Abstract
Knowledge of the inherited risk for cancer is an important component of preventive oncology. In addition to well-established syndromes of cancer predisposition, much remains to be discovered about the genetic variation underlying susceptibility to common malignancies. Increased knowledge about the human genome and advances in genotyping technology have made possible genome-wide association studies (GWAS) of human diseases. These studies have identified many important regions of genetic variation associated with an increased risk for human traits and diseases including cancer. Understanding the principles, major findings, and limitations of GWAS is becoming increasingly important for oncologists as dissemination of genomic risk tests directly to consumers is already occurring through commercial companies. GWAS have contributed to our understanding of the genetic basis of cancer and will shed light on biologic pathways and possible new strategies for targeted prevention. To date, however, the clinical utility of GWAS-derived risk markers remains limited.
INTRODUCTION
Over the past two decades, research advances in cancer genetics have identified and characterized mutated cancer predisposition genes that account for a subset of cancers with a Mendelian pattern of inheritance.1 Examples of cancer predisposition genes are BRCA1 and BRCA2 in hereditary breast and ovarian cancer, the mismatch repair genes in Lynch syndrome, the APC gene in familial adenomatous polyposis (FAP), and the TP53 gene in Li-Fraumeni syndrome. Individuals with mutations in these genes have a much higher risk of developing cancer than those in the general population. Guidelines for genetic testing for these genes and strategies for cancer surveillance and prevention have been developed and incorporated into oncologic practice.2–5
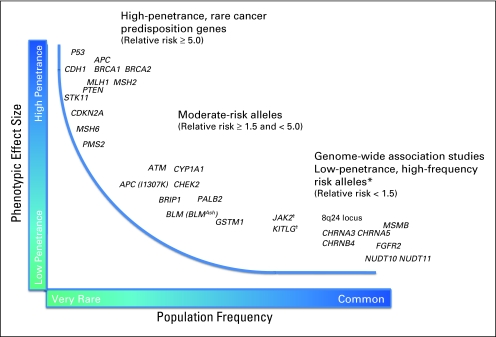
In recent years, genome-wide association studies (GWAS) have identified genetic variants, or susceptibility loci, for a variety of human diseases, including cancer. As methods for sequencing entire personal genomes become less costly, the number of such genetic variants discovered will continue to increase. While the majority of susceptibility loci found by GWAS confer only a modest risk of disease (Fig 1), for-profit companies have made personal genomic profiles available directly to consumers. Although clinicians and scientists have articulated concerns regarding the premature commercial dissemination of personal genomics6 and need for continued research,7 oncologists and other cancer care professionals will be asked by patients to explain results of tests and advise on desired interventions or screening. A recent survey8 indicated that 42% of physicians were aware of direct-to-consumer genomic tests for disease risk, and of those physicians aware of direct-to-consumer tests, approximately 40% had been asked by patients to interpret the results of such testing. It is therefore important for oncologists to be aware of current data regarding the ranges of risk, the clinical validity (accuracy of a test in predicting the clinical outcome), and clinical utility (risks and benefits resulting from the use of a test) associated with genetic variants predisposing to cancer.9,10
Fig 1.
Phenotypic effect size and frequency of occurrence. (*) Named genes only reflect the most likely candidate genes to be implicated by the marker single nucleotide polymorphisms (SNPs) identified from the genome-wide association studies. (†) The marker SNPs mapping to JAK2 in myeloproliferative neoplasms and KITLG in testicular germ cell tumors have odds ratios of approximately 3.0, with allele frequencies ranging from 20% to 40%.
GENETIC SUSCEPTIBILITY TO CANCER
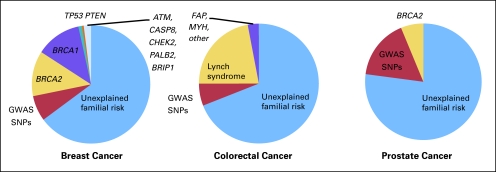
Mutations of more than 50 genes have been associated with high-penetrance cancer susceptibility syndromes (risk of cancer increased approximately 5- to 50-fold). However, these syndromes account for only a small fraction of the familial risk of cancer (Fig 2).11–16 Using breast cancer as an example, mutations in BRCA1 and BRCA2 appear to account for fewer than 20% of the familial risk of breast cancer, with other rare genes (eg, TP53, PTEN) accounting for no more than 5% of the risk.13 Other cancer types are similar in that high-penetrance cancer susceptibility genes only explain a small fraction of the familial risk of cancer.
Fig 2.
Familial risk of common cancers. Most common cancers, including breast, colorectal, and prostate cancer, exhibit familial aggregation, with the disease being more common in family members than in the general population. While familial aggregation may be secondary to genetic or environmental factors, evidence from monozygotic twins of patients suggests that genetic factors are mainly responsible.11,12 As demonstrated in this figure, known high-penetrance cancer predisposition syndromes explain only a fraction of the familial risk of cancer.13–16 While genome-wide association studies have identified numerous low-penetrance loci for each of these three common cancers, their contribution to the familial risk of cancer remains limited and a large portion of the familial risk of these cancers remains unexplained. GWAS, genome-wide association study; SNP, single nucleotide polymorphisms.
Studies of monozygotic twins and inbred populations have provided strong evidence that a large fraction of cancers are mediated by genetic susceptibility.11,12 Surprisingly, in these studies, even seemingly environmental cancers, such as lung cancer, demonstrate familial clustering and are likely to be mediated by genetic susceptibility to shared exposures. A scientific debate over the past decade has centered on whether genetic susceptibility to common diseases is a result of the joint action of several common variants each with low relative risk of disease, or the result of genetic variants with low population frequency but moderate to high risk of disease.17
Examples of rare genetic mutations of moderate effect size emerged from studies of individual candidate genes. For example, genes in a common pathway (ie, DNA damage response), such as ATM18–20, CHEK221–23, BRIP1, 24 and PALB2,25 have been associated with increased risk for breast cancer. However, based on estimates of the risk allele frequencies ranging from 0.1% to 0.5% (ie, 0.2 to one individual in 100 would carry at least one copy of the risk allele) and the modest 2.0-fold increase in relative risk associated with each of these genes, the contribution of mutations in these genes to the familial aggregation of breast cancer is limited. As shown in Figure 2, together with the known high-penetrance genes, all known breast cancer predisposition genes account for only approximately 25% of the familial risk of breast cancer in outbred populations.13
Recently, genetic association studies have been used to discover common genetic variants or risk alleles (minor allele frequency > 10%) with small to moderate (approximately two-fold) risks of cancer.26 Association studies compare the frequency of a genetic variant in disease-affected patients and healthy controls. Recent knowledge gained from the Human Genome Project and the International Hap Map Project together with technical advances in high throughput genotyping technology has resulted in GWAS for a variety of complex diseases including cancer.
GWAS
Study Design
Roughly 99.9% of DNA sequence is identical across different individuals. Given the vast size (3.2 billion base pairs) of the human genome, even this small discrepancy results in millions of potential variations.27 The most common variations are single-base pair changes called single-nucleotide polymorphisms (SNPs). GWAS compare allele frequencies between individuals with a disease (patients) to individuals without disease (controls). By determining which SNPs occur more (or less) frequently in individuals with disease, genomic regions associated with a disease state can be identified and a statistical estimate of the level of increased (or diminished) risk associated with each SNP can be made. GWAS take advantage of the fact that stretches of DNA tend to be inherited together and adjacent alleles sort together nonindependently from generation to generation. This nonrandom association of alleles at nearby loci (linkage disequilibrium) allows certain SNPs to serve as proxies, or tagSNPs, for other nearby SNPs. The use of such marker SNPs allows one to reduce the number of SNPs that need to be genotyped to characterize individual genomic variation to about 500,000.28
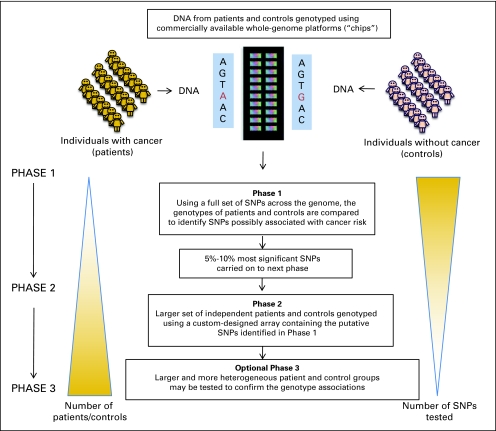
The design of a typical GWAS for cancer susceptibility is shown in Figure 3. DNA from hundreds or thousands of patients and controls is analyzed (genotyped) using commercially available oligonucleotide microarray chips, which allow high throughput analysis of up to a million SNPs in one reaction. Even with per SNP genotyping costs of a fraction of 1 cent, the total cost for genotyping a large sample size using 500,000 or more SNPs can be prohibitive. Cost-effective multistage designs, such as the one shown in Figure 3, retain most of the power of the optimum design often at less than half the cost.29,30 For study designs with three or more phases, the significant SNPs are included for replication testing in different patient-control sample sets.
Fig 3.
Genome-wide association study (GWAS) design. A typical cancer GWAS starts with the selection of a large number of individuals affected with a specific type of cancer and a suitable comparison group without cancer. DNA is isolated from each individual and genotyped using commercially available genome platforms (ie, chips) that assess for common genetic variations in the form of single nucleotide polymorphisms (SNPs) across the entire human genome. Data are reviewed to ensure appropriate genotyping quality. In phase I, stringent statistical methods are used to assess for associations between SNPs passing quality thresholds and cancer risk. In phase II, SNPs found to be significantly associated with cancer risk (approximately 5% to 10% of all SNPs) are tested in a larger set of independent patients and controls using arrays containing the putative SNPs of interest. Some studies proceed to further evaluation of putative SNPs in a third phase using larger and more heterogeneous populations to confirm the genotype associations.
Interpreting GWAS
Initiatives, such as the Strengthening the Reporting of Genetic Association Studies (STREGA),31 have provided guidelines for reporting of genetic association studies and helped standardize GWAS in order to avoid upwardly biased odds ratios (ORs).32 Interpretations of GWAS must take into account possible effects of disease and population heterogeneity, involvement of multiple genetic and environmental factors, and possible gene-gene and gene-environment interactions.
Because power is a function of sample size, minor allele frequency, and the presumed genetic effect size, the detection of modest genetic effects with ORs of 1.3 or lower and minor allele frequencies under 10%, may require more than 10,000 patients and 10,000 controls for adequate statistical power.33 Although large study consortia may approach this size, such efforts are often limited by cost. In addition to ensuring adequate statistical power, patients and controls must be well-matched to avoid population stratification, wherein misleading associations may result from differences in race or ethnicity of those with and without the disease of interest.34–37 Family-based designs can minimize population stratification bias, but also compromise the power of associations.38 As population stratification can lead to significant type I error rates (ie, false positives), principal component analysis39,40 and other methods41,42 have been developed that estimate the population ancestry of patients and controls based on genotypes of a panel of SNPs that are not associated with the disease of interest. Misleading associations may also result from genetic heterogeneity, wherein what appears to be a single disease is in fact a result of genetically separate phenomenon. For example, given the known phenotypic variation in breast cancer, combining all breast cancers into one group may obscure potential genetic associations.
Given multiple comparisons, stringent statistical thresholds are necessary to avoid spurious false-positive associations. The most widely used adjustment for multiple comparisons is the Bonferroni correction, in which the threshold P value (usually 5 × 10−2) is divided by the number of tests performed (approximately 500,000 depending on array used) resulting in an acceptable P value on the order of lower than 1 × 10−7. Even with statistically significant associations, positive results have to be interpreted with caution owing to “winner's curse,” wherein an artificial inflation in measures of association can result from the discovered loci having to pass through stringent significance thresholds.43,44 Regardless of study design and statistical considerations, GWAS are prone to publication bias, as strongly positive associations are more likely to be reported than negative studies.
Interpretation and Clinical Relevance of Results
Genetic associations from GWAS are reported either as relative risks (RR) for cohort studies or as ORs for case-control studies. To date, the majority of cancer GWAS have found risk alleles with only modest associations with cancer risk (OR, 1.1 to 1.5). For example, a risk allele in the fibroblast growth factor receptor 2 (FGR2) gene has been found to be associated with a 1.26-fold increased risk of breast cancer, comparable in magnitude to the modest increased risk of breast cancer conferred by delaying first pregnancy to age older than 35.45 However, a 1.26 risk may be reported to a patient by stating “your risk is increased 26%,” which may seem high, but is far less than the 100-fold increases in breast cancer risk associated with, for example, BRCA mutations. In addition to being misleading, the use of ORs also generally relate to risks over a lifetime, and not over a specific period of time. The very low level of risk associated with most variants found by GWAS remains the greatest barrier to the clinical application of these markers in cancer prevention.
It may also be possible to combine a set of risk alleles into a genomic risk profile for risk estimation of a particular cancer. In a multiplicative model, one simply multiplies one risk factor by the other (eg, 1.2 × 1.5 = 1.8) to derive the combined risk. Such genomic risk panels can then be compared to models that incorporate typical clinical variables (eg, Gail model for breast cancer risk). To date, such comparisons have revealed a very modest, if any, added value to genomic risk profiles.46–48 In addition, more research is needed regarding epistasis (gene-gene interactions) that may confound results of genomic risk profiles. It is also possible that as more variants in the same pathway are discovered, there will be supra-multiplicative interactions that may lead to much higher relative risks. To date such supra-multiplicative interactions have not been found.
It may be common for individuals receiving commercial testing for cancer risk to be told that a certain genetic variant they were found to have is associated with a very high proportion of a type of cancer. The reporting of this type of population attributable risk % (PAR%) of a SNP may be misleading.49 Because of the high allele frequencies of most susceptibility loci identified, the PAR% can be quite high even with very modest elevations in relative risk. In fact, the joint PAR% of common risk variants in some cancers approaches (and may exceed) 100%. These same risk loci, however, may explain only a small fraction of the excess familial relative risk of cancer.14 For example, for the dozen or so SNPs associated with breast cancer risk, the joint PAR% is higher than 70% while the risk loci explain only approximately 8% of the familial relative risk of breast cancer. A large PAR% does not exclude the possibility that other risk alleles exist, that multiple risk factors may occur in the same individual, or that environmental factors may also contribute to disease development.
When clinicians interpret results of GWAS SNPs, it is also important to note that many of the low-risk variants derived by GWAS and used in some commercial panels have not been replicated; a systematic review of 260 meta-analyses of 160 polymorphism gene associations concluded that there is insufficient scientific evidence to conclude that genomic profiles are useful in measuring genetic risk for common diseases or in developing personalized recommendations for disease prevention.50 Finally, since the frequency of risk alleles and the incidence of types of cancer vary within populations, these values may not be generalizable and prospective studies are necessary to directly measure absolute risk, and to judge the accuracy of risk estimates calculated using retrospective methods.
GWAS OF CANCER
In recent years, more than 50 cancer GWAS have been published incorporating at least 15 different malignancies. Nearly all the cancer susceptibility loci identified to date are associated with modest increases in disease risk, with ORs generally below 1.5. Exceptions to this are the risk variants identified in JAK2 in myeloproliferative neoplasms and in KITLG in testicular cancer which are each associated with nearly a three-fold increased risk of disease.51–53 The contribution of the identified susceptibility loci in explaining the genetic basis (heritability) of cancer and the potential clinical implications of GWAS findings is illustrated below in three common malignancies.
Breast Cancer
Breast cancer has been at the forefront of cancer GWAS with at least 13 independent loci implicated in disease risk (Table 2).45,54–81 Of identified susceptibility loci, the most strongly associated risk SNP, with an OR of 1.26, was in FGFR2. The protein encoded by FGFR2 is a member of the FGFR family and is overexpressed in 5% to 10% of breast tumors.82,83 While the precise mechanism(s) of FGFR2 deregulation in breast cancer etiology remains unknown, fine mapping of the region suggests that the causative variants lie in intron 2 of FGFR2. The 10q26 locus mapping to FGFR2 was implicated in a number of breast cancer GWAS using different patient populations and appears to be strongest in estrogen receptor–positive breast cancers.84–86 Some of the other susceptibility loci identified in gene-containing regions have not been implicated in cancer previously (eg, TOX3, LSP1, STXBP4) and are being evaluated for their potential role in carcinogenesis. Other SNPs lie in regions devoid of genes (ie, the 8q24 region) where research into their impact on near-by genes is under investigation. As most GWAS-based associations have correlated with estrogen receptor–positive breast cancers, efforts to identify risk SNPs predicting for estrogen receptor–negative or triple-negative breast cancers are ongoing.
Table 2.
Genome-Wide Association Studies for Breast, Prostate and Colorectal Cancers
| Locus | Implicated Gene | SNP | Per Allele OR Ranges* | Reference |
|---|---|---|---|---|
| Breast | ||||
| 1p11.2 | Pericentric | rs11249433 | 1.16 | 54 |
| 2q35 | Intergenic | rs13387042 | 1.2-1.25 | 54,55 |
| 3p24.1 | SLC4A7 | rs4973768 | 1.11 | 56 |
| 5p12 | Intergenic (MRPS30) | rs4415084 | 1.16-1.19 | 57 |
| rs10941679 | ||||
| 5q11.2 | MAP3K1, MIER3, C5orf35 | rs889312 | 1.13 | 45 |
| 6q22.33 | ECHDC1, RNF146 | rs2180341 | 1.41 | 58 |
| 6q25.1 | ESR1 | rs2046210 | 1.29 | 59 |
| 8q24 | Intergenic | rs13281615 | 1.08 | 45 |
| 10q26 | FGFR2 | rs2981582 | 1.20-1.29 | 45 |
| rs1219648 | 57,60 | |||
| rs1078806 | 58 | |||
| 11p15.5 | LSP1 | rs3817198 | 1.07 | 45 |
| 14q24.1 | RAD51L1 | rs999737 | 1.06 | 54 |
| 16q12 | TNRC9 (TOX3), LOC643714 | rs3803662 | 1.16-1.28 | 45,54,55 |
| 17q23 | STXBP4 | rs6504950 | 1.05 | 56 |
| Prostate | ||||
| 2p15 | EHBP1 | rs721048 | 1.15 | 61 |
| 2p21 | THADA | rs1465618 | 1.08 | 62 |
| 2q31 | ITGA6 | rs12621278 | 1.33 | 62 |
| 3p12 | Intergenic | rs2660753 | 1.18 | 64 |
| 3q21 | Intergenic | rs10934853 | 1.12 | 63 |
| 4q22 | PDLIM5 | rs17021918 | 1.11 | 62 |
| rs12500426 | 1.08 | |||
| 4q24 | TET2 | rs7679673 | 1.10 | 62 |
| 6q25 | SLC22A3 | rs9364554 | 1.17 | 64 |
| 7p15 | Intergenic | rs12155172 | 1.05 | 62 |
| 7p15.2-15.1 | JAZF1 | rs10486567 | 1.12-1.35 | 65 |
| 7q21.3 | LMTK2 | rs6465657 | 1.12 | 64 |
| 8p21 | NKX3-1 | rs2928679 | 1.05 | 62 |
| rs1512268 | 1.18 | |||
| 8q24 | Intergenic | HapC 14 SNPs | 2.10 | 66 |
| rs16901979 | 1.79-1.80 | 63,66 | ||
| DG8S737 | 1.64 | 67 | ||
| rs1447295 | 1.36-1.60 | 63,66–69 | ||
| rs1016343 | 1.37 | 64 | ||
| rs6983267 | 1.26-1.42 | 64,65,69 | ||
| rs4242382 | 1.41-1.87 | 64,65 | ||
| rs1006908 | 1.15 | 70 | ||
| rs620861 | 1.17 | 71 | ||
| rs16902094 | 1.14 | 63 | ||
| 10q11.2 | MSMB | rs10993994 | 1.16-1.25 | 64,65 |
| 10q26.13 | CTBP2 | rs4962416 | 1.17-1.20 | 65 |
| 11p15 | IGF2, IGF2AS, INS, TH | rs7127900 | 1.22 | 62 |
| 11q13.2 | Intergenic | rs10896449 | 1.10-1.28 | 65 |
| rs7931342 | 1.19 | 64 | ||
| rs11228565 | 1.23 | 63 | ||
| 17q12 | TCF2 (HNF1B) | rs4430796 | 1.18-1.38 | 63,65,72 |
| rs7501939 | 1.41 | 64 | ||
| 17q24.3 | Intergenic | rs1859962 | 1.20-1.26 | 64,72 |
| 19q13.2 | PPP1R14A | rs8102476 | 1.12 | 63 |
| 19q13.41 | KLK2, KLK3 | rs2735839 | 1.20 | 64 |
| 22q13 | TTLL1, BIK, MCAT, PACSIN2 | rs5759167 | 1.20 | 62 |
| 22q13 | TNRC6B | rs9623117 | 1.18 | 73 |
| Xp11.23-p11.22 | NUDT10, NUDT11, | rs5945619 | 1.19 | 64 |
| LOC340602, GSPT2, MAGED1 | rs5945572 | 1.23 | 61 | |
| Colorectal | ||||
| 8q23.3 | EIF3H | rs16892766 | 1.25 | 74 |
| 8q24.21 | LOC727677, POU5F1P1 | rs10505477 | 1.17 | 75 |
| Intergenic | rs6983267 | 1.17-1.27 | 74,76 | |
| rs7014346 | 1.19 | 78 | ||
| 10p14 | Intergenic | rs10795668 | 1.11 | 74 |
| 11q23 | Intergenic | rs3802842 | 1.12 | 78 |
| 14q22-q23 | BMP4 | rs4444235 | 1.11 | 79 |
| 15q13 | Intergenic | rs4779584 | 1.23-1.26 | 74,80 |
| GREM1 | rs10318 | 1.19 | 80 | |
| 16q22.11 | CDH1 | rs9929218 | 1.10 | 79 |
| 18q21.1 | SMAD7 | rs4939827 | 1.16-1.20 | 78,81 |
| 19q13.11 | RHPN2 | rs10411210 | 1.15 | 79 |
| 20p12.3 | Intergenic | rs961253 | 1.12 | 79 |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio.
ORs < 1 in the original publication have been converted to ORs > 1 for the alternate allele.
Table 1.
Definition of Terms
| Term | Definition |
|---|---|
| High-penetrance cancer susceptibility syndrome | A cancer predisposition syndrome wherein a mutation in the implicated gene produces a phenotype in a high proportion of individuals who carry the mutation |
| SNP | A variation in DNA sequence where a single nucleotide is replaced by another; SNPs are thought to represent the most common form of genetic variation in the genome |
| Clinical validity | The accuracy with which a genetic test can identify or predict the presence or absence of a particular clinical condition taking into account the specificity, sensitivity as well as the penetrance of the genetic variation |
| Clinical utility | The degree to which the use of a test informs clinical decision making and leads to improved health outcomes |
| Candidate gene studies | A study that identifies genetic associations by assessing genetic variants that are suspected of being involved in the expression of a particular trait or disease |
| GWAS | A systematic hypothesis-free search for genetic variations, in the form of SNPs, across the genome to identify genetic associations with a disease or trait |
| Minor allele frequency | Frequency of the less common allele of a polymorphic locus |
| Risk allele | Any one of several variants of a gene that occupy the same position locus on a chromosome and is associated with risk of a particular trait or disease |
| Linkage disequilibrium | The non-random association of alleles of different polymorphisms in a population |
| Population stratification | Genetic differences between cases and controls not due to the trait or disease being studied but rather due to sampling of populations with different ethnicities or ancestries; population stratification can lead to erroneous genetic associations |
| Genetic heterogeneity | Multiple genetic mutations can result in the same disease phenotype |
| Bonferroni correction | A multiple-comparison correction used when several statistical tests are being performed simultaneously; in GWAS, using the Bonferroni correction helps to avoid spurious genetic associations |
| Winner's curse | Results of GWAS may be subject to varying degrees of upward bias in effect size estimates due to having to pass through stringent statistical thresholds |
| Epistasis | Gene-to-gene interactions where the effects of one gene are modified by one or several other genes |
| PAR% | The reduction in incidence of a particular disease that would be observed if the population was entirely unexposed, compared with its actual exposure; in genetic epidemiology, combining knowledge of risk allele frequency and genotypic relative risk, the attributable fraction of cases that would not occur if no one in the population had the risk allele can be determined |
| FRR | The ratio of disease risk in biological relatives of affected individuals compared with disease risk in the general population; in general, the higher the FRR, the stronger the genetic effect |
| Publication bias | A type of reporting bias wherein statistically significant or positive results are more likely to be published |
Abbreviations: SNP, single nucleotide polymorphism; GWAS, genome-wide association studies; PAR, population attributable risk; FRR, familial relative risk.
Importantly, as compared to the high-penetrance breast cancer susceptibility genes, the magnitude of risk associated with each of the risk SNPs identified in breast cancer GWAS is modest, with ORs largely ranging from 1.1 to 1.4 (Fig 1). The contribution of these loci to the familial risk of breast cancer is no more than approximately 8%14 thereby still leaving the majority of the familial risk of breast cancer unexplained. Modeling studies have predicted that together the seven most common breast cancer–associated SNPs would add little in terms of improved discriminatory accuracy when compared to, or when used in conjunction with, a standard clinical breast cancer risk model (eg, the Gail model).46,47 The addition of information on 10 breast cancer risk SNPs to the Gail model predicted the risk of breast cancer only slightly better than the clinical model alone.48 Similarly, in a cohort of BRCA mutation–negative women with and without breast cancer, incorporation of a genotypic risk score had limited discriminatory accuracy; however, the potential for reclassification of a clinically relevant proportion of women for altered recommendations for chemoprevention or magnetic resonance imaging screening has not been excluded.87 While these clinical studies demonstrate the potential use of SNPs in cancer risk prediction, to date, breast cancer risk stratification based on SNPs remains premature.
Colon Cancer
Although familial susceptibility accounts for as much as 35% of colorectal cancer (CRC)17, only approximately 6% of all CRCs occur in the setting of a known genetic predisposition syndrome. Hereditary nonpolyposis colorectal cancer (Lynch syndrome) and FAP account for the majority of cases, while rare inherited syndromes, such as Peutz-Jeghers syndrome, juvenile polyposis, attenuated FAP, and MYH-associated polyposis, explain only 1% of CRCs.
The seven GWAS in CRC have identified 10 susceptibility loci (Table 2). Multiple CRC GWAS identified the 8q24 locus, containing the rs6983267 SNP, with an associated approximately 1.2-fold increased risk of disease.74–76,78 This SNP was also associated with adenoma risk with an OR of 1.16.88 While the 8q24 region is devoid of known genes, two recent publications suggest that the rs6983267 SNP may be connected to enhanced Wnt signaling and subsequent MYC regulation, known pathways in carcinogenesis.89,90
Nearly half of the susceptibility loci in CRC are in linkage disequilibrium or are nearby genes of the transforming growth factor beta (TGF-ß) signaling pathway previously implicated in carcinogenesis.91,92 Increased TGF-ß1 expression has been linked to tumor progression and recurrence in CRC and germline mutations in components of the TGF-ß signaling pathway, namely SMAD4 and BMPR1A, are responsible for the high-penetrance juvenile polyposis syndrome. Genes implicated from CRC GWAS along the TGF-ß pathway include: SMAD7, RHPN2, BMP4, BMP2, and GREM1.
Overall, the 10 risk loci identified are associated with modest 1.1- to 1.25-fold increases in the relative risk of CRC and account for only approximately 6% of the excess familial risk of CRC.79 There is currently no evidence that individual SNPs or panels of SNPs add to the discriminatory accuracy of current clinical risk criteria based on age, personal and family history of adenomas or CRC, and pre-existing inflammatory bowel disease. Nor is there convincing evidence that these SNPs correlate with survival, early-age at onset, site of tumor, or a histologically more aggressive subset of disease.79,93 By way of comparison, the relative risk for CRC for an individual carrying the 8q24 risk SNP is approximately 1.2- versus a 1.8-fold increased risk for the first-degree relatives of individuals with an adenoma94 and a 2.5-fold increased risk for individuals with a first-degree relative with CRC.95 At this time, recommendations for CRC screening would not be altered from that of the general population based solely on the presence of a CRC-associated risk allele.
Prostate Cancer
There is strong evidence for genetic predisposition to prostate cancer from family studies, including a two- to three-fold increased risk of disease in first-degree relatives of affected men.96,97 Germline mutations in genes such as BRCA2 have been found to be associated with prostate cancer risk, however, such mutations explain less than 10% of the familial risk of prostate cancer.15
GWAS have identified more than a dozen prostate cancer risk loci with ORs mostly ranging from 1.2 to 2.0 (Table 2). As in CRC, bladder, and breast cancer, a number of prostate cancer GWAS identified risk SNPs mapping to the 8q24 locus. In a multiethnic study, Haiman et al77 identified seven independent risk variants in the 8q24 region and, interestingly, observed that the risk variants were most common in the African-American population, possibly suggesting a partial explanation for the higher incidence of prostate cancer in African-American men. Risk SNPs at or near genes (ie, MSMB, KLK3 and KLK2) with plausible roles in prostate carcinogenesis have also been found.64,65
The joint contribution of identified loci to the familial risk of prostate cancer approaches 20%. However, the contribution of the risk SNPs in improving the discriminatory accuracy of clinical models (eg, those based on prostate-specific antigen [PSA]) is likely to be modest, although in at least in one instance a kallikrein-linked SNP is highly correlated with serum levels of proteins associated with prostate cancer risk.98 Studies correlating individual risk SNPs to more aggressive prostate cancer subsets have largely been unrevealing69,99–101 and the risk SNPs have not been associated with survival after diagnosis.102,103 In addition, a family history of prostate cancer still confers a greater risk than the presence of any individual risk allele.101–103
The impact of carrying multiple risk SNPs on prostate cancer risk was assessed by Zheng et al102 with results demonstrating that men who carried ≥ four of five possible risk SNPs had a 4.5-fold increased risk of disease. Importantly, there was no evidence that the risk SNPs were associated with disease aggressiveness, earlier age at diagnosis or presence or absence of family history. A subsequent analysis demonstrated that these five risk alleles do not improve prediction models for disease risk or disease-specific mortality once known risk factors (ie, age, PSA, family history) or prognostic factors (ie, Gleason score, diagnostic PSA, stage, age, primary treatment) are taken into account.103 To date, the validity and clinical utility of using individual or multiple SNP panels as a screening test for prostate cancer have not been demonstrated. Additional studies seeking to identify genetic variants that predict for early-onset or more aggressive disease and may be used for risk stratification are in progress.
Other Malignancies
As presented in Table 3,51–53,104–132 dozens of other SNPs have been associated with an increased risk of cancers of the lung, skin, thyroid, ovaries, pancreas, and other sites. With the exception of SNPs for testicular germ cell tumors and myeloproliferative neoplasms, none of these SNPs has been found to have a risk greater than 2.0.
Table 3.
Genome-Wide Association Studies for Other Cancers
| Locus (entrez gene) | Implicated Gene | SNP | Per Allele OR Ranges* | Reference |
|---|---|---|---|---|
| Lung | ||||
| 5pter-p15.33 | CLPTM1L, TERT | rs401681 | 1.16 | 104 |
| rs402710 | 1.18 | 105 | ||
| rs2736100 | 1.14 | 105 | ||
| 1.23 (adenocarcinoma) | 106 | |||
| 5p15.33 | rs4975616 | 1.16 | 107 | |
| 6p21.33-p21.3 | BAT3, MSH5 | rs3117582 | 1.24 | 104,107 |
| 15q24-q25 | CHRNA3, CHRNA5, CHRNB4, | rs8034191 | 1.30-1.38 | 108–110 |
| PSMA4, LOC123688 | ||||
| rs1051730 | 1.31-1.35 | 105,108,110 | ||
| rs8042374 | 1.33, NR | 104,107 | ||
| 15q25.1 | CHRNA3 | rs938682 | 1.33 | 107 |
| rs12914385 | 1.29 | 107 | ||
| BCC and CM | ||||
| 1p36.13 | PADI4, PADI6, RCC2, AHRGEF10L | rs7538876 | 1.28 BCC | 111 |
| 1q42.11-q42.3 | RHOU | rs801114 | 1.28 BCC | 111 |
| 7q32 | KLF14 | rs157935 | 1.23 BCC | 112 |
| 9p21 | MTAP, CDKN2A | rs7023329 | 1.18 CM | 113 |
| 9p21 | CDKN2A, CDKN2B | rs2151280 | 1.19 BCC | 112 |
| 9p23 | TYRP1 | rs1408799 | 1.15 CM | 114 |
| 11q14-q21 | TYR | rs1126809 | 1.21 CM | 114 |
| 1.14 BCC | ||||
| rs1393350 | 1.29 CM | 113 | ||
| 12q12-q13 | KRT5 | rs11170164 | 1.35 BCC | 112 |
| 16q24 | MC1R | rs258322 | 1.67 CM | 113 |
| 20q11.2-q12 | ASIP | Hap rs1015362G and rs4911414T | 1.45 CM | 114 |
| 1.35 BCC | ||||
| 20q11.22 | CDC91L1 (PIGU) | rs910873 and rs1885120 | 1.75 CM | 115 |
| Urinary bladder | ||||
| 3q28 | TP63 | rs710521 | 1.19 | 116 |
| 8q24.21 | MYC, BC042052 | rs9642880 | 1.22 | 116 |
| 8q24.2 | PSCA | rs2294008 | 1.15 | 117 |
| Neuroblastoma | ||||
| 2q35 | BARD1 | rs3768716 | 1.68 | 118 |
| rs6435862 | 1.68 | |||
| 6p22.3 | FLJ22536, FLJ44180 | rs4712653 | 1.35 | 119 |
| rs9295536 | 1.32 | |||
| rs6939340 | 1.37 | |||
| Glioma | ||||
| 5p15.33 | TERT | rs2736100 | 1.27 | 120 |
| 8q24.21 | CCDC26 | rs4295627 | 1.36 | 120 |
| 9p21.3 | CDKN2B | rs1412829 | 1.42 | 121 |
| rs4977756 | 1.24 | 120 | ||
| 11q23.3 | PHLDB1 | rs498872 | 1.18 | 120 |
| 20q13.33 | RTEL1 | rs6010620 | 1.51 | 121 |
| 1.28 | 120 | |||
| Acute lymphoblastic leukemia (childhood) | ||||
| 7p12.2 | IKZF1 | rs4132601 | 1.69 | 122 |
| 10q21.2 | ARID5B | rs10994982 | 1.62 | 123 |
| Maps to 10q11.22 by HGNC | rs10821936 | 1.91 | 123 | |
| rs7089424 | 1.65 | 122 | ||
| 14q11.2 | CEBPE | rs2239633 | 1.34 | 122 |
| Chronic lymphocytic leukemia | ||||
| 2q13 | ACOXL, BCL2L11 | rs17483466 | 1.39 | 124 |
| 2q37.1 | SP140 | rs13397985 | 1.41 | 124 |
| 2q37.3 | FARP2 | rs757978 | 1.39 | 125 |
| 6p25-p23 | IRF4 | rs872071 | 1.54 | 124 |
| 8q24.21 | Intergenic | rs2456449 | 1.26 | 125 |
| 11q24.1 | GRAMD1B | rs735665 | 1.45 | 124 |
| 15q21.3 | NEDD4, RFX7 | rs7169431 | 1.36 | 125 |
| 15q23 | Intergenic | rs7176508 | 1.37 | 124 |
| 16q24.1 | IRF8 | rs305061 | 1.22 | 125 |
| 19q13.2-q13.3 | PRKD2, STRN4 | rs11083846 | 1.35 | 124 |
| Follicular lymphoma | ||||
| 6p21.33 | STG, PSORS1 | rs6457327 | 1.69 | 126 |
| Thyroid (papillary and follicular) | ||||
| 9q22.33 | Intergenic | rs965513 | 1.75 | 127 |
| 14q13.3 | Intergenic | rs944289 | 1.37 | 127 |
| Myeloproliferative neoplasms | ||||
| 9p24.1 | JAK2 | rs10974944 | 3.10 | 52 |
| Testicular germ cell cancer | ||||
| 4q24 | Intergenic | rs4699052 | 1.21 | 53 |
| 5q31.3 | SPRY4 | rs4324715 | 1.37 | 51 |
| rs4624820 | 1.37 | 53 | ||
| rs6897876 | 1.39 | 51 | ||
| 6p21.3 | BAK1 | rs210138 | 1.50 | 53 |
| 12q22 | KITLG | rs995030 | 2.55 | 53 |
| rs3782179 | 3.08 | 51 | ||
| rs4474514 | 3.07 | 51 | ||
| rs1508595 | 2.69 | 53 | ||
| Pancreatic | ||||
| 1q32.1 | NR5A2 | rs3790844 | 1.30 | 127a |
| 5p15.33 | TERT-CLPTM1L | rs401681 | 1.19 | 127a |
| 9q34 | ABO | rs505922 | 1.20 | 128 |
| 13q22.1 | Intergenic | rs9543325 | 1.26 | 127a |
| Ovarian | ||||
| 9p22 | BNC2, CNTLN, LOC648570 | rs3814113 | 1.22 | 129 |
| Gastric (diffuse) | ||||
| 8q24.3 | PSCA | rs2976392 | 1.62 (Japan) | 130 |
| 1.90 (Korea) | ||||
| Esophageal (squamous cell) | ||||
| 4q21-23 | ADH1B | rs1229984 | 1.79 | 131 |
| 12q24 | ALDH2 | rs671 | 1.67 | 131 |
| Single locus 5p15.33 Multiple cancers | TERT-CLPTM1L | rs401681 | 132 | |
| Prostate | 1.07 | |||
| Lung | 1.15 | |||
| BCC | 1.25 | |||
| Urinary bladder | 1.12 | |||
| Cervical | 1.31 |
Abbreviations: OR, odds ratio; NR, not reported; BCC, basal cell carcinoma; CM, cutaneous melanoma.
ORs < 1 in the original publications have been converted to ORs > 1 for the alternate allele.
CHALLENGES AND FUTURE DIRECTIONS
Identification of Functional Gene Variants and Targeted Prevention
Perhaps one of the most important advantages of GWAS is the use of an agnostic approach wherein genes previously not implicated in cancer susceptibility may be identified. For example, genes such as FGFR2, overexpressed in 5% to 10% of breast tumors, and MSMB, a gene coding for PSP94, an immunoglobulin binding factor synthesized by prostate epithelial cells, seem particularly plausible breast and prostate candidate cancer susceptibility genes, respectively. Variants in genes previously linked to cancer pathogenesis, such as KITLG in testicular cancer, may also help to explain cancer predisposition. A challenge to date has been that the majority of risk SNPs implicated in cancer susceptibility have not been associated with functional changes in the genes residing near the loci of these SNPs. For some of the identified loci, fine-scale genetic mapping and deep sequencing of the implicated regions is ongoing and will hopefully lead to identification of the causal genetic change. For loci that map to genes or nearby genes, functional analyses will be crucial in helping to tailor targeted preventive approaches, such as the potential use of tyrosine kinase inhibitors to prevent breast cancer in women with FGFR2 pathway deregulation.
Incorporation into Prospective Trials
Before incorporating risk variants into individualized cancer risk assessment, the hypothesis-generating GWAS results should be validated in prospective studies in heterogeneous populations to demonstrate the efficacy of these variants in predicting an individual's risk of disease and associated disease outcome. The importance of prospective clinical studies cannot be underestimated for several reasons. First, prospective studies can take into account environmental factors, and can also identify associations with disease-specific mortality or more aggressive clinical phenotypes. Second, prospective studies allow better estimates of absolute risk, sensitivity, specificity as well as positive and negative predictive value that take into account the specific cancer incidences in the target population. These measures are more clinically useful than ORs from retrospective studies. Other measures may include derivation of the discriminatory accuracy of genomic tests using receiver operating characteristic curves that plot graphically the true-positive rate (sensitivity) against the false-positive rate (1 − specificity). Such measures have only recently been applied to GWAS-derived risk markers,46–48,133 and analysis from initial receiver operating characteristic curves indicate that even with strong genetic associations, effective discrimination between patients and controls is not guaranteed.133 In addition, using methods of calibration testing, panels of risk variants should be compared to the predictive power of already existing cancer risk models that incorporate a variety of clinical variables, such as the predictive models available for breast and prostate cancer and the more recently developed CRC risk model for those older than 50 years. Finally, because experience with communication of high-penetrance cancer risk information has shown that people have difficulty understanding probabilistic information134,135 and tend to persist in inaccurate estimates of their risk even in the context of specialized counseling,136–139 the appropriate delivery mode of genetic risk information to individuals must be determined.
Current Challenges
At present, the modest increased risks of cancer associated with the known genetic variants are, for the most part, not medically actionable. There is a lack of data to justify use of individual SNPs or panels of risk variants as independent risk predictors over known clinical variables. Increased population screening based on genomic risk profiles may be harmful and lead to increased cancer screening tests associated with significant false-positive rates (eg, breast magnetic resonance imaging, serum PSA), to invasive tests (eg, colonoscopy), to unnecessary subsequent medical interventions (eg, biopsy), and to overall increased medical costs. Results of genomic risk profiles may also provide false reassurance to individuals who may, in fact, be at high-risk for a particular cancer based on other clinical variables.
Despite the paucity of evidence supporting the use of GWAS results for clinically useful cancer risk prediction, personal genomic testing is currently being offered directly to consumers outside the context of the health care system. Such genome scans may include cancer-specific scans for breast, prostate, colorectal, thyroid, skin, lung and urinary bladder cancer risk. Patients who obtain such tests through nontraditional channels, may nonetheless expect that their physicians will assist them with the interpretations of such data.140,141 Indeed, physicians, including oncologists, have already been asked to explain results of genomic profiles to their patients. Despite the paucity of evidence for clinical utility, 52% to 75% of physicians in an initial survey felt that genomic tests had the potential to impact clinical management.8 In order to inform both patients and physicians, there has been a call for increased regulation or guidance with respect to the provision of such tests. However, such intervention has been limited as many genetic and genomic tests fall outside of the purview of the US Food and Drug Administration and other regulatory agencies.9 Such regulations will clearly be required for pharmacogenomic applications of inherited genetic variants which predict response and toxicity of preventive and therapeutic pharmacologic interventions. Recognizing the scientific merits as well as the limitations of recent discoveries of genetic variants, a recent update by the American Society of Clinical Oncology emphasizes the importance of further research to demonstrate the validity and clinical utility of genomic profiles.142
Future Perspectives
GWAS are powerful tools that have enhanced our understanding of cancer genetics and will inevitably lead to the identification of novel pathways of carcinogenesis. Research regarding other forms of genetic variation in the human genome, including genomic structural changes and sequence variation, is just beginning to emerge and will provide further insight into the genetic basis of complex diseases. As our knowledge of the human genome continues to rapidly expand, health care providers must also be aware of the evidence base required for genomic profiles of cancer risk to be effectively incorporated into the practice of preventive oncology.
Footnotes
Supported in part by The Robert and Kate Niehaus Clinical Cancer Genetics Initiative, The Lymphoma Foundation, the Breast Cancer Research Foundation, and funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views of the Department of Health and Human Services, nor does its mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Mark E. Robson, Pfizer (C); Noah D. Kauff, Wyeth (C) Stock Ownership: Bert Gold, GlaxoSmithKline, Quest Diagnostics Honoraria: Karen E. Hurley, Genzyme Research Funding: Mark E. Robson, Kudos Pharmaceuticals; Robert J. Klein, sanofi-aventis Expert Testimony: Noah D. Kauff, Wyeth (C) Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Zsofia K. Stadler, Peter Thom, Kenneth Offit
Financial support: Kenneth Offit
Administrative support: Kenneth Offit
Collection and assembly of data: Zsofia K. Stadler, Peter Thom, Vincent Devlin
Data analysis and interpretation: Zsofia K. Stadler, Peter Thom, Kenneth Offit
Manuscript writing: Zsofia K. Stadler, Peter Thom, Mark E. Robson, Jeffrey N. Weitzel, Noah D. Kauff, Karen E. Hurley, Vincent Devlin, Bert Gold, Robert J. Klein, Kenneth Offit
Final approval of manuscript: Zsofia K. Stadler, Peter Thom, Mark E. Robson, Jeffrey N. Weitzel, Noah D. Kauff, Karen E. Hurley, Vincent Devlin, Bert Gold, Robert J. Klein, Kenneth Offit
REFERENCES
- 1.Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23:276–292. doi: 10.1200/JCO.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 2.National Comprehensive Cancer Network. Clinical practice guidelines in oncology. http://www.nccn.org/professionals/physician_gls/f_guidelines.asp.
- 3.Robson M, Offit K. Clinical practice: Management of an inherited predisposition to breast cancer. N Engl J Med. 2007;357:154–162. doi: 10.1056/NEJMcp071286. [DOI] [PubMed] [Google Scholar]
- 4.Kauff ND, Domchek SM, Friebel TM, et al. Risk-reducing salpingo-oophorectomy for the prevention of BRCA1- and BRCA2-associated breast and gynecologic cancer: A multicenter, prospective study. J Clin Oncol. 2008;26:1331–1337. doi: 10.1200/JCO.2007.13.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: A systematic review. JAMA. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 6.Offit K. Genomic profiles for disease risk: Predictive or premature? JAMA. 2008;299:1353–1355. doi: 10.1001/jama.299.11.1353. [DOI] [PubMed] [Google Scholar]
- 7.Khoury MJ, McBride C, Schully SD, et al. The Scientific Foundation for Personal Genomics: Recommendations from a National Institutes of Health Centers for Disease Control and Prevention Multidisciplinary Workshop. Genet Med. 2009;9:559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolor K, Liu T, St Pierre J, et al. Health care provider and consumer awareness, perceptions, and use of direct-to-consumer personal genomic tests, United States, 2008. Genet Med. 2009;11:595. doi: 10.1097/GIM.0b013e3181b1cc2c. [DOI] [PubMed] [Google Scholar]
- 9.Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–450. doi: 10.1097/01.gim.0000227935.26763.c6. [DOI] [PubMed] [Google Scholar]
- 10.Burke W. Clinical validity and clinical utility of genetic tests. Curr Protoc Hum Genet. 2009;60:9.15.1–9.15.7. doi: 10.1002/0471142905.hg0915s60. [DOI] [PubMed] [Google Scholar]
- 11.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer: Analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 12.Goldgar DE, Easton DF, Cannon-Albright LA, et al. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 13.Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Res. 1999;1:14–17. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemminki K, Försti A, Lorenzo Bermejo J. New cancer susceptibility loci: Population and familial risks. Int J Cancer. 2008;123:1726–1729. doi: 10.1002/ijc.23702. [DOI] [PubMed] [Google Scholar]
- 15.Edwards SM, Kote-Jarai Z, Meitz J, et al. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet. 2003;72:1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pharoah P, Antoniou AC, Easton DF, et al. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 17.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Athma P, Rappaport R, Swift M. Molecular genotyping shows that ataxia-telangiectasia heterozygotes are predisposed to breast cancer. Cancer Genet Cytogenet. 1996;92:130–134. doi: 10.1016/s0165-4608(96)00328-7. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein JL, Bernstein L, Thompson WD, et al. ATM variants 7271T>G and IVS10-6T>G among women with unilateral and bilateral breast cancer. Br J Cancer. 2003;89:1513–1516. doi: 10.1038/sj.bjc.6601289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bretsky P, Haiman CA, Gilad S, et al. The relationship between twenty missense ATM variants and breast cancer risk: The multiethnic cohort. Cancer Epidemiol Biomarkers Prev. 2003;12:733–738. [PubMed] [Google Scholar]
- 21.CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 23.Thompson D, Seal S, Schutte M, et al. A multicenter study of cancer incidence in CHEK2 1100delC mutation carriers. Cancer Epidemiol Biomarkers Prev. 2006;15:2542–2545. doi: 10.1158/1055-9965.EPI-06-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 25.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 27.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 28.Carlson CS, Eberle MA, Kruglyak L, et al. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 29.Elston RC, Lin D, Zheng G. Multistage sampling for genetic studies. Annu Rev Genomics Hum Genet. 2007;8:327–342. doi: 10.1146/annurev.genom.8.080706.092357. [DOI] [PubMed] [Google Scholar]
- 30.Satagopan JM, Elston RC. Optimal two-stage genotyping in population-based association studies. Genet Epidemiol. 2003;25:149–157. doi: 10.1002/gepi.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the Reporting of Genetic Association studies (STREGA): An extension of the STROBE statement. Ann Intern Med. 2009;150:206–215. doi: 10.7326/0003-4819-150-3-200902030-00011. [DOI] [PubMed] [Google Scholar]
- 32.Garner C. Upward bias in odds ratio estimates from genome-wide association studies. Genet Epidemiol. 2007;31:288–295. doi: 10.1002/gepi.20209. [DOI] [PubMed] [Google Scholar]
- 33.Wang WY, Barratt BJ, Clayton DG, et al. Genome-wide association studies: Theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 34.Wacholder S, Rothman N, Caporaso N. Counterpoint: Bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:513–520. [PubMed] [Google Scholar]
- 35.Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. Am J Hum Genet. 2002;71:304–311. doi: 10.1086/341719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Cardon LR, Phillips MS, et al. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 37.Freedman ML, Reich D, Penney KL, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 38.Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: The sib transmission/disequilibrium test. Am J Hum Genet. 1998;62:450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Li Q, Yu K. Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet Epidemiol. 2008;32:215–226. doi: 10.1002/gepi.20296. [DOI] [PubMed] [Google Scholar]
- 41.Satten GA, Flanders WD, Yang Q. Accounting for unmeasured population substructure in case-control studies of genetic association using a novel latent-class model. Am J Hum Genet. 2001;68:466–477. doi: 10.1086/318195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao R, Boehnke M. Quantifying and correcting for the winner's curse in genetic association studies. Genet Epidemiol. 2009;33:453–462. doi: 10.1002/gepi.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zollner S, Pritchard JK. Overcoming the winner's curse: Estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100:1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101:959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Offit K. Breast cancer single-nucleotide polymorphisms: Statistical significance and clinical utility. J Natl Cancer Inst. 2009;101:973–975. doi: 10.1093/jnci/djp184. [DOI] [PubMed] [Google Scholar]
- 50.Janssens AC, Gwinn M, Bradley LA, et al. A critical appraisal of the scientific basis of commercial genomic profiles used to assess health risks and personalize health interventions. Am J Hum Genet. 2008;82:593–599. doi: 10.1016/j.ajhg.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanetsky PA, Mitra N, Vardhanabhuti S, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilpivaara O, Mukherjee S, Schram AM, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–459. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapley EA, Turnbull C, Al Olama AA, et al. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 56.Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 58.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eeles RA, Kote-Jarai Z, Al Olama AA, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 65.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 66.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 67.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 68.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 70.Al Olama AA, Kote-Jarai Z, Giles GG, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 71.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 73.Sun J, Zheng SL, Wiklund F, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69:10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 75.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 76.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 77.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaeger E, Webb E, Howarth K, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 81.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 82.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:Reviews 3005.1–3005.12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rebbeck TR, DeMichele A, Tran TV, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30:269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia-Closas M, Chanock S. Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin Cancer Res. 2008;14:8000–8009. doi: 10.1158/1078-0432.CCR-08-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garcia-Closas M, Hall P, Nevanlinna H, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Comen E, Balistreri L, Kirchhoff T, et al. Breast cancer risk single nucleotide polymorphism (SNP) genotypes do not correlate with risk estimates from existing prediction models. 32nd Annual Meeting of the San Antonio Breast Cancer Symposium December 9-13, 2009, San Antonio, TX (abstr 901) [Google Scholar]
- 88.Berndt SI, Potter JD, Hazra A, et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. 2008;17:2665–2672. doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 91.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 93.Curtin K, Lin WY, George R, et al. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev. 2009;18:616–621. doi: 10.1158/1055-9965.EPI-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Winawer SJ, Zauber AG, Gerdes H, et al. Risk of colorectal cancer in the families of patients with adenomatous polyps” National Polyp Study Workgroup. N Engl J Med. 1996;334:82–87. doi: 10.1056/NEJM199601113340204. [DOI] [PubMed] [Google Scholar]
- 95.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001;96:2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 96.Zeegers MP, Jellema A, Ostrer H. Empiric risk of prostate carcinoma for relatives of patients with prostate carcinoma: A meta-analysis. Cancer. 2003;97:1894–1903. doi: 10.1002/cncr.11262. [DOI] [PubMed] [Google Scholar]
- 97.Bruner DW, Moore D, Parlanti A, et al. Relative risk of prostate cancer for men with affected relatives: Systematic review and meta-analysis. Int J Cancer. 2003;107:797–803. doi: 10.1002/ijc.11466. [DOI] [PubMed] [Google Scholar]
- 98.Klein RJ, Hallden C, Cronin AM, et al. Blood biomarker levels to aid discovery of cancer-related single nucleotide polymorphisms: Kallikreins and prostate cancer. Cancer Prev Res (Phila PA) 2010;3:611–619. doi: 10.1158/1940-6207.CAPR-09-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 100.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: Association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fitzgerald LM, Kwon EM, Koopmeiners JS, et al. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: Associations with family history and clinical features. Clin Cancer Res. 2009;15:3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 103.Salinas CA, Koopmeiners JS, Kwon EM, et al. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–372. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: A genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 110.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stacey SN, Gudbjartsson DF, Sulem P, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 112.Stacey SN, Sulem P, Masson G, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 115.Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Treviño LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 125.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Skibola CF, Bracci PM, Halperin E, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127a.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Song H, Ramus SJ, Tyrer J, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Study Group of Millennium Genome Project for Cancer. Sakamoto H, Yoshimura K, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 131.Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768–1775. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 132.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jakobsdottir J, Gorin MB, Conley YP, et al. Interpretation of genetic association studies: Markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lipkus IM, Samsa G, Rimer BK. General performance on a numeracy scale among highly educated samples. Med Decis Making. 2001;21:37–44. doi: 10.1177/0272989X0102100105. [DOI] [PubMed] [Google Scholar]
- 135.Han PK, Lehman TC, Massett H, et al. Conceptual problems in laypersons' understanding of individualized cancer risk: A qualitative study. Health Expect. 2009;12:4–17. doi: 10.1111/j.1369-7625.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cull A, Anderson ED, Campbell S, et al. The impact of genetic counselling about breast cancer risk on women's risk perceptions and levels of distress. Br J Cancer. 1999;79:501–508. doi: 10.1038/sj.bjc.6690078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lerman C, Lustbader E, Rimer B, et al. Effects of individualized breast cancer risk counseling: A randomized trial. J Natl Cancer Inst. 1995;87:286–292. doi: 10.1093/jnci/87.4.286. [DOI] [PubMed] [Google Scholar]
- 138.Lidén A, Berglund G, Hansson MG, et al. Genetic counseling for cancer and risk perception. Acta Oncol. 2003;42:726–734. doi: 10.1080/02841860310011023. [DOI] [PubMed] [Google Scholar]
- 139.Finch A, Metcalfe K, Lui J, et al. Breast and ovarian cancer risk perception after prophylactic salpingo-oophorectomy due to an inherited mutation in the BRCA1 or BRCA2 gene. Clin Genet. 2009;75:220–224. doi: 10.1111/j.1399-0004.2008.01117.x. [DOI] [PubMed] [Google Scholar]
- 140.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle–will we get our wish? N Engl J Med. 2008;358:105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 141.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: Raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robson ME, Storm CD, Weitzel J, et al. American Society of Clinical Oncology policy statement update: Genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2010;28:893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]