Abstract
In Singapore, after a major outbreak of dengue in 2005, another outbreak occurred in 2007. Laboratory-based surveillance detected a switch from dengue virus serotype 1 (DENV-1) to DENV-2. Phylogenetic analysis showed a clade replacement within DENV-2 cosmopolitan genotype, which accompanied the predominant serotype switch, and cocirculation of multiple genotypes of DENV-3.
Keywords: Dengue, surveillance, PCR, sequencing, phylogenetic, viruses, dispatch
Dengue poses a threat to public health in >100 countries worldwide. Despite improvements in diagnostics and clinical management, the number of dengue cases continues to rise globally; 2.5 billion persons are at risk for infection (1). Dengue virus (DENV) belongs to the genus Flavivirus and contains a positive-strand RNA genome that encodes 3 structural proteins—core protein, membrane-associated protein, envelope protein—and 7 nonstructural proteins. DENV consists of 4 genetically and antigenically distinct serotypes, 1–4.
Singapore has seen a resurgence of dengue cases since the late 1980s, after 2 decades of successful control that relied mostly on an integrated vector-control program (2,3). The recent epidemiology of dengue in Singapore is characterized by a 5–6-year cycle; incidence rates increase within each cycle before collapsing into 1 or 2 lull years. During an unprecedented dengue outbreak in 2005, a total of 14,006 cases and 27 deaths were reported (4). The outbreak was associated with a switch in predominant serotype, from DENV-2 to DENV-1, in 2004 (5,6). In 2007, Singapore experienced another dengue outbreak after a lull in 2006. We report the laboratory and surveillance findings that assisted vector-control operations during the 2007 dengue outbreak in Singapore.
The Study
In Singapore in 2005, as part of an integrated vector-control program, laboratory-based dengue virus surveillance was established for close monitoring and investigation of the circulating dengue virus serotypes. Samples were sent to the Environmental Health Institute from Tan Tock Seng Hospital, which cares for ≈40% of all reported dengue patients in Singapore, and from a network of participating general practitioners throughout the country. PCR to detect dengue virus RNA and serotyping were performed at the Environmental Health Institute according to its in-house real-time PCR protocol (7). The numbers of dengue-positive samples serotyped were 186 in 2006, 889 in 2007, and 918 in 2008, and represent ≈10% of the total dengue cases reported each year by the Ministry of Health.
The envelope protein gene of DENV (≈1,480 nt) was amplified by reverse transcription–PCR and directly sequenced by using an automated DNA sequencer (ABI 3100; Applied Biosystems, Foster City, CA, USA). Sequences were aligned and submitted to GenBank (accession nos. GQ357666–892). Phylogenetic analysis of DENV sequences was conducted by using the maximum-likelihood method as implemented in PAUP* software, version 4.0b10 (8), and compared with sequence data obtained from GenBank.
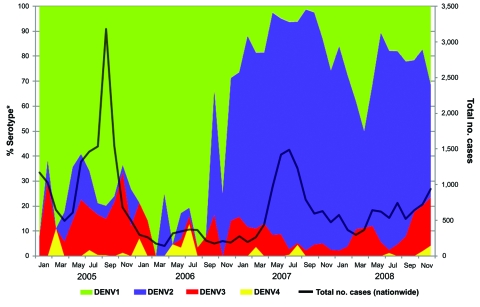
During 2006–2008, all 4 DENV serotypes were detected (Figure 1). DENV-1 (21.7%) and DENV-2 (69.3%) were the predominant serotypes throughout the study period; DENV-3 (7.8%) and DENV-4 (1.2%) were less prevalent. In 2006, the number of DENV cases was relatively low, and DENV-1 remained the predominant serotype after the major 2004–2005 outbreak. During January–September 2006, 75%–100% of samples collected each month contained DENV-1. In early January 2007, the predominant circulating serotype switched from DENV-1 to DENV-2. Early detection of this switch warned of a possible upcoming dengue outbreak. In response, an enhanced vector-control program was activated in February 2007. The proportion of DENV-2–positive samples detected by PCR rose from 57.9% in January 2007 to a peak of 91.0% in July 2007. This increase was accompanied by an increase in the total number of dengue cases reported by the Ministry of Health; cases peaked at 432 in the first week of July 2007. By late August, the number of dengue cases fell to below the warning level (warning level = 256 cases/epidemiologic week) as reported by the Ministry of Health (9). During the switch in predominant serotype, fatality rates (0.32% in 2006 and 0.27% in 2007) and dengue hemorrhagic fever rates (2.4% in 2006 and 2.1% in 2007) did not differ substantially among the reported cases. During this same period of extensive surveillance, 5.2% of the samples in 2007 and 10.8% in 2008 were detected as DENV-3. Our spatial analysis indicated localized emergence of DENV-3 in the eastern region of the country in 2007 and in the central region in 2008. Enhanced control was also attempted in these areas to prevent the spread of the serotype that had been uncommon in Singapore.
Figure 1.
Trends of monthly dengue cases in Singapore, 2005–2008, showing a switch in predominant serotype from dengue virus serotype 1 (DENV-1) to DENV-2 in January 2007 and cocirculation of all 4 serotypes with general dominance of DENV-1 and DENV-2 and lesser circulation of DENV-3 and DENV-4. *From ≈10% of all dengue cases.
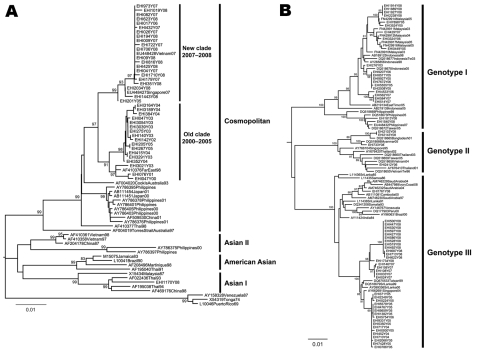
Phylogenetic analysis of DENV-2 envelope gene sequences showed that the switch in predominant serotype in early 2007 coincided with a clade replacement within DENV-2. During 2000–2008, 2 distinct subclades, with strong temporal topology, were found within the cosmopolitan genotype (Figure 2). Specifically, DENV-2 isolates obtained before 2007 formed the subclade herein referred to as the old clade, whereas isolates obtained in 2007 and later formed the new clade with strong bootstrap support. Because 1 of the DENV-2 isolates sampled in 2005 clustered with the new clade but fell closer to the root of that clade, in situ evolution giving rise to DENV-2 viruses that subsequently replaced the old clade viruses is highly likely. A GenBank sequence that belonged to DENV-2, sampled in 2007 in Vietnam, grouped within the new clade, indicating that this virus strain was not restricted to Singapore and may have been circulating in this region.
Figure 2.
Maximum-likelihood tree showing the phylogenetic relationship of A) dengue virus serotype 2 (DENV-2) and B) DENV-3 from Singapore and global isolates based on the envelope protein gene. EHI, sequence data generated at Environmental Health Institute; new clade, isolates obtained in 2007 and later; old clade, isolates obtained before 2007. Numbers on branches represent bootstrap percentages; only those >80% are shown. Scale bars indicate substitutions per site.
Our dengue surveillance also indicated sporadic emergence of DENV-3 from localized areas throughout the country (6,10). Phylogenetic analysis of isolates from Singapore from 2006 through 2008 identified 3 genotypes of DENV-3. These isolates were closely related to those found in Indonesia, Malaysia, Philippines, Thailand, Saudi Arabia, and Côte d’Ivoire (Figure 2), which suggests multiple importations of DENV-3 viruses into Singapore. Analysis of DENV-1 sequences showed that all except 3 belonged to genotype I and were similar to those responsible for the 2005 outbreak (data not shown).
Conclusions
Our dengue surveillance provided early warning of the outbreak in 2007 and contributed to early activation of enhanced vector control. Although we were unable to assess the effectiveness of the control measures, considering the regional situation in 2007 (11,12), we believe that without these measures the dengue situation in Singapore in 2007 would have been worse than or comparable to that in 2004–2005. After a lull year in 2006, dengue cases were expected to rise for a few years. The integrated vector control program has interrupted the dengue trend, with 7,032 cases reported in 2008 and 4,498 in 2009.
As a travel hub, Singapore experiences continuous importation of dengue viruses. Although some become established at various levels, some develop into outbreaks and subsequently get replaced. Our study demonstrates how rapidly dengue virus serotypes can be replaced within a population. It also highlights the complexity of the disease and the challenges faced by affected states that seek to understand the epidemiology for purposes of disease control. To shed further light on the complex interplay among the various factors that affect dengue transmission, studies are being conducted on complete genome sequences of dengue viruses, vectorial capacity of local Aedes spp. mosquitoes, and cross-reactive immune responses to different dengue serotypes.
Acknowledgments
We thank the general practitioners for sending samples for testing and serotyping, the Ministry of Health for approving the use of blood samples from hospitals for viral surveillance, and Jennifer Yap for collating and generating the graph.
Biography
Dr Lee is a research scientist at the Environmental Health Institute. His research interests include the molecular epidemiology of vector-borne diseases.
Footnotes
Suggested citation for this article: Lee K-S, Lai Y-L, Lo S, Barkham T, Aw P, Ooi P-L, et al. Dengue virus surveillance for early warning, Singapore. Emerg Infect Dis [serial on the Internet]. 2010 May [date cited]. http://www.cdc.gov/EID/content/16/5/847.htm
References
- 1.World Health Organization. Strengthening implementation of the global strategy for dengue fever/dengue haemorrhagic fever. 1999. [cited 2009 Jun 2]. http://www.who.int/csr/resources/publications/dengue/WHO_CDS_D_EN_IC_2000_1/en/
- 2.Burattini MN, Chen M, Chow A, Coutinho FA, Goh KT, Lopez LF, et al. Modelling the control strategies against dengue in Singapore. Epidemiol Infect. 2008;136:309–19. 10.1017/S0950268807008667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ooi EE, Goh KT, Gubler DJ. Dengue prevention and 35 years of vector control in Singapore. Emerg Infect Dis. 2006;12:887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh BK, Ng LC, Kita Y, Tang CS, Ang LW, Wong KY, et al. The 2005 dengue epidemic in Singapore: epidemiology, prevention and control. Ann Acad Med Singapore. 2008;37:538–45. [PubMed] [Google Scholar]
- 5.Chow A, Ye T, Ang LW, Foong BH, Kita Y. An update on the dengue situation in Singapore. Ministry of Health Singapore, Epidemiological News Bulletin. 2005;31:54–61 [cited 2009 Mar 10]. http://www.moh.gov.sg/mohcorp/uploadedfiles/Publications/Epidemiological_News_Bulletin/2005/ENB03Q_05.pdf
- 6.Ng LC, Lai YL, Cheng C, Barkham T. Spatial and temporal distribution of dengue serotypes in Singapore. Ministry of Health Singapore, Epidemiological News Bulletin. 2007;33:1–3 [cited 2009 Mar 10]. http://www.moh.gov.sg/mohcorp/uploadedFiles/Statistics/Epidemiological_News_Bulletin/2007/ENB01Q_07.pdf
- 7.Lai YL, Chung YK, Tan HC, Yap HF, Yap G, Ooi EE, et al. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. J Clin Microbiol. 2007;45:935–41. 10.1128/JCM.01258-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland (MA): Sinauer Associates; 1998. [Google Scholar]
- 9.Ministry of Health Singapore. Weekly infectious disease bulletin, epidemiological week 35, 26 Aug–1 Sep, 2007. 2007;4:4 [cited 2009 Mar 10]. http://www.moh.gov.sg/mohcorp/uploadedFiles/Statistics/Infectious_Diseases_Bulletin/2007/2007_week_35.pdf
- 10.Schreiber MJ, Holmes EC, Ong SH, Soh HS, Liu W, Tanner L, et al. Genomic epidemiology of a dengue virus epidemic in urban Singapore. J Virol. 2009;83:4163–73. 10.1128/JVI.02445-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization South-East Asia Region. Situation of dengue/dengue haemorrhagic fever in South-East Asia Region. 2007. [cited 2009 Jun 2]. http://www.searo.who.int/en/Section10/Section332_1098.htm
- 12.World Health organization Regional Office for the Western Pacific. Dengue in the Western Pacific Region [cited 2009 Jun 2]. http://www.wpro.who.int/health_topics/dengue/