Abstract
High-density lipoproteins (HDLs) play an important role in human health through the metabolism and trafficking of cholesterol as well as providing the feedstocks for steroid hormone biosynthesis. These particles contain proteins, primarily Apo-AI and phospholipid and progress through various structural forms including ‘lipid-poor’, ‘discoidal’ and ‘spherical’ entities as cholesterol esters and lipid are incorporated. The discoidal form of HDL is stabilized in solution by two encircling belts of Apo-AI. Previous protein engineering of the Apo-AI sequence has led to a series of amphipathic helical proteins, termed membrane scaffold proteins (MSPs), which have shown great value in assembling nanoscale soluble membrane bilayers, termed Nanodiscs, of homogeneous size and composition and in the assembly of numerous integral membrane proteins for biophysical and biochemical investigations. In this communication we document a protein engineering approach to generate and optimize an extended polypeptide MSP, which will self-assemble phospholipids into larger Nanodiscs with diameters of 16–17 nm. We extensively characterize these structures by size exclusion chromatography and solution X-ray scattering.
Keywords: membrane protein, membrane scaffold protein, nanodiscs
Introduction
Self-assembly is the cornerstone of functioning biological systems, resulting in the synthesis of complex macromolecular assemblies containing protein, nucleic acid, lipid and carbohydrate components. The resulting cellular machines provide for the synthesis, transport and processing of information and energy. One example, which plays a critical role in human health, is the collection of lipid and protein particles which transport cholesterol and cholesterol esters between the vasculature and organs such as liver and steroidogenic tissues. Important players in this process are the high-density lipoproteins. Consisting of primarily a single protein, Apo-AI, and lipid, these nanoscale particles transform from a lipid optimized discoidal form to a spherical ball as cholesterol ester is loaded (Koppaka, 2001; Marcel and Kiss, 2003; Zannis et al., 2006). While important to human diseases such as atherosclerosis, it was noted nearly a decade ago that the discoidal form of HDL particles could be utilized to provide a soluble supported phospholipid bilayer for the incorporation of membrane proteins (Bayburt et al., 1998; 2000; Bayburt and Sligar, 2002; 2003; Civjan et al., 2003). This process results in the membrane protein target being in its native-like lipid environment with ‘naturalistic presentation’, yet soluble in aqueous solution (Sligar, 2003; Nath et al., 2007). Realizing homogeneous and monodisperse particles as an endpoint required extensive protein engineering of the encircling Apo-AI protein, resulting in a class of amphipathic peptides termed membrane scaffold proteins (MSPs) (Bayburt et al., 2002) and the corresponding discoidal bilayer particles with 10–12 nm diameter and 4.5–5.5 nm thickness, termed Nanodiscs (Bayburt et al., 2002; Denisov et al., 2004). Modification of the MSP sequence subsequently allowed incorporation of affinity tags (Histidine, FLAG, etc.), which could be removed by protease treatment (Factor X, TEV) as well as provisions for site-specific labeling of engineered unique amino acid types. Use of synthetic gene technology resulted in the ability to produce MSPs in E. coli at the gram level. As recently reviewed (Nath et al., 2007; Bayburt and Sligar, 2010; Denisov and Sligar, 2010), successful incorporation of fully functional membrane proteins, such as monomeric or trimeric bacteriorhodopsin, monomeric and dimeric bovine rhodopsin, mammalian cytochromes P450 and their complexes with cytochrome P450 reductase, and various transmembrane receptors can be achieved using self-assembly from the mixture of cholate solubilized MSP, lipids and target protein.
A biophysical understanding of the self-assembly process is emerging through the combination of solution X-ray scattering and computational methods (Shih et al., 2007a, b). Even for nanoparticles without incorporated membrane protein targets, this involves the assembly of two distinct MSPs together with hundreds of phospholipids, the precise number determined by the phospholipid type (Bayburt et al., 2002; Denisov et al., 2004; Denisov et al., 2005). In this communication we document a protein engineering approach to generate Nanodisc with larger sizes, 16–17 nm. Earlier efforts at simply fusing two copies of a single MSP resulted in less monodisperse preparations and low yield of large lipoprotein particles (Bayburt et al., 2002). Our current results incorporate knowledge as to precisely which helices of the parent Apo-AI sequence are actually forming contact with the lipid acyl chains, and are based on the previous experience in engineering fusions of these scaffold proteins (Bayburt et al., 2002; Denisov et al., 2004) and on optimization of conditions for the generation of discs with diameters up to 17 nm. This discovery as to how to produce Nanodiscs of increasing size is of great value to the incorporation of larger molecular complexes, such as the enzymes involved in cellular energy metabolism, oligomers of integral membrane receptors, ion channels and transporter proteins.
Experimental section
Chemicals
The lipids 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) were purchased in Avanti Polar Lipids (Alabaster, AL), 3H-DPPC was from American Radiolabeled Chemicals (St. Louis, MO). All other chemicals were from Sigma-Aldrich or Fisher. Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, MA) or Invitrogen (Carlsbad, CA). Water was purified with a Milli-Q purification system (Millipore, Billerica, MA).
Cloning and mutagenesis
Plasmids for expression of MSP1, MSP2, MSP1D1, MSP1D2 and extended MSPs were described previously (Bayburt et al., 2002; Denisov et al., 2004). Plasmids for expression of new MSP2 proteins were produced as follows: first, MSP1D1 plasmid was modified using site-directed mutagenesis to introduce DNA sequence GGTACC (Kpn I restriction site), coding for a dipeptide glycine-threonine linker, between the last amino acid of MSP and Stop codon. The DNA for the second half of the MSP2N was generated by PCR using the MSP1D1 plasmid as a template. Primers were designed to include Kpn I and Hind III restriction sites. The PCR product was digested with these restriction enzymes and cloned into the modified MSP1D1 plasmid from the first step. Presence of the desired insertions or deletions and absence of PCR-induced mutations were verified by DNA sequencing.
Protein expression and purification
MSP expression and purification was carried out as described previously (Bayburt et al., 2002; Denisov et al., 2004). Protein purity was characterized by SDS-PAGE and electrospray mass spectrometry and was found to be greater than 95%.
Preparation of the disc samples
The general procedure for self-assembly of Nanodiscs is the same as described (Bayburt et al., 2002; Denisov et al., 2004). Briefly, the solution of purified MSP at 0.15–0.3 mM concentration was combined with cholate solubilized DPPC to yield an optimal molar ratio, previously estimated from small-scale experiments. After incubation at 38°C, the self-assembly process is initiated by dialysis against 1000-fold excess of buffer at the same temperature using 10 000 MW cut-off membranes, or by incubation with adsorbent beads (Bio-beads SM-2, Biorad or Amberlite XAD-2; Sigma-Aldrich). Assembly of Nanodiscs with POPC is performed in a similar way, but at 4°C instead of 38°C.
Analytical procedures
Concentrations of lipids were measured using tritiated lipids of the same chemical structure together with scintillation counting of the column fractions, as described (Denisov et al., 2004). The specific activity of each batch of radioactive lipids was determined by phosphate quantitation (Chen, 1956) and scintillation counting using standard procedures. For calibration we used standard phosphorus solutions (Sigma, P3869) and [1,2-3H] hexadecanol (Moravec Biochemicals, Bria, CA). Concentrations of scaffold proteins were determined using molar absorption coefficients calculated for the known amino acid sequences according to published methods (Pace et al., 1995). Assembled Nanodiscs were size separated by HPLC (Millenium System, Waters, Milford, MA) on a calibrated size exclusion chromatography (SEC) column with Superdex 200 HR 10/30 (GE Healthcare, Piscataway, NJ).
Small angle X-ray scattering measurements and analysis
Small angle X-ray scattering (SAXS) was measured at the Advanced Photon Source (Argonne National Lab, IL) at 2.012 m distance from the sample to the 2D detector, and a nominal photon energy of 15 keV (wavelength of 0.826 Å). The Nanodisc solutions were sealed in glass capillaries with 1.5 mm diameter (Charles Supper Company, Natick, MA) and placed into an aluminum sample holder with temperature controlled by Peltier elements. Vacuum chambers with Mylar windows were used along the beam path before and after the sample holder to minimize scattering by air. Silver behenate with spacing 58.38 Å (Huang et al., 1993) was used for calibration, and reference buffer solvents for background correction. The data collection was executed after equilibration of the sample at each temperature for 3 min; usually two data frames with 100 s exposure were collected at each temperature to ensure complete equilibration of the sample. After warming to temperatures above the lipid main phase transition, each sample was cooled to 22°C, and the SAXS data were collected again to check for reversibility. In all cases the main lipid transition in Nanodiscs monitored by SAXS was fully reversible. The raw data were processed using the program FIT2D (Hammersley et al., 1996, 1998) to give the scattering curves in the form log(I/I0) vs. Q = 4πsin(θ)/λ. Analysis of the SAXS data used the program GNOM (Svergun, 1992) using the option for homogeneous globular particles.
Results
The assembly of Nanodiscs was initiated by detergent removal as described in the section Methods and in previous publications (Bayburt et al., 2002; Denisov et al., 2004). The correct ratio of MSP to phospholipids is necessary for the generation of a homogeneous size distribution in the resultant phospholipids nanoparticles. The new MSP sequences used in this work, as well as those for generating smaller Nanodiscs are summarized in Table 1. Overall, the main difference between MSP2 (2, Table 1) (Bayburt et al., 2002) and new MSP2N1, MSP2N2 and MSP2N3 proteins (8–10, Table 1) is imposed by various truncations of the helix 1 in the fused proteins 8–10.
Table I.
Labels and amino acid sequences of MSPs used for self-assembly of Nanodiscs
| Abbreviation | Description | Amino acid sequence |
|---|---|---|
| H1 | Helix 1 | LKLLDNWDSVTSTFSKLREQLG |
| H1Δ(1–11) | Truncated Helix 1 | STFSKLREQLG |
| H1Δ(1–17) | Truncated helix 1 | REQLG |
| H2 | Helix 2 | PVTQEFWDNLEKETEGLRQEMS |
| H3 | Helix 3 | KDLEEVKAKVQ |
| H4 | Helix 4 | PYLDDFQKKWQEEMELYRQKVE |
| H5 | Helix 5 | PLRAELQEGARQKLHELQEKLS |
| H6 | Helix 6 | PLGEEMRDRARAHVDALRTHLA |
| H7 | Helix 7 | PYSDELRQRLAARLEALKENGG |
| H8 | Helix 8 | ARLAEYHAKATEHLSTLSEKAK |
| H9 | Helix 9 | PALEDLRQGLL |
| H10 | Helix 10 | PVLESFKVSFLSALEEYTKKLNTQ |
| FX | Original N-terminus | MGHHHHHHIEGR |
| TEV | Modified N-terminus | MGHHHHHHH DYDIPTTENLYFQG |
| Scaffold protein variant | Abbreviated namea | Composition |
| 1 | MSP1 | FX-H1-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 2 | MSP2 | FX-H1- H2-H3-H4-H5-H6-H7-H8-H9-H10-GT-H1-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 3 | MSP1E1 | FX-H1-H2-H3-H4-H4-H5-H6-H7-H8-H9-H10 |
| 4 | MSP1E2 | FX-H1-H2-H3-H4-H5-H4-H5-H6-H7-H8-H9-H10 |
| 5 | MSP1E3 | FX-H1-H2-H3-H4-H5-H6-H4-H5-H6-H7-H8-H9-H10 |
| 6 | MSP1D1 | TEV-H1Δ(1–11)-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 7 | MSP1D2 | TEV-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 8 | MSP2N1 | TEV-H1Δ(1–11)-H2-H3-H4-H5-H6-H7-H8-H9-H10-GT-H1Δ(1–11)-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 9 | MSP2N2 | TEV-H1Δ(1–11)-H2-H3-H4-H5-H6-H7-H8-H9-H10-GT-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
| 10 | MSP2N3 | TEV-H1Δ(1–11)-H2-H3-H4-H5-H6-H7-H8-H9-H10-GT-H1Δ(1–17)-H2-H3-H4-H5-H6-H7-H8-H9-H10 |
Design of the MSPs. a(1) MSP1, membrane scaffold protein 1 with N-terminal hexahistidine tag, followed by Factor X recognition site. (2) MSP2, fusion of two MSP1 molecules with short glycine-threonine linker and N-terminal hexahistidine tag, followed by Factor X recognition site. (3–5) MSP1E1, MSP1E2, MSP1E3, extended membrane scaffold proteins, obtained via insertion of one (residues 56–77 of original MSP1), two (residues 56–99) or three (residues 56–121) extra 22-mer amphipathic helices after the residue Q55 into original MSP1 (1) sequence, as described (JACS 2004). All have N-terminal hexahistidine tag and Factor X recognition site; (6) MSP1D1, MSP1 with the first 11 N-terminal amino acids removed which contains the N-terminal heptahistidine tag and TEV protease recognition site with linker; (7) MSP1D2, MSP1 with the first 22 N-terminal amino acids removed contains the N-terminal heptahistidine tag and TEV protease recognition site with linker; (8–10) New MSP2 proteins based on MSP1 deletion mutants rather than original MSP1. All three have the same first MSP1D1 molecule, but vary in extent of truncation in the second MSP.
Initial characterization of the assembly process is analyzed by SEC (Methods). For instance, Fig. 1 shows the results of a SEC analysis when the assembly utilizes the scaffold protein MSP1. Clearly, the best size homogeneity is obtained at a DPPC:MSP1 ratio of ∼75, corresponding to a Nanodisc with two encircling MSP1 belts and 150 total DPPCs. This ratio is understandable as the optimum packing of lipid inside the solubilizing toroidal amphipathic helix of the scaffold protein to form the cylindrical fragment of Nanodisc bilayer (Bayburt et al., 2002; Denisov et al., 2004).
Fig. 1.
SEC chromatograms of Nanodiscs reconstituted at different lipid:protein ratios for the original MSP1 and MSP2 (Bayburt et al., 2002). (A) DPPC:MSP1 molar ratio: (1) 0; (2) 25; (3) 50; (4) 75; (5) 100; (6) 200; and (7) 300; (B) DPPC:MSP2 molar ratio: (1) 0; (2) 50; (3) 100; (4) 150; (5) 200; (6) 400; and (7) 600. At ratios near optimum (about 90 for MSP1 and all modified MSP1; 180 for MSP2) one predominant form is formed—‘regular’ Nanodisc, ∼10 nm diameter, eluted at 26 min.
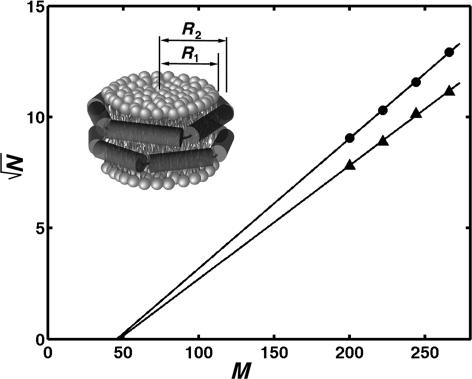
A structural model of the discoidal Nanodisc lipoprotein particles that is consistent with all available experimental data (Denisov et al., 2004, 2005) is represented in Fig. 2. The diameter of the resulting Nanodisc is determined by the length of the encircling MSP, i.e. by the number of amino acids forming the helical belt around the lipid in Nanodiscs, Mh. This number is not known a priori, it may be smaller than the total number of amino acids in a given MSP, M, if not all residues are involved in the formation of amphipathic helix interacting with the lipid bilayer. However, from the data on lipid:protein stoichiometry obtained for the set of extended scaffold proteins with different lengths (Denisov et al., 2004), one can evaluate the actual values of Mh. The following equations connect the structural parameters of the scaffold protein and lipid with radius of Nanodisc R2 and that of lipid bilayer fragment R1 (Denisov et al., 2004):
Fig. 2.
Analysis of lipid:protein stoichiometry in Nanodiscs as a function of the length of scaffold protein. Square root of the number of lipids per protein vs. number of amino acids in MSP is shown for Nanodiscs assembled with DPPC (circles) and POPC (triangles). Slope of linear fit corresponds to the mean surface area per lipid, 52 Å2 for DPPC and 69 Å2 for POPC (Denisov et al., 2004).
Here r is the average radius of the scaffold protein helix, ∼6 Å, Mh is the number of residues in the protein helical belt, L the helical pitch per residue, 1.5 Å, N the number of lipids in Nanodisc per on leaflet of bilayer and S is the mean surface area per lipid. From these equations the following linear connection between the square root of N and Mh can be derived:
| (1) |
Thus, a plot of square root of experimentally determined N vs. M shown in Fig. 2 is linear and allows determining the number of amino acids forming the helical belt around the lipid in Nanodiscs, Mh. As one can see from Equation 1, Mh = 2·π·r/L = 25 at N = 0, however both straight lines cross abscissa axis at M = 46. This means that for all extended scaffold proteins studied earlier, M = Mh + 21, i.e. approximately 21 residues do not participate in formation of amphipathic helical belt around the lipid bilayer. As explained in detail in the original paper (Denisov et al., 2004), the N-terminal 17–20 amino acids in the original MSP1 (similar to the Apo-A1 with removal of 1–43 fragments) apparently do not interact with lipid and can be removed with no effect on the number of lipids encompassed by truncated scaffold protein. This flexibility of the N-terminal fragment in MSP1 may also be necessary for MSP2 to form 10 nm Nanodiscs with only a single scaffold protein, which is twice as long as MSP1. As shown in Fig. 1, particles with 10 nm size are still formed with MSP2 even at higher lipid:protein ratio of 400:1, although significant part of the protein forms larger particles. However, the yield and homogeneity of the larger particles is low when the MSP2 is used, prompting the design of new scaffold proteins that are the subject of this communication.
Size exclusion chromatograms of Nanodiscs formed with new scaffold proteins MSP2N1, MSP2N2 and MSP2N3 at different lipid:protein ratios are shown in Fig. 3. Unlike original MSP2 (Fig. 1), these proteins do not form ‘regular’ disks with high yield at a ratio of MSP:lipid of 1:200. Instead, for two new scaffolds, MSP1N2 and MSP1N3, the proteins with higher degree of truncation in the second MSP molecule, make mostly larger (15 nm Stokes' diameter) disks when assembled at 1:300 and 1:400 protein to lipid ratios. For MSP2N1 the tendency is the same, but population is heterogeneous. In general, the size distribution of the formed complexes is wider for the big discs in comparison with ‘normal’ ones, which results in bigger loss during purification with SEC. The number of DPPC molecules per one scaffold protein in these big discs is 305 ± 8 for MSP2N2 and 307 ± 9 for MSP2N3, almost four times higher than with MSP1, and twice as high as with MSP1E3 (Denisov et al., 2004). To test assembly of larger Nanodiscs with different lipids, we repeated the same set of experiments with DMPC (assembly at 23°C) and POPC (assembly at 4°C) using MSP2N2 as a scaffold protein. Lipid counting for these discs in the main fraction purified by HPLC revealed 323 ± 8 DMPC molecules and 239 ± 8 POPC molecules per Nanodiscs, in a good agreement with the expected mean surface areas per lipid, 50 and 68 Å2 correspondingly.
Fig. 3.
SEC chromatograms of Nanodiscs reconstituted at different lipid:protein ratios for new MSP2s. Lipid:protein ratios are the same in all three panels for DPPC:MSP2N1 (A), DPPC:MSP2N2 (B) and DPPC:MSP2N3 (C): (1) 0; (2) 75; (3) 100; (4) 150; (5) 200; (6) 300; (7) 400; and (8) 600. Unlike original MSP2, these three extended proteins do not form ‘regular’ 10 nm Nanodiscs with high yield at 200:1 ratio. Instead, MSP1N2 and MSP1N3, the proteins with higher degree of truncation in the second MSP molecule, generate mostly big (15 nm) disks at 300:1 and 400:1 ratios eluted at 21 min. For MSP2N1, the tendency is the same, but population is heterogeneous. In general, the size distribution of the formed complexes is more broad for the big discs in comparison with ‘normal’ ones, which results in bigger loss during purification with SEC. The number of DPPC per MSP2 in these big discs determined by scintillation counting of tritiated lipids is 305 ± 8 for MSP2N2 and 307 ± 9 for MSP2N3.
SAXS was used to evaluate the structural parameters of Nanodiscs and to study the thermotropic phase transition of DPPC in nanoscale bilayers. Figure 4 shows the density distribution function calculated from the SAXS curves using program GNOM (Svergun, 1992). For comparison, the density distribution functions of Nanodiscs assembled with other scaffold proteins of different lengths are also shown to illustrate the gradual increase in size with the length of MSP. For all Nanodiscs a characteristic pattern of two peaks at 1.6 nm and 5.6–5.8 nm separated by a minimum at 3.5 nm represents a signature of the lipid bilayer with the thickness of ∼5.5 nm (Denisov et al., 2004, 2005). Both MSP2N2 and MSP2N3 produce Nanodiscs with significantly larger diameters (∼17 nm) than that realized by the first generation MSPs. These sizes are in good agreement with SEC data shown in Fig. 1. Systematic differences between diameters obtained from SAXS measurements in this work as well as in previous studies (Denisov et al., 2004, 2005) and Stokes diameters estimated from SEC, are explained by the anisometric shape of Nanodiscs and by the corresponding difference between real diameter of flat discoidal particle, measured by SAXS, and diameter of the equivalent spherical particle with the same hydrodynamic properties, which is estimated from SEC using globular proteins for calibration (Ortega and Garcia de la Torre, 2003). For instance, the Stokes diameter of a discoidal particle with diameter of 17 nm and thickness of 5.5 nm is approximately 15.8 nm, as calculated using equation (16) in (Ortega and Garcia de la Torre, 2003). The diameters of Nanodiscs formed with MSP2N2 and MSP2N3 correspond to approximately 14.5 nm diameter of the lipid bilayer fragment in these Nanodiscs, and 52 Å2 mean surface area per of lipid molecule, in a good agreement with our earlier results (Denisov et al., 2004, 2005).
Fig. 4.
Density distribution functions calculated using GNOM (Svergun, 1992) from SAXS curves measured at T = 25°C for Nanodiscs formed with DPPC and different scaffold proteins. The increasing size of Nanodiscs is clearly seen from MSP1 (1) to MSP1E1 (2), MSP1E2 (3) and MSP1E3 (4) and finally to the new scaffold proteins MSP2N2 (5) and MSP2N3 (6).
Figure 5 shows the density distribution functions for 17 nm Nanodiscs at different temperatures from 5°C to 55°C. The increase in diameter of Nanodiscs as a function of temperature is plotted in Fig. 6. The main phase transition from gel to liquid crystalline state of DPPC is indicated as a sharp increase of Nanodisc size at 39°C, the position of the transition close to the phase transition documented in multilamellar vesicles (Cevc, 1993). The lateral thermal expansion coefficient CT = (1/A)(dA/dT) for DPPC in large Nanodiscs estimated from Fig. 6 is 0.005 K−1 for the liquid crystalline phase and 0.007 K−1 above phase transition, both values slightly higher than reported earlier for smaller Nanodiscs (Denisov et al., 2005), but similar to those measured for DMPC liposomes (Needham and Evans, 1988). This suggests high elasticity and flexibility of longer scaffold proteins in Nanodiscs, which allows for almost unrestricted expansion of the lipid bilayer with increasing temperature. Phase transition was reversible if Nanodiscs were not incubated at 55°C for more than an hour, and SAXS curves measured for the control at 22°C at the end of experiment are virtually identical to those measured before application of the thermal gradient. This confirms the overall stability of these Nanodiscs in solution and provides additional support to the reported method of generation of large lipoprotein particles suitable for solubilization of functional complexes of membrane proteins.
Fig. 5.
Density distribution functions calculated from the SAXS of MSP2N3–DPPC Nanodiscs as a function of temperature. The lipid packing density gradually decreases, and the size increases with temperature. Phase transition of the lipid bilayer is shown in Fig. 6.
Fig. 6.
Diameter of MSP2N3–DPPC Nanodiscs as a function of temperatures demonstrates sharp phase transition at 39°C, corresponding to the main thermotropic phase transition of the DPPC bilayer. Lipid phase transition in large Nanodiscs is much sharper and less perturbed than the same phase transition in Nanodiscs of the smaller size (Denisov, 2005), indicating the lower fraction of the boundary lipids in MSP2N3 Nanodiscs.
In summary, we have designed, expressed and purified a new class of extended MSPs, which are capable of forming larger Nanodisc particles with high yield. The first extended scaffold protein MSP2, designed as a fusion of two intact copies of MSP1 with a short linker, efficiently forms Nanodiscs of the same size as two copies of the original MSP1, i.e. 10 nm diameter (Bayburt et al., 2002). We later demonstrated that the first 15–20 N-terminal residues do not interact with lipids and are not necessary for the formation of 10 nm Nanodiscs (Denisov et al., 2004). Truncated MSP1 with up to 22 N-terminal residues removed can efficiently form Nanodiscs with the same size and protein–lipid stoichiometry as the original MSP1 (Denisov et al., 2004). This fact suggests that flexibility of the N-terminal fragment in the first MSP2 (Bayburt et al., 2002) was responsible for the formation of the regular 10 nm Nanodiscs with this protein, allowing for the adjustment of amphipathic helices in the single polypeptide chain around the circular fragment of the lipid bilayer. Consistent with this hypothesis, new extended scaffold proteins MSP2N2 and MSP2N3, which do not have the extended flexible fragment in the middle of the designed dimer due to the deletion of the helix 1, do not form small Nanodiscs. The lack of long flexible linker in the sequence of newly designed scaffold proteins MSP2N2 and MSP2N3 favors formation of the long uninterrupted amphipathic helical belt with diameter of 16–17 nm. Using a traditional cholate dialysis method, we optimized the stoichiometry of the protein–lipid ratio and characterized the resulting nanolipoprotein particles using SEC and SAXS. The resulting Nanodiscs provide a planar bilayer domain (∼15 nm) of sufficient size to incorporate large oligomeric membrane proteins and protein complexes. These engineered Nanodiscs are stable at temperatures up to 55°C, as previously demonstrated for the smaller particles (Denisov et al., 2005). This is the first report of the self-assembly of large soluble monodisperse and stable discoidal bilayer fragments, which can be used for the biophysical studies of complex membrane protein assemblies in their native lipid membrane environment.
Funding
This work was supported by grants from the National Institutes of Health [GM33 775 and GM 31 756 to S.G.S]. Portions of this work have been performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center located at Sector 5 of the Advanced Photon Source. DND-CAT is supported by the E.I. DuPont de Nemors & Co., The Dow Chemical Company, the U.S. National Science Foundation [Grant DMR-9304725]; and the State of Illinois through the Department of Commerce and the Board of Higher Education [Grant IBHE HECA NWU 96]. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research [Contract No. W-31-102-Eng-38].
Acknowledgements
We gratefully acknowledge the help and support provided by Dr. J. Quintana, Dr. D. Keane and Dr. S. Weigand while working at Argonne. We thank Dr. M. McLean for design and construction of the Peltier thermostat used in the SAXS experiments and for help in data collection.
Footnotes
Edited by Peter Roepstorff
References
- Bayburt T.H., Sligar S.G. Proc. Natl Acad. Sci. USA. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. doi:10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T.H., Sligar S.G. Prot. Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. doi:10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T.H., Sligar S.G. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. doi:10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T.H., Carlson J.W., Sligar S.G. J. Struct. Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. doi:10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- Bayburt T.H., Carlson J.W., Sligar S.G. Langmuir. 2000;16:5993–5997. doi:10.1021/la991449c. [Google Scholar]
- Bayburt T.H., Grinkova Y.V., Sligar S.G. Nano Lett. 2002;2:853–856. doi:10.1021/nl025623k. [Google Scholar]
- Cevc G. Phospholipids Handbook. New York: Marcel Dekker, Inc; 1993. p. 988. [Google Scholar]
- Chen P.S., Toribara T.Y., Warner H. Anal. Chem. 1956;28:1756–1759. doi:10.1021/ac60119a033. [Google Scholar]
- Civjan N.R., Bayburt T.H., Schuler M.A., Sligar S.G. BioTechniques. 2003;35:556–558. doi: 10.2144/03353rr02. 560,562–563. [DOI] [PubMed] [Google Scholar]
- Denisov I.G., Sligar S.G. Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbapap.2010.05.017. doi:10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov I.G., Grinkova Y.V., Lazarides A.A., Sligar S.G. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. doi:10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Denisov I.G., McLean M.A., Shaw A.W., Grinkova Y.V., Sligar S.G. J. Phys. Chem. B. 2005;109:15580–15588. doi: 10.1021/jp051385g. doi:10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammersley A.P. 1998. http://www.esrf.fr/computing/scientific/FIT2D .
- Hammersley A.P., Svensson S.O., Hanfland M., Fitch A.N., Häusermann D. High Pressure Research. 1996;14:235–248. doi:10.1080/08957959608201408. [Google Scholar]
- Huang T.C., Toraya H., Blanton T.N., Wu Y. J. Appl. Cryst. 1993;26:180–184. doi:10.1107/S0021889892009762. [Google Scholar]
- Koppaka V. Cell Mol. Life Sci. 2001;58:885–893. doi: 10.1007/PL00000908. doi:10.1007/PL00000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel Y.L., Kiss R.S. Curr. Opin. Lipidol. 2003;14:151–157. doi: 10.1097/00041433-200304000-00006. doi:10.1097/00041433-200304000-00006. [DOI] [PubMed] [Google Scholar]
- Nath A., Atkins W.M., Sligar S.G. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. doi:10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- Needham D., Evans E. Biochemistry. 1988;27:8261–8269. doi: 10.1021/bi00421a041. doi:10.1021/bi00421a041. [DOI] [PubMed] [Google Scholar]
- Ortega A., Garcia de la Torre J. J. Chem. Phys. 2003;119:9914–9919. doi:10.1063/1.1615967. [Google Scholar]
- Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. Prot. Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. doi:10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.Y., Arkhipov A., Freddolino P.L., Sligar S.G., Schulten K. J. Phys. Chem. B. 2007a;111:11095–11104. doi: 10.1021/jp072320b. doi:10.1021/jp072320b. [DOI] [PubMed] [Google Scholar]
- Shih A.Y., Freddolino P.L., Sligar S.G., Schulten K. Nano Lett. 2007b;7:1692–1696. doi: 10.1021/nl0706906. doi:10.1021/nl0706906. [DOI] [PubMed] [Google Scholar]
- Sligar S.G. Biochem. Biophys. Res. Comm. 2003;312:115–119. doi: 10.1016/j.bbrc.2003.09.188. doi:10.1016/j.bbrc.2003.09.188. [DOI] [PubMed] [Google Scholar]
- Svergun D.I. J. Appl. Cryst. 1992;25:493–503. [Google Scholar]
- Zannis V.I., Chroni A., Krieger M. J. Mol. Med. 2006;84:276–294. doi: 10.1007/s00109-005-0030-4. doi:10.1007/s00109-005-0030-4. [DOI] [PubMed] [Google Scholar]