Abstract
Stable isotope labeling by amino acids in cell culture (SILAC) is widely used to quantify protein abundance in tissue culture cells. Until now, the only multicellular organism completely labeled at the amino acid level was the laboratory mouse. The fruit fly Drosophila melanogaster is one of the most widely used small animal models in biology. Here, we show that feeding flies with SILAC-labeled yeast leads to almost complete labeling in the first filial generation. We used these “SILAC flies” to investigate sexual dimorphism of protein abundance in D. melanogaster. Quantitative proteome comparison of adult male and female flies revealed distinct biological processes specific for each sex. Using a tudor mutant that is defective for germ cell generation allowed us to differentiate between sex-specific protein expression in the germ line and somatic tissue. We identified many proteins with known sex-specific expression bias. In addition, several new proteins with a potential role in sexual dimorphism were identified. Collectively, our data show that the SILAC fly can be used to accurately quantify protein abundance in vivo. The approach is simple, fast, and cost-effective, making SILAC flies an attractive model system for the emerging field of in vivo quantitative proteomics.
Mass spectrometry-based quantitative proteomics has emerged as a highly successful approach to study biological processes in health and disease (1–3). Most studies have so far been limited to in vitro systems such as cell culture models. Although tremendously useful, these models cannot appropriately reflect relevant regulatory mechanisms of multicellular eukaryotes in vivo. This is particularly relevant for complex processes involving interactions between different cell types such as differentiation and development (4).
Relative changes in protein abundance are most accurately measured by comparing the natural form of a peptide with its stable isotope-labeled analog. Several different approaches enable stable isotope labeling of peptides either by chemical reactions or metabolic incorporation of the label (5, 6). Metabolic labeling has several advantages such as high labeling efficiency and intrinsically higher precision. For example, metabolically labeled samples can be combined before further processing steps so that protein quantification is not affected by differences in sample preparation. Labeling of organisms with stable isotope tracers was pioneered by Rudolf Schoenheimer 75 years ago (7, 8). Since then, several model organisms ranging from prokaryotes to mammals have been labeled metabolically (for an excellent review, see Ref. 9). For example, Caenorhabditis elegans and Drosophila melanogaster have successfully been labeled with 15N (10), and 15N-labeled flies were recently used to study maternal-to-zygotic transition (11) and seminal fluid proteins (sfps)1 transferred at mating (12). 15N has also been used to label entire rats, particularly for quantitative brain proteomics (13, 14). Despite its usefulness, 15N labeling also has several disadvantages. Because most peptides contain dozens of nitrogen atoms, labeling with highly enriched 15N still results in only partial peptide labeling and therefore complex isotope clusters. In addition, the mass shift between the labeled (i.e. heavy) and unlabeled (i.e. light) forms of a peptide depends on the number of nitrogen atoms and therefore varies depending on the peptide sequence. This leads to an increase in the number of candidate masses that need to be considered and therefore complicates peptide identification by search algorithms. Both problems result in smaller identification rates and less accurate quantification that can partially be overcome by computational correction (15, 16).
Stable isotope labeling by amino acids in cell culture (SILAC) is another metabolic labeling approach with several unique advantages (17): because the label is introduced at the amino acid level, mass spectra can easily be interpreted, and peptides can be quantified with high precision. These features have made SILAC a very popular approach for cell culture-based quantitative and functional proteomics (18). As a potential disadvantage, SILAC is generally thought to be restricted to in vitro cell culture experiments. The only SILAC experiments in the fly model were carried out using cell lines cultivated in vitro (19, 20). However, in 2005, Hayter et al. (21) demonstrated that chicken can be partially labeled at the amino acid level by feeding them with a diet containing stable isotope-labeled valine. Three years later, Krüger et al. (22) achieved essentially complete labeling of the laboratory mouse. Until now, this so-called “SILAC mouse” was the only multicellular organism that has been completely labeled with the SILAC approach, and partial labeling was recently achieved in newts (21, 23).
Here, we introduce the fruit fly D. melanogaster in the SILAC zoo. We refer to these animals as SILAC flies because they are obtained by feeding flies on SILAC-labeled yeast. D. melanogaster is one of the best characterized model organisms and has been used to address many fundamental questions in biology (24). Until now, most studies in D. melanogaster have focused on genetic aspects (25). However, proteins are the key actors in most biological processes. It is therefore highly desirable to obtain quantitative information at the protein level in D. melanogaster. We demonstrate in the present study that raising fly larvae on a diet of heavy lysine-labeled yeast cells results in virtually complete heavy labeling in the first filial (F1) generation. Furthermore, we show that the SILAC fly enables proteome-wide quantification with higher precision than a label-free method. In a series of proof-of-principle experiments, we used the SILAC fly to investigate sexually dimorphic protein expression in D. melanogaster, thus providing the first systematic comparison of male and female flies at the protein level.
EXPERIMENTAL PROCEDURES
Culturing of D. melanogaster
D. melanogaster strains w1118 (kindly provided by Dr. Manfred Gossen, Berlin-Brandenburg Center for Regenerative Therapies, Berlin, Germany) and tud1 bw1 sp1/CyO, I(2)DTS5131 (Bloomington Drosophila Stock Center, Bloomington, IN) were raised on a standard culture medium in a 12-h light-dark cycle at 25 °C and 75% relative humidity. Virgin flies were obtained by separating male and female flies within 4 h after eclosion. Flies lacking germ line tissue were generated by crossing homozygous tud1 females with either homozygous or heterozygous tud1 males. Only homozygous tud1 progeny was used for experiments. All flies were aged for 5 days before performing experiments and were collected between 3 and 5 p.m.
Labeling of D. melanogaster
The lysine auxotrophic Saccharomyces cerevisiae strain SUB62 (MATa his3-Δ200 lys2–801 leu2–3, 112 trp1–1 ura3–52) was precultured twice overnight at 30 °C at a dilution of 1:5000 in labeling medium containing 1.7 g/liter yeast nitrogen base (without amino acids and without ammonium sulfate), 20 g/liter d-glucose, 5 g/liter ammonium sulfate, 200 mg/liter adenine hemisulfate, 20 mg/liter uracil, 100 mg/liter Tyr, 10 mg/liter His, 60 mg/liter Leu, 10 mg/liter Met, 60 mg/liter Phe, 40 mg/liter Trp, 100 mg/liter Arg, and 30 mg/liter [12C6,14N2]Lys (Lys0) (all from Sigma-Aldrich) or 30 mg/liter [13C6,15N2]Lys (Lys8) (Sigma Isotec). The second preculture was diluted 1:1000 in labeling medium and incubated at 30 °C until reaching ∼8.0 A600. S. cerevisiae was centrifuged, and the pellets were stored in aliquots at −20 °C. Drosophila embryos were collected on an apple juice-agar plate and subsequently transferred onto a piece of perforated tissue paper on a layer of cotton wool in a 10-cm Petri dish. Cotton was soaked with culture medium containing 60% (w/v) labeled S. cerevisiae (wet mass), 320 mm sucrose (Calbiochem), 0.3 mm ampicillin (Sigma-Aldrich), 6 mm methylparaben, 0.5‰ propionic acid, and 2.5‰ phosphoric acid (all from Merck). Flies were raised in a 12-h light-dark cycle at 25 °C and 75% relative humidity. 10 ml of culture medium was sufficient to breed ∼150 flies. Hatched flies were fed with heavy labeled S. cerevisiae on an apple juice-agar plate.
Sample Preparation
Snap frozen flies were homogenized in ice-cold modified radioimmunoprecipitation assay buffer containing 50 mm Tris-HCl (Carl Roth, Karlsruhe, Germany), pH 7.4, 150 mm NaCl (Merck), 1% Nonidet P-40, 0.25% sodium deoxycholate (both from Sigma-Aldrich), 1 mm EDTA, 0.1% SDS (both from Carl Roth), and 1× Complete protease inhibitor mixture (Roche Applied Science). Homogenates were sonicated and centrifuged, and the concentration of the supernatant containing proteins was determined using the Coomassie Plus Protein Assay kit (Pierce). Total protein extracts of S. cerevisiae were obtained by homogenization with glass beads (Sigma-Aldrich) in ice-cold radioimmunoprecipitation assay buffer. Equal amounts of protein extracts were separated under reducing conditions by SDS-PAGE on a 4–12% NuPAGE gradient gel (Invitrogen) according to the manufacturer's instructions. Proteins were fixed in 50% methanol and 10% acetic acid and stained by colloidal Coomassie Blue (Invitrogen). Gel lanes were cut into 15 slices, and samples were processed essentially as described (26). Lysyl endopeptidase (Lys-C) (Wako, Osaka, Japan) was used for in-gel digestion. Stop and go extraction tips containing C18 Empore disks (3M, Minneapolis, MN) were used to purify and store peptide extracts (27).
LC-MS/MS
On-line LC-MS/MS analysis was performed as described previously (28). In brief, peptide mixtures were separated by reversed phase chromatography using the Eksigent NanoLC-1D Plus system (Eksigent, Dublin, CA) on in-house manufactured 10-cm fritless silica microcolumns with an inner diameter of 75 μm. Columns were packed with ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH, Ammerbuch, Germany) (29). Separation was performed using a 10–60% ACN gradient (155 min) with 0.5% acetic acid at a flow rate of 200 nl/min. Eluting peptides were directly ionized by electrospray ionization and transferred into the orifice of an linear trap quadrupole Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Mass spectrometry was performed in the data-dependent mode with one full scan in the Orbitrap (m/z = 300–1700; resolution = 60,000; target value = 1 × 106). The five most intense ions with a charge state greater than 1 were selected (target value, 5000; monoisotopic precursor selection enabled) and fragmented in the linear trap quadrupole using CID (35% normalized collision energy and wideband activation enabled). Dynamic exclusion for selected precursor ions was 60 s.
Processing of MS Data
The MaxQuant software package (version 1.0.12.36) was used to identify and quantify proteins (30, 31). SILAC duplets were extracted from isotope patterns, recalibrated, and quantified by the Quant module (heavy label Lys8, maximum of three labeled amino acids per peptide, polymer detection enabled, and top six MS/MS peaks per 100 Da). Peak lists were searched on a Mascot search engine (version 2.2, Matrix Science, Boston, MA) against an in-house curated database of D. melanogaster (FlyBase, release 5.13, Indiana University) and/or S. cerevisiae (Saccharomyces Genome Database, ftp://ftp.yeastgenome.org/, database version 060512) plus common contaminants. All protein sequences were also reversed to generate a target-decoy database (32). Carbamidomethylation of cysteine was selected as a fixed modification, and oxidation of methionine and acetylation of the protein N terminus were used as variable modifications. Lys-C was selected as protease (full specificity) with a maximum of two missed cleavages. A mass tolerance of 0.5 Da was selected for fragment ions. A minimum of six amino acids per identified peptide and at least one peptide per protein group were required. The false discovery rate was set to 1% at both the peptide and protein levels. Protein ratios were calculated from the median of all normalized peptide ratios using only unique peptides or peptides assigned to the protein group with the highest number of peptides (“Occam's razor” peptides). Only protein groups with at least three SILAC counts were considered for further analysis.
Cluster Analysis of Gene Ontology (GO) Terms
GO analysis was performed using the DAVID bioinformatics database (DAVID Bioinformatics Resources, National Cancer Institute, Frederick, MD) (33). UniProt entries were binned according to the heavy to light (H/L) protein ratio. Enrichment of GO terms (biological processes, level 4) in each bin was calculated using the entire list of identified proteins as background (threshold count = 2, Expression Analysis Systematic Explorer score = 1). Terms with a p value <0.01 in at least one bin were selected, log-transformed, z-transformed, hierarchically clustered, and plotted as a heat map using an in-house Perl and R script (see below).
Statistical Analysis
Statistical data analysis was done using the R project for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria) or Prism 4.0 (GraphPad Software, San Diego, CA).
Data
The data associated with this study may be downloaded from Proteome Commons (http://proteomecommons.org/) Tranche using the following hash: FE44CxP5/LXJkxuFGCPY40J7mYl6fBm5abGqfZwinzcjANCzLCCyhjSqK8tjtVpPEaJGyt+ZFBFd+ELe11v7cRM+Q+8AAAAAAAAHTg==.
These data include all identified proteins, sequences of identified peptides, and peptide evidences (i.e. SILAC ratio measurements) for all experiments. They are based on the proteinGroups.txt, peptides.txt, and evidence.txt files that were obtained by processing raw files with MaxQuant as described above.
RESULTS
Labeling of D. melanogaster with Heavy Lysine Is Highly Efficient in F1 Generation
D. melanogaster can be raised on a diet consisting exclusively of yeast. Therefore, feeding flies with 15N-labeled yeast yields completely 15N-labeled flies (10). However, in contrast to 15N, SILAC requires that amino acids used for labeling are essential. Typically both lysine and arginine are used for SILAC: because trypsin cleaves C-terminally of these amino acids, all tryptic peptides except for the protein C terminus carry a heavy label and can be used for quantification. Lysine was found to be essential in axenic D. melanogaster culture (34). However, arginine was not required because flies on an arginine-deficient diet survived and laid eggs. Arginine is also problematic because it is converted into proline by some cell lines (35). Therefore, we decided to use heavy lysine as the only label and the endoproteinase Lys-C instead of trypsin to obtain peptides with C-terminal lysine residues.
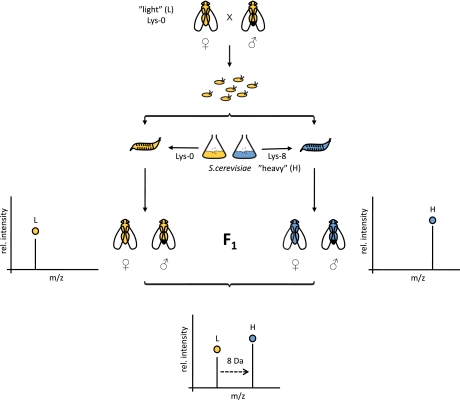
Because cultivating flies axenically is laborious, we tried to label flies in standard culture using SILAC-labeled yeast as food (Fig. 1). First, we generated fly food by cultivating the lysine-auxotrophic yeast strain SUB62 in minimal medium containing either light (12C6,14N2) or heavy (13C6,15N2) lysine (Lys0 or Lys8, respectively). Fly embryos were collected from parental w1118 flies and transferred to dishes containing a suspension of light or heavy SILAC-labeled yeast. The common laboratory strain w1118 features a defective white allele resulting in white eye color. Otherwise w1118 flies are considered to be wild type. Hatched adult flies (F1) were continually fed with heavy or light yeast on an apple juice-agar plate for 5 days. We did not observe any apparent difference between light and heavy flies (phenotype, development time, or number of flies obtained).
Fig. 1.
Work flow for labeling of D. melanogaster with heavy isotope-containing amino acids. Embryos are collected, and hatched larvae are fed with “light” l-[12C6,14N2]lysine (L) or “heavy” l-[13C6,15N2]lysine (H) labeled S. cerevisiae. Different adult F1 subpopulations (male and female) are mixed and analyzed by LC-MS/MS. Pairs of identical peptides with different stable isotope compositions can be distinguished in the mass spectrometer based on their mass difference (8 Da). Consistently, the ratio of peak intensities of heavy versus light peptides reflects differences in protein abundance in vivo. rel., relative.
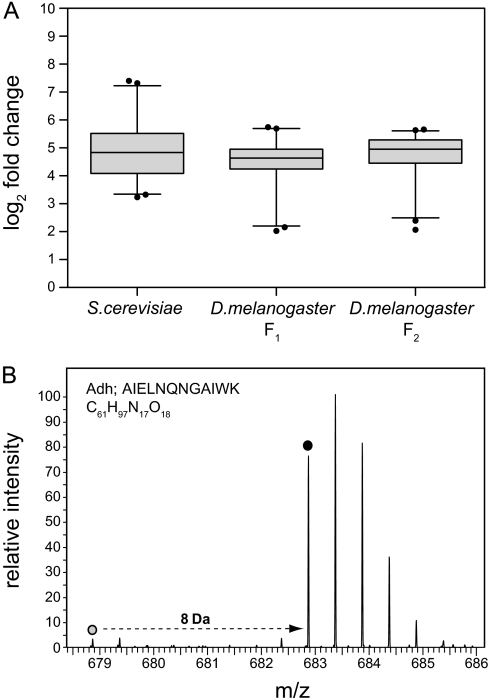
To investigate labeling efficiency, we first analyzed the heavy labeled yeast cells that were used as food source. Yeast samples were lysed, digested with Lys-C, and analyzed by LC-MS/MS. The average labeling efficiency of the 100 most intense proteins was 96.7%, indicating essentially complete labeling (Fig. 2A). Similarly, we investigated label incorporation in F1 of heavy SILAC flies. The average incorporation rate was 96.2% in the F1 generation (Fig. 2A). An exemplary mass spectrum of an alcohol dehydrogenase-derived peptide shows a well defined heavy isotope cluster (Fig. 2B). The corresponding light isotope cluster of this peptide has a much lower intensity with a log2 -fold change of 4.4, corresponding to an incorporation rate of 95.6%. Thus, D. melanogaster can be labeled efficiently with heavy lysine in non-axenic standard culture conditions. To test whether labeling efficiency can be further improved by prolonged metabolic labeling, we collected embryos derived from heavy labeled F1 flies and raised the F2 generation on heavy labeled yeast. Analysis of the same set of proteins revealed only a minor increase of labeling efficiency to 96.9% in F2 (Fig. 2A). Hence, D. melanogaster labeling already is saturated in the first generation. We therefore decided to use F1 flies for further experiments.
Fig. 2.
Effective labeling of D. melanogaster with heavy lysine. A, labeling efficiency of S. cerevisiae and adult D. melanogaster. Total protein extracts of heavy labeled yeast and adult flies from the F1 or F2 generation were analyzed by LC-MS/MS. H/L ratios of the 100 most intense proteins were calculated and expressed as median of log2 -fold changes. Whiskers indicate 2.5 and 97.5 percentiles. Labeling efficiencies were 96.7% (S. cerevisiae), 96.2% (D. melanogaster F1), and 96.9% (D. melanogaster F2). B, representative mass spectrum of an alcohol dehydrogenase (Adh)-derived peptide from heavy adult flies in the F1 generation. The mass shift between the light and the heavy forms of the peptide is 8 Da due to one heavy lysine. Dots mark light and heavy monoisotopic peaks (log2 -fold change is 4.4, corresponding to an incorporation rate of 95.6%).
Labeling with Heavy Lysine Allows for Precise Protein Quantification
Next, we wanted to compare the precision of quantification of the SILAC fly and a label-free approach. Heavy and light male flies were combined, homogenized, processed by in-gel digestion, and analyzed twice by two subsequent LC-MS/MS runs (two technical replicates). 1578 proteins were identified and quantified in both replicates (at least three independent peptide ratios per protein). As we used the same sample in both runs and the same amount of peptides were injected, the ratio of protein abundance comparing replicate 1 and replicate 2 is precisely 1 for all proteins. The spread of the experimentally determined ratios can therefore be used to assess the precision of quantification.
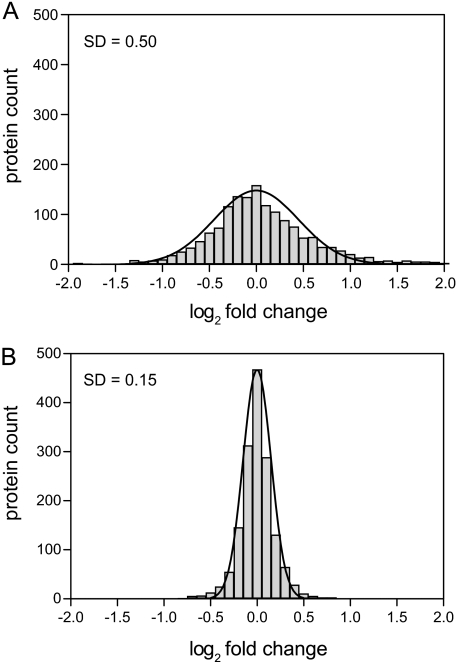
To determine the precision of label-free quantification, we used only light peptide intensities. log2 -fold changes of protein intensities (i.e. summed up peptide intensities calculated in MaxQuant) between both replicates were calculated and plotted in a histogram (Fig. 3A). log2 -fold changes ranged from −6.0 to 4.6 and could be modeled by a normal distribution with a standard deviation of 0.50. Hence, ∼95% of all measured protein ratios do not differ by more than a factor 2 from the correct value using label-free quantification (±2 standard deviations). Next, we performed the same analysis based on SILAC ratios. In this case, we compared the abundance of light proteins between both replicates using the heavy peptides as an internal reference by calculating the ratio of ratios. With this approach, we achieved a considerably higher precision (Fig. 3B). log2 -fold changes ranged from −3.0 to 0.9, and the standard deviation was reduced to 0.15. Accordingly, ∼95% of all measured ratios do not differ by more than 23% from the correct value. These results demonstrate that quantification with the SILAC fly as an internal standard has a ∼4-fold higher precision than label-free quantification in D. melanogaster. This estimated gain in precision is a conservative estimate because we used the same in-gel digest for both technical replicates. Variability in sample processing will increase the error of label-free quantification even further.
Fig. 3.
Precision of label-free versus SILAC-based protein quantification. Heavy and light flies were mixed and analyzed by two different LC-MS/MS runs (n = 1578 quantified proteins). A, for label-free quantification, the ratios of light protein intensities between both replicates were calculated and plotted in a histogram. B, SILAC-based quantification of light proteins between both replicates was performed using the heavy peptides as an internal standard (ratio of ratios). Comparison of standard deviations demonstrates that SILAC is ∼4-fold more precise than label-free quantification.
Identification of Sex-specific Proteins in D. melanogaster
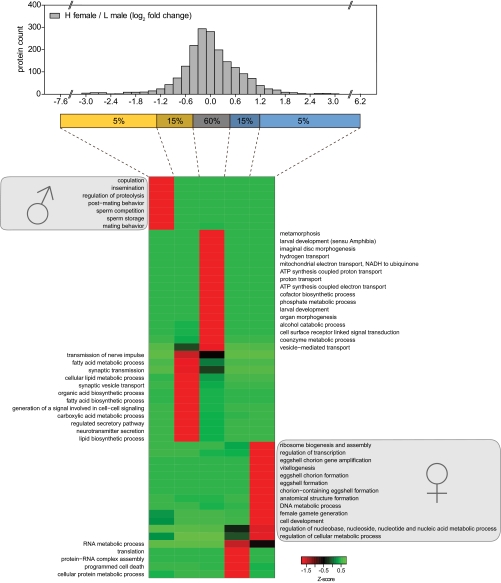
To test whether heavy lysine labeling of D. melanogaster is indeed a suitable method for in vivo quantitative proteomics, we performed a proteome comparison of male and female flies. Equal numbers of heavy w1118 female and light w1118 male flies (10 each) were mixed, and 140 μg of total protein extract was separated by SDS-PAGE, in-gel digested, and analyzed by LC-MS/MS. In total, 1913 proteins were identified and quantified with very high confidence (false discovery rate ≤1% and at least three peptide ratio counts). We sorted all proteins into five bins according to their log2 -fold change (Fig. 4). Next, we investigated whether the proteins in each bin are significantly enriched in GO terms relative to the entire list of identified proteins (36). z-transformed p values of GO terms were clustered and plotted as a heat map. Bins with the 5% highest or 5% lowest H/L ratios show pronounced overrepresentation of proteins belonging to sex-specific GO terms. Male-specific GO terms included copulation, insemination, and sperm storage. The GO term regulation of proteolysis was enriched based on proteins of the male accessory glands such as accessory gland peptide 62F and serine protease inhibitors 2 and 3. Interestingly, proteins moderately up-regulated in males (log2 -fold change between −1.3 and −0.5) were enriched in GO terms associated with neuronal activity (e.g. synaptic transmission and neurotransmitter secretion) and organic acid metabolism (e.g. fatty acid biosynthetic process), suggesting that male flies might potentially have increased neuronal activity and lipid metabolism compared with female flies. Conversely, proteins highly up-regulated in females were enriched in known female-specific terms belonging to oogenesis and the female reproductive system (e.g. female gamete generation and eggshell formation). Several proteins moderately up-regulated in females are involved in protein translation, a process particularly active during oogenesis. Specifically, 68 of 97 identified ribosomal proteins were within the top 20% of female-enriched proteins. Finally, the term programmed cell death was associated with females based on several proteins involved in autophagy, a process that has recently been reported to be involved in early oogenesis (37, 38). Together, these results demonstrate that the SILAC fly enables discrimination of sex-specific protein expression on a global scale and enables assignment of distinct biological processes to male and female flies.
Fig. 4.
Identification of sex-specific protein clusters by GO analysis. Total protein extracts of mixed heavy female and light male flies were analyzed by LC-MS/MS, and log2 -fold changes of H/L protein ratios were calculated (n = 1913). Proteins were divided into five bins according to their H/L ratio. p values of GO terms that were significantly enriched (p < 0.01) in at least in one bin were log-transformed, z-transformed, hierarchically clustered, and plotted as a heat map.
Identification of Sex-specific, Somatically Expressed Proteins in D. melanogaster
Tissues in sexually reproducing organisms can be classified as somatic or germ line. The tudor (tud) gene is essential for assembly of the germ plasm but dispensable for somatic posterior patterning during fly embryogenesis (39, 40). Therefore, progeny derived from female flies homozygous for the tud1 allele lacks germ line tissue. Tud1 progeny has already been used to compare male and female somatic mRNA expression in D. melanogaster (41, 42).
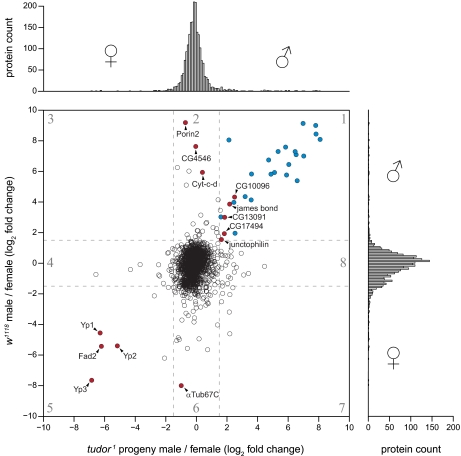
To differentiate between sex-specific proteins in the germ line and soma, we raised tud1 progeny. Sterility of adult tud1 progeny was confirmed by crossing females with w1118 male flies and males with virginal w1118 female flies. In addition, several tud1 progeny male and female flies were dissected to confirm lack of testes and ovaries, respectively (data not shown). To compare the proteome of male and female flies lacking germ line tissue, we combined either male or female tud1 progeny with an internal reference of mixed sex w1118 flies. Likewise, we combined either light male or female w1118 flies with the same mixed sex reference sample. Proteins in all four samples were separated into 15 slices by SDS-PAGE, digested, and analyzed by LC-MS/MS. From the obtained lists of quantified proteins, we calculated relative changes in protein abundance between male and female tud1 progeny for 2079 proteins as the ratio of ratios (i.e. (male tud1 progeny/internal standard)/(female tud1 progeny/internal standard)). Similarly, changes in protein abundance between w1118 males and females (male w1118/internal standard)/(female w1118/internal standard) were calculated (n = 2106). The distribution of log2 -fold changes between male and female w1118 flies is considerably broader than the corresponding distribution for male and female tud1 progeny (Fig. 5, vertical and horizontal histograms, respectively). Hence, the proteome of males and females becomes more similar in the absence of germ line tissue.
Fig. 5.
Identification of sex-specific germ line and somatic protein expression in adult D. melanogaster. The histogram on the right shows male versus female protein ratios in the w1118 strain (n = 2106). The histogram on top shows ratios of male versus female protein ratios in tud1 progeny (n = 2079). Proteins quantified in all experiments are shown in the central scatter plot. The numbered regions in the scatter plot contain different sets of proteins (supplemental Table 1). Known male accessory gland proteins in region 1 are shown in blue. Selected proteins that are mentioned in the main text have labels and are shown in red. Dashed lines indicate log2 -fold change of 1.5 and −1.5, respectively.
We went on to compare log2 -fold changes in both experiments more systematically. As expected, the majority of proteins did not exhibit a sex-specific expression pattern either in w1118 flies or in the tud1 background (Fig. 5, central region in the scatter plot). Closer inspection of proteins in different regions of the scatter plot provides several instructive insights. The proteins in each region are listed in supplemental Table 1 together with corresponding tissue-specific gene expression data retrieved from FlyAtlas (43). A number of proteins were up-regulated in both w1118 and tud1 males (Fig. 5, region 1). These proteins are expressed in a male-specific but germ line-independent manner. Consistently, most of the proteins in this subset (highlighted in blue) were identified in previous studies as accessory gland proteins (acps) or sfps (12, 44, 45). Conversely, when analyzing all previously identified acps and sfps in our data set (w1118 male versus female), we found that they had a strong male-specific expression bias (supplemental Fig. S1). Accessory glands are part of the somatic tissue in the male reproductive system and are involved in the production of seminal fluid. Moreover, we identified several additional proteins not previously known to be expressed in male somatic tissue: the protein encoded by CG10096 has not been identified in male flies, but tissue-specific mRNA expression data confirm that the gene is indeed specifically expressed in male somatic tissue (46). Interestingly, this protein has sequence homology to Arabidopsis thaliana male sterility 2, a protein involved in male gametogenesis in plants (47). The protein James bond catalyzes elongation of very-long-chain fatty acids and plays a crucial role in cytokinesis during male meiosis (48). CG17494 is a protein of unknown function containing a Forkhead-associated domain. The mRNA of CG13091 is enriched in fat body and heart, and junctophilin mRNA is mainly enriched in brain (43). Hence, the latter two proteins appear to be male-specific somatic proteins expressed outside the reproductive system.
Proteins up-regulated in w1118 males but not regulated in tud1 progeny are expected to be expressed in the male germ line (Fig. 5, region 2). The most strongly up-regulated protein in this group is porin 2, a voltage-dependent anion-selective channel located in mitochondria of fly spermatozoa (49). Another mitochondrial protein in this subset is Cyt-c-d, one of the two cytochrome c proteins specifically involved in the apoptosis-like process occurring during terminal differentiation of sperms in D. melanogaster (50). The protein encoded by CG4546 is a predicted arginine kinase. Arginine kinases maintain ATP levels by producing arginine phosphate, which serves as a high energy source from which ATP can be rapidly replenished (51). Importantly, CG4546 is not identical with ArgK, the arginine kinase of the D. melanogaster flight muscle (52). Our data therefore suggest that spermatozoa contain a specific arginine kinase enzyme that may help to buffer ATP levels.
The three yolk proteins in D. melanogaster (Yp1, Yp2, and Yp3) were up-regulated in both w1118 and tud1 females, suggesting that this protein family is mainly expressed in somatic tissue of females (Fig. 5, region 5). Indeed, Yp1, Yp2, and Yp3 are synthesized in the fat body and the ovarian follicular epithelium of females throughout adulthood and are secreted as vitellogenin (53). Furthermore, the fat body-specific fatty acid desaturase (Fad2), which has been shown to be involved in pheromone biosynthesis and courtship behavior, has also been found to be somatically overrepresented in females (54). In contrast, the maternal tubulin isotype α-tubulin67C (αTub67C) was enriched in w1118 but not in tud1 progeny females, indicating that αTub67C is mainly localized in female germ line tissue (Fig. 5, region 6). Indeed, αTub67C is found in unfertilized eggs and embryos, and synthesis of αTub67C protein is restricted to the ovary (55). Finally, a couple of proteins were regulated in tud1 progeny but not in w1118 flies, suggesting that tudor affects gene expression in a sex-dependent way. Collectively, these results demonstrate that the SILAC fly allows for discrimination of tissue-specific protein expression in males and females on a large scale.
DISCUSSION
The fruit fly D. melanogaster is one of the most popular model organisms in biomedical research (24). Advantages of the fly model include the ease of cultivation, rapid development, and a huge number of visible traits. Importantly, 77% of human disease genes with at least one mutant allele in the Online Mendelian Inheritance in Man (OMIM) database have well conserved homologs in D. melanogaster (56). These features render flies a very attractive model system in biomedical research. Until now most research focused on genetic aspects and transcriptomics (25). However, complex biological systems are unlikely to be understood without comprehensive information at the protein level.
A recent large scale project used different cell types, developmental states, multiple fractionation techniques, and many mass spectrometry runs to provide a first high quality catalog of the fly proteome (57). However, functional proteomics also requires quantitative information at the proteomic scale. Metabolic stable isotope labeling is particularly attractive because it enables quantification with high precision. Pioneering experiments established metabolic labeling with 15N in D. melanogaster (10, 11). Since then 15N flies have been used in a further study demonstrating the utility of this approach (12). Here, we establish SILAC as a novel metabolic labeling technique in D. melanogaster.
SILAC as well as 15N labeling have particular strengths and weaknesses. Both methods require special media that do not allow additives containing sources of lysine or nitrogen, respectively, which can be a disadvantage compared with label-free methods. On the other hand, we show that label-free methods have lower precision than SILAC-based quantification (Fig. 3). Advantages of 15N labeling over SILAC are that 15N does not require auxotrophic yeast strains and that the problem of arginine to proline conversion is circumvented. In addition, all peptides in 15N flies are labeled and can be used for quantification. In SILAC, only lysine-containing peptides carry the isotope label. For this reason, we used the protease Lys-C to generate peptides with C-terminal lysine. However, Lys-C is not as efficient as trypsin and produces longer peptides, which typically result in fewer protein identifications.
On the other hand, SILAC has several advantages over 15N. Most importantly, SILAC-labeled peptides have well defined isotope clusters and constant mass shifts (supplemental Fig. S2, A and B). These features greatly facilitate manual and computational data analysis. Indeed, in a comparison of SILAC- and 15N-labeled flies, we identified ∼30% more proteins with SILAC than with 15N at the same false discovery rate (supplemental Fig. S2C). In addition, SILAC does not require computational correction of peptide ratios that are critical for accurate quantification using 15N (15, 16). Perhaps surprisingly, SILAC is also less expensive than 15N: although 1 g of Lys8 is ∼100 times more expensive than 1 g of [15N]ammonium sulfate, only 30 mg instead of 5 g are needed for 1 liter of yeast medium. This makes the SILAC fly a very cost-effective model organism (less than ∼$0.10 per adult fly).
We quantified in the SILAC fly changes in abundance of ∼2000 proteins from 15 gel slices (at least three ratio counts). When performing proteome profiling experiments with tissue culture cells, we typically quantify a higher number of proteins with the same setup (28). A likely reason for this observation is the correlation between protein abundance and tissue-specific expression: most abundant proteins such as histones and ribosomal and cytoskeletal proteins are ubiquitously expressed. Conversely, many low abundance proteins like transcription factors are expressed in a cell-type specific manner. Thus, combining different cell types in an organism increases the dynamic range of protein abundance. Consistent with this explanation, analyzing individual D. melanogaster cell lines enables a deeper coverage of the fly proteome (19, 57). Thus, the SILAC fly will be particularly useful for more targeted experiments focusing on specific body parts (brain, eyes, imaginal disks, etc.).
In a series of proof-of-principle experiments, we used the SILAC fly to study sexual dimorphism in D. melanogaster. Although the molecular mechanisms leading to sexual dimorphism are quite well understood (58, 59), less is known about proteins that determine the dimorphic state in D. melanogaster. Studies have been carried out using microarray technology to analyze sexual dimorphism at the transcript level (41, 42, 46). These studies provide a catalog of genes specifically expressed in somatic and germ line tissues. However, there was no comparative study at the proteome level in D. melanogaster so far. The SILAC fly revealed that the abundance of many proteins changes in a sex-dependent way. We identified several male-specific proteins that were also found in a recent study where sfps were detected in mated female flies (12) and in a study listing known acps (45), suggesting that they are expressed in the male sex organs. GO analysis of differentially regulated proteins revealed a clear picture of processes most relevant for males (copulation and mating behavior) and females (eggshell formation and female gamete generation). The sex bias of several other biological processes was not necessarily expected. For example, we found that several male-specific proteins play a role in neuronal activity and fatty acid biosynthesis, raising the hypothesis that males have higher neuronal activity and lipid metabolism. Females had higher expression levels of proteins involved in translation such as ribosomal proteins. This is most likely due to increased translational activity during oogenesis and has recently been observed in female-like C. elegans (60).
A major advantage of the fly model is the huge collection of available mutant strains. We took advantage of this resource to differentiate between germ line and somatic tissue. Specifically, we used the offspring of tud1 females. Wild-type tudor encodes a 285-kDa protein containing 11 Tudor domains that are essential for germ line development. Tudor belongs to a set of maternally expressed genes (so-called grandchildless or posterior group genes) that are essential for primordial germ cell specification (61). It was recently shown that Tudor domains bind to symmetrically dimethylated arginines in Piwi proteins in mice and flies (62, 63). Our proteomics comparison of w1118 and tud1 males and females enabled us to assess whether sex-specific protein expression originates in the germ line and in somatic tissue. Yolk and seminal fluid proteins were found to be differentially regulated in both w1118 and tud1 flies, consistent with their synthesis in somatic tissue. Conversely, we identified several proteins known to be expressed in the male and female germ line. In addition, we extended the list of germ line and somatic tissue-specific proteins. Hence, our data set can yield valuable information about proteins with so far unknown biological functions.
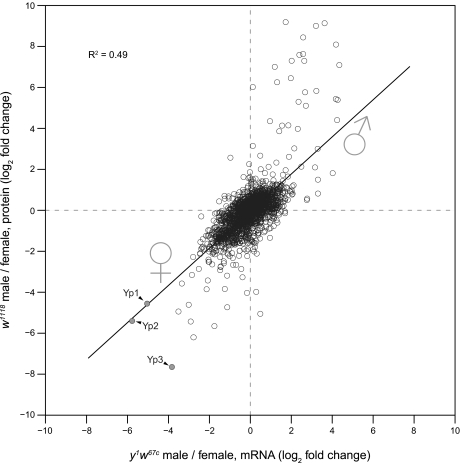
We also compared our quantitative proteome comparison of male and female w1118 flies with a published mRNA data set in a similar genetic background (42). Overall, fold-changes in protein and mRNA abundances (n = 1816) correlated remarkably well (Fig. 6). The slope of the trend line is close to 1, indicating that the direction and magnitude of sex-specific change in expression are similar at the mRNA and protein levels for most detected genes. This observation suggests that sex-specific gene expression is mainly controlled at the mRNA level. However, several proteins strongly up-regulated in males had smaller changes at the mRNA level. Conversely, several female-specific proteins also showed a bigger change at the protein level. This indicates that strongly regulated genes are regulated by a combination of transcriptional and posttranscriptional mechanisms. Interestingly, a large fraction of these genes encode secreted proteins. We previously found a similar overrepresentation of secreted proteins in genes that are regulated by microRNAs at the level of translation (28). It is tempting to speculate that proteins synthesized at endoplasmic reticulum-associated ribosomes (i.e. secreted proteins) generally tend to show a higher degree of posttranscriptional regulation. However, we cannot rule out that the dynamic range of measured mRNA ratios is compressed for technical reasons (64). Of note, although two detected yolk proteins, Yp1 and Yp2, had similar changes at the mRNA and protein levels, regulation of Yp3 was ∼16 times stronger than the corresponding mRNA. The yp3 gene is separated from yp1 and yp2 on the X chromosome, and transcriptional regulation of both loci differs (65, 66). In line with these results, our data indicate that posttranscriptional regulation of yp3 is also different from that of yp1 and yp2.
Fig. 6.
Correlation between male versus female protein -fold change and previously published mRNA data (n = 1816). The trend line indicates a good overall correlation between mRNA and protein -fold changes (slope, 0.9; R2 = 0.5). Strongly regulated genes tend to change more at the protein level than at the mRNA level.
SILAC-labeled model organisms bear great potential. Thus far, the only metazoan species completely labeled at the amino acid level is the laboratory mouse (22). Here, we have introduced the SILAC fly as another animal in the SILAC zoo. This zoo is likely to grow further and will probably contain all popular animal models in the future. In fact, partial labeling of newt and chicken using a pulsed approach has already been reported (21, 23). Our data show that the SILAC fly can provide new insights into biological processes in vivo. The approach is generic and can be used to study countless D. melanogaster mutants available. Importantly, SILAC labeling of flies is fast, simple, and cost-effective. With these features, SILAC flies are particularly attractive model organisms for the emerging field of in vivo quantitative proteomics.
Supplementary Material
Acknowledgments
We thank Dr. Manfred Gossen for providing the D. melanogaster strain w1118 and Dr. Gunnar Dittmar for the S. cerevisiae strain SUB62. Furthermore, we acknowledge Christian Sommer for excellent technical assistance and Jana Richter for preparing Drosophila culture medium.
* This work was supported by the Helmholtz Association and the Swiss National Science Foundation.
 This article contains supplemental Figs. S1 and S2 and Table 1.
This article contains supplemental Figs. S1 and S2 and Table 1.
1 The abbreviations used are:
- sfps
- seminal fluid proteins
- acps
- accessory gland proteins
- GO
- gene ontology
- H/L
- heavy to light
- Lys-C
- lysyl endopeptidase
- SILAC
- stable isotope labeling by amino acids in cell culture
- DAVID
- Database for Annotation, Visualization and Integrated Discovery.
REFERENCES
- 1.Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 2.Cox J., Mann M. (2007) Is proteomics the new genomics? Cell 130, 395–398 [DOI] [PubMed] [Google Scholar]
- 3.Gstaiger M., Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 4.Vermeulen M., Selbach M. (2009) Quantitative proteomics: a tool to assess cell differentiation. Curr. Opin. Cell Biol. 21, 761–766 [DOI] [PubMed] [Google Scholar]
- 5.Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 6.Gevaert K., Impens F., Ghesquière B., Van Damme P., Lambrechts A., Vandekerckhove J. (2008) Stable isotopic labeling in proteomics. Proteomics 8, 4873–4885 [DOI] [PubMed] [Google Scholar]
- 7.Schoenheimer R., Rittenberg D. (1935) Deuterium as an indicator in the study of intermediary metabolism. Science 82, 156–157 [DOI] [PubMed] [Google Scholar]
- 8.Simoni R. D., Hill R. L., Vaughan M. (2002) The use of isotope tracers to study intermediary metabolism: Rudolf Schoenheimer. J. Biol. Chem. 277, e31 [Google Scholar]
- 9.Gouw J. W., Krijgsveld J., Heck A. J. (2010) Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krijgsveld J., Ketting R. F., Mahmoudi T., Johansen J., Artal-Sanz M., Verrijzer C. P., Plasterk R. H., Heck A. J. (2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 11.Gouw J. W., Pinkse M. W., Vos H. R., Moshkin Y., Verrijzer C. P., Heck A. J., Krijgsveld J. (2009) In vivo stable isotope labeling of fruit flies reveals post-transcriptional regulation in the maternal-to-zygotic transition. Mol. Cell. Proteomics 8, 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findlay G. D., Yi X., Maccoss M. J., Swanson W. J. (2008) Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClatchy D. B., Dong M. Q., Wu C. C., Venable J. D., Yates J. R., 3rd (2007) 15N metabolic labeling of mammalian tissue with slow protein turnover. J. Proteome Res. 6, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao L., McClatchy D. B., Park S. K., Xu T., Lu B., Yates J. R., 3rd (2008) Quantitative analysis of brain nuclear phosphoproteins identifies developmentally regulated phosphorylation events. J. Proteome Res. 7, 4743–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouw J. W., Tops B. B., Mortensen P., Heck A. J., Krijgsveld J. (2008) Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal. Chem. 80, 7796–7803 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Webhofer C., Reckow S., Filiou M. D., Maccarrone G., Turck C. W. (2009) A MS data search method for improved 15N-labeled protein identification. Proteomics 9, 4265–4270 [DOI] [PubMed] [Google Scholar]
- 17.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 18.Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 19.Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. (2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762–772 [DOI] [PubMed] [Google Scholar]
- 20.Hilger M., Bonaldi T., Gnad F., Mann M. (2009) Systems-wide analysis of a phosphatase knock-down by quantitative proteomics and phosphoproteomics. Mol. Cell. Proteomics 8, 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayter J. R., Doherty M. K., Whitehead C., McCormack H., Gaskell S. J., Beynon R. J. (2005) The subunit structure and dynamics of the 20S proteasome in chicken skeletal muscle. Mol. Cell. Proteomics 4, 1370–1381 [DOI] [PubMed] [Google Scholar]
- 22.Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 23.Looso M., Borchardt T., Krüger M., Braun T. (2010) Advanced identification of proteins in uncharacterized proteomes by pulsed in vivo SILAC. Mol. Cell. Proteomics 9, 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin G. M., Lewis E. B. (2000) A brief history of Drosophila's contributions to genome research. Science 287, 2216–2218 [DOI] [PubMed] [Google Scholar]
- 25.St Johnston D. (2002) The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3, 176–188 [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 27.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 28.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 29.Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 30.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 31.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 32.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 33.Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 34.Sang J. H., King R. C. (1961) Nutritional requirements of axenically cultured Drosophila melanogaster adults. J. Exp. Biol. 38, 793–809 [Google Scholar]
- 35.Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 36.Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou Y. C., Chittaranjan S., Barbosa S. G., McCall K., Gorski S. M. (2008) Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182, p. 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nezis I. P., Lamark T., Velentzas A. D., Rusten T. E., Bjørkøy G., Johansen T., Papassideri I. S., Stravopodis D. J., Margaritis L. H., Stenmark H., Brech A. (2009) Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 5, 298–302 [DOI] [PubMed] [Google Scholar]
- 39.Thomson T., Lasko P. (2004) Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40, 164–170 [DOI] [PubMed] [Google Scholar]
- 40.Boswell R. E., Mahowald A. P. (1985) tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97–104 [DOI] [PubMed] [Google Scholar]
- 41.Arbeitman M. N., Furlong E. E., Imam F., Johnson E., Null B. H., Baker B. S., Krasnow M. A., Scott M. P., Davis R. W., White K. P. (2002) Gene expression during the life cycle of Drosophila melanogaster. Science 297, 2270–2275 [DOI] [PubMed] [Google Scholar]
- 42.Parisi M., Nuttall R., Edwards P., Minor J., Naiman D., Lü J., Doctolero M., Vainer M., Chan C., Malley J., Eastman S., Oliver B. (2004) A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chintapalli V. R., Wang J., Dow J. A. (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 [DOI] [PubMed] [Google Scholar]
- 44.Takemori N., Yamamoto M. T. (2009) Proteome mapping of the Drosophila melanogaster male reproductive system. Proteomics 9, 2484–2493 [DOI] [PubMed] [Google Scholar]
- 45.Ram K. R., Wolfner M. F. (2007) Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47, 427–445 [DOI] [PubMed] [Google Scholar]
- 46.Arbeitman M. N., Fleming A. A., Siegal M. L., Null B. H., Baker B. S. (2004) A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131, 2007–2021 [DOI] [PubMed] [Google Scholar]
- 47.Aarts M. G., Hodge R., Kalantidis K., Florack D., Wilson Z. A., Mulligan B. J., Stiekema W. J., Scott R., Pereira A. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12, 615–623 [DOI] [PubMed] [Google Scholar]
- 48.Szafer-Glusman E., Giansanti M. G., Nishihama R., Bolival B., Pringle J., Gatti M., Fuller M. T. (2008) A role for very-long-chain fatty acids in furrow ingression during cytokinesis in Drosophila spermatocytes. Curr. Biol. 18, 1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guarino F., Specchia V., Zapparoli G., Messina A., Aiello R., Bozzetti M. P., De Pinto V. (2006) Expression and localization in spermatozoa of the mitochondrial porin isoform 2 in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 346, 665–670 [DOI] [PubMed] [Google Scholar]
- 50.Arama E., Bader M., Srivastava M., Bergmann A., Steller H. (2006) The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 25, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellington W. R. (2001) Evolution and physiological roles of phosphagen systems. Annu. Rev. Physiol. 63, 289–325 [DOI] [PubMed] [Google Scholar]
- 52.Wyss M., Maughan D., Wallimann T. (1995) Re-evaluation of the structure and physiological function of guanidino kinases in fruitfly (Drosophila), sea urchin (Psammechinus miliaris) and man. Biochem. J. 309, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belote J. M., Handler A. M., Wolfner M. F., Livak K. J., Baker B. S. (1985) Sex-specific regulation of yolk protein gene expression in Drosophila. Cell 40, 339–348 [DOI] [PubMed] [Google Scholar]
- 54.Chertemps T., Duportets L., Labeur C., Ueyama M., Wicker-Thomas C. (2006) A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behaviour in Drosophila melanogaster. Insect Mol. Biol. 15, 465–473 [DOI] [PubMed] [Google Scholar]
- 55.Matthews K. A., Miller D. F., Kaufman T. C. (1989) Developmental distribution of RNA and protein products of the Drosophila alpha-tubulin gene family. Dev. Biol. 132, 45–61 [DOI] [PubMed] [Google Scholar]
- 56.Bier E. (2005) Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6, 9–23 [DOI] [PubMed] [Google Scholar]
- 57.Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 58.Hodgkin J. (1990) Sex determination compared in Drosophila and Caenorhabditis. Nature 344, 721–728 [DOI] [PubMed] [Google Scholar]
- 59.Cline T. W., Meyer B. J. (1996) Vive la difference: males vs females in flies vs worms. Annu Rev. Genet. 30, 637–702 [DOI] [PubMed] [Google Scholar]
- 60.Tops B. B., Gauci S., Heck A. J., Krijgsveld J. (2010) Worms from venus and mars: proteomics profiling of sexual differences in Caenorhabditis elegans using in vivo 15N isotope labeling. J. Proteome Res. 9, 341–351 [DOI] [PubMed] [Google Scholar]
- 61.Schupbach T., Wieschaus E. (1986) Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev. Biol. 113, 443–448 [DOI] [PubMed] [Google Scholar]
- 62.Kirino Y., Vourekas A., Sayed N., de Lima Alves F., Thomson T., Lasko P., Rappsilber J., Jongens T. A., Mourelatos Z. (2010) Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16, 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vagin V. V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G. J., Aravin A. A. (2009) Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naef F., Socci N. D., Magnasco M. (2003) A study of accuracy and precision in oligonucleotide arrays: extracting more signal at large concentrations. Bioinformatics 19, 178–184 [DOI] [PubMed] [Google Scholar]
- 65.Barnett T., Pachl C., Gergen J. P., Wensink P. C. (1980) The isolation and characterization of Drosophila yolk protein genes. Cell 21, 729–738 [DOI] [PubMed] [Google Scholar]
- 66.Hutson S. F., Bownes M. (2003) The regulation of yp3 expression in the Drosophila melanogaster fat body. Dev. Genes Evol. 213, 1–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.