitochondrial citrate synthase (protein number 11), and short chain mitochondrial enoyl-CoA hydratase 1 (protein number 16b), deviated considerably from the extended regression line (Fig. 3B, large red triangles) similar to the soluble proteins of the high molecular weight kit (Fig. 2C, dashed black line). These proteins are commonly regarded as water-soluble proteins, although TMH prediction programs only partially support this view (Table II). Seven additional proteins (Fig. 3B, small red triangles) that are also classified as soluble proteins did not fit the appropriate regression line for soluble proteins but fit that for membrane proteins (see “Discussion”). Finally, three very small proteins (protein numbers 1–3) with masses in the range of 15–20 kDa migrated with the dye front and were not resolved by BNE. We conclude that the regression line for mitochondrial complexes I–V can be extended and used for membrane proteins of smaller masses. Mass estimation of soluble proteins seems more complicated because one can hardly predict which soluble proteins will fit the calibration with membrane protein markers and which ones will behave like the soluble proteins of the high molecular weight kit. This problem would be fully relieved if special gel types and electrophoresis conditions were found for which one uniform calibration line was valid for both membrane and soluble proteins.
Searching for Optimal Gel Type with Uniform Calibration for Membrane and Soluble Proteins
Because we planned to compare the migration of respiratory complexes and even larger complexes like pyruvate dehydrogenase complex (13) over a mass range from 100 kDa to 10 MDa in one and the same gel, we searched for the optimal gel by testing gradient gels that start with an acrylamide concentration as low as 3.5% and go up to final acrylamide concentrations of 12–16%. BNE was always stopped shortly before the Coomassie dye reached the gel front, e.g. after 4 h of electrophoresis at a constant power of 5 watts using 14 × 14 × 0.15-cm gels. The 14-cm total gel length was divided into 11.5 cm for the acrylamide gradient, 1.7 cm for a uniform 3.5% acrylamide layer on top, and 0.8 cm for the sample gel.
For 3.5–16% acrylamide gradient gels (as exemplified before in Figs. 2 and 3) the slopes of the regression lines for DDM-solubilized membrane proteins were steeper than those of soluble proteins except after a very short electrophoresis time. In contrast for 3.5–12% acrylamide gradient gels the regression line of soluble proteins was the steeper line (supplemental Fig. S3D). Coincidence or at least a parallel arrangement of the two regression lines was therefore expected at an intermediate acrylamide concentration. In fact, almost superimposable and parallel regression lines were achieved using a linear 3.5–13% acrylamide gradient gel (Fig. 4D), which we consider as the optimal gel type to estimate the mass of DDM- or Triton-solubilized membrane proteins. To apply the calibration for soluble proteins to a membrane protein just a correction factor of 0.8 had to be applied. For example DDM-solubilized respiratory complex II (RC II; 123 kDa), a membrane protein complex, migrated the same distance as the soluble protein lactate dehydrogenase with a mass of 146 kDa (see supplemental Table S3). The mass of RC II was therefore calculated from the apparent mass as calibrated by soluble proteins (146 kDa) and then corrected by multiplying by 0.8. The obtained corrected mass of 117 kDa was close to the actual mass of 123 kDa as calculated by adding the masses of the individual subunits of RC II. No further correction, e.g. for the unknown amount of bound components like detergents, lipids, and dyes, was required. A slightly different correction factor of 0.7 was required to estimate the native masses of digitonin-solubilized membrane proteins when using soluble proteins as standards in the same 3.5–13% gel type (Fig. 4E). The optimal gel type for mass estimation of digitonin-solubilized membrane proteins, however, was a 3.5–12% acrylamide gel (supplemental Fig. S3E) because the two regression lines were almost identical, and no correction factor was required.
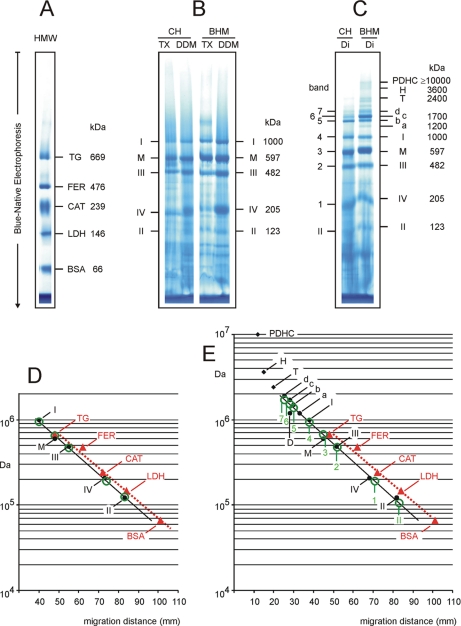
Fig. 4.
Mass calibration by membrane and soluble proteins is almost identical using 3.5–13% acrylamide gradient gels for BNE. A, separation of soluble proteins of the high molecular weight kit. B, separation of TX- and DDM-solubilized mitochondrial complexes from chicken heart (CH) and from BHM. C, separation of digitonin (Di)-solubilized mitochondrial complexes and supercomplexes from chicken and bovine heart. Assignment of proteins and complexes is as in Fig. 1. Additional supercomplexes a–d from BHM comprise respiratory complexes I, III, and IV in different stoichiometric ratio as detailed in supplemental Table S3. PDHC, pyruvate dehydrogenase complex; T and H, tetrameric and hexameric forms of ATP synthase; CAT, catalase; FER, ferritin; LDH, lactate dehydrogenase; TG, thyroglobulin. Bands 1–7 from chicken heart show almost identical migration as bovine complexes IV, III, monomeric ATP synthase (M), I, and supercomplexes b–d, the bovine counterparts. D, mass calibration by soluble proteins of the high molecular weight kit (red triangles) and by DDM-solubilized complexes from bovine (black dots) and chicken heart (green circles). E, mass calibration by soluble proteins (red triangles) and by digitonin-solubilized complexes from bovine (black dots) and chicken heart (green circles). Mass/migration data were taken from supplemental Table S3. Regression lines for calibration by membrane and soluble proteins were almost parallel and close together. A conversion factor of 0.8 was required to estimate the masses of DDM-solubilized membrane proteins after calibration by soluble proteins of the high molecular weight kit (see D). Similarly, a conversion factor around 0.7 was required for digitonin-solubilized membrane proteins (see E). D, dimeric ATP synthase.
High Molecular Mass Markers from Local Supermarket
To validate chicken heart from the local supermarket as a readily available source of membrane protein complexes for mass calibration in BNE we extracted the mitochondrial complexes from chicken heart using the detergents Triton X-100, dodecylmaltoside, and digitonin as described under “Experimental Procedures.” The high molecular weight kit (Fig. 4A) and detergent extracts from chicken heart and from isolated BHM as references were loaded on the same 3.5–13% acrylamide gradient gel for BNE (Fig. 4, B and C).
The bands from TX- and DDM-extracted chicken heart (Fig. 4B, lanes CH/TX and CH/DDM) were assigned to respiratory complexes I–IV and monomeric ATP synthase because the migration distances in the BN gel were almost the same as for the well characterized bovine complexes (10, 14–19) (Fig. 4B, lanes BHM/TX and BHM/DDM). Moreover, the expected characteristic columns of subunits were observed in two-dimensional BN/SDS gels (supplemental Fig. S4A). Mass/migration data of the DDM-solubilized bovine complexes were then plotted in Fig. 4D and used to calculate a regression line (black dots on solid line), which was also valid for the chicken heart mitochondrial complexes (green circles) and very close to that for soluble proteins (red triangles on dotted line).
Mass calibration using TX and DDM extracts must not be mixed with mass/migration data for digitonin-solubilized complexes because digitonin-solubilized complexes (Fig. 4C) showed shorter migration in the same gel compared with TX and DDM extracts (Fig. 4B). This indicated that the migration of membrane protein complexes in BNE was detergent-dependent to a certain degree. Mass/migration data of digitonin-solubilized mitochondrial complexes from BHM (Fig. 4E, black dots) deviated more strongly from the calibration by the high molecular weight kit (red triangles) than the mass/migration data of DDM-solubilized complexes shown before (Fig. 4D). Deviations from linearity in the high mass range, e.g. with tetrameric and hexameric ATP synthase and pyruvate dehydrogenase complex (10 MDa), presumably were due to the 3.5% acrylamide layer on top of the linear acrylamide gradient because minimal friction in this low acrylamide area may enhance migration velocity and migration distance of proteins.
Bands 1–7 from digitonin-solubilized chicken heart much resembled the bovine complexes except that oligomeric forms of ATP synthase (tetrameric and hexameric) were not observed in the one-dimensional BN gel. Chicken heart complexes were assigned according to the apparent masses in the BN gel relative to the bovine complexes and the characteristic polypeptide patterns on a two-dimensional BN/SDS gel (supplemental Fig. S4B) that show high similarity to the bovine mitochondrial complexes (10, 14–22). Chicken heart complexes fitted nicely to the bovine regression line, demonstrating the usefulness of chicken heart homogenates also for the mass estimation of digitonin-solubilized membrane proteins by BNE.
Membrane Protein Markers from Mouse and Rat Heart
Mouse and rat heart homogenates have been resolved many times by BNE for various scientific purposes (14, 23–25), but to our knowledge they have not been used for mass calibration in BNE so far. Preparation and solubilization of the heart homogenates are described under “Experimental Procedures.” Two-dimensional BN/SDS separations and corresponding mass calibrations are exemplified in supplemental Table S4 and supplemental Fig. S5.
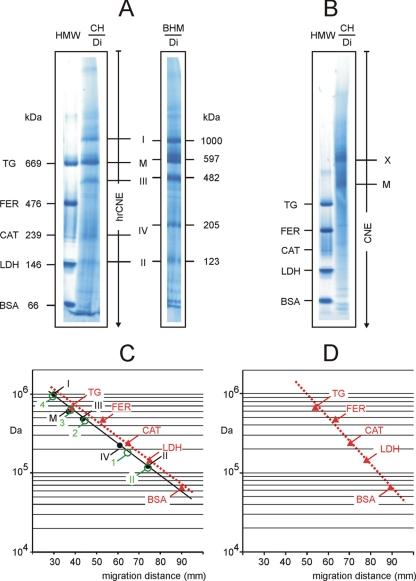
Protein Mass Estimation by High Resolution Clear Native Electrophoresis
High resolution clear native electrophoresis (5) is primarily used to study fluorescently labeled proteins and catalytic activities directly in the native gel. Here we also studied the potential of hrCNE to estimate masses and oligomeric states of proteins and protein complexes. The high molecular weight marker kit and digitonin extracts from BHM and from chicken heart homogenates were loaded on the same 3–12% acrylamide gradient gel for hrCNE (Fig. 5A and supplemental Table S5). The gel was stopped before the Ponceau S dye that was applied with the sample reached the gel front. The identity of the chicken heart complexes was analyzed by two-dimensional hrCNE/SDS-PAGE (supplemental Fig. S4C). Calibration lines for the membrane protein complexes from BHM (Fig. 5C, black dots on solid line) and chicken heart (green circles) were almost parallel and nearly matched that of the soluble proteins of the high molecular weight kit (Fig. 5C, red triangles on dashed line). The error between membrane or soluble protein calibration was around 20%. We conclude that digitonin-solubilized complexes from chicken heart are useful for mass calibration on hrCNE gels. When the high molecular weight kit is used to calibrate the estimated masses of digitonin-solubilized membrane proteins need to be corrected by a factor of 0.8 (80-kDa membrane proteins migrate at the same distance as soluble proteins with a mass of 100 kDa).
Fig. 5.
Mass calibration by membrane and soluble proteins on 3–12% acrylamide gradient gels for hrCNE and CNE. A, hrCNE variant 1 (hrCNE-1; Ref. 5) was used to separate the soluble proteins of the high molecular weight kit and the digitonin-solubilized mitochondrial membrane protein complexes from chicken heart (CH/Di) and from bovine heart mitochondria (BHM/Di). B, CNE could separate the proteins of the high molecular weight kit but failed to separate the digitonin-solubilized mitochondrial membrane protein complexes from chicken heart (CH/Di) sufficiently. Band M, monomeric ATP synthase. Band X was identified as a mixture of complex I and other respiratory complexes. C, mass calibration on hrCNE gel by soluble proteins of the high molecular weight kit (red triangles) and by digitonin-solubilized proteins from bovine heart mitochondria (black dots) and from chicken heart (green circles). D, mass calibration on a CNE gel by soluble proteins of the high molecular weight kit (red triangles). Mass/migration data were taken from supplemental Table S5. CAT, catalase; FER, ferritin; LDH, lactate dehydrogenase; TG, thyroglobulin.
For a complete analysis of the features of mass estimation by various native electrophoresis techniques the performance of CNE was also addressed here. Resolution of proteins of the high molecular weight kit by CNE was of acceptable quality, but the separation of chicken heart mitochondrial complexes was poor (Fig. 5B). Mass estimation by CNE is essentially restricted to acidic water-soluble proteins in many cases (Fig. 5D) because no charge shift is imposed on the native proteins, and most membrane proteins are resolved insufficiently.
DISCUSSION
Estimation of native protein masses by BNE was first described in 1994 (2). It was recognized in this work that some commercially available water-soluble proteins fitted the calibration curve of membrane protein markers. Therefore it was suggested that these special proteins can be used as mass standards for the mass estimation of membrane proteins. Other soluble proteins like bovine cytochrome c, trypsinogen from bovine pancreas, and carbonic anhydrase from bovine erythrocytes did not fit the calibration curve. Basic proteins like cytochrome c and trypsinogen presumably migrated too slowly compared with other marker proteins of comparable size because the number of bound anionic Coomassie dye molecules probably was not sufficient to fully override the protein intrinsic positive charges. Other soluble proteins like carbonic anhydrase (measured pI 5.9) that did not bind Coomassie dye at all also migrated too slowly presumably because their isoelectric points were too close to the native gel pH (around 7.5), and the negative charge excess was not sufficient. Already in 1994 it was obvious that many water-soluble proteins are not useful as protein standards to estimate the masses of native membrane proteins (2).
Introducing novel soluble proteins as mass markers therefore needs comparison with validated mass markers to make mass calibration reliable. A commonly used set of five soluble standard proteins, the high molecular weight kit (GE Healthcare), contains the validated proteins ferritin, catalase, and albumin (2) and also two other protein components, thyroglobulin and lactate dehydrogenase. Thyroglobulin from porcine thyroid is not ideal but sufficiently fits to the calibration line of the high molecular weight kit (Figs. 2 and 4). The observed deviation might be explained by the numerous glycosylations and disulfide linkages of thyroglobulin that potentially affect its overall shape. A second component of the high molecular weight kit, lactate dehydrogenase from bovine heart, should not be confused with the previously analyzed lactate dehydrogenase from rat muscle (2) because the pI and Coomassie binding capability of the heart and muscle isoforms differ considerably. The previously used muscle isoform (pI 8.6) binds Coomassie dye, enhancing its anodic migration velocity (2). In contrast the heart isoform from the high molecular weight kit used here hardly binds the dye, and migration velocity therefore depends on the protein intrinsic charge only. The measured isoelectric point (pI ∼5.0) of lactate dehydrogenase from bovine heart differed clearly from the gel pH of 7.5, and the excess negative charge apparently enhanced electrophoretic mobility sufficiently so that this protein fitted the general calibration line of the high molecular weight kit (Figs. 2 and 4). We conclude that both proteins fit reasonably to the high molecular weight kit proteins; this is in accordance with a previous analysis by Poolman and co-workers (8) performed under different electrophoresis conditions.
Next we asked why the slope of the calibration line of membrane protein markers was steeper than that of soluble markers under some conditions and why the order was sometimes inverted. After very short electrophoresis times (Fig. 2A) we observed that the slope of the calibration line for the high molecular weight kit appeared steeper than that of DDM-solubilized membrane proteins (Fig. 2F, green lines), but the situation was inverted after longer runs. To find a plausible explanation for this phenomenon we considered different parameters that define protein migration in native gels. (i) The charge/mass ratio of a protein defines its maximum electrophoretic mobility in a medium of negligible friction. This situation is approached only during initial electrophoresis when proteins migrate through the sample well and the low acrylamide sample gel. Because quantitative data on the charge/mass ratios are not available, we instead discuss electrophoretic results in terms of the isoelectric point and Coomassie dye binding of the proteins. (ii) As electrophoresis goes on, the molecular sieving effect gains importance. Decreasing pore size in the gel progressively reduces the initial electrophoretic mobility of the proteins. Protein size and pore size of the gel eventually define the maximum migration distance a protein can travel. When we first considered very short electrophoresis runs (Fig. 2, A and F, green lines) it seemed reasonable to assume comparable initial electrophoretic mobilities for all membrane proteins because complexes I, III, IV, and V were all found to bind Coomassie dye at 0.35–0.59 g/g of protein (7). Neglecting potential extra mass contributions from lipids and detergents (see below), the initial electrophoretic mobilities of the mitochondrial complexes should therefore differ by less than a factor of 2, which was confirmed by a reasonably straight regression line. Considering the excess negative charge introduced by several hundreds of bound dye molecules, we expected that the high initial electrophoretic mobility of the mitochondrial complexes could only be beaten by some smaller membrane proteins exhibiting an even higher dye/protein ratio (8). Because BSA showed a lower initial electrophoretic mobility in relation to the regression line of membrane proteins, we assumed that dye binding to BSA was less than to membrane proteins. In fact we could verify low Coomassie binding to BSA (0.10 ± 0.02 g/g) using protein extracts from BN gels and spectral quantification of the dye. An even lower initial electrophoretic mobility was expected for lactate dehydrogenase because no detectable amounts of Coomassie dye were bound to the native protein during electrophoresis. Migration velocity of this protein must therefore rely only on the protein intrinsic negative charge indicated by its measured isoelectric point (pI ∼5.0). Protein-bound detergent cannot contribute to the apparent mass of a protein estimated by BNE because Poolman and co-workers (8) have shown that all detergent that is initially protein-bound is replaced by Coomassie dye during BNE.
Lipid binding to proteins after BNE, a parameter contributing to the apparent mass of a protein, has not been measured yet. At present we can only speculate that lipid binding to membrane proteins after BNE is comparable with or higher than lipid binding after chromatographic isolation for the following reasons. (i) The same detergents (dodecylmaltoside and Triton X-100) and similar detergent/protein ratios (2–5 g/g) have commonly been used to solubilize biological membranes for BNE and for chromatography. (ii) Less lipid should be removed during BNE because the detergent is added only once (for solubilization), whereas chromatographic protocols comprise further delipidating steps. The phospholipid content of chromatographically isolated mitochondrial complexes (28–31) ranged from a few lipids bound to isolated respiratory complex IV (28) to around 100 lipids per dimeric respiratory complex III (31) corresponding to a complete phospholipid bilayer around the formerly membrane-covered part. We speculate that a complete lipid annulus might also be preserved around most membrane proteins following solubilization by dodecylmaltoside or Triton X-100 and separation by BNE. For example, dimeric complex III from BN gels comprises a protein mass of 483 kDa, contains 169-kDa Coomassie dye (0.35 g/g of protein), and may contain 75-kDa lipid (assuming a complete lipid annulus; i.e. 100 phospholipid molecules with an average mass of 750 Da). The lipid content of membrane proteins separated by BNE seems to contribute less than bound Coomassie dye to errors in mass estimation.
Even less or nothing is known on the lipid binding of membrane proteins after solubilization by digitonin. We speculate that the lipid content of digitonin-solubilized membrane proteins is larger than that after solubilization by dodecylmaltoside or Triton X-100 because the apparent masses of digitonin-solubilized proteins in BNE are always larger than those of membrane proteins solubilized by dodecylmaltoside or Triton X-100. In line with this speculation mass/migration data of digitonin-solubilized mitochondrial complexes from BHM (Fig. 4E) deviated more strongly from the calibration by the high molecular weight kit than the mass/migration data of DDM-solubilized complexes (Fig. 4D).
Differences between the calibrations by membrane and soluble markers were most pronounced after extended electrophoresis, i.e. when the proteins had approached their theoretical end points of migration. The differences have been explained by the fact that components like lipids and Coomassie dye bind to membrane proteins but not to the soluble proteins studied (8). Depending on the gel and the electrophoresis conditions, the slope of the regression lines can diverge toward the low mass range as observed here with linear 3.5–16% acrylamide gels, or the slopes can run in parallel as with linear 5–17% acrylamide gels under the conditions described by Poolman and co-workers (8). Provided that the regression lines are parallel, the actual mass of a membrane protein can be calculated from the mass estimated by BNE based on a calibration with the high molecular weight kit by introducing a conversion factor that depends on the offset between the regression lines.
The possibility to invert the order of the slopes of the regression lines for membrane and soluble proteins by variation of electrophoresis times and especially by moving from 3.5–12% (supplemental Fig. S3 and supplemental Table S2) to 3.5–16% acrylamide gels (Figs. 2 and 3) offered the chance to find gels that are characterized by identical slopes of the regression lines for membrane and soluble proteins. An intermediate acrylamide gradient gel type (3.5–13%) was found to exhibit nearly identical slopes for DDM- and Triton-solubilized membrane proteins. Still a certain offset between the regression lines occurred that could be corrected by a conversion factor. For example a conversion factor of 0.8 with DDM- and Triton-solubilized membrane proteins and a conversion factor of 0.7 in the case of digitonin-solubilized membrane proteins had to be used to estimate their masses based on a calibration with the soluble high molecular weight kit. Comparing 3.5–13 and 3.5–12% acrylamide gels (Fig. 4 and supplemental Fig. S3, respectively) indicated that a relatively small variation of the gels can have remarkable effects. Therefore, under “Experimental Procedures” and in supplemental Fig. S6 we describe in detail how we cast the gels. Furthermore, because intended and unintended experimental variations occur between different laboratories and experimentalists, it also seems important that each laboratory first verifies the properties of the gel type used by comparing the regression lines of membrane and soluble protein markers. If non-parallel regression lines are obtained, adjustment of the final acrylamide concentration of the gradient gel will be necessary. However, considering the simplicity and robustness of the protocols to extract mitochondrial complexes from chicken, rat, mouse, or bovine heart, one may set water-soluble markers aside completely and refer generally to membrane protein markers for the mass estimation of membrane proteins. In this case the gel type can be chosen freely to achieve optimal resolution for a given problem. Extending the calibration by larger membrane proteins to the low mass range as shown here seems a very simple option to also estimate the masses of smaller membrane proteins.
In contrast to mass estimation of native membrane proteins by BNE water-soluble proteins cause a lot more difficulties and potential pitfalls. As discussed above the electrophoretic mobilities of mass markers and analyzed proteins must be comparable so that the protein of interest fits to the calibration curve of the markers. This is no problem with membrane proteins because all membrane proteins analyzed so far were found to bind considerable amounts of Coomassie dye and thus showed high electrophoretic mobility. The situation is different and more complicated with soluble proteins because it is difficult to predict whether a specific soluble protein can or cannot bind Coomassie dye. In principle dye binding can be assessed by searching for migrating blue bands during BNE. However, around 5 μg of a relatively pure protein is needed, and negative results do not exclude factual dye binding. Only in rare cases will it be possible to decide whether a specific soluble protein migrates toward the anode because it binds Coomassie dye or because of its intrinsic negative charges. A large excess of negative charges, either intrinsic or introduced with the Coomassie dye, confers high electrophoretic mobility to the analyzed protein so that it can fit the calibration curve. Proteins containing a smaller number of excess negative charges, however, are prone to moderate or large errors in mass estimation.
Because the charge/mass ratio and also the number of excess negative charges commonly are not known, we asked whether the isoelectric points of soluble proteins can be used as an indicator of whether mass estimation by BNE is possible or not allowed. First we compared the calculated isoelectric points using the pI/Mw tool from the ExPASy proteomics server (www.expasy.ch) and the experimentally determined actual isoelectric points (see Table I). We noticed considerable differences and concluded that calculated isoelectric points should be regarded with caution. Two water-soluble proteins with experimentally determined pI ≤5.0 that do not bind Coomassie dye are part of the high molecular weight kit, namely ferritin and lactate dehydrogenase. Their electrophoretic mobilities were sufficient to fit the calibration of the high molecular weight kit indicating that a pI ≤5.0 was sufficient to substitute for the missing Coomassie binding. Either pI ≤5.0 or Coomassie binding of a soluble protein thus seem to assure correct mass estimation with the help of the common soluble mass markers.
Surprisingly, the migration behavior of another group of seven presumably soluble proteins resembled that of membrane proteins rather than that of the soluble proteins of the high molecular weight kit. We hypothesize that these seven proteins (Fig. 3B, small red triangles) migrate as short a distance as membrane proteins of comparable protein mass because they contain, similar to membrane proteins, a large number of bound dye molecules, which increases the protein size. This assumption is supported by the relatively high pI values 8.6, 8.5, and 8.4 for protein numbers 10, 12, and 14, respectively, suggesting that these proteins can only migrate to the anode because a negative charge shift is introduced. This concept is still valid if the actual pI values were 1 unit lower than the calculated pI as also observed for the proteins of the high molecular weight kit (see Table I). Prediction of transmembrane helices for proteins that are commonly regarded as soluble proteins by at least two prediction programs (protein numbers 6, 7, 10, and 16b) may suggest that the prediction programs are not completely reliable, but it may also be taken as an indication that these sequence stretches are rather hydrophobic and may therefore have good dye binding properties. Because it is presently impossible to decide which soluble protein will fit the calibration of the high molecular weight kit and which will fit the calibration of the membrane protein markers, it seems wise to select a gel type with minimal difference between the two calibrations, i.e. to use 3.5–13% acrylamide gradient gels to estimate the native masses of soluble proteins.
Similar to the situation in BNE, mass estimation by hrCNE of soluble basic or uncharged proteins is precluded or difficult except if these proteins bind the mixed anionic/neutral micelles that were added to the cathode buffer for hrCNE. Because it is not easily possible to test for the binding of the mixed micelles, mass estimation of soluble proteins by hrCNE remains rather ambiguous. Basic proteins show cathodic migration and are lost, and proteins with pI close to neutral migrate too slowly toward the anode so that large errors must be expected. The accuracy of mass estimation of membrane proteins by hrCNE, however, seems comparable with that of BNE.
Supplementary Material
* This work was supported by Deutsche Forschungsgemeinschaft Grant SCHA 615/2-1 (to H. S.), by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 815, Project Z1 (Redox-Proteomics), and by Bundesministerium für Bildung und Forschung Grant BMBF 01GM0863 (mitoNET-Deutsches Netzwerk für mitochondriale Erkrankungen).
 This article contains supplemental methods, Figs. S1–S23, and Tables S1–S9.
This article contains supplemental methods, Figs. S1–S23, and Tables S1–S9.
1 The abbreviations used are:
- BNE
- blue native electrophoresis
- BN
- blue native
- BHM
- bovine heart mitochondria
- CNE
- clear native electrophoresis
- Coomassie dye
- Coomassie Blue G-250
- DDM or dodecylmaltoside
- dodecyl-β-d-maltoside
- hrCNE
- high resolution clear native electrophoresis
- RC
- respiratory complex
- TMH
- transmembrane helix
- TX or Triton
- Triton X-100
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 2.Schägger H., Cramer W. A., von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 3.Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 4.Wittig I., Schägger H. (2005) Advantages and limitations of clear native polyacrylamide gel electrophoresis. Proteomics 5, 4338–4346 [DOI] [PubMed] [Google Scholar]
- 5.Wittig I., Karas M., Schägger H. (2007) High resolution clear-native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 6.Wittig I., Carrozzo R., Santorelli F. M., Schägger H. (2007) Functional assays in high resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell-lines. Electrophoresis 28, 3811–3820 [DOI] [PubMed] [Google Scholar]
- 7.Wittig I., Schägger H. (2008) Features and applications of blue-native and clear-native electrophoresis. Proteomics 8, 3974–3990 [DOI] [PubMed] [Google Scholar]
- 8.Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H., Poolman B. (2002) Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317, 591–600 [DOI] [PubMed] [Google Scholar]
- 9.Edelhoch H. (1960) The properties of thyroglobulin. I. The effects of alkali. J. Biol. Chem. 235, 1326–1334 [PubMed] [Google Scholar]
- 10.Berry E. A., Guergova-Kuras M., Huang L. S., Crofts A. R. (2000) Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 69, 1005–1075 [DOI] [PubMed] [Google Scholar]
- 11.Klingenberg M. (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 12.Pedersen A., Karlsson G. B., Rydström J. (2008) Proton-translocating transhydrogenase: an update of unsolved and controversial issues. J. Bioenerg. Biomembr. 40, 463–473 [DOI] [PubMed] [Google Scholar]
- 13.Sanderson S. J., Khan S. S., McCartney R. G., Miller C., Lindsay J. G. (1996) Reconstitution of mammalian pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes: analysis of protein X involvement and interaction of homologous and heterologous dihydrolipoamide dehydrogenases. Biochem. J. 319, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer B., Wittig I., Trifilieff E., Karas M., Schägger H. (2007) Identification of two proteins associated with mammalian ATP synthase. Mol. Cell. Proteomics 6, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 15.Hatefi Y. (1985) The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54, 1015–1069 [DOI] [PubMed] [Google Scholar]
- 16.Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 17.Cecchini G. (2003) Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72, 77–109 [DOI] [PubMed] [Google Scholar]
- 18.Hosler J. P., Ferguson-Miller S., Mills D. A. (2006) Energy transduction: proton transfer through the respiratory complexes. Annu. Rev. Biochem. 75, 165–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schägger H., Brandt U., Gencic S., von Jagow G. (1995) Ubiquinol-cytochrome c-reductase from human and bovine mitochondria. Methods Enzymol. 260, 82–96 [DOI] [PubMed] [Google Scholar]
- 20.Wittig I., Schägger H. (2009) Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim. Biophys. Acta 1787, 672–680 [DOI] [PubMed] [Google Scholar]
- 21.Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schägger H., Pfeiffer K. (2001) The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 276, 37861–37867 [DOI] [PubMed] [Google Scholar]
- 23.Schägger H., Noack H., Halangk W., Brandt U., von Jagow G. (1995) Cytochrome c oxidase in developing rat heart: enzymatic properties and amino-terminal sequences suggest identity of the fetal heart and the adult liver isoform. Eur. J. Biochem. 230, 235–241 [PubMed] [Google Scholar]
- 24.Radford N. B., Wan B., Richman A., Szczepaniak L. S., Li J. L., Li K., Pfeiffer K., Schägger H., Garry D. J., Moreadith R. W. (2002) Cardiac dysfunction in mice lacking cytochrome-c oxidase subunit VIaH. Am. J. Physiol. Heart Circ. Physiol. 282, H726–H733 [DOI] [PubMed] [Google Scholar]
- 25.Vahsen N., Candé C., Brière J. J., Bénit P., Joza N., Larochette N., Mastroberardino P. G., Pequignot M. O., Casares N., Lazar V., Feraud O., Debili N., Wissing S., Engelhardt S., Madeo F., Piacentini M., Penninger J. M., Schägger H., Rustin P., Kroemer G. (2004) AIF deficiency compromises oxidative phosphorylation. EMBO J. 23, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Righetti P. G., Caravaggio T. (1976) Isoelectric points and molecular weights of proteins. J. Chromatogr. 127, 1–28 [DOI] [PubMed] [Google Scholar]
- 27.Samejima T., Kamata M., Shibata K. (1962) Dissociation of bovine liver catalase at low pH. J. Biochem. 51, 181–187 [DOI] [PubMed] [Google Scholar]
- 28.Brandt U., Schägger H., von Jagow G. (1989) Purification of cytochrome-c oxidase retaining its pulsed form. Eur. J. Biochem. 182, 705–711 [DOI] [PubMed] [Google Scholar]
- 29.Sharpley M. S., Shannon R. J., Draghi F., Hirst J. (2006) Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry 45, 241–248 [DOI] [PubMed] [Google Scholar]
- 30.Dröse S., Zwicker K., Brandt U. (2002) Full recovery of the NADH:ubiquinone activity of complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica by the addition of phospholipids. Biochim. Biophys. Acta 1556, 65–72 [DOI] [PubMed] [Google Scholar]
- 31.Schägger H, Hagen T, Roth B, Brandt U, Link TA, von Jagow G. (1990) Phospholipid specificity of bovine heart bc1 complex. Eur. J. Biochem. 190, 123–130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.