Abstract
Respiratory complex I (NADH:quinone oxidoreductase) is an entry point to the electron transport chain in the mitochondria of many eukaryotes. It is a large, multisubunit enzyme with a hydrophilic domain in the matrix and a hydrophobic domain in the mitochondrial inner membrane. Here we present a comprehensive analysis of the protein composition and post-translational modifications of complex I from Pichia pastoris, using a combination of proteomic and bioinformatic approaches. Forty-one subunits were identified in P. pastoris complex I, comprising the 14 core (conserved) subunits and 27 supernumerary subunits; seven of the core subunits are mitochondrial encoded. Three of the supernumerary subunits (named NUSM, NUTM, and NUUM) have not been observed previously in any species of complex I. However, homologues to all three of them are present in either Yarrowia lipolytica or Pichia angusta complex I. P. pastoris complex I has 39 subunits in common with Y. lipolytica complex I, 37 in common with N. crassa complex I, and 35 in common with the bovine enzyme. The mitochondrial encoded subunits (translated by the mold mitochondrial genetic code) retain their N-α-formyl methionine residues. At least eight subunits are N-α-acetylated, but the N-terminal modifications of the nuclear encoded subunits are not well-conserved. A combination of two methods of protein separation (SDS-PAGE and HPLC) and three different mass spectrometry techniques (peptide mass fingerprinting, tandem MS and molecular mass measurements) were required to define the protein complement of P. pastoris complex I. This requirement highlights the need for inclusive and comprehensive strategies for the characterization of challenging membrane-bound protein complexes containing both hydrophilic and hydrophobic components.
Respiratory complex I (NADH:quinone oxidoreductase) is an entry point to the electron transport chain in mitochondria and many aerobic bacteria. It couples NADH oxidation and quinone reduction to proton translocation across the inner mitochondrial (or plasma) membrane, so it is central to energy transduction. Consequently, complex I dysfunctions are linked to an increasing number of neuromuscular and neurodegenerative diseases, including Parkinson's disease, as well as to oxidative stress and aging (1).
Complex I is an l-shaped assembly, with a hydrophobic “arm” in the inner mitochondrial (or plasma) membrane, and a hydrophilic arm extending into the mitochondrial matrix (or cytoplasm) (2). The 14 core subunits of complex I are sufficient for catalysis and conserved in all complex I–encoding species; they comprise two sets of seven subunits that correspond to the two domains (3, 4). The seven hydrophobic subunits are encoded by the mitochondrial genome in eukaryotes, and the seven nuclear-encoded hydrophilic subunits ligate the flavin mononucleotide (which catalyzes NADH oxidation), and the eight (or nine) iron-sulfur clusters (which transfer electrons from the flavin to quinone). The structure of the hydrophilic domain of complex I from Thermus thermophilus, comprising the seven hydrophilic core subunits and a frataxin-like supernumerary subunit, shows how the subunits and cofactors are arranged (5). In addition to the 14 core subunits, eukaryotic complexes I contain a variable number of supernumerary subunits also. The roles of most of the supernumerary subunits are not well defined, although specific functions for some of them have been proposed (6). Twenty-one supernumerary subunits are common to all eukaryotic complexes I, but additional subunits vary between fungi, plants, and animals, as well as between individual species (7). The most intensively characterized eukaryotic complex I is the 1 MDa complex from Bos taurus that contains 45 dissimilar subunits, all with homologues in the human genome (6, 8–10).
The subunit composition of the complexes I from two fungi, Neurospora crassa and Yarrowia lipolytica, have been analyzed previously. Thirty-nine subunits have been identified in N. crassa complex I (11), and four of them do not have mammalian homologues. The mitochondrial genome sequence of Y. lipolytica was published in 2001 (12) and the first assessment of the subunit composition of complex I from Y. lipolytica in 2002 described 37 subunits, identified by a combination of SDS-PAGE, MALDI-TOF mass spectrometry and Edman degradation (13). Subsequently, three additional subunits were described, and, in 2008, the masses of a set of subunits were measured using laser-induced liquid bead ion desorption MS. The accuracy of the molecular mass data was variable, but observed masses were paired with predicted masses within 130 Da (14). Thus, 40 subunits have been identified in Y. lipolytica complex I; 37 of them are found in N. crassa, and 35 in B. taurus.
Pichia pastoris is a methylotrophic ascomycete, which is a commonly used over-expression host for recombinant proteins. The purification and characterization of complex I from P. pastoris, and the related species Pichia angusta, have been described previously (15). Here we present a comprehensive analysis of the protein composition and post-translational modifications of complex I from P. pastoris, using a combination of proteomic and bioinformatic approaches. Proteomic techniques provide a rapid and efficient means of defining the composition of protein complexes. Here the subunits of P. pastoris complex I were resolved either by reverse-phase HPLC or by SDS-PAGE, and then analyzed by peptide mass fingerprinting and tandem mass spectrometry of tryptic peptides, by MALDI-TOF mass spectrometry. The analysis was complicated by the presence of hydrophobic membrane proteins, and by the presence of small subunits which produce few or no proteolytic fragments. After reverse-phase separation, the subunits were also analyzed by ESI-MS, and their molecular masses provided additional information on the subunit composition and amino acid sequences, and on stable post-translational modifications. Forty-one different proteins have been identified in complex I from P. pastoris, including three proteins (named NUSM, NUTM, and NUUM) that have not been detected previously in any species of complex I.
EXPERIMENTAL PROCEDURES
Complex I Purification
Complex I was isolated from P. pastoris strain X-33, and from Y. lipolytica strain GB10, as described previously (15–17).
Separation of the Subunits of Complex I by SDS-PAGE and Reverse-Phase HPLC
SDS-PAGE of 10-20 μg of protein was performed on 18-22% acrylamide gradient Laemmli-style gels (18) for 4 h at 300 mV, and stained with 0.2% Coomassie Blue R250. For reverse-phase HPLC, complex I was precipitated by 20 volumes of cold ethanol, centrifuged at 16,000 × g for 3 min, and the supernatant, containing detergents and lipids, was discarded. The pellet was dissolved in 60% (v/v) formic acid, 15% (v/v) trifluoroethanol, and 1% (v/v) hexafluoroisopropanol, and injected onto a PLRP-S reverse phase HPLC column (1 mm i.d. × 75 mm, Varian Inc., Palo Alto, CA). Proteins were eluted at 50 μl min−1 in a linear gradient of 0-70% propan-2-ol and 15-20% trifluoroethanol, in 1% hexafluoroisopropanol and either 50 mm ammonium formate (pH 3.1) or 0.1% trifluoroacetic acid (pH 1.8) (19).
Analysis of SDS-PAGE and HPLC Fractions by Peptide Mass Fingerprinting and Tandem MS
Protein bands were excised from the SDS-PAGE gels, diced into small cubes (approximately 1 mm3), and digested by trypsin as described previously (20). The tryptic peptides were analyzed by peptide mass fingerprinting and tandem MS using an Applied Biosystems/MDX SCIEX 4800 Plus MALDI TOF/TOF mass spectrometer, with α-cyano-4-hydroxy cinnamic acid as the matrix. Spectra were calibrated using the trypsin autolysis peptides at 2163.057 and 2273.160 Da, and a calcium-associated matrix ion at 1060.048 Da. Peptide sequences were obtained using the same spectrometer, by collision-induced dissociation, at a collision energy of 1 kV. Typically, tandem MS spectra contained data accumulated from 2,500 laser pulses. Fractions from HPLC were dried in a speed-vac centrifuge, then dissolved in 0.2% (w/v) ammonium bicarbonate, 0.5 mm CaCl2, and 12 ng/ml trypsin, and digested overnight at 37 °C.
MALDI-TOF mass spectra were smoothed and monoisotopically labeled using the 4000 Data Explorer software (version 3.5.3, Applied Biosystems). Monoisotopic peak lists were generated using the peaks-to-mascot feature, with a minimum signal-to-noise ratio of 10, and a peak density of 10 (occasionally 15) peaks per 200 Da. Known peaks from human keratin, trypsin autolysis, and matrix associated ions were excluded (see Supplementary Information). Then, the peak lists were compared with the in-house P. pastoris protein sequence database (5,209 sequences, see below), or to the online National Center for Biotechnology Information (NCBInr) database (version 20080912 comprising 7,031,513 sequences) using the Mascot (Matrix Science Ltd., London, UK) search algorithm, version 2.1.0 (21). The significance threshold for peptide identification was p < 0.05, and a single missed cleavage was allowed. The mass tolerances were 70 ppm for peptide mass fingerprinting and 0.8 Da and 120 ppm for tandem MS. N-terminal acetylation and formylation, cysteine propionamidation, and methionine oxidation were selected as variable modifications in initial searches. Error tolerant searches were also performed to identify additional matches that rely on amino acid substitutions or further artifactual or unusual modifications.
Analysis of HPLC Fractions by ESI-MS
Fractions from reverse-phase HPLC were analyzed by ESI-MS in positive ion mode, either online with a Quattro Ultima triple quadrupole mass spectrometer (Waters-Micromass, Waters Corp., Milford, MA) scanning 700–2100 m/z every 5 s, or offline using a Q-TOF1 mass spectrometer (Waters-Micromass). Samples were loaded into the Q-TOF1 by flow injection into 50% isopropanol, and both spectrometers were calibrated with myoglobin and trypsinogen over the mass/charge range of 616 to 2181. Protein molecular masses were determined from series of multiply charged ions, using the component analysis function of MassLynx, version 3.4 (Waters-Micromass).
RESULTS AND DISCUSSION
Prediction of P. pastoris Complex I Subunit Sequences by Comparison to Known Sequences in Other Organisms
The genome sequence of P. pastoris strain X-33 was obtained from Integrated Genomics Inc. (Chicago, IL) in flat file format, and comprised 13 contiguous segments (contigs): 12 nuclear contigs and the mitochondrial genome sequence. A set of approximately 5,000 ORFs and their translated protein sequences were obtained also. Consequently, a genomic DNA sequence database and a protein sequence database were assembled in FASTA format, and tblastn and blastp were used, respectively, to identify the sequences of known complex I subunits (NCBI BLAST v 2.2.16, statistical E-value threshold 1 × 10−10) (22). Comparisons were performed with the amino acid sequences of the subunits of Y. lipolytica complex I plus two subunits (10.4 kDa and NURM) specific to N. crassa complex I (14), and 10 additional subunits from B. taurus complex I (8). Twenty-seven putative complex I subunits, homologous to known nuclear-encoded subunits from Y. lipolytica, were identified in the P. pastoris protein sequence database (although the ORF lengths for NUEM and NUKM required adjusting to produce proteins similar in size to their Y. lipolytica homologues, and a single base deletion was corrected in the C-terminal region of NUAM). A further five putative nuclear-encoded sequences were identified in the DNA sequence database, and were translated, along with their flanking regions, using the NCBInr ORF finder tool (ORFFINDER) (23). ORFs for proteins of similar size to the Y. lipolytica homologues were identified for two sequences, and single introns were predicted in the remaining three (NI9M, NB5M, and NIDM) using the prediction program AUGUSTUS (24) with the organism set as Debaryomyces hansenii. The five additional sequences were added to the in-house P. pastoris protein sequence database. No homologues to the Y. lipolytica NUNM subunit, the N. crassa 10.4 kDa and NURM subunits, or the 10 subunits considered specific to mammalian complex I were identified. Finally, ORFFINDER was used to translate the P. pastoris mitochondrial genome, using both the “yeast” and the “mold, protozoan and coelenterate” codes (the yeast code is used by, for example, Saccharomyces cerevisiae (25) and Kluveromyces thermotolerans (26) and the mold code by, for example, N. crassa (27) and Y. lipolytica (12)). Both codes produced viable sequences for the seven expected subunits of complex I, which were identified by comparison with the NCBInr database and added to the in-house P. pastoris database. In summary, 39 putative subunits of P. pastoris complex I (seven nuclear core subunits, seven mitochondrial core subunits, and 25 supernumerary subunits) were identified. All the DNA and protein sequences for the P. pastoris complex I subunits have been deposited in the European Molecular Biology Laboratory (EMBL) database and the respective accession numbers, along with the amino acid sequences, are presented in the Supplementary Information.
Separation of the Subunits by SDS-PAGE and Their Identification by Mass Spectrometry
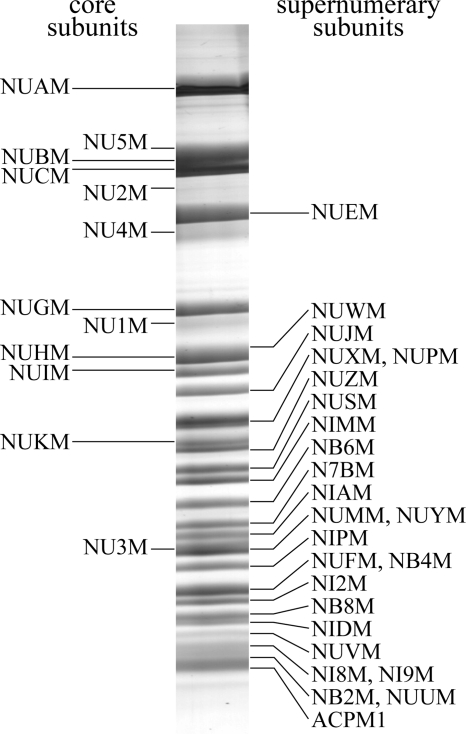
Fig. 1 shows an analysis of complex I from P. pastoris by SDS-PAGE. Each band was excised from the gel, digested with trypsin, and the products were analyzed by peptide mass fingerprinting and tandem mass spectrometry. The unstained regions between the bands were analyzed also because the hydrophobic subunits of complex I do not stain intensively with Coomassie Blue. The data were compared against the in-house P. pastoris protein sequence database and the data are included in Table I and presented fully in Table S1. Thirty-four of the 39 predicted subunits were detected by peptide mass fingerprinting, and 35 were detected by tandem MS. All seven nuclear encoded core subunits were detected, and five of the seven mitochondrial encoded subunits; NU6M and NULM were not detected because, irrespective of the mitochondrial translation code used, their sequences do not contain any tryptic peptides with masses between 700 and 3,000 Da. Significantly more peptides were matched to sequences produced by translation with the mold mitochondrial genetic code (see Table I) than with the yeast mitochondrial code. In particular, subunit NU2M was only identified when the mold genetic code was used, and only the mold code explains a peptide mass and sequence data from subunit NU5M, corresponding to the peptide LIYYTFLNNPNSPK (ATA is translated as Ile by the mold code (underlined), and as Met by the yeast code). Twenty-three of the 25 predicted supernumerary subunits were detected; the ST1 and ACPM2 subunits were not. Two additional proteins, that have been named NUSM and NUUM and which are not similar to any known subunits of complex I, were detected also. Note that the preparation of complex I described here is highly pure: peptide mass fingerprinting and tandem MS analyses sporadically detected only four additional proteins: plasma membrane H+– adenosine triphosphatase, band 7 stomatin protein, 40s ribosomal subunit and a GatB/YeqY superfamily protein (genbank entries 254565045, 254569368, 254566987, and 254566543, respectively, from P. pastoris GS115). All four proteins already have known functions, are present in only low amounts (they were not visible in SDS-PAGE) and were not detected by molecular mass measurements, so they are considered low-level impurities.
Fig. 1.
The subunit composition of complex I from P. pastoris. The subunits of complex I were separated by SDS-PAGE, using a 12-22% acrylamide gradient, and stained with Coomassie Blue R250. The bands were excised from the gel, digested with trypsin, and identified by peptide mass fingerprinting and tandem mass spectrometry. The subunits are labeled according to the SwissProt nomenclature.
Table I. A summary of peptide mass fingerprinting and tandem MS data for the 41 proteins identified in complex I from P. pastoris.
| Subunit | Number of residues | Peptide mass fingerprinting |
Tandem MS |
||||
|---|---|---|---|---|---|---|---|
| Number of peptides1 | Sequence coverage | Score | Number of peptides | Sequence coverage | Score | ||
| 1. NUAM/75 kDa | 702 | 54 (98) | 77% | 298 | 22 | 52% | 1795 |
| 2. NUBM/51 kDa | 462 | 17 (11) | 47% | 175 | 14 | 37% | 767 |
| 3. NUCM/49 kDa | 451 | 20 (11) | 58% | 250 | 19 | 57% | 1328 |
| 4. NUGM/30 kDa | 251 | 18 (15) | 80% | 246 | 18 | 75% | 1213 |
| 5. NUHM/24 kDa | 215 | 9 (14) | 41% | 109 | 9 | 42% | 467 |
| 6. NUIM/TYKY2 | 222 | 11 (23) | 41% | 109 | 12 | 53% | 745 |
| 7. NUKM/PSST2 | 204 | 12 (8) | 45% | 132 | 6 | 32% | 495 |
| 8. NU1M/ND1 | 353 | 4 (28) | 7% | 17 | 2 | 8% | 86 |
| 9. NU2M/ND2 | 523 | 3 (24) | 6% | 18 | 2 | 5% | 132 |
| 10. NU3M/ND3 | 141 | 2 (0) | 17% | 43 | 2 | 17% | 83 |
| 11. NU4M/ND43 | 491 | 3 (24) | 7% | 15 | 1 | 5% | 48 |
| 12. NU5M/ND5 | 642 | 6 (15) | 10% | 35 | 6 | 10% | 265 |
| 13. NU6M/ND64 | 161 | – | – | – | – | – | – |
| 14. NULM/ND4L4 | 82 | – | – | – | – | – | – |
| 15. NUEM/39 kDa | 377 | 22 (16) | 71% | 291 | 18 | 62% | 1526 |
| 16. NESM/ESSS | 215 | 4 (15) | 27% | 23 | 8 | 48% | 870 |
| 17. NUJM/B14.7 | 215 | 12 (45) | 55% | 114 | 7 | 36% | 358 |
| 18. NUXM | 192 | 12 (13) | 65% | 178 | 10 | 63% | 560 |
| 19. NUPM/PGIV | 183 | 9 (8) | 57% | 107 | 8 | 46% | 878 |
| 20. NUZM | 181 | 19 (12) | 84% | 277 | 12 | 69% | 911 |
| 21. NUSM | 154 | 12 (17) | 76% | 173 | 7 | 61% | 521 |
| 22. NIMM/MWFE | 150 | 16 (14) | 62% | 196 | 10 | 52% | 593 |
| 23. NB6M/B16.6 | 146 | 10 (16) | 63% | 141 | 11 | 58% | 650 |
| 24. N7BM/B17.2 | 139 | 9 (16) | 53% | 147 | 6 | 45% | 513 |
| 25. NUYM/18 kDa | 139 | 9 (20) | 67% | 108 | 8 | 62% | 644 |
| 26. NIAM/ASHI | 132 | 7 (17) | 45% | 75 | 6 | 43% | 300 |
| 27. NUMM/13 kDa | 128 | 8 (22) | 71% | 111 | 9 | 66% | 519 |
| 28. NUFM/B13 | 126 | 6 (15) | 49% | 78 | 6 | 58% | 616 |
| 29. NB4M/B14 | 121 | 7 (21) | 61% | 106 | 6 | 56% | 431 |
| 30. NI2M/B22 | 110 | 11 (19) | 78% | 165 | 7 | 59% | 393 |
| 31. NIPM/15 kDa | 105 | 11 (17) | 75% | 145 | 8 | 63% | 506 |
| 32. ACPM1/SDAP15,6 | 92 | 3 | 27% | – | 3 | 27% | 332 |
| 33. NIDM/PDSW | 91 | 5 (24) | 59% | 84 | 4 | 48% | 273 |
| 34. NI8M/B8 | 89 | 6 (24) | 61% | 85 | 5 | 56% | 397 |
| 35. ACPM2/SDAP26,7 | 88 | 2 | 20% | – | 2 | 20% | 100 |
| 36. NB8M/B18 | 86 | 6 (18) | 65% | 67 | 6 | 58% | 334 |
| 37. NUTM5 | 85 | 2 | 28% | 20 | 3 | 27% | 58 |
| 38. NB5M/B15 | 80 | 8 (22) | 62% | 153 | 5 | 36% | 268 |
| 39. NI9M/B9 | 78 | 5 (17) | 43% | 64 | 3 | 26% | 336 |
| 40. NUUM | 73 | 3 (18) | 54% | 40 | 3 | 60% | 165 |
| 41. NB2M/B12 | 60 | 5 (17) | 31% | 68 | 3 | 45% | 147 |
- The number of peptides is given alongside the number of unmatched masses (in brackets).
- Mature protein sequence is not known, number of residues and sequence coverage are based on the complete sequence.
- See Supplementary Information for peptide mass fingerprints and tandem MS spectra.
- No peptides were identified.
- Only observed following protein separation by reverse phase HPLC.
- Peptide mass fingerprinting data alone were insufficient for protein identification.
- Length of mature protein inferred from homology with other species.
To evaluate the completeness of our analysis, complex I from Y. lipolytica was analyzed alongside that from P. pastoris, by comparison of mass spectrometry data with the online NCBInr database. Thirty-six of the 40 known subunits were detected. The second acyl carrier protein (ACPM2) and the mitochondrial encoded subunits NU2M, NU6M, and NULM were not detected; NU6M and NULM do not produce tryptic peptides with masses within the range analyzed.
The Molecular Masses of the Subunits
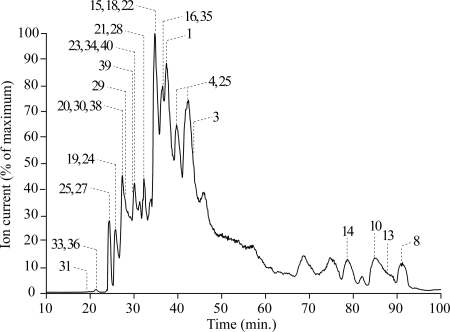
Fig. 2 shows a typical reverse-phase chromatogram for the fractionation of the subunits of P. pastoris complex I. The reverse-phase procedure is compatible with the recovery of intrinsic membrane proteins, and separates both hydrophilic and hydrophobic components in a form suitable for ESI MS (19). The hydrophilic proteins elute first, and the highly hydrophobic proteins elute toward the end of the gradient. The eluting proteins were either collected manually and their intact molecular masses were analyzed by ESI MS, or the eluant from the column was analyzed online using ESI MS. In some experiments, portions of each fraction were digested with trypsin and analyzed using peptide mass fingerprinting and tandem MS to determine the order in which the proteins elute, and to correlate molecular masses with specific subunits. The combined data from all ESI-MS experiments are summarized in Table II. Note that, for some of the larger proteins, the peaks in the ESI spectra were broad; therefore, precise measurements were difficult.
Fig. 2.
Reverse-phase HPLC trace showing the separation of the subunits of complex I from P. pastoris. Proteins were eluted from a reverse-phase HPLC column, at 50 μl min−1, in a 100-min linear gradient of 0-70% propan-2-ol and 15-20% trifluoroethanol, in 1% hexafluoroisopropanol and 50 mm ammonium formate (pH 3.1). The eluant was analyzed by an online ESI mass spectrometer (see Experimental Methods), so the elution trace is described by the total ion current. The labels denote detected molecular masses that could be assigned to subunits of P. pastoris complex I; the numbers refer to the subunits as listed in Table II.
Table II. Calculated and observed protein masses of complex I subunits from P. pastoris and their corresponding post-translational modifications.
| Subunit | Mass |
Δ mass | ppm | Modifications | |
|---|---|---|---|---|---|
| Calc. | Obs. | ||||
| 1. NUAM/75 kDa1 | 76746.3 | 76760.6 | 14.3 | 186.0 | ΔImport(1–24) |
| 2. NUBM/51 kDa1 | 50852.3 | 50867.3 | 15.0 | 295.0 | ΔImport(1–11) |
| 3. NUCM/49 kDa1 | 51347.5 | 51382.9 (3.4) | 35.4 | 689.4 | ΔImport(1–31) |
| 4. NUGM/30 kDa2 | 29046.9 | 29048.3 (1.6) | 1.4 | 48.2 | ΔImport(1–38) |
| 5. NUHM/24 kDa1 | 24344.0 | 24349.6 | 5.6 | 230.0 | ΔImport(1–26) |
| 6. NUIM/TYKY | – | – | – | – | – |
| 7. NUKM/PSST | – | – | – | – | – |
| 8. NU1M/ND13 | 39681.3 | 39717.3 (16.3) | 36.0 | 907.2 | N-α-Formyl |
| 9. NU2M/ND23 | 59074.7 | 59086.1 | 11.4 | 193.0 | N-α-Formyl |
| 10. NU3M/ND33 | 16011.0 | 16014.8 (7.4) | 3.8 | 237.3 | N-α-Formyl |
| 11. NU4M/ND43 | 54756.9 | 54773.2 (8.7) | 16.3 | 297.7 | N-α-Formyl |
| 12. NU5M/ND53 | 72396.9 | – | – | – | – |
| 13. NU6M/ND63 | 18561.6 | 18561.6 (1.3) | 0.0 | 0.0 | N-α-Formyl |
| 14. NULM/ND4L3 | 9116.9 | 9117.0 (0.3) | 0.1 | 11.0 | N-α-Formyl |
| 15. NUEM/39 kDa1 | 42356.0 | 42358.8 (1.9) | 2.8 | 66.1 | ΔImport(1–7) |
| 16. NESM/ESSS2 | 24577.2 | 24579.5 (9.8) | 2.3 | 93.6 | ΔImport(1–24) |
| 17. NUJM/B14.72 | 23393.6 | 23394.1 (1.3) | 0.5 | 21.4 | -Met, N-α-Acetyl |
| 18. NUXM2 | 22254.2 | 22255.2 (1.4) | 1.0 | 44.9 | N-α-Acetyl |
| 19. NUPM/PGIV1,4 | 20704.5 | 20697.4 | 7.1 | 343.0 | -Met, N-α-Acetyl |
| 20. NUZM2 | 20057.0 | 20058.4 | 1.4 | 69.8 | -Met |
| 20099.0 | 20100.4 | 1.4 | 69.7 | -Met, N-α-Acetyl | |
| 21. NUSM2 | 17987.8 | 17988.6 (1.6) | 0.8 | 44.5 | ΔImport(1–28) |
| 22. NIMM/MWFE2 | 17418.7 | 17419.3 (1.0) | 0.6 | 34.4 | none |
| 23. NB6M/B16.61 | 17191.7 | 17192.2 (1.3) | 0.5 | 29.1 | -Met |
| 17233.7 | 17234.6 | 0.9 | 52.2 | -Met, N-α-Acetyl | |
| 24. N7BM/B17.23 | 16323.4 | 16323.4 | 0.0 | 0.0 | -Met, N-α-Acetyl |
| 25. NUYM/18 kDa3 | 15994.0 | 15995.4 (1.9) | 1.4 | 87.5 | ΔImport(1–20) |
| 26. NIAM/ASHI2 | 15226.1 | 15226.6 (0.8) | 0.5 | 32.8 | ΔImport(1–24) |
| 27. NUMM/13 kDa2 | 14339.2 | 14338.0 (0.8) | −1.2 | −83.7 | ΔImport(1–11) |
| 28. NUFM/B132 | 14531.6 | 14533.2 (1.0) | 1.6 | 110.1 | ΔImport(1–8) |
| 29. NB4M/B141 | 14628.8 | 14629.4 | 0.6 | 41.0 | -Met |
| 30. NI2M/B222 | 12733.6 | 12735.0 (1.2) | 1.4 | 109.9 | -Met, N-α-Acetyl |
| 31. NIPM/15 kDa1 | 12097.8 | 12094.4 | −3.4 | −282.5 | -Met |
| 32. ACPM1/SDAP13 | 10659.6 | 10656.4 (0.1) | −3.1 | −290.9 | ΔImport(1–46) + PP5 |
| 33. NIDM/PDSW1 | 10639.9 | 10637.4 (1.5) | −2.5 | −235.0 | -Met, S23L6 |
| 34. NI8M/B82 | 9860.5 | 9861.4 (0.5) | 0.9 | 91.3 | -Met, N-α-Acetyl |
| 35. ACPM2/SDAP2 | 10032.0 | – | – | – | ΔImport(1–42) + PP5 |
| 36. NB8M/B183,4 | 10323.9 | 10320.1 (1.0) | 3.8 | 368.2 | -Met |
| 37. NUTM2 | 9721.2 | 9721.3 (0.7) | 0.2 | 20.5 | -Met |
| 38. NB5M/B152 | 9291.6 | 9292.1 (0.6) | 0.5 | 53.8 | -Met |
| 39. NI9M/B92 | 9037.4 | 9038.3 (0.7) | 0.9 | 99.6 | -Met |
| 40. NUUM3 | 8192.3 | 8192.4 | 0.1 | 12.2 | -Met |
| 41. NB2M/B123 | 6996.8 | 6997.7 (1.4) | 0.9 | 128.6 | -Met, S49L6 |
- Assigned by indirect correlation of a protein mass with tryptic peptide data from a duplicate sample.
- Assigned by direct correlation of a protein mass with tryptic peptide data from the same sample.
- Assigned only by correlation of a measured mass with a predicted candidate mass.
- The mass discrepancies for subunits NUPM/PGIV and NB8M/B18 may be due to the formation of protein disulfides.
- PP, pantetheine-4′-phosphate.
- Amino acid substitution identified by tandem MS (see Table S1).
Molecular masses (within 100 ppm) were obtained for 21 of the 39 putative subunits of P. pastoris complex I, and for the two additional proteins, NUSM and NUUM (see Table II). The data obtained for core hydrophilic subunits NUAM, NUBM, NUCM, and NUHM were less accurate and no data were obtained for either NUIM or NUKM. Of the mitochondrial-encoded subunits, corresponding molecular masses (within 100 ppm) were obtained for NU6M and NULM (when the sequences were translated with the mold mitochondrial code). The masses for four further mitochondrial encoded subunits differed by 3.8 to 36 Da from their predictions, and no mass was assigned for NU5M. Reliable molecular mass assignments were made for most supernumerary subunits, although the masses obtained for NUFM, NI2M, NIPM, ACPM1, NIDM, and NB2M were outside the accepted 100 ppm tolerance. Satisfactory explanations for the differences are lacking. They may result from errors or variations in the sequence data (as for NIDM and NB2M, see Table II), protein isoforms, or protein modifications. No mass data were obtained for ACPM2 (with or without a pantetheine-4′-phosphate modification, with or without an acyl group). Putative subunit ST1 remained undetected (therefore, it is not considered present in complex I from P. pastoris); however, a mass similar to that predicted for ACPM1 (with a pantetheine-4′-phosphate modification) was detected, and tandem MS analysis of peptides from the digest of a reverse-phase HPLC fraction detected two corresponding peptides (see Table II and Table S1). Finally, the molecular mass of a third unknown protein component corresponded to a P. pastoris protein named NUTM (the identification was subsequently supported by peptide mass fingerprinting and tandem MS data, see Table S1). In summary, the reverse-phase separation and associated molecular mass data led to the identification of four subunits (NULM, NU6M, NUTM, and ACPM2) which were not detected by analysis following SDS-PAGE, and accurate molecular masses for 24 subunits.
Post-Translational Modifications and Introns in the Nuclear-Encoded Subunits
Table II summarizes the post-translational modifications of the subunits of P. pastoris complex I. Ten subunits are known to be unmodified, either with (one) or without (nine) the initiator methionine. Fourteen subunits are known to be modified by the removal of N-terminal mitochondrial import sequences. Eight subunits are known to be N-α-acetylated (seven of them lack the initiator methionine, and two of them (NUZM and NB6M) are partially acetylated). In B. taurus complex I, 18 subunits have mitochondrial import sequences (6) and only one conserved subunit, B13, differs in this respect between P. pastoris and B. taurus. In B. taurus complex I, 13 subunits are N-α-acetylated (6), but the acetylation pattern is not strongly conserved, and only five subunits are N-α-acetylated in both species. Two mitochondrial-encoded subunits, NU6M and NULM, were confirmed to retain their N-α-formyl groups in P. pastoris: previous measurements on the B. taurus ND subunits showed that all seven of them retain the N-α-formyl group (19). Finally, all the N-terminal modifications reported in Table II are fully consistent with both peptide mass fingerprinting and tandem MS data, and for 12 subunits the molecular masses of the N-terminal peptides of the processed proteins were measured directly (see Table S1).
Three further post-translational modifications have been observed in B. taurus and N. crassa complexes I. In B. taurus, the histidine-rich N-terminal peptide of B12 (NB2M) is modified by the methylation of one, two, or three histidine residues (28). The B. taurus N-terminal peptide is not conserved in the shorter P. pastoris subunit (or in those of N. crassa or Y. lipolytica) and no evidence for histidine methylation was observed here (see Table II). In B. taurus, subunit B18 (NB8M) is N-α-myristoylated (29), but subunit NB8M is unmodified in both P. pastoris (see Table II) and Y. lipolytica (14) (the N terminus of NB8M is truncated in P. pastoris and Y. lipolytica, relative to B. taurus). A myristoylated lysine has been identified in NU5M of N. crassa complex I (30); it is not present in B. taurus (19), but cannot be either excluded or confirmed in P. pastoris because the mass of NU5M is not known. The SDAP (ACPM) subunits of B. taurus, N. crassa, and Y. lipolytica complex I are all modified by covalent linkages between a serine and a pantetheine-4′-phosphate group, and then by an acyl group. Here, the molecular mass data indicates that the ACPM1 subunit of P. pastoris complex I is modified by a pantetheine-4′-phosphate group also, but no acyl group has been detected.
The genomic sequences of six nuclear-encoded subunits of P. pastoris contain introns (see Supplementary Information). The introns are 62 to 140 bases long, and they all start with the common 5′ consensus hexamer, GTAAGT (31). In comparison, all the supernumerary subunits of Y. lipolytica complex I except NUEM, NESM, and ST1 have been reported to contain at least one intron (32), but, as for P. pastoris, none of the core hydrophilic subunits contain any. In contrast, all the nuclear-encoded subunits of N. crassa complex I have been reported to contain introns (11), revealing P. pastoris as a relatively simple model organism for the genetic manipulation of complex I.
The Mitochondrial-Encoded Subunits of Complex I in P. pastoris
Several lines of evidence show clearly that P. pastoris, like Y. lipolytica and N. crassa, uses the mold mitochondrial genetic code, rather than the yeast code. The molecular masses of NU3M, NU6M, and NULM correspond closely to masses predicted only by the mold code, and tryptic peptides were identified for NU2M only following translation by the mold code. In addition, a peptide from NU5M (see above) analyzed by tandem MS clearly matches a peptide sequence predicted by the mold code, but not by the yeast code.
The mitochondrial genomes of fungi often contain introns; they are difficult to predict, and may either encode endonucleases or other functional genes, or be noncoding (33). Introns have been identified in the ND subunit genes in N. crassa (one in each of NU1M, NULM, and two in NU5M (34, 35)) and Y. lipolytica (one in NU1M and two in NU5M (12)). None of the ND subunits from other fungal complexes I have been characterized sufficiently, but the genes encoding the ND subunits in Debaryomyces hansenii, Pichia Canadensis, and Candida albicans (which are closely related to P. pastoris) are not predicted to contain introns (36). The number of introns present in complete fungal mitochondrial genomes varies widely, from approximately 35 in Podospora anserina (37), to only two in P. canadensis (38). Therefore, there may be introns in the genes for the ND subunits of P. pastoris. The masses of NU6M and NULM correspond closely to the masses predicted by direct translation; therefore, these sequences do not contain introns. For NU1M, NU2M, NU3M, and NU4M, masses were observed that are close to the masses predicted by direct translation, and the translated proteins are homologous to proteins from other species, suggesting that there are no introns in these sequences. Two observed masses (59,086.1 (S.D. 5.0) and 62,012.5 (S.D. 7.3) Da) could be assigned to N-α-formylated NU2M, corresponding to alternative initiation codons (the first corresponds more closely to its calculated mass), and no mass from any putative sequence for NU5M was identified. Our data suggests it is unlikely that there are introns present in the genes for the NU1M, NU3M, NU4M, NU6M or NULM subunits in P. pastoris; however, it is possible that there are introns in the genes for NU2M and NU5M.
Three Putative New Subunits of Complex I Identified in P. pastoris
Our analyses provide strong evidence for two previously unknown subunits in complex I from P. pastoris. A molecular mass (Table II) together with high sequence coverage (60%) from peptide masses and tandem MS data for seven tryptic peptides provide unequivocal evidence for protein NUSM. The data for protein NUUM include a molecular mass and high confidence identification of three tryptic peptides by tandem MS (Table I). The presence of a third protein, NUTM, is less certain. A molecular mass corresponding to the sequence NUTM was obtained by ESI MS, although only a single tryptic fragment was identified with high confidence by tandem MS (Table S1).
The genomes of related organisms were searched for homologues to the three putative new subunits using tblastn (see Fig. 3). Homologues to NUSM and NUTM were identified in P. angusta and Dekkera bruxellensis, but not in Y. lipolytica or N. crassa. Homologues to NUUM were identified in Glomerella graminicola, Aspergillus fumigatus, N. crassa, Magnaporthe grisea, D. bruxellensis, and Y. lipolytica. No P. angusta homologue was identified, but the publically available genome sequence is incomplete. Re-analysis of peptide mass fingerprinting and tandem MS data for complex I from P. angusta (15) subsequently revealed the homologues to NUSM and NUTM. Significant Mascot scores were obtained for both proteins with peptide mass fingerprinting and tandem MS data obtained from SDS-PAGE bands consistent with the expected masses of 21 kDa and 10 kDa (Table S2). In addition, a molecular mass obtained from ESI analysis following reverse-phase fractionation of the P. angusta enzyme corresponds to the predicted sequence of NUTM (predicted 8890.3, measured 8891.1 Da). Re-analysis of the peptide mass finger printing and tandem MS data from complex I from Y. lipolytica revealed two peptides from the homologue to NUUM (see Table S2) from a protein with an apparent mass of approximately 8 kDa in SDS-PAGE. The identification of the three proteins in the purified complexes I from other species increases the likelihood that they are true subunits of the enzyme. None of the three proteins display significant sequence similarity to any previously known complex I subunits, and all three are predicted to contain a single transmembrane helix (using ConPred II (39)).
Fig. 3.
Sequence alignments of the three putative new complex I subunits from P. pastoris. Residues in bold are identical to the P. pastoris sequence. Underlined residues were not present in the mature P. pastoris proteins. NUSM: P. angusta partial sequence 12217898 and D. bruxellensis sequence 145383982. NUTM: P. angusta sequence 12218785 and D. bruxellensis sequence 145373365. NUUM: N. crassa sequence 85108140, Y. lipolytica sequence 49648093, D. bruxellensis 145381781.
The Subunit Composition of Complex I From P. pastoris and Comparison to Other Species
Forty-one proteins have been detected in complex I from P. pastoris, comprising the 14 conserved core subunits, and 27 supernumerary subunits, three of which have not been detected previously in any other species of complex I, and two of which, NUXM and NUZM, are specific to fungal complexes I (7). Forty-one proteins have also been detected in complex I from Y. lipolytica (including a NUTM homologue), and Y. lipolytica and P. pastoris complex I have 39 subunits in common; NUSM and NUTM have been detected in P. pastoris but not Y. lipolytica, and NUNM and ST1 in Y. lipolytica but not P. pastoris. Thirty-nine proteins have been detected in complex I from N. crassa (11), and N. crassa and P. pastoris complex I have 37 subunits in common. NUSM, NUTM, NUUM, and ACPM2 have been detected in P. pastoris but not N. crassa, and the 17.8/NURM and 10.4 kDa subunits in N. crassa but not P. pastoris. It is interesting that most of the variable subunits (NUSM, NUTM, NUUM, NUNM, NURM, and 10.4 kDa) contain a single transmembrane helix, as do four of the subunits found in B. taurus complex I but not in the three fungal species (KFYI, MLRQ, AGGG, and SGDH) suggesting that they may substitute for one another.
The sequences of the seven core hydrophilic subunits of P. pastoris complex I were compared with those of a number of other fungi using pair-wise alignment with ClustalW (40) (see Fig. S8). Of the species compared, P. pastoris complex I is most closely related to the enzymes from Pichia stipitis and D. hansenii, and least closely related to the enzyme from Ustilago maydis. In general, the comparison is consistent with the classification of P. pastoris in the family of Saccharomycetaceae (see Fig. S8).
The sequences of the subunits of P. pastoris complex I were compared with those from Paracoccus denitrificans, Y. lipolytica, and B. taurus (as examples of bacteria, fungi, and mammals) using pair-wise alignment. The percentage identities and similarities, insertions or extensions, and deletions or truncations, are presented in Table S3. As expected, P. pastoris complex I is most closely related to the enzyme from Y. lipolytica. P. denitrificans complex I contains significantly fewer subunits, and they are generally less similar to the P. pastoris proteins than the B. taurus subunits are. The core hydrophilic subunits are best conserved between species with 47-87% similarity (32-75% identity). The core hydrophobic subunits are notably less conserved, with 40-77% similarity (17-49% identity); subunits NU2M and NU6M are the least conserved, and there are significant differences in length in several cases. In particular, P. pastoris NU2M is significantly longer than the B. taurus homologue. Among P. pastoris and B. taurus or Y. lipolytica, the supernumerary subunits are generally less conserved than the core subunits, with 27-82% similarity (13-63% identity). The best conserved supernumerary subunits (>50% similarity for B. taurus and >60% for Y. lipolytica) are NUEM (39 kDa), NB6M (B16.6), NB4M (B14), and NI8M (B8); the least conserved (<35% similarity for B. taurus and < 50% for Y. lipolytica) are NUJM (B14.7), NIMM (MWFE), NIPM (15 kDa), and NB5M (B15). There are significant differences in the sizes of a number of subunits between P. pastoris and B. taurus (notably NESM (ESSS), NIMM (MWFE), NI2M (B22), NIDM (PDSW), and NB5M (B15)). NIMM is particularly interesting because the P. pastoris protein is significantly longer than the Y. lipolytica and B. taurus proteins because of an internal insertion of 22 amino acids, and C-terminal extensions of 34 or 53 amino acid residues, respectively.
Supplementary Material
Footnotes
 This article contains supplemental material.
This article contains supplemental material.
REFERENCES
- 1.DiMauro S., Schon E. A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348, 2656–2668 [DOI] [PubMed] [Google Scholar]
- 2.Guénebaut V., Schlitt A., Weiss H., Leonard K., Friedrich T. (1998) Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 276, 105–112 [DOI] [PubMed] [Google Scholar]
- 3.Walker J. E. (1992) The NADH-ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25, 253–324 [DOI] [PubMed] [Google Scholar]
- 4.Hirst J. (2010) Towards the molecular mechanism of respiratory complex I. Biochem. J. 425, 327–339 [DOI] [PubMed] [Google Scholar]
- 5.Sazanov L. A., Hinchliffe P. (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 6.Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E. (2003) The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604, 135–150 [DOI] [PubMed] [Google Scholar]
- 7.Gabaldón T., Rainey D., Huynen M. A. (2005) Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348, 857–870 [DOI] [PubMed] [Google Scholar]
- 8.Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 9.Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. (2003) Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2, 117–126 [DOI] [PubMed] [Google Scholar]
- 10.Murray J., Zhang B., Taylor S. W., Oglesbee D., Fahy E., Marusich M. F., Ghosh S. S., Capaldi R. A. (2003) The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunoprecipitation. J. Biol. Chem. 278, 13619–13622 [DOI] [PubMed] [Google Scholar]
- 11.Marques I., Duarte M., Assunção J., Ushakova A. V., Videira A. (2005) Composition of complex I from Neurospora crassa and disruption of two “accessory” subunits. Biochim. Biophys. Acta. 1707, 211–220 [DOI] [PubMed] [Google Scholar]
- 12.Kerscher S., Durstewitz G., Casaregola S., Gaillardin C., Brandt U. (2001) The complete mitochondrial genome of Yarrowia lipolytica. Comp. Funct. Genom. 2, 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdrakhmanova A., Zickermann V., Bostina M., Radermacher M., Schägger H., Kerscher S., Brandt U. (2004) Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1658, 148–156 [DOI] [PubMed] [Google Scholar]
- 14.Morgner N., Zickermann V., Kerscher S., Wittig I., Abdrakhmanova A., Barth H.-D., Brutschy B., Brandt U. (2008) Subunit mass fingerprinting of mitochondrial complex I. Biochim. Biophys. Acta 1777, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 15.Bridges H. R., Grgic L., Harbour M. E., Hirst J. (2009) The respiratory complexes I from the mitochondria of two Pichia species. Biochem. J. 422, 151–159 [DOI] [PubMed] [Google Scholar]
- 16.Kashani-Poor N., Kerscher S., Zickermann V., Brandt U. (2001) Efficient large scale purification of his-tagged proton translocating NADH:ubiquinone oxidoreductase (complex I) from the strictly aerobic yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1504, 363–370 [DOI] [PubMed] [Google Scholar]
- 17.Dröse S., Galkin A., Brandt U. (2005) Proton pumping by complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica reconstituted into proteoliposomes. Biochim. Biophys. Acta 1710, 87–95 [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 19.Carroll J., Fearnley I. M., Wang Q., Walker J. E. (2009) Measurement of the molecular masses of hydrophilic and hydrophobic subunits of ATP synthase and complex I in a single experiment. Anal. Biochem. 395, 249–255 [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 21.Perkins D. N., Pappin D. J. C., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 22.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayers E. W., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., DiCuccio M., Edgar R., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Miller V., Mizrachi Il, Ostell J., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Yaschenko E., Ye J. (2009) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. (1980) Codon recognition rules in yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 77, 3167–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talla E., Anthouard V., Bouchier C., Frangeul L., Dujon B. (2005) The complete mitochondrial genome of the yeast Kluyveromyces thermotolerans. FEBS Lett. 579, 30–40 [DOI] [PubMed] [Google Scholar]
- 27.Browning K. S., RajBhandary U. L. (1982) Cytochrome oxidase subunit III gene in Neurospora crassa mitochondria. J. Biol. Chem. 257, 5253–5256 [PubMed] [Google Scholar]
- 28.Carroll J., Fearnley I. M., Skehel J. M., Runswick M. J., Shannon R. J., Hirst J., Walker J. E. (2005) The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 4, 693–699 [DOI] [PubMed] [Google Scholar]
- 29.Walker J. E., Arizmendi J. M., Dupuis A., Fearnley I. M., Finel M., Medd S. M., Pilkington S. J., Runswick M. J., Skehel J. M. (1992) Sequences of 20 subunits of NADH:ubiquinone oxidoreductase from bovine heart mitochondria. J. Mol. Biol. 226, 1051–1072 [DOI] [PubMed] [Google Scholar]
- 30.Plesofsky N., Gardner N., Videira A., Brambl R. (2000) NADH dehydrogenase in Neurospora crassa contains myristic acid covalently linked to the ND5 subunit peptide. Biochim. Biophys. Acta 1495, 223–230 [DOI] [PubMed] [Google Scholar]
- 31.Irimia M., Roy S. W. (2008) Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 4, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman D. J., Martin T., Nikolski M., Cayla C., Souciet J. L., Durrens P. (2009) Génolevures: protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 37, D550–D554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang B. F., Laforest M.-J., Burger G. (2007) Mitochondrial introns: a critical view. Trends Genet. 23, 119–125 [DOI] [PubMed] [Google Scholar]
- 34.Burger G., Werner S. (1985) The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J. Mol. Biol. 186, 231–242 [DOI] [PubMed] [Google Scholar]
- 35.Nelson M. A., Macino G. (1987) Three class I introns in the ND4L/ND5 transcriptional unit of Neurospora crassa mitochondria. Mol. Gen. Genet. 206, 318–325 [DOI] [PubMed] [Google Scholar]
- 36.Santamaria M., Vicario S., Pappadà G., Scioscia G., Scazzocchio C., Saccone C. (2009) Towards barcode markers in fungi: an intron map of Ascomycota mitochondria. BMC Bioinformatics 10, S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummings D. J., McNally K. L., Domenico J. M., Matsuura E. T. (1990) The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr. Genet. 17, 375–402 [DOI] [PubMed] [Google Scholar]
- 38.Sekito T., Okamoto K., Kitano H., Yoshida K. (1995) The complete mitochondrial DNA sequence of Hansenula wingei reveals new characteristics of yeast mitochondria. Curr. Genet. 28, 39–53 [DOI] [PubMed] [Google Scholar]
- 39.Arai M., Mitsuke H., Ikeda M., Xia J.-X., Kikuchi T., Satake M., Shimizu T. (2004) ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32, W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.