Abstract
Nuclear pore complexes (NPCs) gate the only conduits for nucleocytoplasmic transport in eukaryotes. Their gate is formed by nucleoporins containing large intrinsically disordered domains with multiple phenylalanine-glycine repeats (FG domains). In combination, these are hypothesized to form a structurally and chemically homogeneous network of random coils at the NPC center, which sorts macromolecules by size and hydrophobicity. Instead, we found that FG domains are structurally and chemically heterogeneous. They adopt distinct categories of intrinsically disordered structures in non-random distributions. Some adopt globular, collapsed coil configurations and are characterized by a low charge content. Others are highly charged and adopt more dynamic, extended coil conformations. Interestingly, several FG nucleoporins feature both types of structures in a bimodal distribution along their polypeptide chain. This distribution functionally correlates with the attractive or repulsive character of their interactions with collapsed coil FG domains displaying cohesion toward one another and extended coil FG domains displaying repulsion. Topologically, these bipartite FG domains may resemble sticky molten globules connected to the tip of relaxed or extended coils. Within the NPC, the crowding of FG nucleoporins and the segregation of their disordered structures based on their topology, dimensions, and cohesive character could force the FG domains to form a tubular gate structure or transporter at the NPC center featuring two separate zones of traffic with distinct physicochemical properties.
Molecular exchange between the cytoplasm and nucleoplasm of cells is confined to pores in the envelope, which are formed and gated by a proteinaceous structure termed the nuclear pore complex (NPC)1 (1, 2). Metabolites and small proteins diffuse freely through the NPC (3), but the diffusion of larger proteins and RNA is more selective and requires transport signals and mobile receptors termed karyopherins (kaps; importins, exportins, and transportins) (4). The molecular architecture of the NPC is similar in all eukaryotes examined. It features a ring-shaped scaffold that forms a central ∼50 nm transport conduit, eight short fibers extending from the scaffold into the cytoplasm, and a fibrous basket structure extending from the scaffold into the nucleoplasm (5, 6). It also features a poorly defined structure in the center of the conduit (i.e. the transporter structure or central plug structure), which contains kap-cargo complexes in transit (2, 7–10).
Evidence suggests that the NPC passive diffusion conduit is juxtaposed on the facilitated transport conduit (3, 11, 12). This conduit must be flexible enough to accommodate kap-cargo complexes of different shapes and sizes while simultaneously maintaining a barrier against non-karyophilic proteins. Up to 60% of the channel capacity appears occluded at any given time by passing kap-cargo molecules (13). The conduit may also be occluded by NPC components extending into the conduit. The NPC of yeast and mammals is composed of ∼30 proteins called nucleoporins (nups) in multiple copies for a total of ∼450 nups per NPC (14, 15). Half of these nups (the non-FG nups) have structures that resemble membrane coat proteins and together form a ring scaffold that functions as a “stent” to keep the pore membrane open (16, 17). A few pore membrane nups have transmembrane domains and link the ring scaffold to the pore membrane (18–20). The rest of the nups contain multiple copies of phenylalanine-glycine (FG) motifs dispersed over 150–700-amino acid (AA) domains that are intrinsically disordered (i.e. natively unfolded FG domains) (see Fig. 1) (21). These disordered FG domains populate the transport conduit, but are anchored to the NPC ring scaffold by structured domains (22) (see Fig. 1). Despite their structural disorder and functional redundancy, FG domains are essential for the survival of yeast (23) and presumably all eukaryotes.
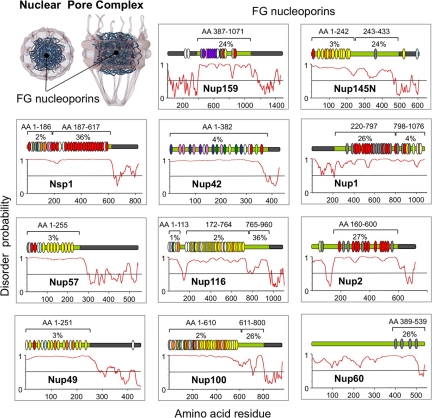
Fig. 1.
Diagram of the NPC and the intrinsically disordered FG nups that line its conduit. Each panel shows one FG nup as a green rectangle (N terminus at left) and the exact location of FG motifs (vertical ovals) in each protein. As defined (22), GLFG motifs are colored yellow, FXFG motifs are red, SPFG motifs are dark green, FXFX motifs are light gray, SAFG motifs are dark blue, PSFG motifs are green, NXFG motifs are light blue, SLFG motifs are orange, XXFG motifs are white, FXXFG motifs are light green, double FG motifs (SAFGXPSFG) motifs are pink, and the triple FG motifs are purple. The plots shown below each nup were generated using PONDR and predict the location of disordered structures (values >0.5) and ordered structures (values <0.5). The brackets above each nup mark the boundaries of intrinsically disordered domains that have undergone rapid evolution (22). Below each bracket is the corresponding percent content of charged AAs. The known and/or predicted NPC anchor domain for each nup (22) is highlighted with a gray box within the green nup rectangle.
It is generally thought that a homogeneous network of random coils provided by intrinsically disordered FG domains forms the NPC permeability barrier (21, 24, 25). In Aspergillus nidulans for example, a 5-min disruption of the NPC diffusion barrier coincides with the cell cycle-dependent dissociation of FG nups from the NPC (26). Also, some yeast strains lacking nup FG domains have a compromised permeability barrier (27), although the effect is subtle and not always observed (23). Whereas in vivo analyses have been confounded by the functional redundancy of FG nups, reductionist approaches carried out in vitro with purified components have provided some insight. Indeed, the selective properties of the NPC toward kaps have been reconstituted in vitro using isolated FG domains on beads (27, 28), FG domain hydrogels (29), and FG domains attached to holes in membranes (30), highlighting the inherent capability of these domains to form selective diffusion barriers that can be specifically permeated by kaps. The exact configuration of FG domains within the NPC and the mechanism of kap movement across the NPC are the subject of much speculation (31), but it seems clear that kaps and passing macromolecules must overcome a hydrophobic barrier imposed by FG domains (24, 32, 33).
Elucidating the dynamic structure of individual FG domains and the intra- and intermolecular interactions they make is key to understanding NPC architecture. The FG domains represent ∼12% of the NPC mass, or 6.5 MDa of unresolved protein structure at the center of the NPC, controlling all nucleocytoplasmic traffic of macromolecules. The tertiary structure of only two FG domains has been characterized in detail, one from the vertebrate Nup153 and the other from yeast Nup116. The Nup153 FG domain adopts extended coil configurations that appear to compact upon binding a kap, giving rise to the proposal that kaps “collapse” the FG domain from extended to compact shapes to gain access across the NPC (34). Consistently, the intrinsically disordered yeast nucleoporin Nup2 adopts extended coil configurations in purified form, but becomes more compact (i.e. to a smaller Stokes radius (RS)) upon kap binding (35). In contrast, the Nup116 FG domain naturally adopts compact, collapsed coil configurations on average in the absence of kaps due in part to its intramolecular cohesion of coils mediated by FG motifs (36). Given these two seemingly disparate findings (collapsed coils or extended coils?), it became necessary to examine all FG nups to get a better picture of how their disordered structures are configured in their native state. For example, there are 11 FG nups in Saccharomyces cerevisiae (Fig. 1), and in principle, their FG domains could adopt any of several categories of intrinsically disordered structures such as molten globules, premolten globules, relaxed coils, or extended coils (37–39). These structures are distinguished from each other by their intramolecular packing density, which is defined by the molecular mass of the polypeptide chain and the hydrodynamic volume it occupies (40). Currently, there are no computer-based structure prediction algorithms that differentiate between these different categories of disordered structures. Hence, we had to purify all FG domains from yeast nups to determine their Stokes radii to make structural assignments based on mass and hydrodynamic volume. In the process, we discovered that yeast FG nups are structurally heterogeneous and adopt distinct categories of disordered structures with separate functions in non-random distributions along their polypeptide chain. The implications of these findings to NPC architecture and function are discussed.
EXPERIMENTAL PROCEDURES
FG Domain Synthesis, Expression, Purification, and Interactivity Assay
Coding sequences for FG domains listed in Table I were PCR-amplified from S. cerevisiae DNA or synthesized de novo by GenScript and were cloned into pGEX-2TK in frame with the 3′ end of the glutathione S-transferase (GST) gene. Where indicated, codons encoding six His residues and one Trp residue were added at the 3′ end. FG domains were expressed as GST fusions in Escherichia coli BL21 strain, and glutathione-coated beads were used to isolate them from bacterial extracts. FG domains were released by thrombolysis from their GST tag, and in some cases, nickel-coated beads were used to recapture the FG domain via its C-terminal His tag. These were eluted from beads with 50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 250 mm imidazole, 0.1% Tween 20 and concentrated (Centricon 3) when necessary. Bead halo assays were performed as described (27).
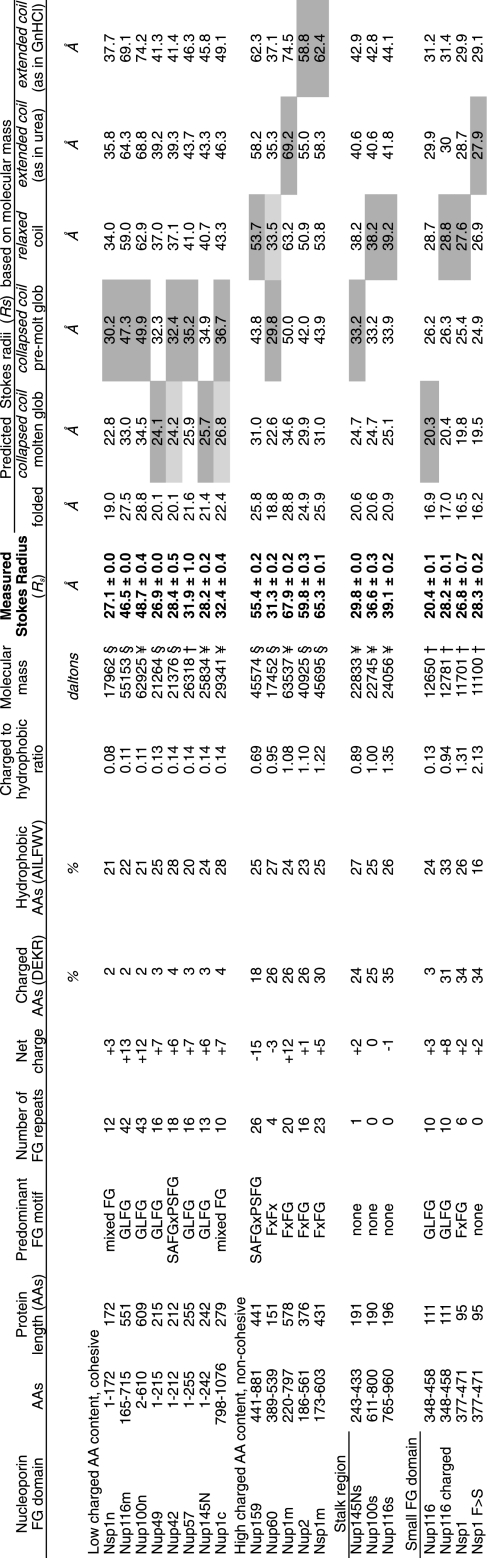
Table I. The hydrodynamic dimensions of intrinsically disordered nup domains in sieving columns at 30 °C.
Domains are ranked by ascending ratio of charged to hydrophobic AAs. Gray shaded boxes highlight the closest match between predicted and measured RS; light gray boxes mark cases where a value falls between categories.
§ Has a 9-AA tag at the N terminus (GSRRASVGS = 876 Da) in addition to the nup sequence.
† Has a 9-AA tag at the N terminus (GSRRASVGS = 876 Da) and a His6 tag (HHHHHH = 841 Da) at the C terminus in addition to the nup sequence.
¥ Has a 9-AA tag at the N terminus (GSRRASVGS = 876 Da) and a His6 tag with a tryptophan (WHHHHHH = 1027 Da) at the C terminus in addition to the nup sequence.
Determination of Stokes Radii
Purified FG domains were subjected to size fractionation in FPLC Superose 6 or Superdex 75 sizing columns. Proteins were injected at a flow rate of 0.3 or 0.5 ml/min into columns equilibrated at 30 °C in 20 mm Hepes, pH 6.8, 150 mm KOAc, 2 mm Mg(OAc)2. The protein elution profiles were monitored by UV absorbance and by SDS-PAGE analysis of collected fractions. Nup elution profiles were compared with those of carbonic anhydrase (29 kDa, RS = 23.5 Å), ovalbumin (45 kDa, RS = 29.8 Å), BSA (68 kDa, RS = 35.6 Å), aldolase (161 kDa, RS = 4.81 Å), catalase (232 kDa, RS = 5.22 Å), β-galactosidase (465 kDa, RS = 6.9 Å), and thyroglobulin (670 kDa, RS = 8.5 Å). ATP (0.25 mm) and plasmid pUC19 (4 μg) were included in the runs to mark the included and excluded volumes, respectively; their presence did not alter the mobility of nups. Protein elution volumes were determined according to the formula Kd = (Ve − V0)/(Vc − V0). Standards were plotted in relation to their known Stokes radii, allowing for RS calculation of unknowns using a linear regression formula.
MD Simulations
Simulations were performed using AMBER (Assisted Model Building with Energy Refinement, versions 7 and 8). FG domains were started from a fully extended structure with the φ and ψ angles set at 180° except for proline residues. Implicit solvent MD simulations were performed using the generalized Born/surface area model using Bondi atomic radii for the atoms. The Amber99 force field parameters were used. Each system was energy-minimized using 100 cycles of steepest descents and conjugate gradients. Constant temperature simulations were performed for 5 ns at 300 K by weakly coupling the system (using a 2 ps coupling constant) to an external heat bath. The salt concentration (Debye-Huckel screening) was set at 0.15 m, and a non-bonded cutoff of 250 Å was used (equivalent to infinity for these systems). A time step of 2 fs was used, and bonds containing hydrogen were constrained to their equilibrium distance using SHAKE. Replicate simulations were performed using different random seed values to generate different initial starting velocities. The radius of gyration (Rg) was calculated directly from the trajectories using the α carbons in the protein backbone (using CARNAL). Additional high temperature simulations were performed for all replicates by restarting each simulation for 3 additional ns coupled to an external heat bath at 350 K.
NMR Experiments
NMR experiments were performed on purified FG domains (0.5 mm) in 50 mm potassium phosphate, pH 6.4 using a Varian INOVA 600-MHz spectrometer with a 5 mm probe with single axis (along z) shielded magnetic field gradients. One-dimensional 1H NMR experiments were obtained using the water suppression scheme 1-3-3-1 Watergate (41). Self-diffusion coefficient measurements were obtained using a BPP-SED (bipolar gradient pulse pair selective echo dephasing) sequence (42). Translational diffusion tensor values were calculated based on the bead model approximation method (43) used successfully to calculate translational and rotational diffusion tensors of proteins (44, 45). All atoms were considered as beads of equal size (σ = 5.1 Å). The overall isotropic translational self-diffusion coefficient was calculated by taking the average of the principal values of the diffusion tensor.
Protein Composition Profiling
The AA compositions of disordered nup domains were analyzed using an approach developed for intrinsically disordered proteins (46). The fractional difference in composition between FG domains (or a set of disordered proteins from the DisProt database (47)) and a set of ordered proteins was calculated as (CX − Corder)/Corder and plotted for each AA residue where CX is the content of a given AA in a given protein or protein set and Corder is the corresponding content in a set of ordered proteins. In corresponding plots, the AAs were arranged from the most “order-promoting” to the most “disorder-promoting” according to the AA distribution in the DisProt database (47).
RESULTS
Distinct Categories of Intrinsically Disordered Structures in FG Nups: from Collapsed Coils to Extended Coils
We recently characterized the dynamic structure of a portion of the intrinsically disordered FG domain of Nup116 and found that it adopts an ensemble of collapsed coil conformations (36). To investigate whether other FG domains of yeast nups adopt similar collapsed coil configurations in contrast to extended coil configurations as reported for the human Nup153 FG domain (34), we purified the FG domains of S. cerevisiae nups (supplemental Fig. S1), subjected each to size fractionation in sieving columns to measure their RS (Table I), and compared the measured RS values with the predicted RS values for proteins of equal mass in various hypothetical structural configurations such as folded, molten globule, premolten globule, relaxed coil, extended coil (as in urea), or very extended coil (as in guanidinium hydrochloride). The latter values were obtained using the scaling relations reported (40). The sieving experiments were conducted at 30 °C, which is a physiologically relevant temperature for yeast growth.
Some FG domains of nups such as Nup145N and Nup49 (see Fig. 1) had measured Stokes radii that best matched the dimensions predicted for proteins of equal mass in the molten globular configurations (Table I). Others such as the FG domains of Nup116, Nup100, Nup57, Nup42, and Nup60 and the N- and C-terminal portions of the Nsp1 (Nsp1n) and Nup1 (Nup1c) FG domains, respectively, had dimensions that best matched the dimensions predicted for proteins in the premolten globular configurations, although some values fell between categories such as the Nup60 FG domain. In contrast, the FG domain of Nup159 best matched a protein in the relaxed coil configuration, and the FXFG-rich regions of Nup1 (Nup1m), Nup2, and Nsp1 (Nsp1m) had dimensions that best matched the dimensions predicted for proteins in extended coil configurations (i.e. akin to conformations adopted by proteins in chemical denaturants such as urea or guanidinium hydrochloride).
The Category of Intrinsically Disordered Structure in FG Nups Is Related to AA Composition
We next sought to identify AA determinants that influence which category of intrinsically disordered structure is adopted by an FG domain. We found that neither the total number of FG motifs, the type of FG motif, nor the length of the FG domain could predict the category adopted (see Table I), although these variables may have minor effects. Instead, it seemed that a difference in the content of charged AAs (i.e. the charge content) and, more specifically, the ratio of charged to hydrophobic AAs could best predict the different structural categories. According to that measure, the FG domains segregated into two distinct categories based on their AA composition and their measured Stokes radii (Fig. 2). One category (Fig. 2, left bottom group), comprising the Nsp1n, Nup116m, Nup100n, Nup57, Nup42, Nup1c, Nup49, and Nup145N FG domains, was characterized by a low charge content. These adopted collapsed coil configurations on average. A second category (Fig. 2, top right group), comprising the Nsp1m, Nup2, Nup1m, Nup159, and Nup60 FG domains plus the Nup116 and Nup100 stalk regions (defined below), was characterized by a high content of charged AAs. These adopted relaxed or extended coil configurations on average. Lastly, the Nup145N stalk region had a high charge content, but adopted collapsed coil configurations, making it an outlier from either group.
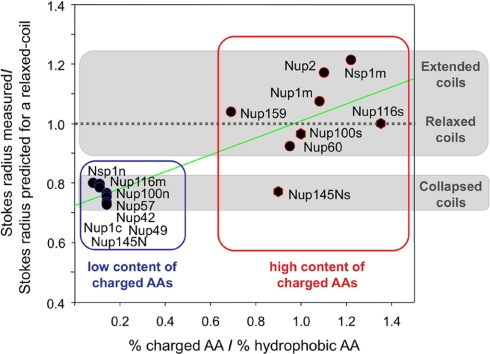
Fig. 2.
Distinct categories of intrinsically disordered structures adopted by FG nups. Stokes radii values obtained for the purified FG domains (Table I) were divided by the values predicted for each nup based on its mass assuming a relaxed coil configuration (y axis). The AA composition of each domain was expressed as the percent content of charged AAs (i.e. DEKR) divided by the percent content of hydrophobic AAs (i.e. AILFWV) (x axis). Gray boxes highlight the two main categories of intrinsically disordered structures. Nup domains with a low or high charge content are highlighted in blue or red boxes, respectively. The green line highlights the correlation between category of intrinsically disordered structure and charged-to-hydrophobic AA ratio.
To learn more about the AA composition differences between disordered domains that adopt collapsed coil configurations versus those that adopt relaxed or extended coil configurations, we compared the average abundance of each AA residue between these two structurally distinct groups. As an added comparison, the values were plotted relative to the average AA composition of proteins that fold (assigned a value of 0 in the y axis) and in comparison with intrinsically disordered proteins in general. As expected, all of the intrinsically disordered domains were depleted of order-promoting AAs and were variably enriched in disorder-promoting AAs (Fig. 3A). As a notable exception, Phe residues were enriched in the disordered FG domains despite being considered order-promoting. This was expected because phenyl rings in FG domains are used as key binding determinants for kaps (48, 49) and between FG nups (27). More importantly, among all the disordered domains examined, those that adopted relaxed or extended coil configurations had a high content of charged and chain-bending AAs such as Asp, Lys, Glu, and Pro with the charged residues being the dominant feature (Fig. 3B). We refer to these as having a high charge content and depict them and their preferred AAs in red in the figures. In contrast, all of the domains that adopted collapsed coil configurations had a high content of uncharged polar residues such as Asn, Gly, Gln, and Thr (Fig. 3A) with Asn and Gly residues being the dominant feature (Fig. 3B). We refer to these domains as having a low charge content and depict them and their preferred AAs in blue in the figures. Overall, the results implied that an enrichment in Asp, Lys, Glu, and Pro residues and a depletion of Asn, Gly, Gln, and Thr residues could convert a collapsed coil domain into a relaxed or extended coil domain. To test this hypothesis, a mutant version of the small Nup116 FG domain (AAs 348–458) that normally adopts collapsed coil configurations in the wild type form (36) was created where the intervening sequences between its FG motifs (which are otherwise rich in Asn, Gly, Gln, and Thr residues) were replaced by Asp-, Lys-, Glu-, and Pro-containing sequences in a way that resembled the Nsp1m FG domain (Fig. 3C). We refer to this mutant as the Nup116 FG domain charged mutant. A second mutant was created by removing Phe residues from the FXFG motifs of a small Nsp1 FG domain (AAs 377–471) and replacing them with Ser; we refer to this mutant as the FXFG → SXSG mutant or Phe → Ser mutant (Fig. 3C). It was designed to test the notion that hydrophobic FXFG motifs in a relaxed or extended coil FG domain can mediate intramolecular cohesion of coils similarly to what was observed for GLFG motifs in the collapsed coil Nup116 FG domain (36).
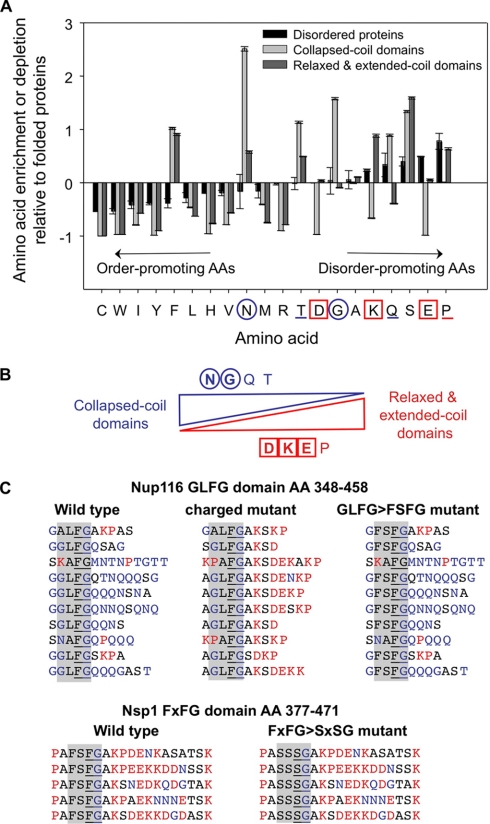
Fig. 3.
AA composition comparison between distinct structural categories of disordered domains. A, the AA composition of all FG domains that adopt collapsed coil configurations was compared with the composition of all domains that adopt relaxed or extended coil configurations (see Fig. 2). The outlier Nup145Ns domain was excluded from the analysis. The AA compositions were plotted relative to the AA composition of folded proteins (46), which sets the 0 value in the y axis for each AA. AA residues are listed from left to right according to their classification as order- or disorder-promoting (82). AA residues highlighted with blue circles are enriched in collapsed coil FG domains, and those highlighted by red squares are enriched in relaxed or extended coil FG domains. Error bars represent non-parametric estimations of the confidence intervals calculated by bootstrapping (10,000 interactions). B, a diagram summarizing the significant differences in AA composition between the two main categories of intrinsically disordered structures found in the FG nups. The diagram also highlights the proposed AA changes needed to convert a collapsed coil FG domain to an extended coil FG domain. C, AA sequences of small FG domains analyzed. Residues common in collapsed coil FG domains (blue text) were substituted with residues common in extended coil FG domains (red text) in the Nup116 FG domain (AAs 348–458) to create the Nup116 charged mutant. A Nup116 “swap” mutant featuring FXFG motifs instead of GLFG motifs was also created. Lastly, a small Nsp1 FG domain (AAs 377–471) was mutated by conversion of its FXFG motifs to SXSG motifs. The Nsp1 FG domain used in the MD simulations (AAs 375–479) was very similar to the one shown here (AAs 377–471), but contained six instead of five FG motifs. FG motifs are underlined. The category of FG motif (i.e. FXFG, GLFG, etc.) is shaded in gray.
The small FG domains mentioned above were purified (supplemental Fig. S1B), and their hydrodynamic dimensions were determined by measuring their Stokes radii (Table I) and their NMR diffusion coefficients. As predicted, the mutant Nup116 FG domain with the charged intervening sequences (the Nup116 charged mutant) displayed a larger Stokes radius (RS = 28.2 Å) and a smaller NMR diffusion coefficient (Ds = 13.04 ± 0.07 × 10−11 m2 s−1) than the wild type FG domain (RS = 20.4 Å; Ds = 13.27 ± 0.14 × 10−11 m2 s−1), indicating that the hydrodynamic volume of the mutant was bigger due to a molecular decompaction. Indeed, the dimensions of the mutant now best matched the dimensions predicted for a protein of equal mass in relaxed coil configurations rather than the collapsed coil conformations preferred by the wild type version (Table I).
The wild type version of the small Nsp1 FG domain adopted relaxed coil configurations on average (Table I); this was expected given its high charge content. However, substitution of its FXFG motifs for SXSG motifs caused it to decompact (i.e. increasing its RS value from 26.8 ± 0.7 to 28.3 ± 0.2 Å), converting it into an extended coil FG domain (Table I). The NMR experiments, however, were unable to detect this small increase in size because of poor resolution. The diffusion coefficient increased slightly for the mutant from 12.62 ± 0.09 to 12.71 ± 0.01 × 10−11 m2 s−1, but the difference in values was not statistically significant. Altogether, the results highlighted two key parameters influencing the category of intrinsically disordered structure adopted by FG domains: the charge content and the intramolecular cohesion of coils mediated by FG motifs.
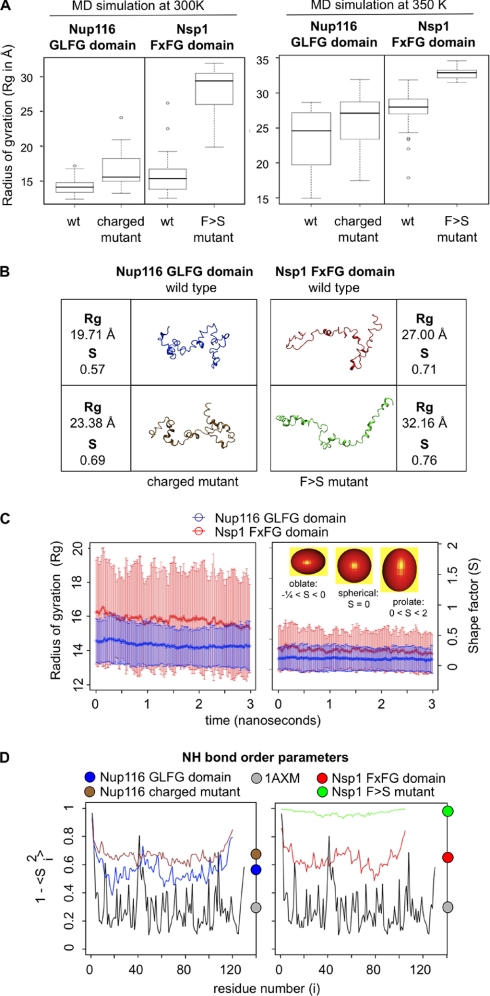
Relaxed and Extended Coil FG Domains Are More Dynamic than Collapsed Coil FG Domains
Molecular dynamics modeling can assist in characterizing the structural dynamics of intrinsically disordered proteins, which can be measured as time-dependent fluctuations in the molecular shape and/or the Rg and as fluctuations in the φ-ψ bond angles along the polypeptide backbone. To explore FG domain dynamics, 40 independent MD simulations were performed on each of the small FG domains (Nup116, Nup116 charged mutant, Nsp1, and Nsp1 FXFG → SXSG mutant; Fig. 3C), starting from a fully stretched conformation. The simulations were conducted at 300 K for 5 ns and were extended for an additional 3 ns at 350 K. As soon as the simulations started, the maximally stretched FG domains relaxed into more compact configurations with small patches of unstable secondary structure. Because these are disordered proteins, the resulting end structures for the replicates did not resemble one another as expected. However, and despite the fact that the nup structures were ever changing, the ensemble of structures for each ended up sampling a similar range of sizes and shapes during the last 3 ns of the 300 K simulations and during the last 2 ns of the 350 K simulations; this was according to various metrics of size, which changed little during these periods (data not shown). Notwithstanding, root mean square deviation versus time plots showed that the FG domains continued to sample structural change, albeit with similar dimensions, even after equilibration (supplemental Fig. S2). Thus, the simulated FG domain trajectories were saved at 1-ps intervals during the last 3 ns of all replicate simulations at 300 K to generate a total of 120,000 structures for each FG domain and during the last 2 ns of the 350 K simulations to generate 80,000 structures. The structures at 300 K were used to extract dynamical data because of a better resolution of dynamical differences between FG domains at that temperature. The structures generated at 350 K were used to calculate molecular shape parameters (Rg and shape parameter (S) values) because we established previously (36) and confirmed here that simulated structures at this temperature best match the magnitude of size differences detected for purified FG domains in sieving columns. Regardless of simulation temperature, all conclusions drawn below were derived from averaging properties across all 40 replicates for each FG domain.
Using the thousands of structures generated for each FG domain, we first calculated the average Rg values for each protein. At first glance, the pattern of differences in the Rg values between the four simulated FG domains matched the pattern of differences measured in hydrodynamic radius (Rh) values from the sieving columns (Fig. 4A and Table I). Specifically, the mutant Nup116 FG domain with a high charge content had larger dimensions (i.e. larger Rg) than its wild type version with low charge. This suggested that charged AAs in inter-FG motif regions can promote decompaction of an FG domain. Likewise, the Nsp1 FXFG → SXSG mutant exhibited larger dimensions than its wild type version, indicating that Phe residues in FXFG motifs mediated some intramolecular cohesion of coils (i.e. compaction) in the Nsp1 FG domain.
Fig. 4.
Structural dynamics of nup FG domains. A, box plots of the Rg for the FG domain structures simulated. The data represent the mean Rg obtained for 40 independent simulations of each domain at 300 or 350 K. The plots show a box around the central half of the data points enclosing the interquartile range. The error bars (‘whiskers’) enclose the remaining data points except for any outlier points (shown as individual points) that are more than 1.5 times the interquartile range outside the box. B, a representative snapshot of average structures obtained during 350 K simulations. The selected structures were those whose Rg value best matched the measured Stokes radius value and the average S value. The overall best match was with structures in the top of the first quartile in Rg values (i.e. an Rg > 25% of sampled structures; see A). C, plot of the mean and standard deviation (error bars) of the Rg and S values over time for FG domain simulations in implicit solvent at 300 K. Rg provides a measure of compactness for each protein, whereas S provides a statistical measure of its shape. The inset shows ellipsoids with shapes that match sample S parameters. D, order parameter (1 − S2) calculations for FG domains simulated at 300 K. Values were calculated for the amide N–H bond on each residue across all 300 K implicit solvent simulations. High or low values indicate larger or smaller fluctuations, respectively, in the N–H bond angle due to amplitude motions. Values for a folded protein (Protein Data Bank code 1AXM) are shown for comparison. The color circles on the right mark the average value for each protein.
The shape of a disordered protein can be characterized in terms of the eigenvalues of the Rg, which can be used to derive S, which can tell whether the protein resembles a sphere, an ellipsoid, or a rod (50). A value of S = 0 represents a sphere, whereas a value of S > 0 represents a prolate object, and a value of S < 0 represents an oblate object (see Fig. 4C, inset). The average shape of each small FG domain was calculated using its ensemble of 80,000 structures obtained at 350 K. Consistent with the greater Rg and RS values, the mutant FG domains adopted more prolate configurations than their wild type versions with an average S value of 0.68 versus 0.58 for the mutant and wild type Nup116 FG domains, respectively, and 0.76 versus 0.71 for the mutant and wild type Nsp1 FG domains, respectively. Finally, to provide a visual image of the different FG domain structures or rather a snapshot of their ever changing ensemble of conformations, we selected one structure in each data set whose S value was close to the average value for its ensemble of structures and whose Rg value best matched the RS/Rh values measured in the sizing columns (Fig. 4B). These structure snapshots clearly showed the difference in compaction between the different categories of FG domains.
The structural dynamics of the small FG domains were analyzed by plotting the fluctuations in the Rg and shape over time (Fig. 4C). The plots showed that the collapsed coil Nup116 FG domain with a low charge content adopted loose but compact conformations that although dynamic did not change dramatically in radius over time (Fig. 4C, blue lines) (also see supplemental Movie S1). This was contrasted by the relaxed coil Nsp1 FG domain containing a high charge content that continued to change widely in conformation over time (Fig. 4C, red lines) (also see supplemental Movie S2). As expected, each individual FG domain replicate did not exhaustively search all available conformation space in such a short time frame, hence the reason why we conducted 40 independent 5 ns replicates for each FG domain (i.e. to ensure good overall sampling). To determine whether good sampling was achieved, the secondary structure content of each FG domain replicate was analyzed, and the results were plotted as an average for the whole FG domain using all 40 replicates in combination (supplemental Fig. S3A) and individually for each FG domain, matching the secondary structure to its location along the polypeptide chain (supplemental Fig. S3B). Consistent with our previous CD spectroscopic analysis of FG domains (21), there was little secondary structure (i.e. β sheet and α helix) content in the simulated FG domains, but the presence of helical structures was notable for 310 helices because these structures are rarely found in folded proteins. However, examination of the per residue contribution to these data (supplemental Fig. S3B) revealed that the helical structures are very transient in nature, appearing and disappearing intermittently. Finally, when the secondary structure content was plotted relative to its location along the polypeptide chain, it showed that every FG domain simulation replicate adopted similar patterns of secondary structure along its chain (supplemental Fig. S3B) despite the fact that all end structures for the replicates looked different. This reproducibility in secondary structure distribution among the replicates gave us confidence that robust conformational sampling was achieved.
The structural dynamics of the FG domains were also analyzed by calculating the backbone N–H order parameter (S2) for each AA residue (Fig. 4D). Values of 1 − S2 close to 0 indicate highly constrained motions, and values close to 1 indicate unrestricted motions. The results indicated that the backbones for these proteins are highly flexible. The 1 − S2 values for all the FG domains were greater than the values obtained for a folded protein of similar size (e.g. fibroblast growth factor 1; Protein Data Bank code 1AXM; Fig. 4D, gray) and were consistent with a lack of stable secondary structure. The analysis also showed that the Nup116 charged mutant (Fig. 4D, brown) is more flexible than the wild type version (blue) and that the FXFG → SXSG mutant of Nsp1 (green) displays almost unrestricted motions and was significantly more flexible than its wild type version (red).
A main goal here was to identify the physical and/or dynamical properties that best distinguish the different categories of structures adopted by the FG domains. Hence, we used Tukey “honest significant difference” tests (51) to perform statistical analyses of nup properties calculated from the MD trajectories. These included the average Rg value (over each 3 ns simulation), the standard deviation of the Rg (over each 3 ns simulation), the average S value, the standard deviation of the S value, and the order parameter (S2) (averaged over all residues). Because a characteristic feature of the FG domains was their dynamics, we also analyzed the frequency spectrum of the Rg and S time series for each of the simulations. Specifically, we calculated the spectral density (in decibels) at frequencies with periods ranging from 2 ps to 1.5 ns and identified the frequency range that best distinguished the different FG domains and their characteristic structures. Overall, the frequency analysis of the Rg values yielded the best overall pairwise distinction between the FG domains with a p value <0.05 for all pairs (supplemental Table S1).
Some FG Nups Display A Bimodal Distribution of Charged AAs in Their Disordered Domains
In the experiments above, the ratio of charged to hydrophobic AAs correlated with the distinct category of intrinsically disordered structure that each FG domain adopted (Table I and Fig. 2). Given this relationship, we examined closely the distribution of charged AAs in the intrinsically disordered region of every FG nup. Interestingly, a subset of nups had disordered domains with only a low content of charged AAs (2–4%) (e.g. Nup42, Nup49, and Nup57), whereas others had disordered domains with only a high charge content (24–35%) (e.g. Nup159, Nup60, and Nup2) (Fig. 1 and Table II). In contrast, two FG nups (e.g. Nsp1 and Nup1) had a bimodal distribution of charged residues within their disordered domain (Fig. 1 and Table II). For example, the Nsp1 N terminus (AAs 1–172; Nsp1n) had a low charge content (2%) and adopted an ensemble of collapsed coil configurations. In contrast, the remainder of the Nsp1 FG domain (AAs 173–603; Nsp1m) had a high content of charged AAs (36%) and adopted extended coil configurations (Table I). Such bimodal distribution of charged AAs and of distinct categories of intrinsically disordered structures predicted a bipartite topology in these FG domains where one portion adopts collapsed coil configurations while the other adopts relaxed or extended coil configurations. Topologically, these would loosely resemble the canopy and trunk (or stalk) of a “tree,” respectively (see Fig. 6).
Table II. Properties of intrinsically disordered domains in S. cerevisiae FG nucleoporins.
MG, molten globule; PMG, premolten globule.
| Nucleoporin | Disordered domain AAsa | FG motifs | Net charge | pI | Charged AAs (DEKR) | Hydrophobic AAs (AILFWV) | Charged to hydrophobic ratio | Molecular mass | Configuration at 30 °C | Predicted Rh at 30 °Cb | Predicted diameter | Volume | Number per NPC | Volume × number | FG domain interactionc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | daltons | nm | nm | nm3 | nm3 | |||||||||
| Nup159 | 387–1071 | 16 | −38 | 4.7 | 24 | 22 | 1.1 | 71,748 | Relaxed coil | 6.7 | 13.4 | 1,259 | 8 | 10,074 | Repulsive |
| Nup42 | 1–382 | 20 | +11 | 10.7 | 4 | 25 | 0.2 | 37,577 | PMG | 4.1 | 8.2 | 289 | 8 | 2,309 | Cohesive |
| Nup116 | 172–764 | 42 | +11 | 10.7 | 2 | 22 | 0.1 | 59,006 | PMG | 4.9 | 9.8 | 492 | 8 | 3,940 | Cohesive |
| Nup116 | 765–960 | 0 | −1 | 6.3 | 35 | 26 | 1.4 | 22,152 | Relaxed coil | 3.8 | 7.6 | 230 | 8 | 1,838 | Repulsive |
| Nup100 | 1–610 | 43 | +12 | 11.4 | 2 | 21 | 0.1 | 61,153 | PMG | 4.9 | 9.8 | 493 | 8 | 3,941 | Cohesive |
| Nup100 | 611–800 | 0 | 0 | 7.2 | 26 | 25 | 1.0 | 20,842 | Relaxed coil | 3.7 | 7.4 | 212 | 8 | 1,697 | Repulsive |
| Nsp1 | 1–186 | 13 | +4 | 10.5 | 2 | 22 | 0.1 | 18,507 | PMG | 3.1 | 6.2 | 125 | 32 | 3,991 | Cohesive |
| Nsp1 | 187–617 | 22 | +4 | 8.7 | 36 | 24 | 1.5 | 44,749 | Extended coil | 6.2 | 12.4 | 998 | 32 | 31,930 | Repulsive |
| Nup49 | 1–251 | 17 | +7 | 10.7 | 3 | 24 | 0.1 | 24,320 | MG | 2.5 | 5.0 | 65 | 16 | 1,047 | Cohesive |
| Nup57 | 1–255 | 15 | +7 | 10.8 | 3 | 21 | 0.1 | 24,601 | PMG | 3.4 | 6.8 | 165 | 16 | 2,633 | Cohesive |
| Nup145N | 1–242 | 13 | +6 | 10.4 | 3 | 17 | 0.2 | 23,931 | MG | 2.5 | 5.0 | 65 | 16 | 1,047 | Cohesive |
| Nup145N | 243–433 | 1 | +2 | 8.8 | 24 | 27 | 0.9 | 20,930 | PMG | 3.2 | 6.4 | 137 | 16 | 2,195 | Repulsive |
| Nup1 | 220–797 | 19 | +12 | 9.3 | 26 | 24 | 1.1 | 61,634 | Extended coil | 6.8 | 13.6 | 1,316 | 8 | 10,531 | Repulsive |
| Nup1 | 798–1076 | 10 | +7 | 11.8 | 4 | 26 | 0.2 | 27,525 | PMG | 3.6 | 7.2 | 195 | 8 | 1,563 | Cohesive |
| Nup60 | 389–539 | 4 | −3 | 5.3 | 26 | 27 | 1.0 | 16,576 | Relaxed coil | 3.3 | 6.6 | 151 | 8 | 1,204 | Repulsive |
| Nup2 | 160–600 | 15 | −8 | 5.2 | 27 | 23 | 1.2 | 46,960 | Extended coil | 6.3 | 12.6 | 1,047 | 8 | 8,375 | Repulsive |
a Specific boundaries for the intrinsically disordered domains were defined in Denning and Rexach (22) according to a bimodal AA substitution rate in these nups during evolution. The boundaries for Nup1 and Nup2 were refined here according to the PONDR predictions shown in Fig. 1.
b The predicted Rh for each domain was derived from its molecular weight and its best match structural category using the scaling equations defined in Table I.
c These assignments are based on the interactions reported here or in Patel et al. (27); the assignments are consistent with the rules of cohesion discussed in the text.
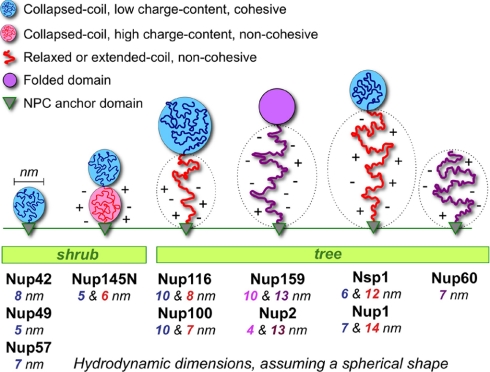
Fig. 6.
Hydrodynamic dimensions and topology of intrinsically disordered domains in S. cerevisiae FG nups. Polypeptide chains with a net positive charge are depicted as blue or red wiggly lines depending on their high (red) or low (blue) content of charged AAs. Nup domains with a net negative charge greater than 1 are shown as purple lines. The NPC anchor domain for each nup is depicted as a small gray triangle. Nups were categorized as shrubs if their FG domain adopts collapsed coil configurations on average and is adjacent to the NPC anchor domain. FG nups were categorized as trees if they feature a relaxed or extended coil domain separating a collapsed coil or folded globule from the NPC anchor domain. The Nup60 FG domain tree is shown topless because it lacks an attached globular domain. The color-matched numbers listed under each nup correspond to the calculated hydrodynamic diameter of each domain according to RS and mass (Table II) and assuming a spherical shape. Domains with a high charge content are decorated by charge symbols that reflect the net charge.
An additional subset of FG nups (Nup116, Nup100, and Nup145N) also exhibited a bimodal distribution of charged AAs along their disordered domain except it included a region without FG motifs that separates the FG domain from the folded NPC anchor domain (Fig. 1 and Table II); we termed these the stalk regions. For example, the Nup100 FG domain (AAs 1–610) has a low charge content and is contiguous with a predicted disordered region (Nup100s; AAs 611–800) that features a 26% charge content (Fig. 1). To test whether these stalk regions were indeed intrinsically disordered, we purified them and measured their hydrodynamic radii and protease sensitivity. When exposed to a very mild proteinase K digestion, each was degraded quickly as was the FG domain of Nup145N (supplemental Fig. S4A) and all other FG nups (21); this was consistent with their categorization as intrinsically disordered proteins. Also, far-UV CD spectroscopy analyses showed that these regions were devoid of secondary structure as was the FG domain of Nup145N (supplemental Fig. S4B) and all other FG domains tested (21). Finally, to categorize the subtype of intrinsically disordered structure adopted by these stalk regions, their RS/Rh was measured and compared with the hydrodynamic dimensions predicted for proteins of equal mass in different intrinsically disordered configurations (Table I). The RS obtained for Nup116s and Nup100s best matched the dimensions predicted for a protein in relaxed coil configurations, and the RS for the Nup145N stalk region matched best the dimension predicted for a premolten globule.
Exploring FG Domain Function in Vitro: Charge Content Determines the Attractive or Repulsive Character of Intermolecular FG Domain Interactions
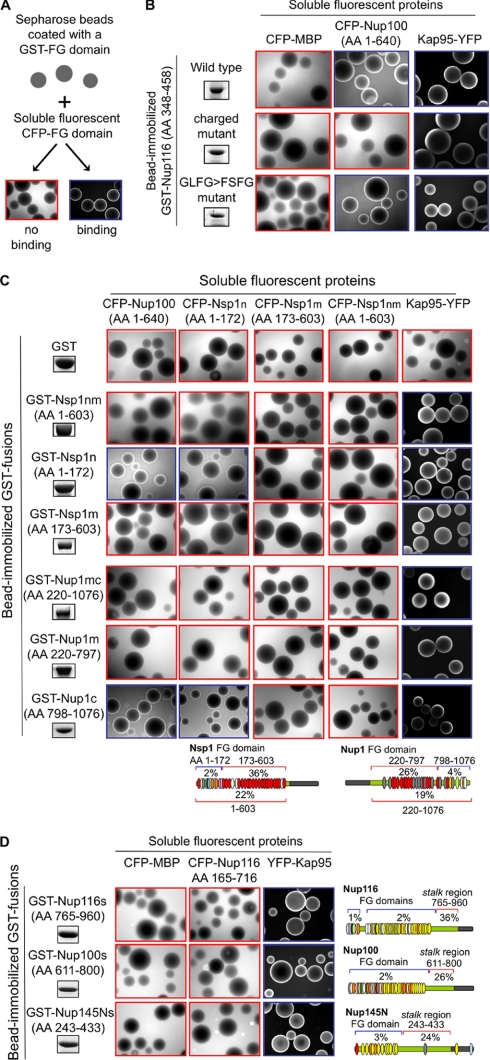
We previously showed that a subset of FG domains displays attraction (i.e. cohesion) toward each other in vitro and in vivo via hydrophobic attraction between their FG motifs. We also showed that other FG domains do not form such cohesions under identical conditions (27). Back then it was noted that the ability to form interactions seemed to correlate with a low charge content in the FG domains, but this was not tested experimentally. To explore these correlations and, more importantly, to determine whether the distinct categories of intrinsically disordered structures are linked to this key FG domain function (e.g. intermolecular cohesion and FG domain oligomerization), we examined the cohesive properties of the small wild type Nup116 FG domain (featuring a low charge content) in comparison with its mutant containing a high charge content (the Nup116 charged mutant) (Fig. 3C). We also tested whether FXFG motifs are as cohesive as GLFG motifs using a GLFG → FXFG swap mutant (Fig. 3C). Because FG domains that interact do so with low affinity (KD ∼ 5–70 μm), we used the bead halo assay (Fig. 5A), which robustly tests low affinity interactions under equilibrium binding conditions (27). In the assay, Sepharose beads coated with GST-FG domain fusions are mixed with another FG domain added as a soluble fluorescent protein, and the mixtures are photographed under a fluorescence microscope. Positive binding interactions between soluble and immobilized FG domains are evident as a halo of fluorescence around beads, whereas negative interactions appear as dark beads surrounded by a fluorescent background. Beads coated with the GST-Nup116 FG domain (AAs 348–458) or with its mutant forms were incubated with soluble CFP-Nup100 FG domain (AAs 1–640) (a cohesive FG domain fused to the cyan fluorescent protein (27)), YFP-Kap95 (a kap that binds FG motifs fused to the yellow fluorescent protein), or CFP-maltose-binding protein (Fig. 5B). As expected, none of the immobilized Nup116 FG domains interacted with the negative control CFP-maltose-binding protein, but all interacted with the positive control YFP-Kap95, confirming the specificity and the functionality of the isolated FG domains. In contrast, the wild type Nup116 FG domain and the GLFG → FXFG swap mutant, but not the Nup116 charged mutant, interacted with the cohesive CFP-Nup100 FG domain. This demonstrated that cohesion between FG domains can be mediated by FXFG motifs as well as GLFG motifs (36) as long as the intervening sequences have a low charge content.
Fig. 5.
Test of low affinity interaction between FG domains. A, diagram of the bead halo assay used to detect low affinity interactions (27). Soluble CFP-FG domain fusions were mixed with bead-immobilized GST-FG domains, and the mixture was visualized (and photographed) directly under a fluorescence microscope at 25 °C. B–D, soluble CFP or YFP fusions (top row) were mixed with bead-immobilized GST fusions (left columns) as indicated, and the mixtures were visualized as in A. An aliquot of the immobilized protein was resolved by SDS-PAGE, was stained with Coomassie Blue, and is shown on the left. Blue or red boxes highlight positive or negative interactions, respectively. The nup diagrams highlight the bimodal distribution of charged AAs in the intrinsically disordered domains of the nups used. MBP, maltose-binding protein.
The observed correlation between charge content and the ability to form intermolecular FG domain interactions was particularly intriguing when considering that some FG nups such as Nsp1, Nup1, Nup116, Nup100, and Nup145N contain a bimodal distribution of high and low charge content along their disordered domains (Fig. 1 and Table II). Based on the lessons learned above, it therefore seemed possible to predict the cohesive properties of each subdomain relying on its content of charged AAs and FG motifs. In the case of Nsp1 for example, the analysis predicted that its N-terminal portion (Nsp1n; AAs 1–186) (Fig. 1) would be cohesive based on its low (2%) charge content (Table I) and that the middle portion (Nsp1m; AAs 187–617) would be repulsive based on its high charge content (36%). As for Nup1, the C-terminal portion (Nup1c; AAs 798–1076) with a low (4%) charge content was predicted to be cohesive, whereas the middle portion (Nup1m; AAs 220–797) was predicted to be repulsive due to its high charge content (26%). Lastly, the stalk regions of Nup100 (AAs 611–800), Nup116 (AAs 765–960), and Nup145N (AAs 243–433) were also predicted to be non-cohesive based on their high charge content (24–35% charged AAs) and their lack of FG motifs (Table II). To test the predictions, we used the bead halo assay using immobilized and soluble FG domains representing the relevant regions of these nups. As expected for the negative control, none of the soluble fluorescent proteins bound to immobilized GST (Fig. 5C, top row). Also as expected, YFP-Kap95 bound to all immobilized FG domains because they all have FG motifs (Fig. 5C, right column). As reported previously (27), the immobilized full-length Nsp1 FG domain (Nsp1nm; AAs 1–603) containing regions of high and low charge content (but a high overall charge content of 22%) did not bind to the soluble FG domains (Fig. 5C, second row) and not even to the cohesive FG domains such as the Nup100 FG domain for example (Fig. 5C, left column). However, when the Nsp1 FG domain was separated into the two distinct regions based on charge content (Nsp1n and Nsp1m) (see Fig. 5C, bottom diagrams), a different picture emerged. The immobilized Nsp1n FG domain interacted with the cohesive CFP-Nup100 FG domain and even with itself when added as a soluble CFP fusion (Fig. 5C). In contrast, the portion of the Nsp1 FG domain with the high charge content (Nsp1m; AAs 173–603) displayed no cohesion toward other FG domains or itself either in its soluble or immobilized form. A similar scenario was observed for Nup1. We previously showed that a Nup1 FG domain (AAs 332–1076) containing separate regions of high and low charge content but an overall high charge content (18%) displays no attraction toward other FG domains (27). The same result was shown here for a similar but larger portion of the Nup1 FG domain (AAs 220–1076) (Fig. 5C). These Nup1 domains included the C terminus (AAs 798–1076), which has a low charge content (4%) and was predicted to be cohesive. Indeed, as predicted, when only the C terminus of Nup1 was immobilized, it bound to other cohesive FG domains (Nup100 AAs 1–640 and Nsp1 AAs 1–172; Fig. 5C), whereas the highly charged middle portion of Nup1 (AAs 220–797) did not. Lastly, none of the immobilized stalk regions of Nup116, Nup100, and Nup145N interacted with soluble cohesive FG domains (e.g. the CFP-Nup116 FG domain AAs 165–716) as expected based on their high charge content and the lack of FG motifs. Yet surprisingly, the stalk regions interacted with Kap95-YFP (Fig. 5D).
Tertiary Structure (Topology) and Hydrodynamic Dimensions of S. cerevisiae FG Nups
Based on the observations above, it became possible to estimate the topology of each intrinsically disordered domain in the FG nups. For the nups with disordered regions of high and low charge content in a bimodal distribution, estimates were determined separately for each subregion (Table II). In general, the FG domains could be described in two shape categories, which we termed “shrubs” and “trees” (Fig. 6) depending on their content and position of collapsed coil versus relaxed or extended coil domains in relation to the NPC anchor domain (the “roots”). For illustration purposes, the FG domains that adopt collapsed coil configurations (Fig. 2) were depicted as elliptical globules filled with coils (Fig. 6), whereas the domains that adopt relaxed or extended coil configurations were depicted just as coils. It was clear from the diagrams that the collapsed coil region of several FG nups is separated from the NPC anchor domain by a relaxed or extended coil region that features a high charge content. This peculiar topology implied a functional significance to having collapsed coil globules anchored to the NPC via more dynamic coils in relaxed or extended configurations (see “Discussion”).
Finally, we used scaling equations (40) to calculate the hydrodynamic volume of all yeast FG domains and stalk regions, adjusting for the mass of the entire domain when necessary (in cases where only a fragment was analyzed here; compare Tables I and II). The derived Rh/RS values were then used to estimate (i) the molecular diameter of each FG domain or stalk assuming it is a sphere, (ii) the hydrodynamic volume it would occupy in isolation, and (iii) the combined volume that all intrinsically disordered nup domains would occupy at the NPC given their stoichiometry there (52). Interestingly, all of the intrinsically disordered nup domains combined would occupy a hydrodynamic volume equivalent to 88,315 nm3 (e.g. 20,471 nm3 occupied by cohesive FG domains and 67,844 nm3 occupied by repulsive FG domains and stalk regions) (Table II). This value is remarkably close to the presumed volume of the NPC transport conduit (i.e. 86,162 nm3) assuming it is a cylinder ∼35 nm high with a ∼56 nm diameter and that FG domains are anchored 28 nm away from the NPC center on average (52) (see Fig. 7). However, because not all FG domains populate the interior of the transport conduit (i.e. some are anchored to the periphery of the conduit) (see Fig. 7), there would be empty space within the conduit.
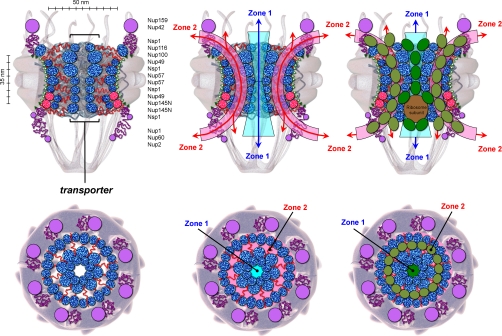
Fig. 7.
Diagram of the forest model of NPC architecture with a central transporter structure and two distinct zones of traffic. This model is based on the topology, dimensions, stoichiometry, interactivity, and approximate anchoring site of yeast FG nups. FG nups are listed from top to bottom according to their stoichiometry and the reported location of their tether sites along the NPC z axis (52). Nup2 does not have an NPC anchor domain but binds Nup60 directly and indirectly via Ran (Gsp1) (83). The disordered structures shown are drawn to scale according to the topology and dimensions delineated for each nup in Fig. 6. For simplicity, only 34 of the 136 FG domains predicted for each NPC are shown in side views (i.e. as two vertical columns of eight total), and only 40 of the 136 FG domains are shown in the top (cytosolic face) views, which feature Nsp1 and Nup116 forming two of the seven octoglobular FG domain rings at the central transporter. Kaps are depicted as dark or light green ovals if traversing Zone 1 or 2, respectively. The kap dimensions shown are those of Kap95 as a representative. See “Discussion” for a detailed explanation of the forest model, its central transporter structure, and the two distinct zones of traffic it creates along the NPC conduit.
DISCUSSION
Defining Two Categories of Intrinsically Disordered Structures in FG Nups
The analysis here of 20 different intrinsically disordered protein domains led to the conclusion that yeast FG nups feature two distinct categories of disordered structures with unique AA composition, dynamics, and function. One category had a low content of charged AAs (2–4%) (Table I), was enriched in Asn and Gly residues (Fig. 3, A and B), and exhibited Stokes radii in physiological buffer equivalent to those of proteins of equal mass in molten or premolten globular configurations (i.e. collapsed coil configurations) (Table I and Fig. 2). Functionally, these FG domains displayed attraction toward one another, forming low affinity binding interactions (Fig. 5, B and C) (27). The second category had a high content of charged AAs (18–35%) (Table I); was rich in Asp, Lys, and Glu residues (Fig. 3, A and B); and adopted relaxed or extended coil conformations (Table I and Fig. 2) that were more dynamic than the collapsed coil conformations (Fig. 4 and supplemental Movies S1 and S2). Functionally, these FG domains avoided interactions with each other (i.e. were repulsive) (Fig. 5C) despite having cohesive (i.e. sticky) FG motifs (27). An important question is whether these two distinct categories of disordered structures are relevant to NPC architecture and/or function in vivo. Indeed, a recent bioinformatics analysis identified key features in the NPC that have been conserved since the last eukaryotic common ancestor. It noted that nup FG domains evolved as two unique groups with distinct AA compositions, those rich in Gly residues and those rich in DEKR residues (53). Remarkably, these are almost the same AA composition differences identified here for the nups when using the two distinct categories of intrinsically disordered structures as a guide. This close correlation provides strong evidence that the two categories of disordered structures in the FG nups evolved as an indispensable feature of NPC architecture and function in eukaryotes.
Exploring the Correlation between AA Composition and Category of Intrinsically Disordered Structure
The AA composition of the intrinsically disordered FG domains, specifically the ratio of charged to hydrophobic AAs, was a key determinant of the structural conformations that each domain preferred on average (Fig. 2). Indeed, all FG domains with a low ratio of charged to hydrophobic AAs (≤0.14) adopted collapsed coil configurations similar to those adopted by premolten and molten globules (Table I). In contrast, the FG domains with a high ratio of charged to hydrophobic AAs (>0.69) adopted relaxed or extended coil configurations on average except for Nup145Ns. This correlation was tested and confirmed by genetic manipulation of the small Nup116 FG domain (AAs 348–458). This collapsed coil FG domain was transformed into a relaxed coil FG domain by increasing its charge content from 3 to 31% at intervening sequences between FG motifs without altering the FG motif spacing (Table I and Fig. 3C). In fact, the relationship observed here between the charged-to-hydrophobic AA ratio and the category of intrinsically disordered structure was evident in 19 of 20 domains analyzed (Table I and Fig. 2). The only exception was the Nup145N stalk region, which had a 0.89 ratio and was predicted to adopt extended coil configurations, but adopted collapsed coil configurations on average (Table I). This outlier may contain an unrecognized structural feature(s) that permits better compaction of its ensemble of structures. In conclusion, the charge-to-hydrophobicity relationship observed here for the two main categories of intrinsically disordered structures in nups was reminiscent of the charge-to-hydrophobicity relationship noted previously between folded and disordered proteins (54). We suggest that within the intrinsically disordered class of proteins the charge-to-hydrophobicity ratio strongly biases the category of intrinsically disordered structures that will form (Table I), extending the charge-to-hydrophobicity paradigm to this peculiar class of proteins with no fixed secondary or tertiary structure.
Exploring the Correlation between Structural Dynamics and Category of Intrinsically Disordered Structure
The MD simulations of FG domains revealed that their dynamics differ according to the category of intrinsically disordered structure (Fig. 4 and supplemental Table S1). The collapsed coil and relaxed coil FG domains examined (Nup116 AAs 348–458 and Nsp1 AAs 377–471, respectively) were much more dynamic than a folded protein (i.e. Protein Data Bank code 1AXM); but comparing the two FG domains, the physical dimensions (Rg and shape factor) and the bond angles along the peptide backbone fluctuated more widely for the Nsp1 FG domain (Fig. 4). This domain was less compact and more dynamic (see supplemental Movies S1 and S2) presumably because it made fewer intramolecular interactions and/or experienced more intramolecular repulsion in comparison with the Nup116 domain. Consistently, the simulated Nsp1 domain had only six FG motifs serving as possible intramolecular cohesion elements compared with ten FG motifs in the Nup116 domain. Also, the Nsp1 domain had a 34% content of charged AAs versus only 3% in the Nup116 domain, implying that the high charge content in Nsp1 caused intramolecular coil repulsion. From the movies, it was evident for the collapsed coil Nup116 GLFG domain that some parts of the polypeptide chain seemed intermittently trapped in low energy wells. This was expected because its FG motifs can mediate intramolecular cohesion of coils (Table I) (36) and can therefore restrict their mobility at least temporarily. Finally, the most dynamic and extended FG domain examined here was the FXFG → SXSG mutant of Nsp1 (Fig. 4), which had no FG motifs that could serve as cohesion elements and featured a >2:1 ratio of charged to hydrophobic AAs, the highest of any domain examined (Table I). In conclusion, these data suggest that the structural dynamics of disordered FG domains are governed by two parameters: the ability of their coils to form intramolecular interactions via GLFG or FXFG motifs (and/or possibly other FG motif types; see Fig. 1) and the charge content of the regions between FG motifs, which interferes with FG motif interactions when it is high. Regardless of category, the disordered structures in the FG domains could constantly fluctuate in orientation and dimensions, permitting any FG domain to extend beyond its average shape with some frequency. The frequency of these events, which was much higher for the relaxed and extended coil structures than for the collapsed coil structures (supplemental Movies S1 and S2), was the dynamical feature that best distinguished the distinct structural categories of disordered FG domains (supplemental Table S1).
Two Categories of Disordered Structures in FG Nups Have Unique and Overlapping Functions
When considering molecular interactions, both categories of intrinsically disordered FG domains bound karyopherins (Fig. 5) (28). However, only the collapsed coil FG domains were able to bind each other to form oligomers (Fig. 5) (27). This was in stark contrast to all of the relaxed or extended coil FG domains, which tend to repel FG domains (Fig. 5, C and D) (27) regardless of their content of sticky FG motifs (Fig. 5B). Hence, there seems to be a functional need in cells to have some FG domains aggregate and others repel. This could be another reason why the two distinct structural categories of FG domains evolved separately. This seems insightful considering that current models of NPC gate architecture differ in exactly that point (i.e. cohesion between FG domains in the hydrogel model versus repulsion in the polymer brush model) (25, 55). We therefore conclude, as we did before (27), that two different gating mechanisms likely operate at the NPC at distinct locations, one acting as a hydrogel and the other acting as an entropic brush. Lastly, it seems relevant to point out that all relaxed and extended coil FG domains (from Nup159, Nup2, Nup60, Nup1, and Nsp1) bind only one or two kaps with high avidity in affinity capture assays (using bead-immobilized FG domains and yeast cytosol), whereas all FG domains that adopt collapsed coil configurations (Nup42, Nup49, Nup57, Nup100, and Nup116) bind many (from seven to 11) different kaps with high avidity (28, 56, 57). It may be that the preclustering of FG motifs and/or other determinants in the collapsed coil FG domains makes them more amenable to high affinity interactions with kaps.
Molecular Features That Influence Interaction between FG Domains
Besides the presence of FG motifs, the overall charge content in the FG domains was the best indicator of their propensity to form intermolecular interactions (i.e. their cohesiveness). In fact, two simple “rules of cohesion” could explain all yeast FG domain interactions detected thus far. First, FG domains with a low content of charged AAs (≤4%) can bind each other, whereas FG domains with a high charge content (>18%) cannot (Table I and Fig. 5) (27). This was tested and confirmed here by introducing charged AAs (Fig. 3C) in the otherwise cohesive Nup116 FG domain, converting it into a non-cohesive FG domain (Fig. 5B). Second, the presence of FG motifs is required for binding between FG domains as mutant domains that lack them cannot interact (27). Previously, we showed that the GLFG motifs of Nup116 function as hydrophobic cohesion elements for inter- and intramolecular FG domain interactions (27, 36). Here, we expanded that observation by showing that the FXFG motifs of Nsp1 can also function as cohesion elements, but only when surrounded by uncharged polar AAs (Fig. 5B). This particular detail is important because human FG nups (except for hNup98) lack the canonical GLFG motifs present in most of the cohesive yeast FG nups. Nonetheless, the human FG nups such as Nup62 (i.e. the Nsp1 ortholog) have FXFG motifs embedded in regions of low charge content that according to our analysis and prediction should display cohesion toward other FG domains with a low charge content.
Bimodal Distribution of Structure and Function in Disordered Regions of Nups
The rules of cohesion between FG domains delineated above were used to identify the N terminus of Nsp1 (Nsp1n; AAs 1–172) and the C terminus of Nup1 FG domains (Nup1c; AAs 798–1076) (Fig. 1 and Fig. 5C) as regions that could bind other cohesive FG domains. This functional prediction based on their low (2–4%) charge content contradicted our previous observation that larger FG domains containing these sequences (Nsp1 AAs 1–603 and Nup1 AAs 332–1076) did not bind to cohesive FG domains (27). However, as we showed here, the termini of these FG domains did interact with each other and with additional FG domains, but only when detached from their highly charged and repulsive neighbor domain (Fig. 5C). This implied that the tips of Nup1 and Nsp1 FG domains can interact with other cohesive FG domains (27) (Fig. 7) as long as their repulsive neighbor domain was physically shielded by steric hindrance or some other means. This observation could in fact explain why a larger Nsp1 FG domain (AAs 1–601) at a concentration of 300 mg/ml is able to form hydrogels in vitro in a FG motif-dependent manner (29) despite having an overall high content of charged AAs (22%). According to the rules above, the charged AAs should have prevented its interaction with other FG domains at least at physiological pH as observed here (Fig. 5C) and previously (27). Therefore, we suggest that during formation of nup hydrogels in vitro, which is done in 0.2% TFA at a pH <2 (29), the negatively charged AAs in the Nsp1 FG domain become protonated, and their charge is neutralized. This significantly increases the hydrophobicity of the domain and decreases its ratio of charged to hydrophobic AAs from a value of 0.9 to a value of 0.5, which more closely resembles the ratio in cohesive FG domains (Table I) (27). In conclusion, only by studying individual segments of intrinsically disordered regions of nups (in isolation rather than studying them strictly as part of the intact protein or always combined with other FG domains) were we able to identify and characterize the two functionally distinct categories of FG domains. Hence, this partitioning approach may be especially important in cases where crowding (such as in a polymer brush scenario) or physical masking by a binding partner could physically isolate one segment of a disordered domain from another.
A bimodal distribution of structure and function was also detected in the intrinsically disordered domains of Nup116, Nup100, and Nup145N. These contained a previously unrecognized region of disordered structure, which we termed the stalk region, located between the FG domain and the NPC anchor domain. The stalk regions lack FG motifs (except for one in Nup145N) and have a high charge content that starkly contrasts with its neighbor FG domain (Fig. 5D). Initially, these stalk regions were predicted to be disordered based on their AA composition (according to PONDR; see Fig. 1) and on their rapid AA substitution rate (22), which is common in disordered domains (58). Here, we confirmed these to be disordered using three additional criteria: (i) by direct measurement of their hydrodynamic dimensions in sizing columns (Table I), which showed that they have hydrodynamic dimensions similar to those of proteins of equal mass in disordered configurations, (ii) by the apparent lack of stable secondary structures as judged by CD spectroscopic analysis (supplemental Fig. S4B), and (iii) by their hypersensitivity to proteases (supplemental Fig. S4A). When examining the function of the stalk regions, we found that unlike their neighboring FG domain they do not interact with the cohesive FG domains (Fig. 5D). This was not surprising given that they lack FG motifs and have a high charge content, which is similar to the repulsive FG domains (see below). What was surprising, however, was that the stalk regions interacted with the importin Kap95 directly despite their lack of FG motifs (Fig. 5D). Hence, Kap95 could use the stalk regions as docking sites during transport or may recognize a targeting motif in these regions for use during NPC biogenesis. Regardless, the ∼200-AA stalk region in Nup116 and Nup100 could function as a dynamic tether or spring connecting the globular FG domain to the NPC anchor domain (Fig. 6) (see below).
Topology of FG Nups within the NPC Transport Conduit
FG nups are likely anchored to the inner walls of the NPC transport conduit and its periphery. Their overall distribution has been envisioned as a “cloud” of polypeptide chains populating the conduit, but leaving a 10 nm hole through the center (1) or as an amorphous hydrogel without a central opening (29). The fact remains, however, that these are only hypothetical models of NPC conduit architecture that are difficult to test in vivo due to the complexity and redundancy of FG nup function. Based on these models for example, it is debated whether individual FG domains can extend their coils sufficiently to reach across the central cavity of the NPC to interact and mingle with other FG nups anchored at the opposing side (as in the hydrogel model) leaving no central hole open. At first glance, our data would indicate that they are not. The distance from their NPC tether sites to the conduit center (∼28 nm on average (1)) is greater than the dimensions reported here for the individual FG domains (i.e. ∼5–14 nm diameter assuming spherical shapes; Tables I and II) and even for the bipartite domains stacked on top of each other in tree configurations (Fig. 6). Importantly, however, FG domains are likely crowded at the NPC because their dimensions (e.g. their hydrodynamic diameter) (Fig. 6) are equal to or greater than the average distance between their anchor sites (∼5–6 nm (52)). Hence, a crowding effect would promote formation of a polymer brush whose entropic effect would force some FG domains to extend upward away from their grafting or tethering sites to become taller (59). In the polymer brush scenario shown here, the FG domain shrubs would fit snugly under the canopy of the FG domain trees for an overall forest-like organization as shown (Fig. 7, left panels). The crowding effect near the grafting sites would preferentially elongate the more dynamic and charged portions of the FG nups (i.e. the stalk regions of Nup116 and Nup100 and the FXFG-rich regions of Nsp1, Nup1, and Nup2) because these prefer extended configurations on average that are repelled by the FG domain shrubs due to charge content (Fig. 5) (27). The collapsed coil shrubs would interact laterally due to their cohesive nature (27), possibly forming a thin hydrogel along the inner wall surface of the ring scaffold. Lastly and most importantly, if the force of attraction between the cohesive tips of the FG domain trees (i.e. their canopy) was greater at the conduit center than the force needed to pull or stretch the neighbor FG domain or stalk region toward the conduit center,2 then a modified picture would emerge. There, the sticky globules at the end of the extended coils from Nsp1, Nup1, Nup116, and Nup100 would coalesce at the NPC center to form a large quaternary structure consisting of ∼56 collapsed coil globules organized as seven octoglobular rings stacked with one another to form a tunnel structure with a central open channel as shown (Fig. 7). It seems possible that this hypothetical structure, which appears “suspended” in the center of the NPC conduit, could take the form of a hydrogel (29) if its cohesive FG domains were to lose their “identity” by coalescing into one metastable network (60) rather than remaining as discrete collapsed coil globules as depicted.
Physical Evidence in Support of a Suspended, Tubular Gate Structure or Transporter Formed by Disordered FG Domains at the Center of the NPC Transport Conduit
The molecular gate structure proposed here formed by disordered FG domains closely matches the description of the transporter structure observed by cryoelectron microscopy and described in great detail by Akey and co-workers over a ten year period (7, 8, 61–64). This is the same structure as the central plug, which was originally observed by tomography of single negatively stained Xenopus NPCs (65). Notably, our model of FG domain distribution within NPCs predicts the existence of a low density protein ring at the NPC center (because of the intrinsically disordered nature of the collapsed coil FG domains forming the transporter) connected directly to the spoke ring scaffold by even lower density “cables” (i.e. the extended coil FG domains of Nsp1 and Nup1 and the FG nup stalk regions of Nup116 and Nup100), overall giving the impression that the transporter structure is suspended as described (7, 8). This suspended gate architecture is also consistent with the most recent high resolution cryoelectron tomography reconstructions of Dictyostelium NPCs (2, 10) that depict the transporter as a cylindrical structure suspended in the center of the NPC conduit (but without the connecting cables). In fact, the proposed transporter tubule or “plug” (the term that may best describe it if it were to form a hydrogel) has been observed in nuclei, nuclear envelopes, and isolated NPCs from a variety of species (e.g. yeast, frog, human, flies, slime mold, and birds) (7, 8, 10, 61, 66, 67) even without the aid of computer-assisted image averaging (63, 65, 68–70). However, its features seemed variable and easily perturbed during physical studies as expected for a structure made of disordered proteins. Notwithstanding and despite the overwhelming morphological evidence supporting its existence, the transporter structure has often been dismissed as representing only kap-cargo complexes caught in transit across the NPC (9, 66).
Although the tubular NPC gate structure proposed here (i.e. the transporter) differs significantly from the hydrogel model of NPC gate architecture or the simple polymer brush gate model, it is consistent with both because it combines key features of the models as instructed by the physical and functional parameters uncovered here for the FG domains. An important and reasonable assumption made in all current models including ours is that FG domains are tethered to the inner walls and to the periphery of the NPC transport conduit. The details regarding the exact position of FG nup anchor sites seem less important to the model proposed because in every arrangement the shrubs would still be shrubs and the trees would still be trees, and the canopies of the trees would still interact with each other at the conduit center to form the transporter. Notwithstanding, the positions selected here for each FG nup anchor site closely match the location reported for them in the yeast NPC by immuno-EM measurements (14, 52).
Two Zones of Traffic across the NPC with Distinct Physicochemical Properties
According to our proposed model, Zone 1 would constitute the interior of the central transporter structure (Fig. 7, center panels). The environment within this zone would be expected to be hydrophobic because of the abundance of FG motifs and the scarcity of charged AAs in the FG domains forming it (i.e. the FG domains of Nup116 and Nup100 and the cohesive tip of the Nsp1 and Nup1 FG domains). Nonetheless, these FG domains have a net positive charge and an average isoelectric focus point of ∼11 (Table II). Possibly, Zone 1 and particularly the FG domains flanking its narrow entrances could act as electrostatic barriers attracting negatively charged macromolecules with hydrophobic surfaces (such as kaps) while repelling positively charged macromolecules. Kaps carrying large cargos would easily traverse the NPC through Zone 1 because its transport tunnel could rapidly deform and expand in diameter to accommodate large mRNPs and ribosomes (Fig. 7, right panels), cargo proteins coupled to gold beads (∼10–20 nm in diameter), and even very large viral particles ∼36 nm in diameter (71). The hydrodynamic dimensions measured here for the collapsed coil FG domains forming the transporter (∼6–10 nm in diameter assuming a spherical shape) (Tables I and II and Fig. 6) are similar to those of kaps (∼5–15 nm) (72). The nups, however, have less than half of the protein mass per unit of volume. Hence, if the FG domains remained as self-contained globules sticking readily to others like them without merging at the NPC, then the kaps would have to move in between FG domain globules rather than going through their hydrodynamic space as in a hydrogel. Alternatively, if the cohesive FG domains merged at the NPC to become one homogeneous meshwork as proposed for a hydrogel, then the kaps would have to break and reseal the meshwork to go through the NPC (29). However, because kaps bind to the same FG motifs that cohesive FG domains use for interaction among themselves (Fig. 5) (27) and within themselves (Table I) (36), then all of these FG motif-dependent interactions would likely compete at the NPC. This is important because kap binding sites on nups are thought to be saturated in vivo (31). Hence, if intramolecular FG domain interactions were to dominate in the collapsed coil FG domains, then these interactions would permit them to retain their globular identity within the NPC at all times, even while interacting with kaps and other FG domains.
According to our proposed model, Zone 2 would contain the extended coil regions of the Nsp1 and Nup1 FG domains and the ∼200-AA relaxed coil stalk regions of Nup116 and Nup100 protruding laterally from the exterior surface of the transporter tubule. Physically, these elongated domains would connect the transporter structure to the ring scaffold, keeping it suspended about the center of the NPC conduit. Because these domains have a high content of charged AAs, they would impart a unique character, a hydrophilic character, to Zone 2, which we define as the space between the exterior surface of the transporter structure and the inner wall surface of the conduit lined by Nup57, Nup49, and Nup145N FG domains in shrub configurations (Fig. 7) (see below). Individually, each of the extended coil FG domains and stalk regions traversing Zone 2 from the ring scaffold to the transporter could function as a dynamic tether connecting the globular collapsed coil FG domain at one end to the folded NPC anchor domain at the other. These tethers could operate as springs that contract and extend to accommodate compressions and extensions of the globular FG domains in Zone 1 as they interact with other FG domains in the transporter or with kaps or are pushed by large cargos in transit such as ribosomes exiting the nucleus (Fig. 7, right panels).
As discussed below, we suggest that unloaded kaps, kaps loaded with small cargos, and small Ntf2-Ran import complexes could move across the NPC via Zone 2 (Fig. 7, right panels). If so, kap binding to the extended coil FG domains and stalks in Zone 2 could cause them to collapse as proposed for the interaction between importin β and the human Nup153 FG domain (34). This could effectively retract the globular FG domains away from Zone 1, causing a widening of the transporter tunnel. Interestingly, the conversion of all relaxed and extended coil structures in Zone 2 to molten globular structures with smaller hydrodynamic dimensions could “liberate” up to ∼56,498 nm3 of space at the NPC, which is sufficient to accommodate ∼150 molecules of unloaded karyopherin Kap95 for example (the Kap95 RS is 4.44 ± 0.04 nm) (Fig. 7, right panels). Specifically, kaps that prefer FXFG motifs such as Ntf2 (48) or those transporting integral membrane proteins across the NPC likely traverse through Zone 2. Lastly, the entrances to Zone 2 are flanked by relaxed or extended coil FG domains from Nup159, Nup60, and Nup2 featuring net negative charges (Table II), which are opposite to the rest of the FG domains in the conduit. These gatekeepers of Zone 2 could therefore function as proposed for entropic bristles that capture kaps to let them through (25).
Evidence in Support of the Existence of Two Distinct Zones of Traffic in the NPC
First, there is ample morphological evidence showing that Zone 1 is utilized for nuclear export of large cargos such as mRNPs and for nuclear import of small cargos conjugated to large 10–20 nm gold particles (2, 67, 73–75). In fact, the single file transport route observed for cargos across the NPC directly supports the existence of the transporter structure because it shows that FG domains organize to form a single centrally confined channel for large cargos. In contrast, there is less evidence supporting the existence of Zone 2 as a route for kap transport, but this may be explained by the fact that all kap-cargo complexes examined to date by electron microscopy have been large in size such as mRNPs (67, 73) and proteins coupled to gold particles such as nucleoplasmin-gold (74, 75), Ran-gold, and nuclear localization signal-GFP-gold (2). Simply, their large size would have prevented entry to Zone 2 and hence visualization in Zone 2. It seems likely, however, that such large kap-cargo complexes could bind FG domains at the entrance of Zone 2 in preparation for transit through Zone 1 as observed (74). Second, the recent finding that small (≤4-nm diameter) cargos imported by the yeast karyopherins Kap104 and Kap121 follow a mostly peripheral route of traffic across the NPC conduit rather than a central route (judging from immunoelectron microscopy studies that use postembedding labeling techniques and antibodies directed to the cargo3) directly supports the existence of Zone 2 as a transport route. Third, it is known that Kap121 binds Nup53 at the NPC (76, 77) at a site in Nup53 (AAs 405–430) that is very close (∼30 AAs away or <10 nm) to its membrane insertion domain (AAs 461–475) (78, 79). Hence, Kap121 must travel through Zone 2 to interact with Nup53 at the NPC. Likewise, Kap95-Kap60 heterodimers must gain access to Zone 2 when engaged in protein import of integral inner nuclear membrane proteins (80). Finally, a genetic study that defined a minimal set of FG domains needed to support Kap104-mediated import and mRNP export in yeast concluded that the FG domain of Nup57 (which is entirely in Zone 2) and the FG domain of Nup116 (which is the major component of Zone 1) define two functionally separate routes for traffic across the NPC (81).
Passive Diffusion of Macromolecules across the NPC
According to our model, both Zones 1 and 2 would permit the diffusion of small proteins and metabolites. Zone 1 would feature one diffusion channel with a highly variable diameter, whereas Zone 2 would contain eight channels with more restricted dimensions (Fig. 7). For Zone 2, a preferred size limit of ∼5–6 nm could be imposed by the average distance between the collapsed coil domains of the shrubs and the collapsed coil domains of the central transporter (Fig. 7). This hypothetical 5–6 nm size limit could also be influenced by the average distance between extended coils spanning Zone 2, which in turn is established by the average distance between FG nup anchor sites (estimated at ∼5–6 nm (52)) (Fig. 7, central panels). In Zone 1, if the transporter adopted a hydrogel or plug configuration, then overlapping channels would form with diameters restricted by the average distance between polypeptide chains (27) or by the physical distance between FG motifs along each FG domain (29). Finally, when the NPC is saturated with kaps (Fig. 7, right panels) as predicted for NPCs in vivo, both zones could become even tighter barriers against the diffusion of small inert proteins simply due to mass action as observed (3).
Concluding Remarks: Finding Order within Disorder
We conclude that the NPC permeability barrier or transport gate formed by intrinsically disordered domains of FG nups is not an amorphous and homogeneous network of random coils as previously imagined. Rather, it is a sophisticated mosaic of intrinsically disordered domains that show organization at multiple levels. At the primary sequence level, the disordered domains display a bimodal distribution of AAs (e.g. NG versus DKE) (Fig. 3B) and a non-random distribution of FG motif types with different spacings (Fig. 1). At the secondary structure level, they display a bimodal distribution of distinct categories of intrinsically disordered structures ranging from molten globules to extended coils (Fig. 2) often next to NPC anchor domains that are rich in secondary structure and fold into defined structures (22). At the tertiary structure level, the bimodal distribution of collapsed coil versus relaxed or extended coil structures, combined with the position of the NPC anchor domain and the crowded conditions of the NPC, could allow the FG domains to form structures whose topology resemble shrubs and trees of distinct charge content and net charge as shown (Fig. 6). Finally, we hypothesize that the distribution of distinct categories of intrinsically disordered nup domains at the NPC is also not random. Instead, it may feature a central cluster of collapsed coils that stick together to form the elusive transporter structure (Fig. 7).
Supplementary Material
* This work was supported, in whole or in part, by National Institutes of Health Grants RO1 GM077520 (to M. F. R., M. E. C., V. V. K. and E. Y. L.) and RO1 LM007688 and GM071714 (to V. N. U.). This work was also supported by the Program of the Russian Academy of Sciences for “Molecular and Cellular Biology” (to V. N. U.) and by the Indiana University-Purdue University Indianapolis Signature Centers Initiative.
 This article contains supplemental Figs. S1–S4, Table S1, and Movies S1 and S2.
This article contains supplemental Figs. S1–S4, Table S1, and Movies S1 and S2.
2 Y. W. Kim, R. Zandi, M. E. Colvin, M. F. Rexach, and A. Gopinathan, manuscript in preparation.
3 M. Goldberg, personal communication.
1 The abbreviations used are:
- NPC
- nuclear pore complex
- nup
- nucleoporin
- FG nup
- nucleoporin with FG motifs
- FG domain
- nup domain with FG motifs
- kap
- karyopherin
- MD
- molecular dynamics
- PONDR
- predictors of natural disordered regions
- AA
- amino acid
- CFP
- cyan fluorescent protein
- mRNP
- messenger ribonucleoprotein.
REFERENCES
- 1.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 2.Beck M., Lucić V., Förster F., Baumeister W., Medalia O. (2007) Snapshots of nuclear pore complexes in action captured by cryo-electon tomography. Nature 449, 611–615 [DOI] [PubMed] [Google Scholar]
- 3.Mohr D., Frey S., Fischer T., Güttler T., Görlich D. (2009) Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28, 2541–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terry L. J., Shows E. B., Wente S. R. (2007) Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 5.Lim R. Y., Aebi U., Fahrenkrog B. (2008) Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 129, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenkiel-Krispin D., Maco B., Aebi U., Medalia O. (2010) Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J. Mol. Biol. 395, 578–586 [DOI] [PubMed] [Google Scholar]
- 7.Akey C. W., Radermacher M. (1993) Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q., Rout M. P., Akey C. W. (1998) Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1, 223–234 [DOI] [PubMed] [Google Scholar]
- 9.Fahrenkrog B., Aebi U. (2003) The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4, 757–766 [DOI] [PubMed] [Google Scholar]
- 10.Beck M., Förster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. (2004) Nuclear pore complex structure and dynamics revealed by cryoelectron microscopy. Science 306, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 11.Feldherr C. M., Akin D. (1997) The location of the transport gate in the nuclear pore complex. J. Cell Sci. 110, 3065–3070 [DOI] [PubMed] [Google Scholar]
- 12.Keminer O., Peters R. (1999) Permeability of single nuclear pores. Biophys. J. 77, 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters R. (1984) Nucleo-cytoplasmic flux and intracellular mobility in single hepatocytes measured by fluorescence microphotolysis. EMBO J. 3, 1831–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. (2002) Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U. (2008) Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsia K. C., Stavropoulos P., Blobel G., Hoelz A. (2007) Architecture of a coat for the nuclear pore membrane. Cell 131, 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rexach M. (2009) Piecing together nuclear pore complex assembly during interphase. J. Cell Biol. 185, 377–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hetzer M. W., Wente S. R. (2009) Border control at the nuclear pore complex: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17, 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Martinez J., Rout M. P. (2009) Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 21, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. (2003) Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. U.S.A. 100, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denning D. P., Rexach M. F. (2007) Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell. Proteomics 6, 272–282 [DOI] [PubMed] [Google Scholar]
- 23.Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. (2004) Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6, 197–206 [DOI] [PubMed] [Google Scholar]
- 24.Ribbeck K., Görlich D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T. (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 13, 622–628 [DOI] [PubMed] [Google Scholar]
- 26.De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A. (2004) Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14, 1973–1984 [DOI] [PubMed] [Google Scholar]
- 27.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. (2007) Natively-unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 [DOI] [PubMed] [Google Scholar]
- 28.Allen N. P., Huang L., Burlingame A., Rexach M. (2001) Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276, 29268–29274 [DOI] [PubMed] [Google Scholar]
- 29.Frey S., Görlich D. (2009) FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 28, 2554–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jovanovic-Talisman T., Tetenbaum-Novatt J., McKenney A. S., Zilman A., Peters R., Rout M. P., Chait B. T. (2009) Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters R. (2009) Translocation through the nuclear pore: Kaps pave the way. BioEssays 31, 466–477 [DOI] [PubMed] [Google Scholar]
- 32.Naim B., Zbaida D., Dagan S., Kapon R., Reich Z. (2009) Cargo surface hydrophobicity is sufficient to overcome the nuclear pore complex selectivity barrier. EMBO J. 28, 2697–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jäkel S., Mingot J. M., Schwarzmaier P., Hartmann E., Görlich D. (2002) Importins fulfill a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim R. Y., Fahrenkrog B., Köser J., Schwarz-Herion K., Deng J., Aebi U. (2007) Nanomechanical basis for selective gating by the nuclear pore complex. Science 318, 640–643 [DOI] [PubMed] [Google Scholar]
- 35.Denning D. P., Uversky V., Patel S. S., Fink A. L., Rexach M. (2002) The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol. Chem. 277, 33447–33455 [DOI] [PubMed] [Google Scholar]
- 36.Krishnan V. V., Lau E. Y., Yamada J., Denning D. P., Patel S. S., Colvin M. E., Rexach M. F. (2008) Intramolecular cohesion of coils mediated by phenylalanine-glycine motifs in the natively unfolded domain of a nucleoporin. PLoS Comput. Biol. 4, e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 38.Uversky V. N. (2003) Protein folding revisited. A polypeptide chain at the folding-misfolding-non-folding crossroads: which way to go? Cell. Mol. Life Sci. 60, 1852–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uversky V. N. (2002) Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tcherkasskaya O., Davidson E. A., Uversky V. N. (2003) Biophysical constraints for protein structure prediction. J. Proteome Res. 2, 37–42 [DOI] [PubMed] [Google Scholar]
- 41.Piotto M., Saudek V., Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 42.Krishnan V. V., Thornton K. H., Cosman M. (1999) An improved experimental scheme to measure self-diffusion coefficients of biomolecules with an advantageous use of radiation damping. Chem. Phys. Lett. 302, 317–323 [Google Scholar]
- 43.Garcia de la Torre J. G., Bloomfield V. A. (1981) Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Q. Rev. Biophys. 14, 81–139 [DOI] [PubMed] [Google Scholar]
- 44.Krishnan V. (1997) Determination of oligomeric state of proteins in solution from pulsed-field-gradient self-diffusion coefficient measurements. A comparison of experimental, theoretical, and hard-sphere approximated values. J. Magn. Reson. 124, 468–473 [Google Scholar]
- 45.Krishnan V. V., Cosman M. (1998) An empirical relationship between rotational correlation time and solvent accessible surface area. J. Biomol. NMR 12, 177–182 [DOI] [PubMed] [Google Scholar]
- 46.Vacic V., Uversky V. N., Dunker A. K., Lonardi S. (2007) Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 8, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sickmeier M., Hamilton J. A., LeGall T., Vacic V., Cortese M. S., Tantos A., Szabo B., Tompa P., Chen J., Uversky V. N., Obradovic Z., Dunker A. K. (2007) DisProt: the database of disordered proteins. Nucleic Acids Res. 35, D786–D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayliss R., Leung S. W., Baker R. P., Quimby B. B., Corbett A. H., Stewart M. (2002) Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21, 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayliss R., Littlewood T., Strawn L. A., Wente S. R., Stewart M. (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277, 50597–50606 [DOI] [PubMed] [Google Scholar]
- 50.Tran H. T., Pappu R. V. (2006) Toward and accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys. J. 91, 1868–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crawley M. (2007) The R book, Wiley and Sons, New Jersey, 482–486 [Google Scholar]
- 52.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 53.DeGrasse J. A., DuBois K. N., Devos D., Siegel T. N., Sali A., Field M. C., Rout M. P., Chait B. T. (2009) Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8, 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uversky V. N., Gillespie J. R., Fink A. L. (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct. Funct. Genet. 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 55.Frey S., Richter R. P., Görlich D. (2006) FG-rich repeats of nuclear pore complex proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 [DOI] [PubMed] [Google Scholar]
- 56.Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. (2002) Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1, 930–946 [DOI] [PubMed] [Google Scholar]
- 57.Huang L., Baldwin M. A., Maltby D. A., Medzihradszky K. F., Baker P. R., Allen N., Rexach M., Edmondson R. D., Campbell J., Juhasz P., Martin S. A., Vestal M. L., Burlingame A. L. (2002) The identification of protein-protein interactions of the nuclear pore complex of Saccharomyces cerevisiae using high throughput matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry. Mol. Cell. Proteomics 1, 434–450 [DOI] [PubMed] [Google Scholar]
- 58.Tompa P. (2003) Intrinsically unstructured proteins evolve by repeat expansions. BioEssays 25, 847–855 [DOI] [PubMed] [Google Scholar]
- 59.Bright J. N., Woolf T. B., Hoh J. H. (2001) Predicting properties of intrinsically unstructured proteins. Prog. Biophys. Mol. Biol. 76, 131–173 [DOI] [PubMed] [Google Scholar]
- 60.Kustanovich T., Rabin Y. (2004) Metastable network model of protein transport through nuclear pores. Biophys. J. 86, 2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiseleva E., Goldberg M. W., Allen T. D., Akey C. W. (1998) Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci. 111, 223–236 [DOI] [PubMed] [Google Scholar]
- 62.Akey C. W. (1995) Structural plasticity of the nuclear pore complex. J. Mol. Biol. 248, 273–293 [DOI] [PubMed] [Google Scholar]
- 63.Akey C. W. (1990) Visualization of transport-related configurations of the nuclear pore transporter. Biophys. J. 58, 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akey C. W. (1989) Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J. Cell Biol. 109, 955–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unwin P. N., Milligan R. A. (1982) A large particle associated with the perimeter of the nuclear pore complex. J. Cell Biol. 93, 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoffler D., Feja B., Fahrenkrog B., Walz J., Typke D., Aebi U. (2003) Cryo-electron tomography provides novel insight into nuclear pore architecture: Implications for nucleocytoplasmic transport. J. Mol. Biol. 328, 119–130 [DOI] [PubMed] [Google Scholar]
- 67.Daneholt B. (1997) A look at messenger RNP moving through the nuclear pore. Cell 88, 585–588 [DOI] [PubMed] [Google Scholar]
- 68.Kiseleva E., Allen T. D., Rutherford S, Bucci M., Wente S. R., Goldberg M. W. (2004) Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145, 272–288 [DOI] [PubMed] [Google Scholar]
- 69.Goldberg M. W., Solovei I., Allen T. D. (1997) Nuclear pore complex structure in birds. J. Struct. Biol. 119, 284–294 [PubMed] [Google Scholar]
- 70.Rutherford S. A, Goldberg M. W., Allen T. D. (1997) Three-dimensional visualization of the route of protein import: the role of nuclear pore complex substructure. Exp. Cell Res. 232, 146–160 [DOI] [PubMed] [Google Scholar]
- 71.Panté N., Kann M. (2002) Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fukuhara N., Fernandez E., Ebert J., Conti E., Svergun D. (2004) Conformational variability of nucleo-cytoplasmic transport receptors. J. Biol. Chem. 279, 2176–2181 [DOI] [PubMed] [Google Scholar]
- 73.Dworetzky S. I., Feldherr C. M. (1988) Translocation of RNA-coated gold particles through the nuclear pores of oocytes. J. Cell Biol. 106, 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akey C. W., Goldfarb D. S. (1989) Protein import through the nuclear pore complex is a multistep process. J. Cell Biol. 109, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feldherr C. M., Kallenbach E., Schultz N. (1984) Movement of a karyophilic protein through the nuclear pores of oocytes. J. Cell Biol. 99, 2216–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marelli M., Aitchison J. D., Wozniak R. W. (1998) Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143, 1813–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makhnevych T., Lusk C. P., Anderson A. M., Aitchison J. D., Wozniak R. W. (2003) Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115, 813–823 [DOI] [PubMed] [Google Scholar]
- 78.Marelli M., Lusk C. P., Chan H., Aitchison J. D., Wozniak R. W. (2001) A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153, 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patel S. S., Rexach M. F. (2008) Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics 7, 121–131 [DOI] [PubMed] [Google Scholar]
- 80.King M. C., Lusk C. P., Blobel G. (2006) Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 81.Terry L. J., Wente S. R. (2007) Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) Intrinsically disordered protein. J. Mol. Graph. Model. 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 83.Denning D., Mykytka B., Allen N. P., Huang L., Burlingame A. L., Rexach M. (2001) The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.