Abstract
Karyotypic analysis and genomic copy number analysis with single nucleotide polymorphism (SNP)-based microarrays were compared with regard to the detection of recurrent genomic imbalances in 20 clear cell renal cell carcinomas (ccRCCs). Genomic imbalances were identified in 19 of 20 tumors by DNA copy number analysis and in 15 tumors by classical cytogenetics. A statistically significant correlation was observed between the number of genomic imbalances and tumor stage. The most common genomic imbalances were loss of 3p and gain of 5q. Other recurrent genomic imbalances seen in at least 15% of tumors included losses of 1p32.3-p33, 6q23.1-qter and 14q and gain of chromosome 7. The SNP-based arrays revealed losses of 3p in 16 of 20 tumors, with the highest frequency being at 3p21.31-p22.1 and 3p24.3-p25.3, the latter encompassing the VHL locus. One other tumor showed uniparental disomy of chromosome 3. Thus, altogether loss of 3p was identified in 17 of 20 (85%) cases. Fourteen tumors showed both overlapping losses of 3p and overlapping gains of 5q, and the karyotypic assessment performed in parallel revealed that these imbalances arose via unbalanced 3;5 translocations. Among the latter, there were common regions of loss at 3p21.3-pter and gain at 5q34-qter. These data suggest that DNA copy number analysis will supplant karyotypic analysis of tumor types such as ccRCC that are characterized by recurrent genomic imbalances, rather than balanced rearrangements. These findings also suggest that the 5q duplication/3p deficiency resulting from unbalanced 3;5 translocations conveys a proliferative advantage of particular importance in ccRCC tumorigenesis.
INTRODUCTION
Epithelial tumors of the kidney represent approximately 3% of all adult malignancies, with an incidence approximating that of all forms of leukemia combined (Uzzo et al., 2003). Histopathologically, clear cell renal cell carcinoma (ccRCC) is the most common kidney neoplasm, accounting for 80% of sporadic RCC. Classical chromosome banding analysis of ccRCC has documented frequent loss of 3p due to deletion or unbalanced translocation, often with chromosome 5. The 3;5 translocations result in duplication of a portion of distal 5q. Loss of chromosome 14q, especially 14q24-q31 and 14q31-q32, represents another recurrent cytogenetic alteration in ccRCC, and a positive correlation has been observed between allelic loss of 14q and a poor prognosis (Gunawan et al., 2001; Kardas et al., 2005; Klatte et al., 2009).
The von Hippel-Lindau gene, VHL, located at 3p25.3, is considered to be a major target of 3p loss in sporadic ccRCC, with intragenic VHL mutations being observed in 50%–60% and allelic loss in 80%–90% of cases (Gnarra et al., 1994; Kondo et al., 2002). Whether duplication of 5q targets a specific gene in ccRCC cases with an unbalanced translocation between chromosomes 3 and 5 is not known. However, patients with 3p loss and gain at distal 5q, due to at (3;5), have been reported to have a significantly better disease-specific prognosis (Nagao et al., 2005).
The recent availability of array-based platforms designed to detect recurring copy number abnormalities (CNAs) at a high-resolution, genome-wide level has facilitated the discovery of tumor suppressor genes and oncogenes involved in various cancers (Greshock et al., 2007). Genomic copy analysis has been proposed as a beneficial tool that is becoming an important part of the clinical cytogenetics laboratory setting, although studies documenting parallel routine karyotypic and microarray-based comparisons are limited. In this investigation, we compared classical cytogenetics to single nucleotide polymorphism (SNP)-based microarrays with regard to detection of recurrent sites of genomic imbalance in a series of 20 sporadic ccRCC samples. The high-density arrays revealed imbalances in three cases that had a normal karyotype and in one case that yielded no metaphases. The alterations identified by these two procedures were similar in other cases, although breakpoints were better defined with arrays, as expected. Among cases with an unbalanced t(3;5), there was a common region of loss at 3p21.3-pter and a common region of gain at 5q34-qter.
MATERIALS AND METHODS
Patients and Tumor Specimens
The specimens were surgically resected primary ccRCCs, except case 10, which was a recurrent tumor. Samples were obtained directly after surgery following institutional review board guidelines, with written consent obtained from all patients as part of our IRB-approved institutional Kidney Cancer Database. To date, all patients are alive. Tumor stage and grade of each case are summarized in Table 1. All cases were diagnosed by an oncologic pathologist with a special interest in urologic pathology (TAS). Fresh specimen was immediately cut in half, with one piece used for cytogenetics and the other used for microarray analysis.
Table 1.
Comparison of Chromosomal Findings Detected by Karyotyping Versus Genomic Copy Number Analysis
| Case No. | Histology | Stage | Grade | Karyotype | Genomic Copy Analysis |
|---|---|---|---|---|---|
| 1a | ccRCC | I | III | 46,XY[20] | Normal |
| 2 | ccRCC | II | II | 35–45,XY,der(3)t(3;5)(p21;q31),−14[cp13] /41~45,sl,+7[cp2]/46,XY[4] | −3p12-pter, +5q21, +5q23.1-qter, +7, −14 |
| 3 | ccRCC | I | II | 46,XY[20] | −8 |
| 4 | ccRCC | I | III | 46,XY,der(3)t(3;5)(p21;q31)[17]/46,XY[3] | −3p13-pter, +5q23.1-qter |
| 5 | ccRCC | I | III | 46–53,XX,+2,−3,add(11)(q23),add(14)(q24)[cp7] | +2p, +2q11-q31, −3p, −3q11-q23 |
| 6 | ccRCC | II | III | 46,XX[2] | −1p31.3-p36.3, −3p, −4, +5q14-qter, −8, −14, −18 |
| 7 | ccRCC | I | III | 45,XY,der(2)t(2;3)(q33;q12),−3[16]/46,XY[4] | −2q32-qter, −3p11-pter |
| 8 | ccRCC (sarcomatoid transformation) | III | IV | 46,XY[19] | −Y, −3p, −5q11-q21, −6, −10q, +16p, −16q, upd(5q) |
| 9 | ccRCC (recurrent to adrenal gland) | R | III | 47,XX,der(3)t(3;5)(p13;q22),+del(14)(q22),+21[17] /46~47,sl,−14[3] | −1p31.3, −3p12-pter, +5q21-qter, +21 |
| 10 | ccRCC | I | I | 46,XX,del(3)(p13 or p14)[6]/45,XX,der(3)del(3)(p13 or p14)inv(3)(q11.2q21),−14[10] /42~51,XX,+7[cp3]/46,XX[1] | −3p14-pter, +5q34-qter |
| 11 | ccRCC (multicystic with papillary features) | I | II | 47,XY,+18[18]/46,XY[4] | +18 |
| 12 | ccRCC | IV | IV | No dividing cells observed. | −1p34.2-pter, +1p31.1-p34.2, +1q, −3p12-pter, +3p12-qter, +5q, +7, −8p, +8q, −14 |
| 13 | ccRCC | II | III | 40~43,XX,der(3)t(3;5)(p14;q13),del(6)(q23),−8,−9, −14,del(16)(q22),−17[cp15]/46,XX[4] | −3p13-pter, +5q21-qter, −6q22.3-qter, −9, −14, −17p |
| 14 | ccRCC | I | III | 45,XY,add(3)(q27),der(3)t(3;5)(p13;q22),−14[cp15] /44,sl,−Y[cp5] | −Y, −3p14.1-pter, +5q23.1-qter |
| 15 | ccRCC | I | II | 31,XX,del(3)(p21p25),der(3)t(3;5)(p13;q13),inc[1] /76~82<4n>,slx2,+2,+2,inc[cp3]/46,XX[4] | +2, −3p21-pter, −3q22, +5q13-qter, +7 |
| 16 | ccRCC | I | II | 46,X,−Y,t(3;5)(p13;p13),+5,+7,−14[13]/46,XY[7] | −Y, +5, +7, −14, upd(3pq) |
| 17 | ccRCC | I | II | 46,XY,der(3)t(3;5)(p13;q15)[21] | −3p12-pter, +5q14-qter |
| 18 | ccRCC | I | II | 46,XX,der(3)t(3;5)(p13;q31),add(4)(q35)[22] | −3p12-pter, +4q26-qter, −5q21, +5q31.1-qter |
| 19 | ccRCC | I | III | 43~45,XX,der(3)t(3;5)(p13;q22),−14[cp13] /46,XX[4] | −3p14.3-pter, +5q23.1-qter, −14 |
| 20 | ccRCC | II | I | 46,XX,der(3)t(3;5)(p13;q31)[1]/47,sl,+X[11] /92,slx2[1]/46,XX[7] | +X, −3p12-pter, +5q23.3-qter, upd(4q) |
The pathology was re-examined, and the diagnosis of ccRCC was confirmed; however, the histopathological reassessment revealed that only 30% of the specimen contained tumor cells.
Karyotypic Analysis
Each cytogenetic sample was minced mechanically using two scalpels and then treated with 0.2% collagenase for 30–60 min at 37°C. The samples were cultured for two to fourteen days in RPMI 1640 medium containing 15% fetal bovine serum. Metaphase slide preparations and G- banding were performed according to our standard method (Miura et al., 1990).
Genomic Copy Number Analysis
Tumor tissue used for genomic copy number analysis was macrodissected by a pathologist (AL) to remove obvious necrotic areas, stroma, and adjacent normal tissue. For each tumor, a Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA), containing more than 906,600 single nucleotide polymorphism (SNP) probes and more than 946,000 probes for detection of copy number variation, was used as the hybridization target. The array probe datasheet can be found at the link shown below: http://www.affymetrix.com/support/technical/datasheets/genomewide_snp6_datasheet.pdf.
Briefly, total genomic DNA (500 ng) from each test sample was digested with NspI and StyI restriction enzymes and ligated to adapters that recognize cohesive four-basepair (bp) overhangs. A generic primer that recognizes the adapter sequence was used to amplify the adapter-ligated DNA fragments. PCR conditions were optimized to preferentially amplify fragments in the 200–1,100-bp size range. PCR amplification products for each restriction enzyme digest were combined and purified using Agencourt SNPClean magnetic beads (Beckman Coulter, MA). The amplified DNA was fragmented, biotin-labeled, and hybridized to a Genome-Wide Human SNP Array 6.0 according to the manufacturer’s recommendations. The intensities of probe hybridization were analyzed using Affymetrix software GCOS 1.4, and genotyping and copy number analysis were performed using Affymetrix Genotyping Console 3.0.2 with default setting, which compares all hybridization intensities against a data set consisting of 270 HapMap normal reference samples. All tiny focal copy number peaks observed were checked against the Database of Genomic Variants (DGV), Hospital for Sick Children, Toronto, and each of these focal peaks was found to be a known or suspected copy number variant (CNV). In addition, the DNA copy number results for our 20 ccRCC samples were interpreted by comparison to results for three benign renal tumors, none of which showed any genomic imbalances.
RESULTS
Karyotypic Findings
Cytogenetic analysis was performed on 20 tumor samples from patients with ccRCC, and the findings are summarized in Table 1. Abnormal karyotypes were found in 15 cases. Normal karyotypes were observed in four samples, presumably due to the outgrowth of normal stroma. No metaphases were found in the remaining sample.
The most frequent cytogenetic abnormality was loss of material from the short arm of chromosome 3. Such losses were documented in 13 of 15 (87%) cytogenetically abnormal samples analyzed. In 10 samples, the loss of 3p and gain of 5q were due to an unbalanced translocation that resulted in a derivative chromosome 3 consisting of a portion of 5q translocated to 3p. Breakpoints in 3p ranged from 3p13 to 3p21. Breakpoints in 5q ranged from 5q13 to 5q31. It is noteworthy that one additional tumor (case 16, see Table 1) had a balanced translocation, t(3;5)(p13;p13), suggesting a propensity for rearrangements between chromosomes 3 and 5 in ccRCC, perhaps due to their proximity in the nucleus in kidney cancer precursor cells. Three samples showed either monosomy 3, deletion of 3p, or loss of the entire 3p due to an unbalanced translocation between 3q and 2q. Less common recurrent abnormalities included monosomy 14 (6 of 15 cases, 40%), trisomy 7 (3 cases), and loss of the Y chromosome (2 cases).
Genomic Copy Number Analysis
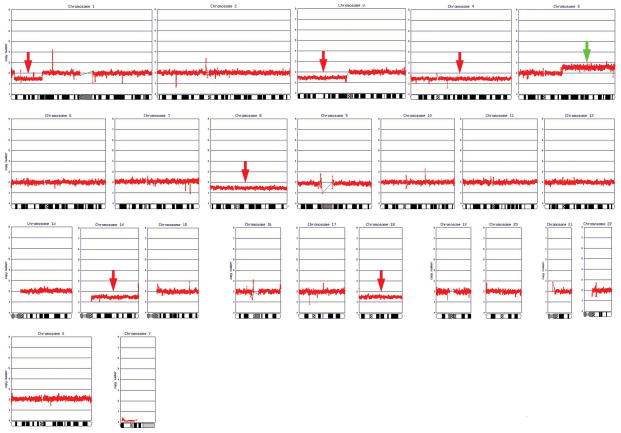
DNA copy number analysis was performed on the same set of 20 ccRCC samples that were examined cytogenetically (Table 1). Nineteen of the 20 tumors (95%) exhibited genomic imbalances, including one case in which no metaphases were found and three cases that had a normal karyotype, likely due to overgrowth of fibroblasts or other contaminating stromal cells in the cell cultures. Genomic alterations were not found in one tumor (case 1) either by karyotypic or genomic copy number analysis. The pathology of this case was re-examined, and the diagnosis of ccRCC was confirmed. However, the histopathological reassessment revealed that only 30% of the specimen represented tumor cells. Thus, the high proportion of stromal cells may have masked the presence of any genomic imbalances in this case. Alternatively, it is possible that a small minority of ccRCCs do not harbor gross genomic alterations. Fig. 1 depicts a genomic copy number analysis profile of a representative ccRCC. A schematic summary of CNAs observed in all tumors is depicted in Fig. 2.
Figure 1.
Genomic copy number analysis profile of a representative primary ccRCC specimen (case 6).
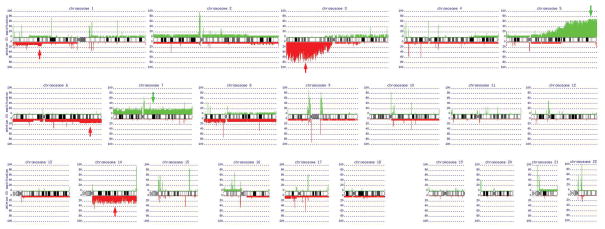
Figure 2.
Schematic summary of copy number abnormalities identified in all 20 ccRCC specimens. In addition to losses of 3p and gains of 5q, other recurrent genomic imbalances seen in at least 15% of tumors include losses of 1p32.3-p33, 6q23.1-qter and 14q and gain of chromosome 7.
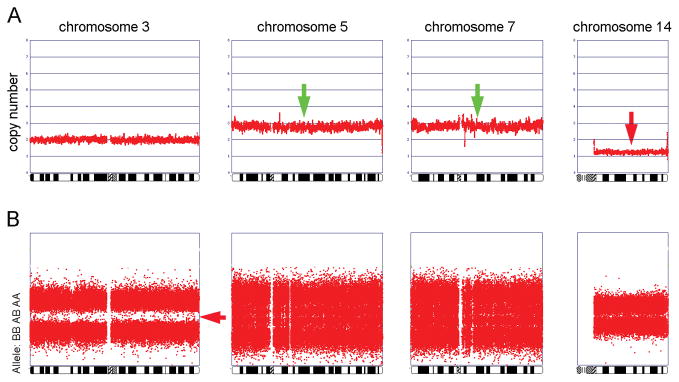
Sixteen of the 20 samples showed losses of 3p, with the highest frequency of loss being at 3p21.31-p22.1 and 3p24.3-p25.3, the latter encompassing the VHL locus. One additional tumor (case 16) did not show genomic loss of 3p based on the DNA copy number profile, but the allelic profile showed LOH for the entire chromosome 3. In this case, there were no heterozygous (AB) genotype calls; instead, all genotype calls appeared to be either AA or BB (Fig. 3). Collectively, this suggests that one entire chromosome 3 is lost and the second copy is duplicated, indicative of uniparental disomy of chromosome 3, upd(3pq). Thus, altogether loss of 3p was identified in 17 of our 20 (85%) ccRCCs.
Figure 3.
Genomic copy number profiles of chromosomes 3, 5, 7, and 14 (panel A) and allelic profile of chromosome 3 (panel B) from case 16. Note that although the copy number profile of chromosome 3 is normal, the allelic profile shows LOH for the entire chromosome, based on the lack of AB genotype calls.
Fourteen tumors showed both overlapping losses of 3p and overlapping gains of 5q. In nearly all of these cases, the karyotypic assessment performed in parallel with the array analysis revealed that an unbalanced t(3;5) was present. Case 10 was an exception in that the genomic copy number analysis revealed loss of 3p14-pter and gain of 5q34-qter, whereas the karyotype showed a 3p deletion but not the gain of distal 5q. Re-review of the karyotype did not uncover a gain of distal 5q, although the quality of the metaphase spreads was insufficient to rule out the possibility of a cryptic translocation of extra 5q material. Therefore, the cell population dividing in culture may not have been representative of the tumor as a whole; alternatively, this tumor may have been polyclonal in origin.
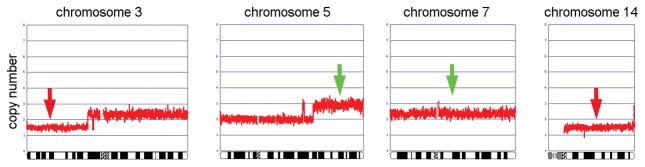
In case 2, the DNA copy number profile revealed distinctive losses of 3p and 14 and gain of 5q, whereas the evidence for a gain of chromosome 7 was less striking (Fig. 4). These findings are consistent with the karyotype, which showed 3p-, 5q+, and −14 in 13 cytogenetically abnormal metaphase spreads, whereas +7 was seen in only 2 metaphases. Thus, gain of chromosome 7 appears to represent a late change in the karyotypic evolution of this tumor.
Figure 4.
Genomic copy number profiles of chromosomes 3, 5, 7, and 14 from case 2. Note distinct losses of 3p and 14 and gain of 5q, whereas the gain of chromosome 7 is less striking, suggesting that this abnormality was present in a subset of the tumor cells and, thus, might be a late change in karyotypic evolution.
Among the cases with both 3p loss and 5q gain, there was a commonly deleted region at 3p21.3-pter and a common region of copy number gain at 5q34-qter. A diagram depicting genomic profiles of cases exhibiting both 3p copy number loss and 5q gain is presented in Fig. 5. Other recurrent genomic imbalances seen in at least 15% of tumors included losses of 1p32.3-p33, 6q23.1-qter and 14q and gain of chromosome 7. In addition to these larger segmental and whole chromosome CNAs, numerous tiny focal gains or losses were observed, many of which were near centromeric regions. All but one of these focal changes could be attributed to a CNV, based on a search of the Database of Genomic Variants. The lone exception was a loss observed at 9p11.1, which was found to reside at a gene “desert”, i.e., a region with no known genes, and could represent a rare CNV.
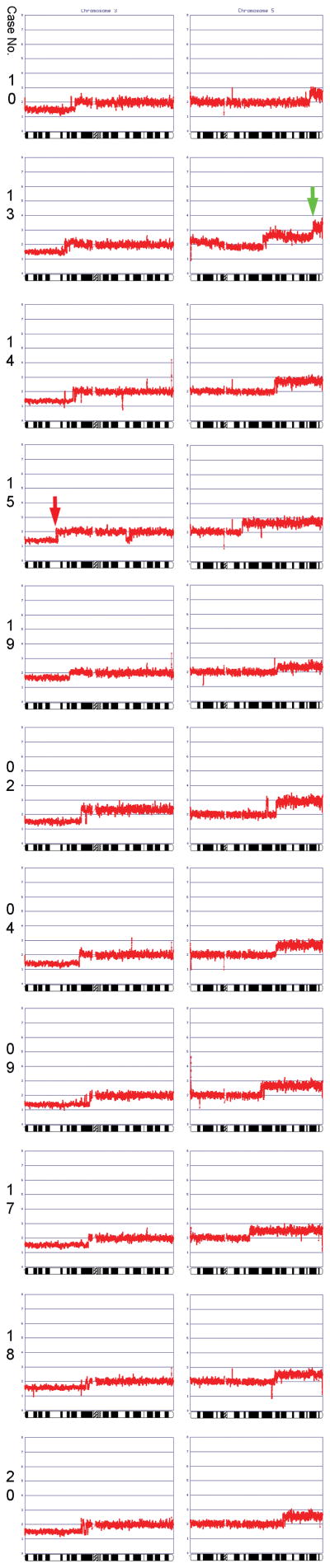
Figure 5.
Summary of all losses of 3p and all gains of 5q observed in 11 ccRCCs.
Although this series was small, the large (stage II, >7 cm) and advanced (stages III/IV and recurrent) tumors together had significantly more genomic alterations (range 4–10, median 6) than the stage I tumors (range 1–5, median 2.5; p-value = 0.006, Wilcoxin test). No statistically significant correlation was observed between the number of genomic imbalances and tumor grade. It is notable, however, that the two tumors with the highest grade (IV) and stage (III or IV), i.e., cases 9 and 12, had either 8 or 10 imbalances, the highest numbers observed in this series. Collectively, these data suggest that by the time a ccRCC becomes larger than 7 cm and/or metastasizes, it acquires more genomic imbalances that may contribute to a more aggressive biology. No specific chromosomal imbalance was found to correlate with stage or grade. Most patients in the series were recently diagnosed and thus could not be analyzed for possible correlations between the genomic findings and survival.
Comparison of Karyotypic and Genomic Copy Number Findings
Among the 15 cases that exhibited chromosomal alterations by both karyotyping and array analysis, the imbalances observed were generally similar with the two detection methods, although the karyotypic interpretation of chromosomal breakpoints often was imprecise compared to the genomic copy number analysis. In two ccRCCs, a single abnormality, −8 in one case and +7 in another, was identified by genomic copy number analysis but not by karyotyping. On the other hand, in five tumors, one or two abnormalities (3p- in one case; −14 in two cases; −8 and 16q- in one case; and rearrangements of 11q and 14q in one case) were detected by karyotyping but not by genomic copy number analysis. In case 9, a 1p- was detected in the genomic copy number analysis only, whereas a 14q- was observed only in the cytogenetic analysis. Similarly, in case 10, a gain of 5q was identified in the microarray analysis only, whereas monosomy 14 was observed only in the karyotypic study. Among the tumors with differential findings based on the two methods used, it is noteworthy that in four tumors (cases 5, 9, 10, and 14) loss of all or part of chromosome 14 was consistently observed in the karyotypic analysis and not in the copy number array analysis. The lone exception was case 6, which had a normal karyotype but had loss of chromosome 14 and six other genomic imbalances. Whether the preferential detection of chromosome 14 losses in the cytogenetic analysis is simply a reflection of the modest size of our series or an indication that a subpopulation of cells with chromosome 14 loss has a proliferative advantage in cell culture is unclear.
DISCUSSION
The cytogenetic findings presented here are consistent with those of several other investigators (Gunawan et al., 2001; Kardas et al., 2005; Klatte et al., 2009). In particular, loss of 3p was the most prevalent abnormality in all three of these studies of ccRCC, with the incidence ranging from 60% to 98%, compared to 87% of the cytogenetically abnormal samples analyzed in our series. As in our study, gain of part of 5q was common, usually due to an unbalanced t(3;5), previously reported to be preferentially involved in rearrangements between chromosome 3p and other chromosomes in RCC (Kovacs et al., 1987). As in our study, Kardas et al. (2005) and Klatte et al. (2009) identified +7, −14, and loss of Y as recurrent abnormalities, although the incidence of −Y was much higher in those studies (48%–55%) than in our series (20%). Our findings also indicate that genomic copy number analysis is a sensitive tool for the detection of chromosomal imbalances in ccRCC and potentially other cancers and hematological malignancies in which copy number alterations, as opposed to balanced rearrangements, have clinical utility. Moreover, as expected, breakpoints could be identified more precisely by the genomic copy number analysis.
Ten of our tumors showed both overlapping losses of 3p and overlapping gains of 5q; in each case, the karyotypic studies revealed an unbalanced t(3;5). Beroukhim and colleagues performed an integrated, genome-wide analysis of copy number changes and gene expression profiles in 90 ccRCCs, including both sporadic and VHL disease-associated tumors (Beroukhim et al., 2009). They identified seven regions of nonrandom copy number gains at chromosomes 1q, 2q, 5q, 7q, 8q, 12p, and 20q and seven regions of deletion (1p, 3p, 4q, 6q, 8p, 9p, and 14q). Overall, 3p loss and 5q gain were statistically the most significant events in ccRCC in this series, with segments 3p25.3 and 5q35.3 identified as the critical regions involved [10]. Our parallel cytogenetic and genomic copy number studies indicate that the finding of 3p loss and 5q gain in the same tumor are typically due to unbalanced translocations between 3p and 5q. Several smaller series of ccRCCs have also been reported, using lower density arrays (Cifola et al., 2008; Matsuda et al., 2008; Toma et al., 2008; Yoshimoto et al., 2007). As in the reports summarized above, these studies also showed a high frequency of 3p loss and 5q gain. While there were considerable variations in the breakpoints identified in 3p and 5q, all showed overlapping losses of 3p and gains of distal 5q. Our study indicates that the smallest region of overlapping loss is at 3p21.3-pter and the common region of gain is at 5q34-qter.
Copy number loss on 3p in ccRCC is associated with mutations of the VHL gene (3p25.3), known to be involved in renal carcinogenesis. Copy number gain of 5q may contribute to tumor progression, as suggested by other studies (Nagao et al., 2005; Toma et al., 2008), although a putative RCC-related oncogene in this region remains unknown. Integration of SNP findings in the distal 5q region with expression data obtained from the public domain to search for genes with elevated copy number as well as high RNA expression revealed a candidate gene, TGFBI (transforming growth factor, beta-induced gene), located at 5q31 (Matsuda et al., 2008). TGFBI encodes a secreted matrix protein with apoptotic and adhesion-related growth activities. TGFBI is up-regulated in various cancers and was proposed to act as an oncogene in clear cell RCC (Matsuda et al., 2008). It is also noteworthy that the colony-stimulating factor 1 receptor gene, CSF1R, is located at 5q33. CSF1R has been implicated as an oncogene and has been shown to be up-regulated in nearly 70% of ccRCCs (Toma et al., 2008). However, the smallest region of overlapping duplications observed in our series of der(3)t(3;5) cases resides at 5q34-qter, i.e., distal to the sites of the TGFBI and CSF1R genes. Thus, whether these or some other gene in the distal 5q region is a ccRCC oncogene will require further study.
Spatial contiguity of chromosomal loci within the interphase nucleus is thought to provide an explanation for the existence of certain nonrandom gene fusions. For example, in prostate cancer, approximately 50% of human prostate cancers exhibit a gene fusion involving TMPRSS2, an androgen-regulated gene, and ERG, which encodes an ETS transcription factor. Interestingly, fluorescence in situ hybridization studies have revealed that androgen signaling induces proximity of the TMPRSS2 and ERG genomic loci specifically in human prostate cancer cells, and subsequent exposure of these cells to gamma irradiation induced TMPRSS2-ERG rearrangements (Mani et al., 2009). This may explain why TMPRSS2-ERG fusions are restricted to the prostate, which is dependent on androgen signaling. Rearrangements between chromosomes 3 and 5 appear to be unusually common in ccRCC, suggesting spatial contiguity of these chromosomes in renal epithelial cells. Instead of a fusion gene, however, the unbalanced 3;5 translocations seen in ccRCC consistently result in loss of 3p, encompassing the VHL locus, and gain of distal 5q, changes that appear to provide a proliferative advantage specific for ccRCC tumorigenesis.
Collectively, our findings validate that genomic copy number analysis is a useful detection tool that can supplant or, at least complement, karyotypic analysis of tumors such as ccRCC that are characterized by genomic imbalances rather than balanced chromosomal rearrangements. In tumor types characterized by recurrent chromosomal losses and/or gains, genomic copy number analysis can provide a higher rate of detection of clinically relevant alterations, with greater precision, than does classical cytogenetics. This is noteworthy, because our findings indicate that the number of CNAs increases in large and more advanced tumors. Our study also indicates that 3p loss is a hallmark of ccRCC, and that this alteration is frequently connected with 5q gain due to an unbalanced translocation with variable breakpoints. Whether the nonrandom involvement of chromosomes 3 and 5 arises due to their proximity within the interphase nucleus of a target kidney cell and/or represents a selective growth advantage due to duplication of a 5q gene that cooperates with VHL loss/inactivation is worthy of future investigation.
Acknowledgments
The following Fox Chase Cancer Center shared core services were used in the course of this work: Genomics and Tumor Bank Facilities.
Supported by: NIH, NCI grant P30 CA006927. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Additional funds were provided by Fox Chase Cancer Center via institutional support of the Kidney Cancer Keystone Program.
References
- Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WGJ, Signoretti S. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifola I, Spinelli R, Beltrame L, Peano C, Fasoli E, Ferrero S, Bosari S, Signorini S, Rocco F, Perego R, Proserpio V, Raimondo F, Mocarelli P, Battaglia C. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, Lubensky I, Duan DR, Florence C, Pozzatti R, Walther MM, Bander NH, Grossman HB, Brauch H, Pomer S, Brooks JD, Isaacs WB, Lerman MI, Zbar B, Linehan WM. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Greshock J, Feng B, Nogueira C, Ivanova E, Perna I, Nathanson K, Protopopov A, Weber BL, Chin L. A comparison of DNA copy number profiling platforms. Cancer Res. 2007;67:10173–10180. doi: 10.1158/0008-5472.CAN-07-2102. [DOI] [PubMed] [Google Scholar]
- Gunawan B, Huber W, Holrup M, von Heydebreck A, Efferth T, Poustka A, Ringert RH, Jakse G, Fuzesi L. Prognostic impacts of cytogenetic findings in clear cell renal cell carcinoma: gain of 5q31-qter predicts a distinct clinical phenotype with favorable prognosis. Cancer Res. 2001;61:7731–7738. [PubMed] [Google Scholar]
- Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, Lerman MI. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardas I, Mrozek K, Babinska M, Krajka K, Hadaczek P, Lubinski J, Roszkiewicz A, Kuziemska E, Limon J. Cytogenetic and molecular findings in 75 clear cell renal cell carcinomas. Oncol Rep. 2005;13:949–956. [PubMed] [Google Scholar]
- Klatte T, Rao PN, de Martino M, LaRochelle J, Shuch B, Zomorodian N, Said J, Kabbinavar FF, Belldegrun AS, Pantuck AJ. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol. 2009;27:746–753. doi: 10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yao M, Yoshida M, Kishida T, Shuin T, Miura T, Moriyama M, Kobayashi K, Sakai N, Kaneko S, Kawakami S, Baba M, Nakaigawa N, Nagashima Y, Nakatani Y, Hosaka M. Comprehensive mutational analysis of the VHL gene in sporadic renal cell carcinoma: relationship to clinicopathological parameters. Genes Chromosomes Cancer. 2002;343:58–68. doi: 10.1002/gcc.10054. [DOI] [PubMed] [Google Scholar]
- Kovacs G, Szücs S, De Riese W, Baumgärtel H. Specific chromosome aberration in human renal cell carcinoma. Int J Cancer. 1987 Aug 15;198740(2):171–8. doi: 10.1002/ijc.2910400208. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Khoo SK, Massie A, Iwamura M, Chen J, Petillo D, Wondergem B, Avallone M, SJK, Tan MH, Koeman J, Zhang Z, Kahnoski RJ, Group FKCS, Baba S, Teh BT. Identification of copy number alterations and its association with pathological features in clear cell and papillary RCC. Cancer Lett. 2008;272:260–267. doi: 10.1016/j.canlet.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Miura I, Siegfried JM, Resau J, Keller SM, Zhou JY, Testa JR. Chromosome alterations in 21 non-small cell lung carcinomas. Genes Chromosomes Cancer. 1990;2:328–338. doi: 10.1002/gcc.2870020411. [DOI] [PubMed] [Google Scholar]
- Nagao K, Yamaguchi S, Matsuyama H, Korenaga Y, Hirata H, Yoshihiro S, Fukunaga K, Oba K, Naito K. Allelic loss of 3p25 associated with alterations of 5q22.3~q23.2 may affect the prognosis of conventional renal cell carcinoma. Cancer Genet Cytogenet. 2005;160:43–48. doi: 10.1016/j.cancergencyto.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Toma MI, Grosser M, Herr A, Aust DE, Meye A, Hoefling C, Fuessel S, Wuttig D, Wirth MP, Baretton GB. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density affymetrix 10K single nucleotide polymorphism mapping array. Neoplasia. 2008;10:634–642. doi: 10.1593/neo.08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzo RG, Cairns P, Al-Saleem T, Hudes G, Haas N, Greenberg RE, Kolenko V. The basic biology and immunobiology of renal cell carcinoma: considerations for the clinician. Urol Clin North Am. 2003;30:423–436. doi: 10.1016/s0094-0143(03)00021-1. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Matsuura K, Karnan S, Tagawa H, Nakada C, Tanigawa M, Tsukamoto Y, Uchida T, Kashima K, Akizuk S, Takeuchi I, Sato F, Mimata H, Seto M, Moriyama M. High-resolution analysis of DNA copy number alterations and gene expression in renal clear cell carcinoma. J Pathol. 2007;213:392–401. doi: 10.1002/path.2239. [DOI] [PubMed] [Google Scholar]