Abstract
KiSS-1 is a metastasis suppressor gene reported to be involved in the progression of several solid neoplasias. The loss of KiSS-1 gene expression has been shown to be inversely correlated with increasing tumor stage, distant metastases, and poor overall survival in bladder tumors. To identify the molecular pathways associated with the metastasis suppressor role of KiSS-1 in bladder cancer, we carried out a proteomics analysis of bladder cancer cells (EJ138) transiently transfected with a vector encompassing the full-length KiSS-1 gene using an iTRAQ (isobaric tags for relative and absolute quantitation) approach. Protein extracts collected after 24- and 48-h transfection were fractionated and cleaved with trypsin, and the resulting peptides were labeled with iTRAQ reagents. The labeled peptides were separated by strong cation exchange and reversed phase LC and analyzed by MALDI-TOF/TOF MS. Three software packages were utilized for data analysis: ProteinPilot for identification and quantification of differentially expressed proteins, Protein Center for gene ontology analysis, and Ingenuity Pathways Analysis to provide insight into biological networks. Comparative analysis among transfected, mock, and empty vector-exposed cells identified 1529 proteins with high confidence (>99%) showing high correlation rates among replicates (70%). The involvement of the identified proteins in biological networks served to characterize molecular pathways associated with KiSS-1 expression and to select critical candidates for verification analyses by Western blot using independent transfected replicates. As part of complementary clinical validation strategies, immunohistochemical analyses of proteins regulated by KiSS-1, such as Filamin A, were performed on bladder tumors spotted onto tissue microarrays (n = 280). In summary, our study not only served to uncover molecular mechanisms associated with the metastasis suppressor role of KiSS-1 in bladder cancer but also to reveal the biomarker role of Filamin A in bladder cancer progression and clinical outcome.
Bladder cancer represents the fourth most common malignancy among men and the eighth cause of male cancer deaths (1). Bladder cancer can be classified based on the depth of invasion. Clinically, ∼75% of transitional cell carcinomas (TCCs)1 are non-muscle-invasive (pTis, pTa, and pT1), 20% are muscle infiltrating (pT2–pT4), and 5% are metastatic at the time of diagnosis (1). Low grade tumors are always papillary and usually non-invasive, whereas high grade tumors can be either papillary or non-papillary and are often invasive. Patients diagnosed with localized TCC have a 5-year relative survival rate over 90%. However, patients presenting with regional and distant metastatic disease spread have 5-year relative survival rates of lower than 50 and 10%, respectively (1). Bladder cancer progression and the development of secondary metastases follow complex sequential steps. The changes at the genetic and/or epigenetic level to the many genes involved in critical cell functions are not completely understood (2).
KiSS-1 has been shown to suppress metastases without affecting tumorigenicity in melanoma and breast cancer cells (3–7). It maps to chromosome 1q32 (8) and is regulated by genes mapping to chromosome 6 (3–7). KiSS-1 encodes a 145-amino acid protein, which is processed into kisspeptins of several sizes (9–11). Kisspeptins have been shown to control the onset of puberty and inhibit cancer metastasis of different tumor types (9–11). Experimental and clinical studies indicate KiSS-1 to be a functionally active metastasis suppressor gene in several solid tumors (12–19). Molecular profiling analysis revealed that KiSS-1 was lost in advanced cell lines and bladder tumors, providing prognostic information for bladder cancer (13). Independent analyses of transcript levels of KiSS-1 using in situ hybridization and real time quantitative PCR (RT-PCR) in large cohorts of bladder tumors showed that low expression of KiSS-1 was significantly associated with increasing histopathologic stage, grade, and poor survival (13, 19). Regulation of events downstream of cell-matrix adhesion involving cytoskeleton reorganization has been attributed to KiSS-1 expression (3–19). However, the mechanism by which KiSS-1 plays a role in bladder cancer progression or is involved in the invasive/metastatic phenotype has not been fully elucidated.
Quantitative proteomics is driving the discovery of disease-specific targets and biomarkers. The challenge of proteomics resides in the complexity of protein chemistry and multiple potential post-translational functional modifications. The design of a proteomics experiment is typically dependent on whether the proteins to be measured are known or unknown. Protein and antibody arrays allow relative differential quantification of known proteins (20). Mass spectrometry techniques have become the dominant means of protein identification (20). The use of isobaric tags for relative and absolute quantitation (iTRAQ) combined with multidimensional liquid chromatography (LC) and tandem MS analysis (21) is emerging as a powerful methodology in the search for disease-specific targets and biomarkers using cell lines (22–30), tissues (31–37), and body fluids (38, 39). The iTRAQ method places tags on primary amines (NH2-terminal or ε-amino group of the lysine side chain), allowing detection of most tryptic peptides. Because of the isobaric mass design of the iTRAQ reagents, differentially labeled peptides appear as single peaks in MS scans, but when subjected to tandem MS (MS/MS) analysis, the mass-balancing carbonyl moiety is released as a neutral loss, thereby liberating isotope-encoded reporter ions that provide relative quantitative information on the proteins from which the peptides originate (21). In this study, we describe a quantitative proteomics analysis of bladder cancer cells (EJ138) transiently transfected with a vector containing the full-length KiSS-1 gene using the multiplex capability of the iTRAQ approach. To the best of our knowledge, the iTRAQ strategy has not been reported to date in bladder cancer. As part of the experimental design (see Fig. 1), some of the proteins identified using the iTRAQ strategy as being regulated by KiSS-1 were validated independently by Western blotting using transfected cell lines. Moreover, their potential biomarker role in bladder cancer progression was evaluated in tissue arrays containing well characterized bladder tumors.
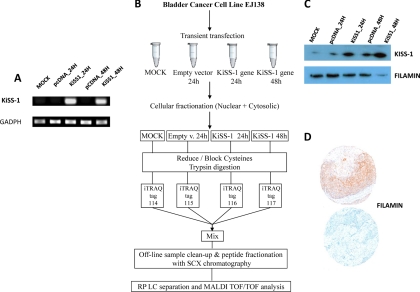
Fig. 1.
Experimental design. A, bladder cancer cells (EJ138) were transiently transfected with a vector encompassing the full-length KiSS-1 gene. Confirmation of efficient transfection was performed by RT-PCR. B, schematic diagram showing the design work flow used for the multiplexed iTRAQ-based experiments. Replicated biological samples were fractionated, the protein extracts were digested using trypsin, and peptides were labeled with iTRAQ reagents. Labeled peptides were combined and fractionated by strong cation exchange chromatography. Collected fractions were separated by nano-LC and analyzed by MALDI-TOF/TOF mass spectrometry. Data analyses were performed with the ProteinPilot software using the Paragon algorithm as the search engine. C, validation of the effect of KiSS-1 overexpression in protein changes of the identified proteins by Western blotting analyses on protein extracts obtained from transfected cells. D, immunohistochemistry on tissue arrays containing bladder tumors served to validate associations of identified proteins regulated by KiSS-1 with clinicopathological variables in bladder cancer. RP, reversed phase; SCX, strong cation exchange chromatography.
EXPERIMENTAL PROCEDURES
Overexpression of KiSS-1 in Bladder Cancer Cell Lines
Cell Culture, Transient Transfection, and RNA Extraction
Bladder cancer cell lines (RT4, 5637, RT112, UM-UC-3, T24, J82, SW780, EJ138, TCCSUP, and ScaBER) were obtained from the American Type Culture Collection and cultured following standard procedures as described previously (40). The differential expression of KiSS-1 was assessed among cell lines by RT-PCR to select a bladder cancer cell line for transfection experiments (data not shown). EJ138, an invasive bladder cancer cell line, was selected based on low KiSS-1 levels and its feasibility to be transiently transfected. Cells utilized were grown no longer than four to six passages and harvested at 75–90% confluence. After harvesting, cell pellets were washed three times in cold PBS and frozen at −20 °C for RNA and protein extraction. Two replicate transient transfections were performed (A and B). Mock cells were transfected with the FuGENE reagent (Roche Applied Science) alone in parallel to transfection of EJ138 with the empty vector (EV) and the vector encompassing the full-length KiSS-1 gene. Transfectants were collected at two time points: 24 h (KiSS-1/24 h) and 48 h (KiSS-1/48 h). The increased expression of KiSS-1 after transfection was confirmed by RT-PCR of the corresponding cDNAs (see Fig. 1).
RNA Analysis of KiSS-1 in Bladder Cancer Cell Lines
Total RNA from transfected cells was isolated in two steps using TRIzol reagent (Invitrogen) followed by RNeasy (Qiagen, Valencia, CA) purification. RNA (1 μg) was reverse transcribed using avian myeloblastosis virus reverse transcriptase (Promega) and amplified for KiSS-1 using specific primers (sense, ACTCACTGGTTTCTTGGCAGC; antisense, ACCTTTTCTAATGGCTCCCCA) and conditions (28 PCR cycles using an annealing temperature of 60 °C). PCR was performed using a final volume of 15 μl containing 1× PCR Ecostart buffer (Ecogen) 1.5 mm MgCl2, 0.2 mm dNTP, 0.25 μm each primer, and 1.5 units of Ecostart Taq polymerase (Ecogen). For PCR amplification, 0.4 μg of cDNA was utilized. GADPH was used as an internal control ensuring cDNA quality and loading accuracy. The amplification product was resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
Functional Analyses of KiSS-1 Overexpression
Cell Cycle Analysis
Cells were seeded in DMEM containing 10% fetal bovine serum (FBS) at a density of 3.5 × 105 cells/well in 6-well plates and incubated at 37 °C for 24 and 48 h after transfection. At these time points, cells were harvested by trypsinization, washed with PBS, and fixed with paraformaldehyde (1%) and cold absolute ethanol for 1 h at 4 °C. Then, cells were incubated with propidium iodide (Sigma) for 30 min at room temperature in the dark. The percentage of cells present in the sub-G0, G0/G1, S, and G2/M phases of the cell cycle was determined by propidium iodide incorporation and flow cytometry analysis (FACSCalibur, BD Biosciences) using CellQuest software (BD Biosciences).
Apoptosis Assay
3.5 × 105 cells/well were seeded in 6-well plates in DMEM containing 10% FBS and incubated at 37 °C for 24 and 48 h after transfection. At these time points, cells were harvested by trypsinization, washed with PBS, and incubated with propidium iodide (Sigma) and Annexin V-allophycocyanin (BD Biosciences) for 20 min at room temperature in the dark. The percentages of live/death cells were determined by flow cytometry analysis (FACSCalibur) using CellQuest software.
Proliferation Assay
1.2 × 104 cells/well were seeded in 96-well plates in triplicate in DMEM containing 10% FBS and incubated for 24 and 48 h after transfection. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide proliferation kit (Roche Diagnostics) was used as recommended by the manufacturer. Cell proliferation was spectrophotometrically measured at 550 and 595 nm using 690 nm as reference.
Wound Healing Assay
The migration wound healing assay was performed by plating 3.5 × 105 cells in 6-well plates. Once cells were attached and reached confluence, a scratch was made through the confluent monolayer using a sterile pipette tip. Photographs of cells invading the scratch were taken at the indicated time points.
Invasion Assay
Cell culture inserts (24-well plates; pore size, 8 μm; BD Biosciences) were seeded with 2.5 × 104 cells after 24 and 48 h of transfection in 500 μl of DMEM with 0.1% FBS in the upper chamber. Medium with 10% FBS (500 μl) was added to the lower chamber and served as a chemotactic agent. BD Biocoat Matrigel invasion chambers (BD Biosciences) were incubated for 24 h in a humidified tissue culture incubator at 37 °C in a 5% CO2 atmosphere. Cells were fixed with 4% paraformaldehyde in both sides of the Matrigel chamber for 10 min and washed with PBS. Then, cells in both sides of the chamber were stained with 1 μg/ml 4′-6-diamidino-2-phenylindole (DAPI) (Sigma) for 10 min. Both sides of the Matrigel chamber were photographed using confocal microscopy (Leica TCS-SP5, Leica Microsystems GmbH, Wetzlar, Germany), and the number of invading cells was analyzed with the Imaris software (Bit Plane AG, Zurich, Switzerland). The percentage of invasion was estimated as follows: (number of invading cells/number of total cells) × 100.
iTRAQ Protein Profiling
Sample Preparation
For protein extraction and subcellular fractionation (41), cell pellets were resuspended in 400 μl of ice-cold Buffer A (10 mm Hepes, pH 7.5, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 1 mm PMSF plus protease and phosphatase inhibitors). Samples were incubated on ice for 15 min, 25 μl of 10% Nonidet P-40 were added, and the samples were centrifuged at 4 °C and 16,100 × g for 15 min. Supernatants containing the cytosolic and other cellular proteins were stored at −80 °C. Nuclear pellets were washed with 400 μl of ice-cold Buffer A and centrifuged at 4 °C and 16,100 × g for 11 min. After discarding the supernatant, pellets were resuspended in 100 μl of ice-cold Buffer B (10 mm Hepes, pH 7.5, 0.4 NaCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 1 mm PMSF plus protease and phosphatase inhibitors). Samples were incubated for 30 min in a shaking incubator, and protein lysates were then clarified by centrifugation at 4 °C and 16,100 × g for 10 min. After repeating this step twice, supernatants were combined and stored at −80 °C prior to sample cleanup.
Peptide Labeling
For the 4-plex iTRAQ experiments, 24-h mock-treated cells and cells transfected with the EV after 24 h were used as controls and compared with cells transiently transfected with a vector encompassing the full-length KiSS-1 gene and collected after 24 h (KiSS-1/24 h) and 48 h (KiSS-1/48 h). Two biological replicates (A and B) were prepared and analyzed by iTRAQ-based LC-MALDI MS/MS. Protein extracts were cleaned up by acetone precipitation with 6 volumes of ice-cold acetone, and pellets were dissolved in 8 m urea, 2% SDS, 0.5 m triethylammonium bicarbonate. Protein concentration was determined with the Bradford assay using BSA as standard (Protein Assay kit, Bio-Rad). iTRAQ labeling of each sample was performed according to the manufacturer's protocol (Applied Biosystems, Framingham, MA). Briefly, a total of 50 μg of protein from each transfectant and control was reduced with 50 mm tris(2-carboxyethyl)phosphine at 60 °C for 1 h, and the cysteine residues were subsequently alkylated with 200 mm methyl methanethiosulfonate at room temperature for 15 min. Protein enzymic cleavage was carried out with trypsin (Promega, Madison, WI; 1:20, w/w) at 37 °C for 16 h. Each tryptic digest was labeled according to the manufacturer's instructions with one isobaric amine-reactive tag as follows: Tag114, mock; Tag115, EV; Tag116, KiSS-1/24 h; and Tag117, KiSS-1/48 h (Fig. 1). After 1-h incubation, labeled samples were pooled and evaporated to dryness in a vacuum centrifuge. Each set of labeled samples was dissolved in 1 ml of loading buffer (15 mm KH2PO4 in 25% acetonitrile, pH <3.0) prior to strong cation exchange fractionation.
Cation Exchange Fractionation
A Coulter Gold HPLC system (Beckman, Fullerton, CA) equipped with a 2.1-mm-inner diameter × 100-mm-long PolySULPHOETHYL-A column packed with 5-μm beads with 300-Å pores (PolyLC, Columbia, MD) was used for cation exchange fractionation. A 2.1-mm-inner diameter × 10-mm-long guard column of the same material was fitted immediately upstream of the analytical column (31). Peptides were loaded onto the column and washed isocratically at 100% eluent A. Peptide fractionation was performed using a linear binary gradient from 0 to 100% B at 0.2 ml/min over 75 min. Finally, the column was washed at 100% B for 5 min and returned to 100% A over 10 min. Eluent A was identical in composition to the loading buffer; eluent B was eluent A containing 350 mm KCl. The UV detector was set at 214 nm, and fractions were collected every 2 min using a Gilson FC 203B fraction collector (Gilson, Middleton, WI) and later pooled together according to variations in peak intensity (27). In total, 24 fractions were pooled for each sample and dried by vacuum centrifuge for subsequent nano-reversed phase liquid chromatography (nano-LC) fractionation.
Nano-LC
Each fraction was resuspended in loading buffer (0.05% heptafluorobutyric acid) and separated using an Ultimate 3000 nano-LC system (Dionex-LC Packings, Amsterdam, The Netherlands) equipped with a ProbotTM MALDI spotting device (Dionex-LC Packings). To preconcentrate and desalt the samples before switching the precolumn in line with the separation column, 20 μl from each strong cation exchange chromatography fraction was loaded onto a reversed phase monolithic polystyrene-divinylbenzene 200-μm-inner diameter × 5-mm peptide trapping cartridge (Dionex-LC Packings) and washed for 8 min at 20 μl/min (24). The peptides were eluted from a reversed phase monolithic polystyrene-divinylbenzene 200-μm-inner diameter × 5-cm analytical column (Dionex-LC Packings) by application of a binary gradient with a flow rate of 2.5 μl/min. Buffer A was 2% ACN in 0.05% TFA; Buffer B was 50% ACN with 0.04% TFA. Peptides were separated using the following gradient: 0–8 min, 0% B; 8–9 min, 0–15% B; 9–39 min, 15–95%; 39–45 min, 95% B; and 45–55 min, 0% B. The column effluent was mixed directly with the MALDI matrix solution (5 mg/ml α-cyano-4-hydroxycinnamic acid in 70% ACN) at a flow rate of 2.5 μl/min through a microtee connection before spotting onto 1664-well stainless steel MALDI target plates (Applied Biosystems, Foster City, CA) using a Probot microfraction collector (Dionex-LC Packings) with a speed of 5 s/well. The matrix contained 10 mm NH4H2PO4 and 30 fmol/μl Fibrinopeptide B as internal standard controls for mass calibration (42).
MALDI-TOF/TOF Mass Spectrometry Analysis
MALDI target plates were analyzed using a 4800 Analyzer equipped with TOF/TOF ion optics (Applied Biosystems, Framingham, MA; MDS-Sciex Concord, Ontario, Canada) and 4000 Series Explorer software, version 3.5.1. The instrument was operated in positive ion mode and externally calibrated using a mass calibration standard kit (Bruker, Madison, WI). The laser power was set to 2800 for MS and 3600 for MS/MS acquisition. Typically, 1000 laser shots were accumulated for each sample well, and MS spectra were acquired from 800 to 4000 Da with a minimum signal-to-noise ratio filter of 50 for precursor ion selection (22, 26). MS/MS analyses were performed for the 10 most abundant precursor ions per well using air as the collisionally activated dissociation gas at a collision energy of 1 kV and a collision gas pressure of 1 × 10−6 torr with an accumulation of 2000 shots for each spectrum. To look for less abundant proteins, reinterrogation of the target plates was carried out to acquire the next 10 most intense peaks (if there were any above the signal-to-noise ratio threshold of 25) (22).
Data Analysis
Relative quantification and protein identification were performed with the ProteinPilotTM software (version 2.0.1; Applied Biosystems; MDS Sciex) using the ParagonTM algorithm as the search engine (43). Each MS/MS spectrum was searched against a database of human protein sequences (NCBInr, released March 2008, downloaded from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/protein/). The search parameters allowed for cysteine modification by methyl methanethiosulfonate and biological modifications programmed in the algorithm (i.e. phosphorylations, amidations, semitryptic fragments, etc.). The detected protein threshold (unused protscore (confidence)) in the software was set to 2.0 to achieve 99% confidence, and identified proteins were grouped by the ProGroup algorithm (Applied Biosystems, Foster City, CA) to minimize redundancy. The bias correction option was executed.
The peptide and protein selection criteria for relative quantitation were performed as follows. Only peptides unique for a given protein were considered for relative quantitation, excluding those common to other isoforms or proteins of the same family. Proteins were identified on the basis of having at least one peptide with an ion score above 99% confidence. Among the identified peptides, some of them were excluded from the quantitative analysis for one of the following reasons. (i) The peaks corresponding to the iTRAQ labels were not detected. (ii) The peptides were identified with low identification confidence (<1.0%). (iii) Either the same peptide sequence was claimed by more than one protein or more than one peptide was fragmented at the same time because of shared MS/MS spectra. (iv) The sum of the signal-to-noise ratio for all of the peak pairs was <6 for the peptide ratios. The protein sequence coverage (95%) was estimated for specific proteins by the percentage of matching amino acids from the identified peptides having confidence greater than or equal to 95% divided by the total number of amino acids in the sequence.
Several quantitative estimates provided for each protein by ProteinPilot were utilized: the -fold change ratios of differential expression between labeled protein extracts; the p value, representing the probability that the observed ratio is different than 1 by chance; and the error factor (EF) as a measure of the error in the average ratio expressing the 95% confidence interval (CI) of the average iTRAQ ratio (EF = 1095% CI where 95% CI = (ratio × EF) − (ratio/EF)). A decoy database search strategy was also used to estimate the false discovery rate (FDR), defined as the percentage of decoy proteins identified against the total protein identification. The FDR was calculated by searching the spectra against the NCBInr Homo sapiens decoy database. The estimated low FDR of 1.6% indicated a high reliability in the proteins identified (44). The results were then exported into Excel for manual data interpretation. Although relative quantification and statistical analysis were provided by the ProteinPilot 2.0 software, an additional 1.3-fold change cutoff for all iTRAQ ratios (ratio <0.77 or >1.3) was selected to classify proteins as up- or down-regulated (24, 44, 45). Proteins with iTRAQ ratios below the low range (0.77) were considered to be underexpressed, whereas those above the high range (1.3) were considered to be overexpressed.
Functional distribution analyses of the identified proteins were initially performed using the Protein Center software (Proxeon, Odense, Denmark; http://www.proteincenter.proxeon.com). The Ingenuity Pathways Analysis software (Ingenuity Systems, Redwood City, CA; http://www.ingenuity.com) was used to identify pathways associated with KiSS-1 metastasis suppressor overexpression. The data sets generated by ProteinPilot from the proteomics analysis containing identifiers and corresponding expression values were uploaded into the Ingenuity application. Each identifier was mapped to its corresponding object (gene/protein) in the Ingenuity Knowledge database. These genes, called focus genes, were overlaid onto a global molecular network developed from information previously reported based on known regulatory relationships, such as protein interactions, modifications, regulation of expression, etc. The filters and general settings for the core analysis considered all molecules, including endogenous chemicals, as well as both direct and indirect relationships. Human was the taxonomy selected. All data sources, tissues, and cell lines were considered, and a stringent filter for molecules and relationships was chosen. Networks of focus genes were then algorithmically generated based on their connectivity and ordered by a score. This score reflects the relevance of the network based on a p value calculation, which calculates the likelihood that the network-eligible molecules that are part of a network are found therein by random chance alone. Networks were also associated to biological functions (and/or diseases) that were most significant to the genes in the network. Fischer's exact test was used to calculate a p value determining the probability that each biological function and/or disease assigned to that network is due to chance alone.
Validation by Western Blotting in Bladder Cancer Cells
Total proteins were extracted from mock and transfected bladder cancer cell lines using radioimmune precipitation assay lysis buffer and quantified with the Bradford assay using BSA as standard (Protein Assay kit, Bio-Rad). Total protein extracts (50 μg) were mixed with 5× SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.005% bromphenol blue) and resolved by SDS-PAGE on 10% acrylamide gels. Proteins were detected immunologically following electrotransfer onto PVDF membranes (Millipore, Bedford, MA) after activation with methanol. The membranes were blocked with 5% nonfat dry milk in PBS and 0.1% Tween 20 for 1 h at room temperature and incubated overnight at 4 °C with the following primary antibodies: anti-KiSS-1 (mouse monoclonal, 18 kDa; generated by our laboratory at the Centro Nacional de Investigaciones Oncológicas), anti-Filamin A (mouse monoclonal, 250 kDa; 1:50 dilution; NCL-FIL, Novocastra, Wetzlar, Germany), anti-Ezrin (mouse monoclonal, 80 kDa; 1:7000 dilution; E-8897, Sigma), MMP-2 (mouse monoclonal, 92 kDa; 1:100 dilution; 550892, BD Pharmingen), MMP-9 (mouse monoclonal, 64 and 72 kDa; 1:100 dilution; 550892, BD Pharmingen), p53 (mouse monoclonal, 53 kDa; 1:50 dilution; PAb1801, Calbiochem), DDX21 (rabbit polyclonal, 87 kDa; 1:3500 dilution; 10528-1-AP, Proteintech Europe, Manchester, UK), p21 (rabbit polyclonal, 21 kDa; 1:50 dilution, sc-397, Santa Cruz Biotechnology, Santa Cruz, CA), and anti-pRB (mouse monoclonal, 106 kDa, 1:200 dilution; Neomarkers, Fremont, CA). Blots were washed three times for 10 min in PBS and 0.1% Tween 20 and incubated with the following horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature: HRP-conjugated anti-mouse (1:1000 dilution), anti-rabbit (1:2000 dilution), and anti-goat IgG (1:2000 dilution) (Dako, Glostrup, Denmark). Blots were developed using a peroxidase reaction with an enhance chemiluminescence immunoblotting detection system (ECL, GE Healthcare). Antibodies were accepted as displaying a single predominant band at the expected molecular weights. α-Tubulin (mouse monoclonal, 50 kDa; 1:4000 dilution; Sigma) was utilized as the loading control.
Clinical Evaluation of Expression of Metastasis-related Biomarkers Related to KiSS-1 Overexpression
Tissue Samples and Microarrays
Several bladder cancer tissue microarrays were constructed at the Spanish National Cancer Center and used in this study. These arrays included primary urothelial cell carcinomas of the bladder belonging to patients recruited under Institutional Review Board-approved protocols at collaborating institutions. Tumor tissues were embedded in paraffin, and 5-μm sections were stained with hematoxylin and eosin to identify viable, morphologically representative areas of the specimen from which needle core samples were taken using a precision instrument (Beecher Instruments, Silver Spring, MD). From each specimen, triplicate cores with diameters of 1.0 mm were punched and arrayed on the recipient paraffin block. Five-micrometer sections of these tissue array blocks were cut and placed on charged polylysine-coated slides and used for immunohistochemistry analysis. We constructed six different bladder cancer tissue microarrays, including a total of 218 bladder tumors for which KiSS-1 transcript levels were also measured as described below. Demographic information indicated the presence of 196 males and 22 females with a median age of 65.0 years (range, 25–81 years). Tumor stage distribution was pTa (25), pT1 (87), pTis (6), pT2 (54), pT3 (28), and pT4 (18). Tumor grade distribution was grade 1 (19), grade 2 (10), and grade 3 (189). Tumor stage and grade were defined according to consensus criteria (46, 47). Three additional tissue microarrays including a total of 62 bladder primary muscle-invasive (pT2, pT3, and pT4) high grade TCC tumors with lymph node metastatic status were also analyzed. Importantly, 40 of these cases were shown to display lymph nodes negative for bladder cancer, whereas 22 had lymph node metastases. Clinicopathologic and annotated follow-up information of the tumors spotted onto the tissue microarrays allowed the evaluation of associations of KiSS-1 and the proteins identified by the proteomics iTRAQ approach among them and with the staging properties and outcome assessment.
Transcript Expression Analysis by RT-PCR in Bladder Tumors
KiSS-1 transcript expression was assessed in paraffin-embedded tumors contained in the tissue arrays described above. RNA was extracted using the TRIzol method. Complementary DNA was synthesized using the ThermoScript RT-PCR system (Invitrogen). Template cDNA was added to Taqman Universal Master Mix (Applied Biosystems, Foster City, CA) in a 15-μl reaction with specific primers and probes acquired from Applied Biosystems (Foster City, CA) for KiSS-1 (Hs_00158486_m1) and the chaperonin containing T-complex protein 1 subunit 6A (ζ1) (CCT6A) (Hs_00798979_s1). Quantification of gene expression was carried out using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Relative gene expression quantification was calculated according to the comparative cycle threshold (Ct) method using CCT6A as endogenous control. Final results were estimated as follows: 2−(ΔCt KiSS-1) where ΔCt KiSS-1 values for each sample were determined by subtracting the Ct value of the target KiSS-1 gene from the value of the CCT6A gene. Only triplicates with standard deviations of the Ct value <0.20 were accepted.
Immunohistochemistry
Protein expression patterns of the differentially expressed proteins were assessed at the microanatomical level using the tissue microarrays outlined above. Standard avidin-biotin immunoperoxidase procedures were used for immunohistochemistry. Antigen retrieval methods (0.01% citric acid for 15 min under microwave treatment) were utilized prior to incubation with primary antibodies overnight at 4 °C. The same primary antibodies used in Western blotting worked for immunohistochemistry under the following condition: Filamin A, mouse monoclonal at 1:100 dilution. Staining conditions were optimized on sections from formalin-fixed, paraffin-embedded tissue controls for each antibody as specified by the manufacturers. The secondary antibody (Vector Laboratories) was biotinylated goat anti-mouse antibody (1:500 dilution). The absence of primary antibody was used as negative control. Skin and skeletal muscle were used as positive controls. Diaminobenzidine was utilized as the final chromogen, and hematoxylin was utilized as the nuclear counterstain (40).
Statistical Analysis
The consensus value of the three representative cores from each tumor sample arrayed was used for statistical analyses. The association of the expression of Filamin A measured by immunohistochemistry on tissue arrays with histopathologic stage and tumor grade was evaluated using the non-parametric Wilcoxon-Mann-Whitney and Kruskall-Wallis tests (48). Associations between biomarkers were analyzed using Kendall's tau test. Filamin A expression was evaluated as a continuous variable based on the number of cells expressing the protein in the cytoplasm. The intensity of the staining was categorized from negative (−) to low (+), intermediate (++), and high (+++). The cutoff of expression for prognostic evaluation was selected based on the median values of expression among the groups under analyses. The associations of this protein with overall survival were also evaluated using the log rank test in those cases for which follow-up information was available. Disease-specific overall survival time was defined as the months elapsed between transurethral resection or cystectomy and death as a result of disease (or the last follow-up date). Patients who were alive at the last follow-up or lost to follow-up were censored. Survival curves were plotted using the standard Kaplan-Meier methodology (48). Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) (version 11.0).
RESULTS
Experimental Design
The present study was based on four major sets of experiments (Fig. 1). First, bladder cancer cell lines were transiently transfected with a vector encoding the full length of the metastasis suppressor gene KiSS-1 (Fig. 1A). Second, an iTRAQ proteomics approach was performed to identify molecular pathways associated with KiSS-1 overexpression in transfected cells (Fig. 1B). Third, immunoblot analyses of protein extracts of transfected cells served to validate the impact of KiSS-1 overexpression on the protein levels of the targets identified by the iTRAQ method (Fig. 1C). Fourth, tissue samples of cohorts of well characterized and follow-up annotated cases contained in tissue microarrays served to evaluate the potential clinical significance of KiSS-1 and related identified targets at the microanatomical level using immunohistochemistry (Fig. 1D). These analyses served to assess the potential involvement of KiSS-1 and novel related proteins in bladder cancer progression.
Functional Analyses of KiSS-1 Overexpression
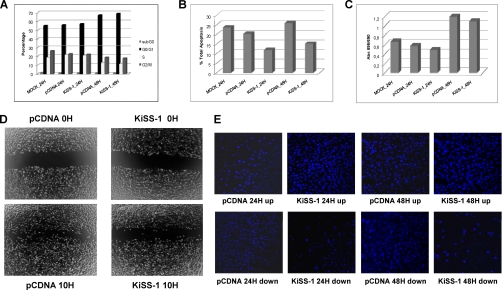
Analyses in vitro were performed to evaluate the phenotype of KiSS-1-transfected cells (Fig. 2). More specifically, the impact of KiSS-1 overexpression on the cell cycle (Fig. 2A), apoptosis (Fig. 2B), proliferation (Fig. 2C), migration (Fig. 2D), and invasion (Fig. 2E) was analyzed. We did not observe changes in cell cycle subpopulations after KiSS-1 transfection (Fig. 2A). However, a significant lower apoptotic rate was observed in KiSS-1 transfectants as compared with the mock and empty vector at both time points (p < 0.05) (Fig. 2B). Subsequent to KiSS-1 overexpression proliferation diminished without reaching statistical significance (Fig. 2C). Importantly, wound healing assays revealed the slower migration rate of KiSS-1 transfectants (Fig. 2D). Moreover, invasion assays indicated that cells overexpressing KiSS-1 were on average 24 and 20% less invasive at 24 and 48 h, respectively, than cells transfected with the empty vector (Fig. 2E). Overall, the overexpression of the KiSS-1 gene provided a less aggressive phenotype as shown in the functional assays, which were performed in duplicate, and average results are presented in Fig. 2.
Fig. 2.
Functional analyses of impact of KiSS-1 overexpression on cell cycle (A), apoptosis (B), proliferation (C), migration (D), and invasion (E). The averages of duplicate experiments of each functional assay of KiSS-1-overexpressing cells at 24 and 48 h after transfection versus the empty vector and mock controls are presented in each panel.
Reproducibility Assessment of Protein Identification among Replicates and Subcellular Fractions
In total, 1529 proteins were identified across both biological replicates using ProteinPilot (1157 in Replicate A and 1173 in Replicate B). The Venn diagram provided in supplemental Fig. 1A shows that 801 of these proteins were common to both sets. There were high correlation rates between both biological replicates because 69% of proteins identified in Replicate A were detected in Replicate B, and 68% of those identified in Replicate B were detected in Replicate A. When comparing the number of proteins identified in each of the subcellular fractions taking both replicates together (supplemental Fig. 1B), it was observed that most of the proteins identified were detected in the nuclear fractions (47.5%), and nearly 30% of all the proteins detected were found simultaneously in nuclear and cytoplasmic fractions. The proteins identified in both biological replicates in each nuclear (supplemental Fig. 1C) and cytoplasmic (supplemental Fig. 1D) fraction were also compared. Around 50% of the proteins were found in both biological replicates in each cellular fraction. In this study, we focused on the 203 proteins that were identified in all the protein fractions for which four data estimations per protein were available. Detailed information of the identified proteins and their respective quantification for each cellular subfraction and each biological replicate is provided in supplemental Tables 1–4 (protein summary) and 5–8 (peptide summary).
As mentioned above, from the 1529 proteins identified across all the fractions analyzed, 203 (13%) proteins were found in all fractions, meaning both in nuclear and cytoplasmic fractions from both biological replicates (four data estimations per protein). Taking the cutoffs of 1.3 and 0.77 to define differentially expressed proteins with p values lower than 0.05, from the 203 proteins identified across the fractions and replicates, 73 were differentially expressed between the mock and the empty vector, 132 were differentially expressed between the 24-h transfectants and the empty vector, and 104 were differentially expressed between the 48-h transfectants and the empty vector. Three data estimations per protein were found for 179 (12%) proteins, 461 (30%) were identified in two fractions (two data estimations per protein), and 686 (45%) were identified in only one fraction. The list for the identified proteins with four and three data estimations are provided in supplemental Tables 9 and 10, respectively. These tables include their abundance ratios, p values, EFs, and iTRAQ modification sites from iTRAQ-based LC-MALDI MS/MS analyses.
Functional Classifications of Identified Proteins
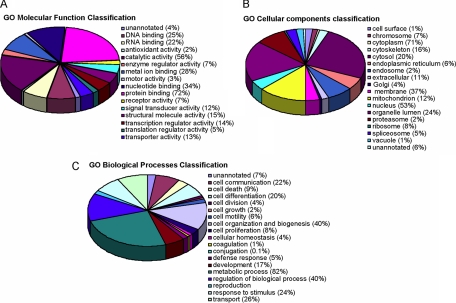
The functional annotation of the 1529 identified proteins was initially assigned using the Protein Center software. Three main types of annotations were obtained from the gene ontology consortium web site: cellular components, molecular functions, and biological distribution. A GOslim approach defined specifically for ProteinCenter served to reduce the multiple gene ontology annotations to a manageable set of ∼20 high level terms that were used for filtering the information into percentage estimations (Fig. 3). The ontology analysis of the identified proteins indicated the relevance and diversity of molecular functions, such as protein binding (72%) or catalytic activity (56%). Many of the identified proteins were involved in metabolism (82%) or cell organization (40%). The data in Fig. 3 supported the functional in vitro evaluation shown above of the impact of KiSS-1 overexpression on the cellular reorganization that affects cellular migration and invasion. Finally, a high number of the identified proteins were found to be localized in the nuclei (71%) and the cytoplasm (53%), consistent with the subcellular fractionation performed. Classifications were redundant (over 100%) because proteins were annotated in more than one compartment (Fig. 3).
Fig. 3.
Classification of identified proteins based on their functional annotations using gene ontology (GO) molecular function (A), biological processes (B), and cellular components (C). These analyses were performed with the 1529 proteins identified in both cytoplasmic and nuclear replicates. When more than one assignment was available for a given protein, all the functional annotations were considered in the analyses.
Molecular Pathways Associated with KiSS-1 Overexpression
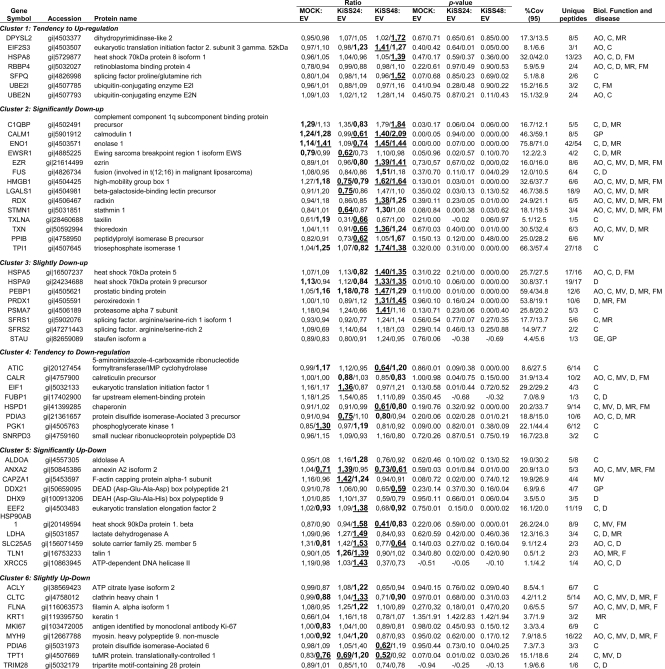
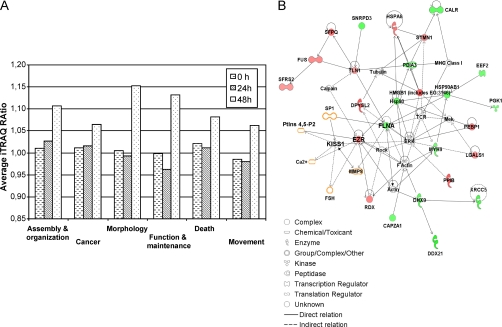
To narrow down the number of protein candidates related to KiSS-1 overexpression, the data set containing the proteins detected in the four cellular subfractions (n = 203) was uploaded into the Ingenuity Pathways Analysis software. Cytoplasmic reorganization of the cytoskeleton is important in the metastatic process. Ezrin and Filamin A were shown to change their levels of expression under the influence of KiSS-1. Because these two cytoskeleton proteins have been reported to be altered in the metastatic process in other neoplasias and antibodies were available, they were selected for validation analyses. Focus was directed to the networks in which Ezrin and Filamin A participated. This functional analysis identified 57 proteins that were biologically related to Ezrin and Filamin A (Table I) as they participated in the following critical neoplasia-related biological function annotations: cellular assembly and organization, cancer, cell movement, cell death, cell morphology, and cell function and maintenance (Fig. 4A).
Table 1. Statistically significant differentially expressed cytoplasmic proteins identified by iTRAQ analysis of EJ138 cells transfected with KiSS-1.
The 57 unique proteins found in both replicates, identified with >99% confidence (corresponding to a protein score cutoff >2.0) and sharing similar biological functions with both Ezrin and Filamin A, were clustered based on similar trends of differential expression over time. The table includes gene name; accession number; ratios and p-values of expression between mock, transfectants at 24 and 48 hours and empty vector, percent coverage (95%) (%Cov(95)); unique peptides; and biological (Biol.) function described for these proteins. Values shown correspond to Replicate A/Replicate B data from cytoplasmic fractions analysis. Mean ratio corresponds to the protein reporter ion intensity originating from mock (114.1), KiSS-1/24 h (KISS24) (116.1) or KiSS-1/48 h (KISS48) (117.1) relative to EV (115.1). Ratios deemed to signify differential expression (p value < 0.05) are bold and shown in a larger font. In a few cases, p values were not available due to single peptide assignment. These are indicated with a dash (–). Additionally, the proteins were considered to show a significant upward or downward trend if their expression ratios were <0.77 or >1.3, respectively; ratios are shown underlined. Biological functions are as follows: cell assembly and organization (AO), cancer (C), cell movement (MV), cell death (D), cell morphology (MR), cell function and maintenance (FM), gene expression (GE), and cell growth and proliferation (GP). The complete set of proteins identified is provided in supplemental Tables 1–4.
Fig. 4.
Functional networks of identified proteins. A, overview of the temporal effects of KiSS-1 overexpression on the various cellular processes. The differential regulation of the proteins from each cellular process is summarized to illustrate the overall temporal effects of KiSS-1 on EJ138 cells. B, biological interaction networking of proteins regulated by KiSS-1. The accession numbers and KiSS-1/48 h:EV ratio values for proteins identified in Table I were imported into Ingenuity Pathways Analysis software, generating different molecular networks. A network where Filamin A and Ezrin were involved was selected. KiSS-1 was connected to Filamin A and Ezrin by creating new pathways through different molecules (labeled in orange). In this network, genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge. All edges are supported by at least one publication from the information stored in the Ingenuity Knowledge database. The intensity of the node color indicates the degree of up- (red) or down (green)-regulation. The legend of the interaction network and the relationships between molecules is summarized on the right of the figure. KiSS-1, Ezrin, and Filamin A are highlighted on the map. CALR, calreticulin; CAPZA1, capping protein (actin filament) muscle Z-line, α1; DDX21, DEAD (Asp-Glu-Ala-Asp) box polypeptide 21; DHX9, DEAH (Asp-Glu-Ala-His) box polypeptide 9; DPYSL2, dihydropyrimidinase-like 2; EEF2, eukaryotic translation elongation factor 2; EZR, Ezrin; FLNA, Filamin A, α (actin-binding protein 280); FSH, follicle-stimulating hormone; FUS, fusion (involved in t(12;16) in malignant liposarcoma); HMGB1 (includes EG:3146), high mobility group box 1; HSP90AB1, heat shock protein 90 kDa α (cytosolic) class B member 1; HSPA8, heat shock 70-kDa protein 8; LGALS1, lectin, galactoside-binding, soluble, 1; MMP9, matrix metalloproteinase 9; MYH9, myosin, heavy chain 9, non-muscle; PDIA3, protein-disulfide isomerase family A, member 3; PEBP1, phosphatidylethanolamine-binding protein 1; PGK1, phosphoglycerate kinase 1; PPIB, peptidyl-prolyl isomerase B (cyclophilin B); RDX, radixin; SFPQ, splicing factor proline/glutamine-rich (polypyrimidine tract-binding protein-associated); SFRS2, splicing factor, arginine/serine-rich 2; SNRPD3, small nuclear ribonucleoprotein D3 polypeptide 18 kDa; STMN1, stathmin 1/oncoprotein 18; TLN1, talin 1; XRCC5, x-ray repair complementing defective repair in Chinese hamster cells 5 (double strand break rejoining).
An independent analysis was performed by importing the ratio values for KiSS-1/48 h:EV for the 57 proteins in Table I into the Ingenuity Pathways Analysis software. The unique molecular network that connected Filamin A and Ezrin selected 23 proteins among the 57 provided in Table I. The top biological functions participating in this network were cell morphology, cellular assembly and organization, and cellular function and maintenance. Addition of KiSS-1 to this molecular network served to generate an interaction map connecting KiSS-1 to Ezrin and Filamin A through the biological interactions previously described for these proteins (Fig. 4B).
Clusters of Differentially Expressed Proteins
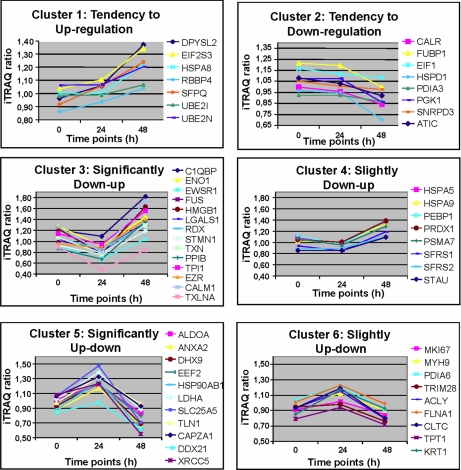
As shown above, from the total number of identified proteins in both biological replicates (four data points per protein; n = 203), we focused on those biologically related to Ezrin and Filamin A (n = 57). Table I provides a detailed description of the identification and quantification data, including accession number, gene symbol, percent coverage (95%), number of unique iTRAQ peptides identified, and the biological annotations related to these 57 proteins. Although the relative quantification analysis given by the ProteinPilot 2.0 software provided statistical analysis, a 1.3-fold change cutoff was applied to all iTRAQ ratios (ratio <0.77 or >1.3) to classify proteins as up- or down-regulated. The 57 proteins displayed in Table I were grouped into different clusters based on their degree of modification and similar trends of differential expression over time as shown in Fig. 5. The represented mean ratio indicates the protein reporter ion intensity derived from the mock (114.1), KiSS-1/24 h (116.1), or KiSS-1/48 h (117.1) relative to EV (115.1) protein extracts. These analyses revealed a diminished expression of Filamin A and an increased expression of Ezrin upon KiSS-1 transient transfection.
Fig. 5.
Identification of protein clusters that showed similar trends of differential expression over time. These proteins exhibit progressive up- or down-regulation on a temporal basis and were clustered into groups based on their degree of modification: clusters 1, 2, 3, 4, 5, and 6. Cluster 1 includes the following: DPYSL2, dihydropyrimidinase-like 2; EIF2S3, eukaryotic translation initiation factor 2, subunit 3γ, 52 kDa; HSPA8, heat shock 70-kDa protein 8 isoform 1; RBBP4, retinoblastoma-binding protein 4; SFPQ, splicing factor proline/glutamine-rich (polypyrimidine tract-binding protein-associated); UBE2I, ubiquitin-conjugating enzyme E2I; UBE2N, ubiquitin-conjugating enzyme E2N. Cluster 2 includes the following: C1QBP, complement component 1 q subcomponent-binding protein precursor; CALM1, calmodulin 1; ENO1, enolase 1; EWSR1, Ewing sarcoma breakpoint region 1 isoform EWS; EZR, Ezrin; FUS, fusion (involved in t(12;16) in malignant liposarcoma); HMGB1, high mobility group box 1; LGALS1, β-galactoside-binding lectin precursor; RDX, radixin; STMN1, stathmin 1; TXLNA, taxilin; TXN, thioredoxin; PPIB, peptidyl-prolyl isomerase B precursor; TPI1, triose-phosphate isomerase 1. Cluster 3 includes the following: HSPA5, heat shock 70-kDa protein 5; HSPA9, heat shock 70-kDa protein 9 precursor; PEBP1, prostatic binding protein; PRDX1, peroxiredoxin 1; PSMA7, proteasome α7 subunit; SFRS1, splicing factor, arginine/serine-rich 1 isoform 1; SFRS2, splicing factor, arginine/serine-rich 2; STAU, staufen isoform a. Cluster 4 includes the following: ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; CALR, calreticulin precursor; EIF1, eukaryotic translation initiation factor 1; FUBP1, far upstream element-binding protein; HSPD1,chaperonin; PDIA3, protein-disulfide isomerase-associated 3 precursor; PGK1, phosphoglycerate kinase 1; SNRPD3, small nuclear ribonucleoprotein polypeptide D3. Cluster 5 includes the following: ALDOA, aldolase A; ANXA2, annexin A2 isoform 2; CAPZA1, F-actin capping protein α-1 subunit; DDX21, DEAD (Asp-Glu-Ala-Asp) box polypeptide 21; DHX9, DEAH (Asp-Glu-Ala-His) box polypeptide 9; EEF2, eukaryotic translation elongation factor 2; HSP90AB1, heat shock 90-kDa protein 1, β; LDHA, lactate dehydrogenase A; SLC25A5, solute carrier family 25, member 5; TLN1, talin 1; XRCC5, ATP-dependent DNA helicase II. Cluster 6 includes the following: ACLY, ATP citrate lyase isoform 2; CLTC, clathrin heavy chain 1; FLNA, filamin A, α isoform 1; KRT1, keratin 1; MKI67, antigen identified by monoclonal antibody Ki-67; MYH9, myosin, heavy polypeptide 9, non-muscle; PDIA6, protein-disulfide isomerase-associated 6; TPT1, tuMR protein, translationally controlled 1; TRIM28, tripartite motif-containing 28 protein.
Protein Identification and Relative Quantification of Filamin A and Ezrin
Two MALDI-derived MS2 spectra of two iTRAQ-tagged peptides of Filamin (supplemental Fig. 2) and Ezrin (supplemental Fig. 3) were selected to illustrate the protein identification and relative quantification process. As shown in Table I, the identification of Ezrin was possible because of eight and six uniquely occurring tryptic peptides detected in Replicate A and Replicate B 4-plex experiments, respectively. Analyses of all uniquely occurring peptides contributed to the quantification statistics revealing that Ezrin was significantly up-regulated after KiSS-1 overexpression (Fig. 4, cluster 3). One of these uniquely occurring peptides, VTTMDAELEFAIQPNTTGK, was fragmented to its constituent iTRAQ reporter and backbone fragment ions (supplemental Fig. 3). These fragments allowed its relative quantification based on the signal intensity values of the reporter ions and amino acid sequence of the peptide based on the b- and y-product ion signal pattern. Filamin A was identified through detection of five and seven uniquely occurring tryptic peptides in Replicate A and Replicate B, respectively. Similarly, analysis of the relative quantification of these peptides showed that Filamin A was slightly down-regulated upon KiSS-1 overexpression (Fig. 4, cluster 6). The fragmentation analysis of one of these peptides, FNEEHIPDSPFVVPVASPSGDAR, to its constituent iTRAQ reporter and backbone fragment ions is shown in supplemental Fig. 2.
Validation of iTRAQ-identified Candidates by Western Blotting in Bladder Cancer Cells Transfected with KiSS-1
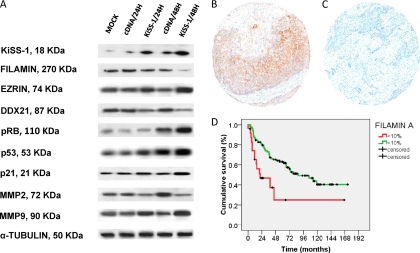
As part of the validation analyses of the potential relevance of the proteins identified in the iTRAQ experiments, immunoblotting analyses were performed on a series of protein extracts from transfected bladder cancer cells (Fig. 6A). The results verified differential regulation of these proteins upon KiSS-1 transient overexpression. Parallel to KiSS-1 transfection, increased expression was observed for Ezrin, and decreased expression was found for DDX21 and Filamin A. Proteins belonging to the pathways identified using the iTRAQ approach and previously reported to be linked to KiSS-1 expression were also tested by immunoblotting (Fig. 6A). KiSS-1 transfection was shown to impact the pattern of metalloproteinases (15, 49,60–51) and the expression of p53, p21, and pRB (13) (supplemental Tables 2, 9, and 10). The results obtained are in good agreement with the expression ratios obtained using the iTRAQ method. Based on these observations, further analyses were performed to assess the clinical relevance in bladder cancer progression of identified proteins, such as Filamin A, in independent sets of human clinical material.
Fig. 6.
Verification analyses of differential expression of identified proteins. A, validation of the iTRAQ results of selected proteins by immunoblotting using protein extracts from transfected bladder cancer cells. The results verified differential regulation of these proteins upon KiSS-1 transient overexpression. Parallel to KiSS-1 overexpression, decreased expression was found for Filamin A, and increased expression was observed for Ezrin. Antibodies were accepted as displaying a single predominant band at the expected molecular weights; α-tubulin was used as the loading control. B and C, representative immunohistochemistry expression patterns of Filamin A in non-invasive and invasive bladder tumors contained in tissue arrays. High expression levels of Filamin A were observed in non-invasive superficial lesions as compared with invasive bladder tumors. There was a significant difference regarding the expression of Filamin A regarding tumor stage (p < 0.0005) (original magnifications in C and D, ×200). D, Kaplan-Mayer curve survival analysis indicating that a decreased protein expression of Filamin A measured by immunohistochemistry on tissue arrays was significantly associated with poor disease-specific survival (p = 0.001).
Filamin A Is Differentially Expressed in Bladder Tumors and Associated with KiSS-1 Expression
The next set of analyses dealt with the optimization and characterization of protein expression patterns of identified proteins, such as Filamin A, by means of immunohistochemistry in bladder tumors. Differential expression was observed for Filamin A among the bladder tumors tested as shown by the representative immunohistochemical analyses displayed in Fig. 6, B and C. The next set of analyses searched for associations with clinicopathologic variables in patients with bladder cancer using an independent series of bladder tumors contained in several tissue arrays. These analyses revealed statistical associations between Filamin A expression and tumor stage when comparing non-invasive versus muscle-invasive bladder tumors (p < 0.0005, n = 218). Moreover, Filamin A expression was associated with poor disease-specific overall survival (log rank, p = 0.001) (Fig. 6D). All primary invasive bladder tumors that developed lymph node metastases showed low transcript levels of KiSS-1. Importantly, Filamin overexpression was more frequently found in patients with lymph node metastases as compared with those with negative lymph nodes (p = 0.036, n = 62). Moreover, an inverse correlation between KiSS-1 expression measured by RT-PCR and Filamin A protein expression (Kendall's tau = −0224, p < 0.0005) was observed in this series. These observations were consistent with the loss of Filamin A after KiSS-1 overexpression observed in transiently transfected bladder cancer cells (that could be associated with the metastatic phenotype). Overall, expression patterns of Filamin A on these independent series of patients with bladder tumors further confirmed the clinical associations of Filamin A with tumor progression and clinical outcome at the protein level in human clinical material.
DISCUSSION
The novelty of this study deals with the application of an iTRAQ approach to dissect the molecular pathways associated with KiSS-1 overexpression in bladder cancer, which provided a less aggressive phenotype as shown in the functional assays by decreased apoptotic, migration, and invasion rates in vitro. This proteomics technique is a powerful method for the identification of the molecular mechanisms associated with bladder cancer metastases and the discovery of potential related targets and biomarkers aiming to address prevention and/or therapy for metastatic disease. Several proteins were found to be differentially expressed in a significant manner in KiSS-1 transfectants and were confirmed to be regulated by KiSS-1 by immunoblotting, an independent analytical method. Immunostainings on tissue arrays containing independent larger series of patients with bladder cancer served to assess the associations of novel KiSS-1-related proteins with clinicopathological variables. Thus, this combination of proteomics approaches has identified novel differentially expressed proteins related to KiSS-1 expression during bladder cancer progression.
To our knowledge, this is the first study on expression of KiSS-1 in bladder cancer cells using a novel proteomics approach and validating the results with in vitro and clinical material. The multisample capability of the iTRAQ technology was considered ideally suited to our study because it allowed comparison of two time points after transfection and two controls. The advantage of iTRAQ was the simultaneous identification and quantification of proteins differentially expressed between experimental (transfected) and control (non-transfected) specimens. The extent of the proteomic profile observed in this study was comparable with those observed in other iTRAQ-two-dimensional LC-MS/MS studies using cell lines of several tumor types such as lung (22), prostate (27, 28), leukemia (29), colorectal (30), and breast (44). On the basis of the identity and biological abundance of the identified proteins, the iTRAQ-two-dimensional LC-MS/MS method exhibited a large dynamic range in profiling both high and low abundance proteins. The broad spectrum of proteins observed with this approach testified to its suitability for proteomics studies of transfected cancer cell lines.
Replicated biological experiments served to provide confidence in the proteomics methodology applied. The use of such independent experimental replicates in which efficient transfection was confirmed by RT-PCR and subfractionation was performed served to assess interassay biological and technical reproducibility. High agreement was observed among experimental replicates and among subcellular fractions. The variability found between these factions could be attributed to biological differences; reproducibility issues in the fractionation method utilized; the potential effect of KiSS-1 overexpression in the cellular translocation or sublocalization of regulated proteins, translocations that may deserve further investigation using immunofluorescence analyses; or the restrictive cutoff selected to consider a change to be over- or underexpressed (24, 44, 45). The limitation of selecting a threshold of expression to consider proteins to be differentially expressed upon KiSS-1 overexpression was followed up with validation analyses for key data. This study focused on describing those proteins detected in both biological replicates and subfractionations. It is likely that other proteins that showed significant differences in three of the four protein extracts could also be potentially relevant in KiSS-1 networks and may deserve further investigation, for example Zyxin (13) (supplemental Table 10). Overall, the verification of the changes induced by KiSS-1 by an independent analytical method in the same cell transfectants provided confidence in the experimental design, allowing validation of significant identified abundance changes.
It is important to point out that in our experimental design subcellular fractionation was carried out to reduce the sample complexity and increase the probability of detecting low abundance proteins. The additional advantage of analyzing subcellular fractions and organelles is the opportunity to track proteins that shuttle between different compartments. The protocol we followed for cellular fractionation used centrifugations designed to isolate nuclear extracts (41). The integrity of the nuclear extracts was confirmed by the presence of Lamin B only in nuclear fractions and not in cytoplasmic fractions. Similar to previously reported studies, higher reproducibility was observed between cytoplasmic replicates than nuclear extracts, which is likely due to the variability of the subfractionation methods (41). Because the cytoplasmic reorganization of the cytoskeleton is important in the metastatic process, Fig. 5 displays the average of the cytoplasmic counterparts.
The mechanism by which KiSS-1 is involved in the invasive/metastatic bladder cancer phenotype has not been completely elucidated. Consistent with the hypothesis that KiSS-1 regulates events downstream of cell-matrix adhesion, independent or not of cytoskeleton reorganization (13, 49,60–51), the results reported here showing reduced migration and invasion rates in vitro revealed the likely involvement of proteins known to be associated with these processes. In bladder cancer, the inactivation of the key regulatory RB1 and TP53 pathways has been shown to be necessary for tumorigenesis and progression (2, 52, 53). Importantly, the proteomics approach undertaken in this study identified the increased expression of p53, p12, and pRB after transfection of KiSS-1 (supplemental Tables 2, 9, and 10) that could be involved in the diminished apoptotic rate under KiSS-1 overexpression. The expression of KiSS-1 was confirmed to be associated with the phosphorylation status of pRB, p53, and its effector p21 (13) and altered patterns of expression of metalloproteinases (15, 49,60–51) by immunoblotting. The link of KiSS-1 with TP53/RB pathways was consistent with a previous report in which the loss of KiSS-1 expression analyzed by in situ hybridization on tissue arrays was significantly associated with protein expression of total retinoblastoma by immunohistochemistry on the same specimens (13). The association found between expression of KiSS-1 and the inactive hyperphosphorylated pRB products (total pRB) and p53 supported the relevance of KiSS-1 in bladder cancer progression because these are critical targets in this disease (2, 13, 52, 53). Overall, this proteomics discovery approach identified molecular networks in which a great number of the aforementioned members are involved. Moreover, the changes induced by KiSS-1 in these proteins were also independently shown by observations from Western blotting analyses supporting such associations.
Although we generated a monoclonal antibody for KiSS-1 allowing Western blotting validation, this antibody did not work for immunohistochemistry. The lack of suitable antibodies is an obstacle to carrying out extensive clinical studies of KiSS-1 at the microanatomical level by immunohistochemistry. To circumvent this problem, the analysis of clinical tumor material was performed by RT-PCR. Because the cytoplasmic reorganization of the cytoskeleton is important in the metastatic process, Ezrin and Filamin A, two cytoskeleton proteins reported to be altered in the metastatic process in other neoplasias and for which antibodies were available, were selected for validation analyses. In this study, we did not pursue Ezrin immunohistochemistry validation because we had recently described how this protein is related to bladder cancer progression. The novelty in this regard is the impact of KiSS-1 transfection on Ezrin expression. In our series, the loss of KiSS-1 expression was observed in those metastatic patients that displayed cytoplasmic Filamin A expression (54). Importantly, this report revealed the regulation of the expression of these proteins by KiSS-1 (54, 55). This observation may indicate a cooperative involvement of KiSS-1 together with Ezrin and Filamin A, among others, in the suppression of metastatic potential of bladder tumors. KiSS-1 was revealed to function as an upstream regulator of adhesion molecules such as Filamin A. This protein belongs to a family of cytoskeletal proteins that organize filamentous actin into networks and stress fibers (56). Filamin A is a non-muscle actin-binding protein, the appropriate functioning of which is essential for development (57, 58). Filamins are essential for mammalian cell locomotion and anchoring of transmembrane proteins, including integrins, and also act as interfaces for protein-protein interaction (57, 59). More than 30 proteins of great functional diversity are known to interact with filamins, which function as a signaling scaffold by connecting and coordinating a large variety of cellular processes (57, 59). This study revealed Filamin A as a target that can be regulated by KiSS-1. Furthermore, this innovative finding was clinically relevant by the link of the expression of this protein to the invasive/metastatic phenotype in human bladder tumors.
Cancer cell lines represent an ideal model to explore the potential biological relevance of targets or biomarker candidates. To our knowledge, this is the first large scale quantitative proteomics profiling of transfected bladder cancer cell lines. Previously reported and novel proteins differentially regulated under KiSS-1 overexpression have been identified. The initial validation set of analyses dealt with confirmatory studies showing increased protein expression of the identified proteins in association with KiSS-1 overexpression in the same replicates utilized for iTRAQ analyses and independent replicates by immunoblotting. To evaluate the relevance of these identified proteins during bladder cancer progression, immunohistochemical stainings were performed on several tissue arrays containing independent larger series of bladder tumors comprising early and advanced stages of the disease with available follow-up. Interestingly, Filamin A was associated with tumor progression (by being correlated with histopathologic tumor stage), lymph node metastases, and clinical outcome in bladder cancer. These observations served to support phenotypically the tumor specificity of the proteins detected by the iTRAQ approach. Furthermore, they supported our experimental design searching for KiSS-1-related bladder progression biomarkers. Overall, immunoblotting analyses of transfected cells confirmed the iTRAQ findings, and these observations are in agreement with the relative concentration trends in the clinical specimens analyzed in this study.
CONCLUSIONS
The iTRAQ proteomics approach identified molecular pathways associated with the overexpression of the metastasis suppressor KiSS-1 in bladder cancer, which provided a less aggressive phenotype as shown in the functional assays and the clinical validation in human samples. Independent in vitro validation by Western blotting confirmed the role of KiSS-1 at regulating novel and critical known proteins involved in bladder cancer progression. Clinical validation analyses by immunohistochemistry on tissue arrays revealed the association of KiSS-1 and related Filamin A expression with pathological correlates of tumor progression, lymph node metastasis, and clinical outcome. These analyses provided mechanistic support for the involvement of KiSS-1 in the progression of the disease. In summary, our study not only has served to reveal molecular mechanisms associated with the metastasis suppressor role of KiSS-1 in bladder cancer but also to identify novel potential metastasis-related biomarkers that may assist in the clinicopathological stratification of patients affected with bladder tumors. Among our concluding remarks, we point out the utility of the iTRAQ proteomics strategy undertaken for the identification of proteins regulated with KiSS-1 expression in bladder cancer. As a result of this analysis, Filamin A was found to be associated with bladder cancer progression and clinical outcome. These observations suggest a role for Filamin A and KiSS-1 as metastasis-related biomarkers for bladder cancer staging and prognosis.
Supplementary Material
Acknowledgments
We thank all members of the Tumor Markers Group and the Proteomics Unit for technical support and constructive suggestions in the preparation of this manuscript. We acknowledge the members of our clinical collaborators at the different institutions involved in this study and the members of the Spanish Oncology Group of Genitourinary Cancer for support in facilitating the tumor specimens and the clinical follow-up of the bladder cancer cases analyzed in this study. We are grateful to Danny Welch for kindly providing us the vector encompassing the full KiSS-1 cDNA for transient transfection experiments.
* This work was supported by Spanish Ministry of Education and Culture Grant SAF2006-08519 to M. S.-C.
 This article contains supplemental Figs. 1–3 and Tables 1–10.
This article contains supplemental Figs. 1–3 and Tables 1–10.
1 The abbreviations used are:
- TCC
- transitional cell carcinoma
- EF
- error factor
- FDR
- false discovery rate
- iTRAQ
- isobaric tags for relative and absolute quantitation
- RB
- retinoblastoma
- TP53
- tumor protein 53
- CI
- confidence interval
- EV
- empty vector
- Ct
- cycle threshold.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) Cancer statistics. CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2.Sánchez-Carbayo M., Cordon-Cardó C. (2007) Molecular alterations associated with bladder cancer progression. Semin. Oncol. 34, 75–84 [DOI] [PubMed] [Google Scholar]
- 3.Welch D. R., Chen P., Miele M. E., McGary C. T., Bower J. M., Stanbridge E. J., Weissman B. E. (1994) Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene 9, 255–262 [PubMed] [Google Scholar]
- 4.Miele M. E., Robertson G., Lee J. H., Coleman A., McGary C. T., Fisher P. B., Lugo T. G., Welch D. R. (1996) Metastasis suppressed, but tumorigenicity and local invasiveness unaffected in the human melanoma cell line MelJuSo after introduction of human chromosomes 1 or 6. Mol. Carcinog. 15, 284–299 [DOI] [PubMed] [Google Scholar]
- 5.Lee J. H., Miele M. E., Hicks D. J., Phillips K. K., Trent J. M., Weissman B. E., Welch D. R. (1996) KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J. Natl. Cancer Inst. 88, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 6.Lee J. H., Welch D. R. (1997) Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int. J. Cancer 71, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 7.Lee J. H., Welch D. R. (1997) Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 57, 2384–2387 [PubMed] [Google Scholar]
- 8.West A., Vojta P. J., Welch D. R., Weissman B. E. (1998) Chromosome localization and genomic structure of the KiSS-1 metastasis suppressor gene. Genomics 54, 145–148 [DOI] [PubMed] [Google Scholar]
- 9.Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., Ishibashi Y., Watanabe T., Asada M., Yamada T., Suenaga M., Kitada C., Usuki S., Kurokawa T., Onda H., Nishimura O., Fujino M. (2001) Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature 411, 613–617 [DOI] [PubMed] [Google Scholar]
- 10.Kotani M., Detheux M., Vandenbogaerde A., Communi D., Vanderwinden J. M., Le Poul E., Brézillon S., Tyldesley R., Suarez-Huerta N., Vandeput F., Blanpain C., Schiffmann S. N., Vassart G., Parmentier M. (2001) The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 276, 34631–34636 [DOI] [PubMed] [Google Scholar]
- 11.Muir A. I., Chamberlain L., Elshourbagy N. A., Michalovich D., Moore D. J., Calamari A., Szekeres P. G., Sarau H. M., Chambers J. K., Murdock P., Steplewski K., Shabon U., Miller J. E., Middleton S. E., Darker J. G., Larminie C. G., Wilson S., Bergsma D. J., Emson P., Faull R., Philpott K. L., Harrison D. C. (2001) AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J. Biol. Chem. 276, 28969–28975 [DOI] [PubMed] [Google Scholar]
- 12.Shirasaki F., Takata M., Hatta N., Takehara K. (2001) Loss of expression of the metastasis suppressor gene KiSS-1 during melanoma progression and its association with LOH of chromosome 6q16.3q23. Cancer Res. 61, 7422–7425 [PubMed] [Google Scholar]
- 13.Sanchez-Carbayo M., Capodieci P., Cordon-Cardo C. (2003) Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. Am. J. Pathol. 162, 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeguchi M., Yamaguchi K., Kaibara N. (2004) Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin. Cancer Res. 10, 1379–1383 [DOI] [PubMed] [Google Scholar]
- 15.Hesling C., D'Incan M., Mansard S., Franck F., Corbin-Duval A., Chèvenet C., Déchelotte P., Madelmont J. C., Veyre A., Souteyrand P., Bignon Y. J. (2004) In vivo and in situ modulation of the expression of genes involved in metastasis and angiogenesis in a patient treated with topical imiquimod for melanoma skin metastasis. Br. J. Dermatol. 150, 761–767 [DOI] [PubMed] [Google Scholar]
- 16.Dhar D. K., Naora H., Kubota H., Maruyama R., Yoshimura H., Tonomoto Y., Tachibana M., Ono T., Otani H., Nagasue N. (2004) Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int. J. Cancer 111, 868–872 [DOI] [PubMed] [Google Scholar]
- 17.Masui T., Doi R., Mori T., Toyoda E., Koizumi M., Kami K., Ito D., Peiper S. C., Broach J. R., Oishi S., Niida A., Fujii N., Imamura M. (2004) Metastin and its variant forms suppress migration of pancreatic cancer cells. Biochem. Biophys. Res. Commun. 315, 85–92 [DOI] [PubMed] [Google Scholar]
- 18.Martin T. A., Watkins G., Jiang W. G. (2005) KiSS-1 expression in human breast cancer. Clin. Exp. Metastasis 22, 503–511 [DOI] [PubMed] [Google Scholar]
- 19.Nicolle G., Comperat E., Nicolaïew N., Cancel-Tassin G., Cussenot O. (2007) Metastin (KISS-1) and metastin-coupled receptor (GPR54) expression in transitional cell carcinoma of the bladder. Ann. Oncol. 18, 605–607 [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Carbayo M. (2006) Antibody arrays: technical considerations and clinical applications in cancer. Clin. Chem. 52, 1651–1659 [DOI] [PubMed] [Google Scholar]
- 21.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 22.Keshamouni V. G., Michailidis G., Grasso C. S., Anthwal S., Strahler J. R., Walker A, Arenberg D. A., Reddy R. C., Akulapalli S., Thannickal V. J., Standiford T. J., Andrews P. C., Omenn G. S. (2006) Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 5, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 23.Overall C. M., Dean R. A. (2006) Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 25, 69–75 [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Choong L. Y., Lin Q., Philp R., Wong C. H., Ang B. K., Tan Y. L., Loh M. C., Hew C. L., Shah N., Druker B. J., Chong P. K., Lim Y. P. (2007) Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol. Cell. Proteomics 6, 2072–2087 [DOI] [PubMed] [Google Scholar]
- 25.Gagné J. P., Ethier C., Gagné P., Mercier G., Bonicalzi M. E., Mes-Masson A. M., Droit A., Winstall E., Isabelle M., Poirier G. G. (2007) Comparative proteome analysis of human epithelial ovarian cancer. Proteome Sci. 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson H., Lengqvist J., Hedlund J., Uhlén K., Orre L. M., Bjellqvist B., Persson B., Lehtiö J., Jakobsson P. J. (2008) Quantitative membrane proteomics applying narrow range peptide isoelectric focusing for studies of small cell lung cancer resistance mechanisms. Proteomics 8, 3008–3018 [DOI] [PubMed] [Google Scholar]
- 27.Glen A., Gan C. S., Hamdy F. C., Eaton C. L., Cross S. S., Catto J. W., Wright P. C., Rehman I. (2008) iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression. J. Proteome Res. 7, 897–907 [DOI] [PubMed] [Google Scholar]
- 28.Sardana G., Jung K., Stephan C., Diamandis E. P. (2008) Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res. 7, 3329–3338 [DOI] [PubMed] [Google Scholar]
- 29.Pierce A., Unwin R. D., Evans C. A., Griffiths S., Carney L., Zhang L., Jaworska E., Lee C. F., Blinco D., Okoniewski M. J., Miller C. J., Bitton D. A., Spooncer E., Whetton A. D. (2008) Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteomics 7, 853–863 [DOI] [PubMed] [Google Scholar]
- 30.Tan H. T., Tan S., Lin Q., Lim T. K., Hew C. L., Chung M. C. (2008) Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Mol. Cell Proteomics 7, 1174–1185 [DOI] [PubMed] [Google Scholar]
- 31.DeSouza L., Diehl G., Rodrigues M. J., Guo J., Romaschin A. D., Colgan T. J., Siu K. W. (2005) Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 4, 377–386 [DOI] [PubMed] [Google Scholar]
- 32.Seshi B. (2006) An integrated approach to mapping the proteome of the human bone marrow stromal cell. Proteomics 6, 5169–5182 [DOI] [PubMed] [Google Scholar]
- 33.Chaerkady R., Harsha H. C., Nalli A., Gucek M., Vivekanandan P., Akhtar J., Cole R. N., Simmers J., Schulick R. D., Singh S., Torbenson M., Pandey A., Thuluvath P. J. (2008) A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J. Proteome Res. 7, 4289–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garbis S. D., Tyritzis S. I., Roumeliotis T., Zerefos P., Giannopoulou E. G., Vlahou A., Kossida S., Diaz J., Vourekas S., Tamvakopoulos C., Pavlakis K., Sanoudou D., Constantinides C. A. (2008) Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (iTRAQ) with two-dimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 7, 3146–3158 [DOI] [PubMed] [Google Scholar]
- 35.Bouchal P., Roumeliotis T., Hrstka R., Nenutil R., Vojtesek B., Garbis S. D. (2009) Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J. Proteome Res. 8, 362–373 [DOI] [PubMed] [Google Scholar]
- 36.Ralhan R., Desouza L. V., Matta A., Chandra Tripathi S., Ghanny S., Datta Gupta S., Bahadur S., Siu K. W. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics 7, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ralhan R., Desouza L. V., Matta A., Chandra Tripathi S., Ghanny S., Dattagupta S., Thakar A., Chauhan S. S., Siu K. W. (2009) iTRAQ-multidimensional liquid chromatography and tandem mass spectrometry-based identification of potential biomarkers of oral epithelial dysplasia and novel networks between inflammation and premalignancy. J. Proteome Res. 8, 300–309 [DOI] [PubMed] [Google Scholar]
- 38.Streckfus C. F., Mayorga-Wark O., Arreola D., Edwards C., Bigler L., Dubinsky W. P. (2008) Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Invest. 26, 159–167 [DOI] [PubMed] [Google Scholar]
- 39.Bijian K., Mlynarek A. M., Balys R. L., Jie S., Xu Y., Hier M. P., Black M. J., Di Falco M. R., LaBoissiere S., Alaoui-Jamali M. A. (2009) Serum proteomic approach for the identification of serum biomarkers contributed by oral squamous cell carcinoma and host tissue microenvironment. J. Proteome Res. 8, 2173–2185 [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Carbayo M., Socci N. D., Charytonowicz E., Lu M., Prystowsky M., Childs G., Cordon-Cardo C. (2002) Molecular profiling of bladder cancer using cDNA microarrays: defining histogenesis and biological phenotypes. Cancer Res. 62, 6973–6980 [PubMed] [Google Scholar]
- 41.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17, 6419–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choe L., D'Ascenzo M., Relkin N. R., Pappin D., Ross P., Williamson B., Guertin S., Pribil P., Lee K. H. (2007) 8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics 7, 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 44.Ho J., Kong J. W., Choong L. Y., Loh M. C., Toy W., Chong P. K., Wong C. H., Wong C. Y., Shah N., Lim Y. P. (2009) Novel breast cancer metastasis-associated proteins. J. Proteome Res. 8, 583–594 [DOI] [PubMed] [Google Scholar]
- 45.Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 46.American Joint Committee on Cancer (1988) Staging of cancer at genitourinary sites, in American Joint Committee on Cancer: Manual for Staging of Cancer, 3rd Ed., pp. 194–195, J. B. Lippincott & Co., Philadelphia [Google Scholar]
- 47.Mostofi F. K. (1973) Histological Typing of Urinary Bladder Tumors, World Health Organization, Geneva [Google Scholar]
- 48.Dawson-Saunders B., Trapp R. G. (1994) Basic and Clinical Biostatistics, 2nd Ed., Appleton & Lange, Norwalk, CT [Google Scholar]
- 49.Ringel M. D., Hardy E., Bernet V. J., Burch H. B., Schuppert F., Burman K. D., Saji M. (2002) Metastin receptor is overexpressed in papillary thyroid cancer and activated MAP kinase in thyroid cancer cells. J. Clin. Endocrinol. Metab. 87, 2399. [DOI] [PubMed] [Google Scholar]
- 50.Hori A., Honda S., Asada M., Ohtaki T., Oda K., Watanabe T., Shintani Y., Yamada T., Suenaga M., Kitada C., Onda H., Kurokawa T., Nishimura O., Fujino M. (2001) Metastin suppresses the motility and growth of CHO cells transfected with its receptor. Biochem. Biophys. Res. Commun. 286, 958–963 [DOI] [PubMed] [Google Scholar]
- 51.Yoshioka K., Ohno Y., Horiguchi Y., Ozu C., Namiki K., Tachibana M. (2008) Effects of a KiSS-1 peptide, a metastasis suppressor gene, on the invasive ability of renal cell carcinoma cells through a modulation of a matrix metalloproteinase 2 expression. Life Sci. 83, 332–338 [DOI] [PubMed] [Google Scholar]
- 52.Markl I. D., Jones P. A. (1998) Presence and location of TP53 mutation determines pattern of CDKN2A/ARF pathway inactivation in bladder cancer. Cancer Res. 58, 5348–5353 [PubMed] [Google Scholar]
- 53.Lu M. L., Wikman F., Orntoft T. F., Charytonowicz E., Rabbani F., Zhang Z., Dalbagni G., Pohar K. S., Yu G., Cordon-Cardo C. (2002) Impact of alterations affecting the p53 pathway in bladder cancer on clinical outcome, assessed by conventional and array-based methods. Clin. Cancer Res. 8, 171–179 [PubMed] [Google Scholar]
- 54.Seraj M. J., Harding M. A., Gildea J. J., Welch D. R., Theodorescu D. (2000) The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin. Exp. Metastasis 18, 519–525 [DOI] [PubMed] [Google Scholar]
- 55.Palou J., Algaba F., Vera I., Rodriguez O., Villavicencio H., Sánchez-Carbayo M. (2009) Protein expression patterns of ezrin are predictors of progression in T1G3 bladder tumours treated with nonmaintenance Bacillus Calmette-Guérin. Eur. Urol. 56, 829–836 [DOI] [PubMed] [Google Scholar]
- 56.Gorlin J. B., Yamin R., Egan S., Stewart M., Stossel T. P., Kwiatkowski D. J., Hartwig J. H. (1990) Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J. Cell Biol. 111, 1089–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y., Walsh C. A. (2004) The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6, 1034–1038 [DOI] [PubMed] [Google Scholar]
- 58.Robertson S. P., Twigg S. R., Sutherland-Smith A. J., Biancalana V., Gorlin R. J., Horn D., Kenwrick S. J., Kim C. A., Morava E., Newbury-Ecob R., Orstavik K. H., Quarrell O. W., Schwartz C. E., Shears D. J., Suri M., Kendrick-Jones J., Wilkie A. O.; OPD-spectrum Disorders Clinical Collaborative Group (2003) Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 33, 487–491 [DOI] [PubMed] [Google Scholar]
- 59.van der Flier A., Sonnenberg A. (2001) Structural and functional aspects of filamins. Biochim. Biophys. Acta 1538, 99–117 [DOI] [PubMed] [Google Scholar]
- 60.Yan C., Wang H., Boyd D. D. (2001) KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha-induced block of p65/p50 nuclear translocation. J. Biol. Chem. 276, 1164–1172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.