Abstract
Signal-induced alternative splicing of the CD45 gene in human T cells is essential for proper immune function. Skipping of the CD45 variable exons is controlled, in large part, by the recruitment of PSF to the pre-mRNA substrate upon T cell activation; however, the signaling cascade leading to exon exclusion has remained elusive. Here we demonstrate that in resting T cells PSF is directly phosphorylated by GSK3 thus promoting interaction of PSF with TRAP150 which prevents PSF from binding CD45 pre-mRNA. Upon T cell activation, reduced GSK3 activity leads to reduced PSF phosphorylation, releasing PSF from TRAP150 and allowing it to bind CD45 splicing regulatory elements and repress exon inclusion. Our data place two new players, GSK3 and TRAP150, in the complex network that regulates CD45 alternative splicing and demonstrate a new paradigm for signal transduction from the cell surface to the RNA processing machinery through the multi-functional protein PSF.
Introduction
Signal-induced alternative splicing is a primary, but poorly understood, mechanism for regulating protein isoform expression in response to changing cellular environments (Lynch, 2007; Shin and Manley, 2004). In humans, examples of signal-induced splicing regulation are known to play essential roles in diverse cellular responses including neuronal depolarization, insulin signaling and T cell activation (An and Grabowski, 2007; Chalfant et al., 1995; Lee et al., 2007; Lynch, 2007). However, surprisingly few cases have been studied in detail (Shin and Manley, 2004; Lynch, 2007). Therefore, despite the functional importance of linking extracellular stimuli to pre-mRNA processing, the mechanisms governing this regulation are mostly unknown.
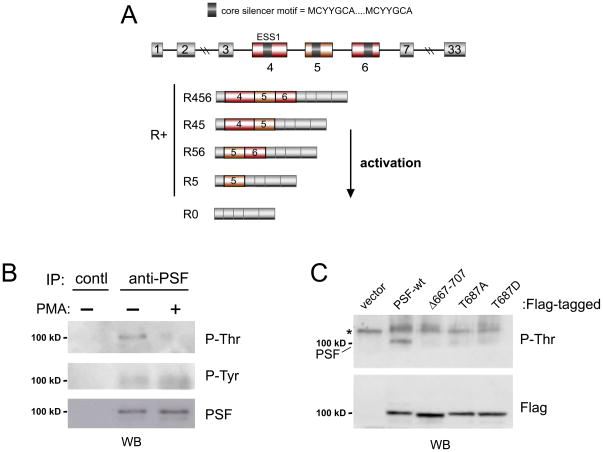
A well-documented example of signal-induced splicing regulation is the transmembrane tyrosine phosphatase CD45, which encodes at least 5 isoforms as a result of tightly controlled alternative splicing (Fig 1A, Hermiston et al., 2002). CD45 is expressed on all nucleated hematopoietic cells and has regulatory functions in a variety of signal transduction pathways, including cytokine-, interferon- and antigen receptor mediated signaling (Hermiston et al., 2002). In T cells, where CD45 plays an essential role in signal transmission from the T cell receptor to the intracellular machinery, antigenic stimulation induces skipping of three exons thereby increasing expression of the smallest CD45 isoform, CD45R0 (Fig 1A, Hermiston et al., 2002). This differential CD45 isoform expression in naïve versus activated and memory T cells has long been used as the defining marker of these T cell states. At a functional level, the activation-induced exon exclusion in CD45 has been suggested to play an important role in the homeostasis of the immune system as the resulting CD45R0 protein forms catalytically inactive dimers that attenuate T cell signaling (Hermiston et al., 2002; Xu and Weiss, 2002). Consistently, a silent point mutation in CD45 exon 4 that disrupts the essential splicing regulatory element ESS1 (exonic splicing silencer 1; Fig 1A) results in aberrant exon inclusion, loss of CD45R0 isoform expression, and increased susceptibility to several autoimmune diseases in humans (Jacobsen et al., 2002; Lynch, 2004).
Figure 1.
Phosphorylation of PSF on T687 is decreased upon T cell stimulation. (A) Schematic of CD45 showing expressed isoforms, ESS1 silencer sequence, and core motif common to ESS1 and other variable exons through which PSF functions. (B) Endogenous PSF immunoprecipitated from nuclear extract from either untreated or PMA stimulated JSL1 cells (72 hrs), or IP with unrelated control antibody, Western blotted for total PSF or with anti-phospho-threonine (P-Thr) or anti-phospho-tyrosine (P-Tyr) antibody. Size of PSF in anti-P blots is as indicated. (C) JSL1 cells stably expressing indicated Flag-tagged PSF variants were lysed, subjected to anti-Flag immunoprecipitation and blotted for P-Thr or Flag. Non-specific band is labeled with an asterisk.
In recent work, we and others have demonstrated that PSF and hnRNP L-like (hnRNP LL) bind to the ESS1 regulatory element in stimulated cells and mediate the increased skipping of the CD45 variable exons observed upon T cell activation (Melton et al., 2007; Oberdoerffer et al., 2008; Topp et al., 2008; Wu et al., 2008; Motta-Mena et al., 2010). Both PSF and hnRNP LL are RNA binding proteins that have been shown to regulate the alternative splicing of many genes in addition to CD45 (Hung et al., 2008; Oberdoerffer et al., 2008; Shav-Tal and Zipori, 2002). Moreover, PSF is a highly abundant nuclear protein that has functions in a range of RNA biogenesis processes from basic splicing catalysis to transcription to nuclear export (Shav-Tall and Zipori, 2002). However, how the various activities of PSF are regulated in cells is not yet clear.
In terms of CD45 alternative splicing, the stimulation-specific activity of PSF and hnRNP LL is due to the fact that these proteins show a marked preference for binding to the ESS1 regulatory sequence in activated versus resting T cells (Melton et al., 2007; Motta-Mena et al., 2010; Topp et al., 2008). In the case of hnRNP LL this activation-induced binding is readily attributed to an increase in protein expression in activated T cells (Topp et al., 2008). By contrast, nuclear PSF expression remains unchanged upon activation (Melton et al., 2007), suggesting that its binding to the CD45 pre-mRNA is regulated at a post-translational level. However, the underlying signaling cascade regulating PSF function has thus far not been characterized.
Glycogen synthase kinase 3 (GSK3) was initially described as an enzyme regulating glucose metabolism but has since attracted much interested due to its involvement in many other cellular processes (Cohen and Frame, 2001). For example, GSK3 has been linked to the innate immune system (Martin et al., 2005) and is involved in regulating neuronal cell fate and development of tauopathies (Plattner et al., 2006). In the acquired immune system, specifically T cells, GSK3 activity is decreased upon antigen stimulation through phosphorylation on serine 9 (Ohteki et al., 2000; Welsh et al., 1996). The reduced GSK3 activity has been shown to be involved in mediating the CD28 costimulatory signal and is thus required for an optimal T cell response (Diehn et al., 2002). In our present work, we have identified an additional role of GSK3, the modulation of alternative splicing. We show that in resting T cells, GSK3 directly phosphorylates the splicing regulatory protein PSF. In this phosphorylated form PSF is sequestered in a complex with TRAP150, precluding it from binding the ESS1 sequence in CD45 alternatively spliced exons. Upon T cell stimulation reduced GSK3 activity leads to reduced PSF phosphorylation, thereby releasing PSF from TRAP150 and allowing it to participate in activation-induced CD45 exon skipping. Thus, we have now identified the complete signaling cascade linking T cell receptor engagement with PSF-mediated exclusion of alternatively spliced CD45 exons and have implicated GSK3 and TRAP150 as two critical regulators of this pathway.
Results
PSF T687 is differentially phosphorylated in resting versus activated T cells
Previously we have demonstrated that PSF is uniquely recruited to the ESS1 regulatory element of CD45 variable exons in response to cell stimulation, even though this protein is present at equal concentration in the nuclei of resting and activated cells (Melton et al., 2007; Motta-Mena et al., 2010). Importantly, PSF purified from activated cells has silencing activity on the CD45 variable exons in in vitro splicing assays, while PSF purified from resting cells does not (Melton et al., 2007). Moreover, using the MS2 system to tether PSF to a model exon, we find that forced recruitment of PSF results in exon exclusion under both resting and activated conditions (data not shown), suggesting that the activity of PSF is primarily regulated at the level of RNA binding. Taken together, these data led us to test the hypothesis that PSF itself is differentially modified in resting versus activated T cells in a manner that controls its ability to bind to CD45 exons and cause repression.
We first compared possible posttranslational modifications of PSF precipitated from resting versus activated JSL1 cells that we and others have demonstrated to be a faithful model for antigen-induced alternative splicing of CD45 when stimulated with the phorbol ester PMA (Lynch and Weiss, 2000; Oberdoerffer et al., 2008). Using an antibody specific for phospho-threonine we observed a substantial decrease in threonine phosphorylation under activated conditions (Fig 1B and see below). In contrast, global tyrosine phosphorylation of PSF is weak and does not change in the presence of PMA (Fig 1B). A database search (www.phosphosite.org) revealed only one site of predicted threonine phosphorylation, namely T687. Consistent with this prediction, either deletion of the C-terminus encompassing T687, or mutation of T687 to either alanine or aspartic acid resulted in an almost complete loss of the anti-phospho-threonine reactive band (Fig 1C). This result is further supported by a previous study which mapped threonine phosphorylation in PSF to the C-terminal third of the protein (Shav-Tal et. al., 2001). Therefore, while we cannot entirely rule out the presence of additional phosphorylated threonine residues which may escape the detection with the antibody, we conclude that T687 is the primary site of threonine phosphorylation in PSF.
PSF specifically associates with TRAP150/BTF in a phosphorylation dependent manner
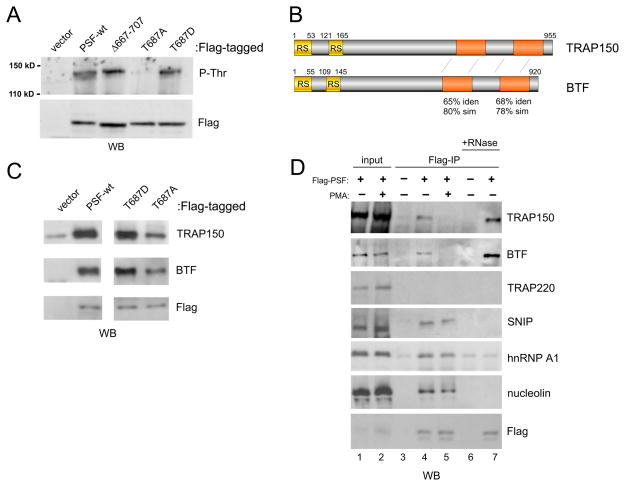
Strikingly, the immunoprecipitations of PSF derivatives also revealed a protein of around 135 kD that co-precipitated with the wildtype PSF and the T687D mutant but was largely absent in the precipitate of the T687A mutant when detected by P-Thr antibody or silver stain (Fig 2A and data not shown). Although a few additional PSF-associated proteins were also observed to react with the P-Thr antibody, only the 135 kD species differs in association between the T687A and T687D mutant (Fig S1A). As the T687A mutant mimics the dephosphorylated state of PSF that is observed in activated cells, we reasoned that differential association with the 135 kD protein could explain differential RNA binding capability in resting and activated cells. We therefore subjected the 135 kD band to mass spectroscopy analysis and identified specific peptides for two nuclear proteins, TRAP150 and BTF (Fig S1B) which co-migrate in SDS-PAGE and share extensive sequence homology (Fig 2B). Using specific antibodies we confirmed that both TRAP150 and BTF co-precipitate with PSF from nuclear extract and that this interaction is reduced in the T687A mutant when compared to either the phosphomimic T687D or wildtype PSF (Fig 2C). Consistent with reduced phosphorylation of T687 upon T cell stimulation, the interaction of PSF with TRAP150 and BTF is also significantly stronger in resting (−PMA) versus activated (+PMA) JSL1 cells (Fig 2D). Importantly, we observe no significant change in the threonine phosphorylation status of TRAP150 itself upon PMA treatment (Fig S1C). Furthermore, the interaction between PSF and TRAP150/BTF in resting cells is independent of RNA, as addition of RNase A and T1 did not decrease the observed co-precipitation (Fig 2D, lanes 4 vs. 7). In fact we observe a reproducible increase in the association of TRAP150 and BTF with PSF following RNase treatment, suggesting that loss of RNA association makes PSF more accessible to bind TRAP150/BTF. Finally, we note that the interaction of TRAP150/BTF with PSF is unlikely to be directly mediated by T687 as deletion of the C-terminal 40 amino acids of PSF, encompassing T687, is also permissive for interaction with TRAP150 and BTF (Figs 2A, S1D). We hypothesize therefore that phosphorylation of T687 drives association with TRAP150 and/or BTF in an allosteric manner as discussed below.
Figure 2.
Phosphorylation of PSF on T687 regulates association with TRAP150/BTF. (A) anti-P-Thr blot of Flag-PSF immunoprecipitates as in Fig 1C, looking at co-precipitated proteins of around 150 kD. (B) Schematic of TRAP150 and BTF showing location of RS (yellow) and homologous (orange) domains. (C) Immunoprecipitates of Flag-PSF variants, similar to Fig 1C, blotted with antibodies specific to TRAP150 or BTF. (D) Immunoprecipitates of Flag-PSF from resting (−PMA) versus stimulated (+PMA, 72 hrs) JSL1 cells, blotted with antibodies specific to the indicated proteins. Samples in lanes 6 and 7 were treated with 20 U RNase T1 and 20 ug RNase A for 15 min at 37°C prior to IP. Lanes 1 and 2 correspond to 30% input. See also Figure S1.
The function of both TRAP150 and BTF remains poorly characterized. TRAP150 was initially cloned as a component of the transcription mediator complex (Fondell et al., 1996). However, consistent with subsequent studies suggesting that TRAP150 is not a primary mediator subunit (Conaway et al., 2005), we do not observe an interaction of PSF with TRAP220, a core component of the mediator complex (Fig 2D). More recent studies have implicated TRAP150 and BTF in RNA processing (Merz et al., 2007), either as components of a complex with SNIP1 involved in mRNA stability (Bracken et al., 2008) or in a loosely-associated complex of splicing regulatory factors including nucleolin and hnRNP A1 (Li et al., 2003). We do detect SNIP1, nucleolin and hnRNP A1 in precipitates with PSF. However, unlike TRAP150 and BTF, these proteins associate with PSF equally in resting and stimulated cells and the addition of RNase completely abolishes this interaction (Fig 2D). Together these data demonstrate a specific and phosphorylation-dependent interaction of TRAP150/BTF with PSF and suggest that this TRAP150-BTF-PSF complex is distinct from other assemblies in which TRAP150 or BTF have been characterized thus far.
Interaction of TRAP150 with PSF inhibits CD45 exon skipping
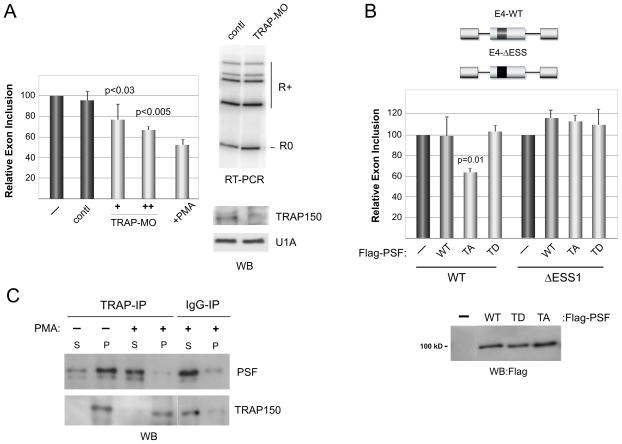
To investigate the functional role of the PSF-TRAP150-BTF interaction with respect to CD45 alternative splicing, we next knocked down TRAP150 and BTF using morpholino oligos. Remarkably, knock down of TRAP150 results in a dose dependent decrease in CD45 variable exon inclusion in resting cells, which at the highest level of TRAP150 depletion (Fig 3A) is similar to that observed upon T cell activation (+PMA). This effect of TRAP150 depletion on CD45 alternative splicing is dependent on the presence of the ESS1 regulatory element through which PSF functions as inclusion of CD45 variable exon 4 from a standard minigene construct is decreased upon depletion of TRAP150, while splicing of an analogous construct lacking the ESS1 regulatory sequence is unaffected by TRAP150 knock down (Fig S2A). Notably, however, this change in splicing occurs without any alterations in the expression of the ESS1 regulatory proteins, PSF, hnRNP L or hnRNP LL (S2B).
Figure 3.
The majority of cellular PSF is bound to TRAP150 to inhibit CD45 exon skipping in resting cells. (A) Representative RT-PCR and quantitation (n=3) of CD45 isoform expression in JSL1 cells under resting (−) or stimulated (+PMA, 72 hrs) conditions or after treatment of resting cells with control or anti-TRAP150 morpholino oligomers. Error bars correspond to standard deviation. Western blot confirmation of TRAP150 knock down shown in lower right. (B) Quantitation (n=4) of splicing of CD45 exon 4-derived minigenes in cells transfected with the WT, T687A or T687D version of Flag-PSF (see Western). Error bars correspond to standard deviation, p-value for difference between WT and TA PSF is shown. Minigene E4-WT contains the complete native CD45 exon 4 sequence, while in E4-ΔESS the ESS1 motif has been replaced by an unrelated sequence of similar length (Melton, et al., 2007). (C) Analysis of PSF in pellet (P) versus supernatant (S) following precipitation with antibody to TRAP150 or control antibody (IgG). See also Figure S2.
To initially investigate if the ESS1-dependent splicing change we observe upon TRAP150 depletion is a result of altered TRAP150-PSF interaction, we also tested the effect of disrupting this interaction by mutating PSF. We predict that the T687A mutant of PSF, which interacts more weakly with TRAP150 (Fig 2A,C), should promote CD45 exon repression in a manner similar to that observed upon knock down of TRAP150. Strikingly, we observe a significant decrease in the inclusion of wildtype CD45 exon 4 upon overexpression of PSF-T687A in resting cells (Fig 3B). In contrast expression of WT PSF had no effect on exon 4 splicing, and neither protein repressed the ΔESS control (Fig 3B). Moreover, the phosphomimic PSF-T687D had no effect on exon 4 splicing in either resting or stimulated cells (Fig 3B and data not shown), consistent with the notion that the T687 phosphorylated pool of PSF is itself non-functional for CD45 repression and does not block the accumulation and activity of the hypophosphorylated form of endogenous PSF upon stimulation (see below). A functional role for the TRAP150-PSF interaction is further supported by the finding that the majority of wildtype, endogenous PSF is associated with TRAP150 in resting cells, as indicated by the efficiency of co-immunoprecipitation (Fig 3C). In contrast, the bulk of PSF is not precipitated by TRAP150 following stimulation (Fig 3C). Taken together these data confirm a significant and regulated interaction between endogenous TRAP150 and PSF, and are consistent with this interaction playing a critical role in regulating PSF function in CD45 splicing.
In contrast to the results with TRAP150, knock down of BTF did not substantially change CD45 alternative splicing either on its own (Fig S2C) or in combination with TRAP150 depletion (data not shown). As BTF and TRAP150 have been shown to associate (Bracken et al., 2008) we reasoned that TRAP150 could be the functionally relevant PSF binding partner, with BTF only recruited to PSF indirectly via TRAP150. Consistent with this prediction we find that upon knock down of TRAP150 BTF no longer co-precipitates with PSF, although total protein levels remain unchanged (Fig S2B, D). In contrast, the association of TRAP150 with PSF does not require BTF as knock down of BTF does not weaken TRAP150 co-precipitation with PSF (Fig S2D). Moreover, we observe a direction interaction in vitro between purified recombinant TRAP150 and PSF (data not shown). Therefore we conclude that TRAP150 is the primary functionally-relevant PSF-associated protein and have focused our subsequent studies on characterizing the regulation and consequence of this TRAP150-PSF interaction.
TRAP150 directly inhibits binding of PSF to the ESS1 RNA
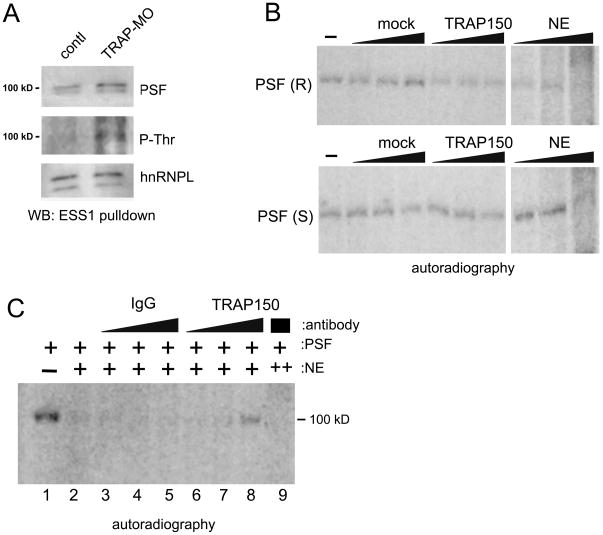
The above results, together with our previous demonstration that T cell activation induces PSF’s association with the ESS1 regulatory sequence in CD45 (Melton et al., 2007; Motta-Mena et al., 2010), suggest a mechanism in which the interaction between TRAP150 and PSF limits exon skipping by blocking the interaction of PSF with ESS1. We therefore assayed the association of PSF with the ESS1 regulatory element in an RNA pull-down assay done from cells depleted of TRAP150 or wildtype controls (Fig 4A). Remarkably, upon TRAP150 knock down we observe an increase in the association of PSF with ESS1 RNA that is highly similar to that observed upon T cell activation (Melton et al., 2007; Motta-Mena et al., 2010). Moreover, PSF associated with ESS1 RNA does not react efficiently with the P-Thr antibody unless TRAP150 is depleted (Fig 4A, P-Thr), suggesting that the phosphorylated population of PSF, bound by TRAP150, is hindered from binding to ESS1. In further support of this model we are unable to detect any TRAP150 itself, or BTF, among the ESS1 associated proteins (Melton et al., 2007 and data not shown).
Figure 4.
TRAP150 directly inhibits RNA binding of PSF. (A) RNA affinity purification using biotinylated 60 nt ESS1 sequence and nuclear extracts of JSL1 cells treated with control or anti-TRAP150 morpholino oligomers, blotted for endogenous PSF, P-Thr or hnRNP L as loading control. (B) PSF was purified from resting or stimulated (PMA, 72 hrs) JSL1 cells stably expressing Flag-PSF. UV crosslinking of recombinant PSF purified from resting (R) or stimulated (S) JSL1 cells to ESS1 RNA alone (−) or in the presence of anti-Flag eluate from control lysate that lacks Flag-TRAP150 (mock), purified TRAP150 or nuclear extract (NE). (C) Addition of anti-TRAP150 or control antibody, to UV crosslinking experiment as in panel B. Lane 9 is the same as 8 except for the addition of more NE, demonstrating that the increase in PSF binding is a consequence of the antibody titrating an inhibiting factor in NE (i.e. TRAP150). See also Figure S3.
We next sought to determine whether TRAP150 directly regulates the binding of PSF to the CD45 ESS1 RNA using purified proteins (Fig. S3A). Purified PSF binds specifically to radiolabeled ESS1 RNA in a UV crosslinking assay, as shown by competition with non-specific versus ESS1 RNA (S3B). Importantly, PSF purified from resting or stimulated cells binds equally well to ESS1 RNA, demonstrating that the RNA binding capability of PSF itself is not altered upon stimulation (Fig 4B). However, the addition of purified TRAP150 specifically inhibits the RNA binding capability of PSF purified from resting cells (Fig 4B, top) with little impact on the RNA binding of PSF purified from stimulated cells (Fig 4B, bottom).
We further observe that nuclear extract differentially inhibits RNA binding of PSF from resting and activated cells (Fig 4B). This result provides an explanation for our previous observation that PSF purified from resting cells cannot repress CD45 exon usage when added to nuclear extract in an in vitro splicing assay (Melton et al., 2007) as the nuclear extract would inhibit recruitment of this “resting” PSF to the substrate. Notably, nuclear extracts from resting or activated T cells are equally effective in inhibiting RNA binding of PSF, demonstrating that stimulation alters the inherent susceptibility of PSF to binding inhibition, without inducing additional activities within the nuclear extract (Fig 4B and S3C). Moreover, the inhibitory effect of nuclear extract is partially relieved by titration of antibody against TRAP150, but not by a variety of control antibodies (Fig 4C and data not shown). Taken together, our data from interaction, knock down and RNA binding studies demonstrate that the phosphorylation dependent TRAP150-PSF interaction directly blocks PSF from binding ESS1 in resting T cells thereby limiting the repression of CD45 exons prior to T cell activation.
GSK3 directly phosphorylates PSF and increases PSF-TRAP150 interaction and CD45 variable exon inclusion
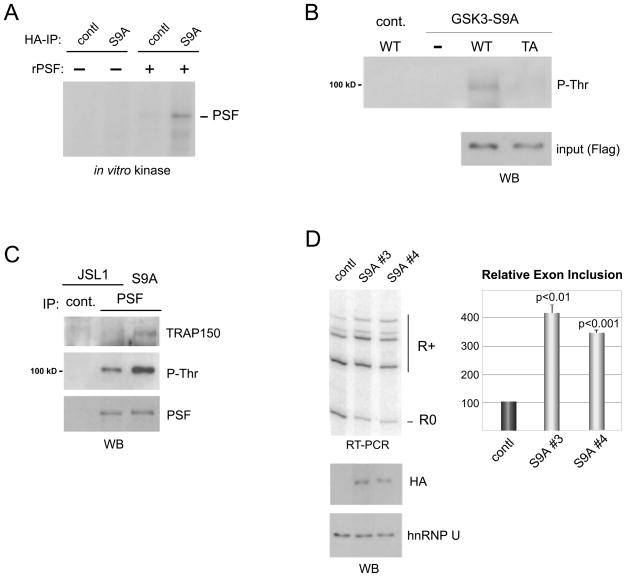
In order to link the TRAP150-dependent regulation of PSF to T cell signaling, we next sought to uncover how T cell stimulation allows for reduced phosphorylation of PSF. The Ser/Thr kinase GSK3 (glycogen synthase kinase 3) has several of the hallmarks expected of a signal-responsive regulator connecting T cell activation to PSF phosphorylation: GSK3 is highly expressed and constitutively active in resting human T cells and the JSL1 cell line (Fig S4A), T cell activation leads to increased phosphorylation of the auto-inhibitory S9 thereby reducing GSK3 activity (Diehn et al., 2002, Fig S4A), and T687 in PSF is surrounded by several prolines, which is a preferred substrate context for GSK3 (Hooper et al., 2008). We therefore purified a HA-tagged version of the constitutively active GSK3beta S9A mutant from HEK293 cells for use in an in vitro kinase assay with PSF. In this assay GSK3-S9A does induce phosphorylation of both bacterially-expressed (Fig 5A) or JSL1-expressed (Fig. S4B) recombinant PSF. Importantly, in vitro phosphorylation of bacterially-expressed recombinant PSF by GSK3-S9A indicates that a priming phosphate is not required for this activity as is sometimes observed for GSK3 substrates. We further show that threonine phosphorylation of PSF by GSK3 primarily occurs on residue T687, as this phosphorylation is detected by the P-Thr antibody and is markedly decreased in the T687A mutant of PSF (Fig 5B).
Figure 5.
GSK3 directly phosphorylates PSF thereby regulating CD45 alternative splicing. (A) SDS-PAGE of in vitro kinase assay using the constitutively active GSK3beta S9A mutant purified by HA-tag from HEK293 cells (or control mock purification) incubated with recombinant PSF in the presence of 32P-gamma-ATP. Radiolabeled protein corresponding to the size of PSF is indicated. (B) In vitro kinase assay as in panel A with PSF (WT) and PSF T687A (TA) purified from transiently transfected HEK293 cells. Reactions were performed in the presence of cold ATP and phosphorylation was detected by anti-phospho-threonine Western blot. (C) JSL1 cells stably expressing HA-GSK3beta S9A blotted for P-Thr and TRAP150 in precipitates of PSF. (D) Representative RT-PCR and quantitation (n=3) of CD45 isoform expression in two representative stable GSK3beta S9A cell lines versus control all under resting conditions. Error bars correspond to standard deviation. Expression of the HA-tagged GSK3beta S9A confirmed by Western blot with hnRNP U as loading control (lower panel). See also Figure S4.
To examine the functional effects of GSK3, we next produced JSL1 cell lines stably overexpressing the activated HA-GSK3-S9A mutant. Consistent with the in vitro kinase results, we observed increased phosphorylation of endogenous PSF in these cell lines (Fig 5C). Remarkably, this increase in PSF phosphorylation correlates with an increased interaction between PSF and TRAP150 and a dramatic decrease in CD45 variable exon skipping (Fig 5C, D). As for the TRAP150 knock down, the effect of GSK3-S9A expression on CD45 splicing is dependent on the presence of the ESS1 element, as determined by minigene experiments (data not shown and see below). Since expression levels of the other known regulators of CD45 alternative splicing were not affected by GSK3-S9A expression (Fig S4C), we conclude that the change in CD45 splicing induced by GSK3-S9A is due to the increased interaction of PSF with TRAP150 sequestering PSF away from the CD45 pre-mRNA.
Reduced GSK3 activity leads to CD45 exon exclusion in JSL1 cells and in primary human T cells
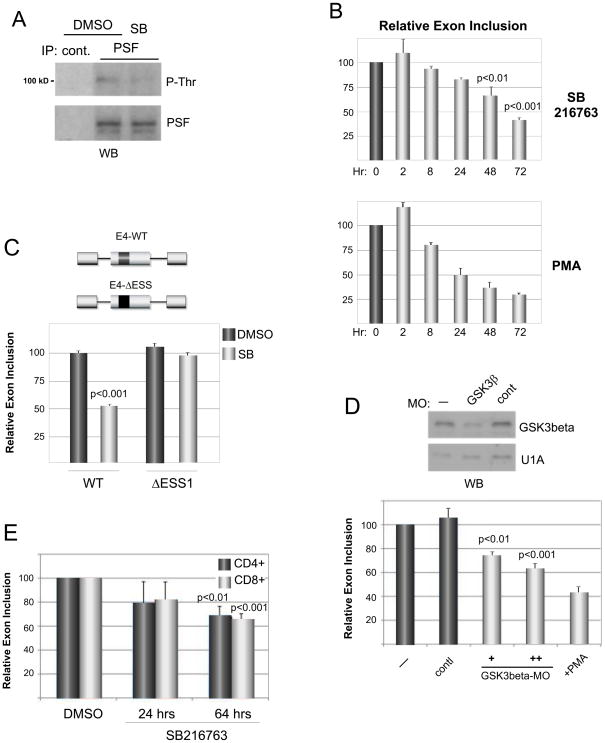
To confirm that GSK3 is in fact an endogenous regulator of PSF we took advantage of the widely used GSK3 inhibitor SB216763. Consistent with the studies above, treatment of JSL1 cells with SB216763 results in a decrease in PSF threonine phosphorylation, with no change in the expression of PSF itself or any other known CD45 regulatory protein (Fig 6A, S5A). Moreover, SB216763 induces a robust increase in CD45 exon skipping in a manner that mimics the effect of PMA stimulation (Fig. 6B). Such exon repression in response to SB216763 is dependent on the presence of the ESS1 regulatory element, as substitution of the ESS1 in a CD45 exon 4 minigene greatly abrogates the influence of this compound (Fig 6C, S5B). Two other independent methods to reduce GSK3 activity in resting T cells also resulted in increased CD45 variable exon skipping, namely knock down of protein expression by morpholino oligos (Fig 6D) or treatment of cells with LiCl, another GSK3 inhibitor (data not shown). Therefore, regardless of method, reduction of GSK3 activity in resting cells closely mimics the effect of PMA stimulation on PSF phosphorylation and CD45 exon skipping, suggesting that GSK3 inhibition is the primary mode by which PMA stimulation causes PSF-mediated changes in CD45 splicing. Consistently, treatment of cells simultaneously with SB216763 and PMA does not substantially increase the magnitude of exon skipping over PMA alone (Fig S5C), suggesting that SB216763 and PMA act on the same pathway in order to achieve CD45 exon exclusion.
Figure 6.
Inhibition of endogenous GSK3 causes a stimulation-like change in CD45 splicing. (A) Western blot of endogenous PSF precipitated from JSL1 cells treated with 10 uM SB216763 (SB) or DMSO vehicle control. (B) Quantitation (n=3) of CD45 isoform expression in cells treated with SB216763 (10uM) or PMA (20ng/ml) for times indicated. (C) Quantitation (n=3) of splicing of minigenes (described in Figure 3) in cells treated with SB216763 or DMSO control. (D) Quantitation (n=3) of endogenous CD45 isoform expression in cells treated with a morpholino oligo against GSK3. Western blot confirmation of knock down relative to U1A loading control shown at top. (E) Quantitation (n=3) of endogenous CD45 isoform expression in primary CD4+ or CD8+ T cells purified from human blood and cultured in the absence (DMSO) or presence of SB216763 for the times indicated. For all panels error bars correspond to standard deviation. See also Figure S5.
Finally, in order to confirm the physiological relevance of our data, we examined whether GSK3 regulates CD45 alternative splicing in primary cells. To this end, we cultured purified CD4+ or CD8+ primary human T cells in the presence of SB216763 or solvent control and analyzed CD45 splicing after 24 or 64 hours. Consistent with our observations in JSL1 cells, SB216763 induces repression of CD45 variable exons in both CD4+ and CD8+ T cells at both time points (Fig 6E). Therefore, we conclude that our data from JSL1 T cells indeed accurately reflect the in vivo pathway from T cell activation to CD45 alternative splicing.
Accumulation of unphosphorylated PSF upon T cell activation requires de novo protein synthesis
The kinetics of the effect of SB216763 on CD45 splicing is remarkably similar to that observed upon PMA stimulation of JSL1 cells or in response to activation of primary T cells (Fig 6B and Lynch and Weiss, 2000). However, we note that this time course is markedly slower than the kinetics with which most signaling events are thought to occur. We previously have demonstrated that the delayed response of CD45 splicing to PMA treatment is due, at least in part, to a requirement for de novo protein synthesis (Lynch and Weiss, 2000). Given that the GSK3 inhibitor displays similar kinetics, we conclude that the rate limiting step in this pathway must be downstream of GSK3 inactivation, which occurs within hours of T cell stimulation (Diehn et al., 2002; Sengupta et al., 2007).
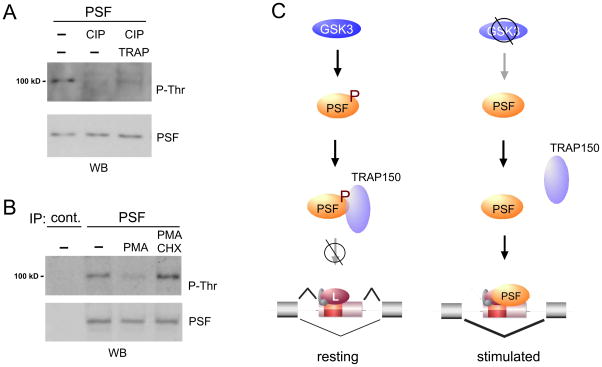
One model to explain the delay in alternative splicing following GSK3 inhibition would be a highly stable Phospho-T687 form of PSF, perhaps aided by being associated with TRAP150. In this model the kinetics of changes in CD45 splicing would be limited by the half-life of PSF, as turnover of PSF in the absence of GSK3 activity would lead to a slow but steady accumulation of PSF unphosphorylated at T687 that is not able to be sequestered by TRAP150. Consistent with this hypothesis, PSF itself, as well as TRAP150, are both highly stable with a half-life of over 24 hours (Fig. S6A; A. A. Melton, F.H. and K.W.L. unpublished), and a strong loss of PSF phosphorylation is not observed until after 24 hours post-stimulation (Fig. S6B). Moreover, although the presence of TRAP150 does not change the overall half-life of PSF (Fig. S6A), it does stabilize the threonine phosphorylation of PSF as pre-incubation with recombinant TRAP150 inhibits the ability of phosphatase to dephosphorylate PSF in vitro (Fig 7A).
Figure 7.
Stability of P-Thr form of PSF and model of pathway from GSK3 activity to CD45 splicing. (A) Western blot of P-Thr form of PSF following in vitro incubation of purified PSF with CIP in the absence or presence of recombinant TRAP150. (B) Western blot of PSF precipitated from cells treated with PMA (20ng/ml), PMA + cycloheximide (CHX, 20uM) or DMSO vehicle control (−) for 48h. Samples were normalized for total PSF in each lane. (C) Schematized model for signal-induced regulation of CD45 in resting and stimulated T cells as described in text. See also Figure S6.
A further prediction of the above model is that the relative amount of phosphorylated PSF would remain high in the absence of protein synthesis following stimulation. Indeed, we find that PMA treatment fails to induce a change in PSF phosphorylation when cells are treated with cycloheximide (Fig 7B, S6B), consistent with the block in activation-induced alternative splicing of CD45 conferred by cycloheximide (Lynch and Weiss, 2000). We have further shown previously that blocking PSF translation with a morpholino oligomer abrogates the ability of PMA to induce CD45 exon skipping (Melton et al., 2007). Thus, taken together, our data suggest that de novo PSF synthesis, in the absence of GSK3 activity, is likely the rate limiting step by which the proper temporal control of CD45 splicing is achieved.
Discussion
In this study, we have identified the complete signaling pathway that leads from T cell receptor engagement to RNA binding of PSF and CD45 exon exclusion (Fig 7C). We show that TRAP150, a protein of largely unknown function, and GSK3, a versatile signaling mediator, are key components of this pathway. Our data also provide a molecular explanation for the built-in “time lag” that is a hallmark of CD45 signal-induced splicing. This study thus reveals substantial new insight into the mechanism for signal induced alternative splicing in T cells, and extends the known cellular functions of TRAP150 and GSK3.
While numerous examples of signal-induced alternative splicing have been described in a variety of cell types, the signaling pathways that connect membrane-bound receptors to the nuclear RNA processing machinery have been identified in only very few cases (Lynch, 2007; Shin and Manley, 2004). Some notable examples in T cells are the Sam68-mediated splicing of CD44 or the TIA-1 regulated Fas alternative splicing (Izquierdo and Valcárcel, 2007; Matter et al., 2002). In these cases, direct phosphorylation of Sam68 or TIA-1 was shown to occur rapidly upon activation of signaling pathways and to directly increase the activity of these proteins bound to RNA. Such a direct effect of phosphorylation on splicing activity is markedly different from the mechanism we demonstrate here in which phosphorylation of PSF indirectly regulates its accessibility to the target RNA via protein-protein interactions. Therefore, the GSK3-dependent regulation of a mutually exclusive interaction between PSF and either TRAP150 or CD45 RNA is a unique paradigm for connecting intracellular signaling pathways to RNA processing events.
In our model, the crucial regulatory point is the phosphorylation dependent interaction of PSF with TRAP150, which prevents PSF from binding to the ESS1 RNA. We were able to map the functionally-relevant phosphorylation in PSF to a single threonine residue, T687, as a T687A point mutant shows substantially reduced interaction with TRAP150 and bypasses the regulatory pathway leading to CD45 exon exclusion in resting cells. We do detect a residual interaction of the PSF T687A mutant with TRAP150, suggesting that phosphorylation strongly facilitates this interaction, but is not an absolute requirement. Furthermore, the finding that deletion of the C-terminal 40 amino acids does not prevent interaction of PSF with TRAP150, argues against direct interaction of TRAP150 with the phosphorylated T687 residue. Rather, our data suggest that phosphorylation of T687 drives conformational changes regulating the interaction with TRAP150. In such a model, a small proportion of the T687A mutant or the non-phosphorylated WT PSF could still be in the conformation permissive to TRAP150 interaction thereby explaining a basal level of interaction in the non-phosphorylated state. Future studies to characterize the interface between TRAP150 and PSF are thus predicted to uncover complex and important intra- and intermolecular interactions.
A further broad implication of the data presented here is in characterizing the regulation of PSF T687 phosphorylation, and defining PSF as a substrate of GSK3. In many cases GSK3-mediated phosphorylation requires a priming phosphate located 4 amino acids towards the C-terminus from the target site (Cohen and Frame, 2001). PSF does contain a tyrosine at position 691, which could potentially serve as a priming phosphate of GSK3-mediated T687 phosphorylation. However, as GSK3 phosphorylates PSF purified from bacteria, we conclude that a priming phosphate is not strictly required, limiting the potential functional relevance of Y691. Finally, as PSF is a mostly nuclear protein and GSK3 resides mostly in the cytoplasm, the simplest model for GSK3-mediated PSF phosphorylation would be that it occurs immediately upon translation, prior to nuclear import, although we cannot rule out other possible models for the location of this activity.
Interestingly, reduced GSK3 activity has been previously shown to regulate the CD28 costimulatory signal in activated T cells (Diehn et al., 2002). This data, combined with our work suggest that a decrease in GSK3 activity is required early during T cell activation to initiate proliferation, but that it later acts on CD45 alternative splicing to increase the threshold for activation, thereby leading to T cell attenuation. Obviously it is important for proper immune function to have the attenuation step temporally delayed with respect to initial activation. In general, direct phosphorylation or dephosphorylation of proteins occurs rapidly upon receptor engagement and are thus not suited to exert a delayed response. In our model the appropriate temporal response is accomplished by a mostly stable phosphorylation of PSF and its interaction with TRAP150, which is only released by de novo synthesis of PSF in conditions with reduced GSK3 activity.
Our data regarding the requirement for de novo protein synthesis in order to induce reduced PSF phosphorylation and CD45 exon skipping is fully consistent with the model we have proposed. We cannot formally rule out that cycloheximide blocks synthesis of a phosphatase required to dephosphorylate PSF under activated conditions. However, as we have demonstrated that changing the activity of GSK3 alone is sufficient to change the phosphorylation state of PSF in the absence of any phosphatase inhibitors (Fig 5, 6), we favor a model in which the predominant factor determining the phosphorylation state of PSF is GSK3. This notion is further supported by our data showing that experimental decrease of GSK3 activity in resting conditions is sufficient to induce a CD45 splicing pattern resembling that of activated cells. In addition, if PMA stimulation would mainly act through increasing expression of a phosphatase, treatment of cells simultaneously with PMA and SB216763 should have an additive effect. The fact that this is not the case again suggests that the contribution of a phosphatase, if any, is minor. Nevertheless, it is possible that there may be alternative models that are consistent with our data and further studies are necessary to work out the complete details of the molecular interactions between GSK3, PSF and TRAP150.
It is interesting to note that we have previously identified a large number of genes that are alternatively splicing in response to T cell activation, several of which are regulated in an ESS1 dependent manner in concert with CD45 (Ip et al., 2007; Rothrock et al., 2003). Consistent with PSF binding specifically to the ESS1 sequence motif, we have shown PSF to be involved in the regulation of several of the ESS1-containing exons studied thus far ((Motta-Mena et al., 2010) and data not shown). PSF is also known to have multiple functions in the nucleus, including roles in transcription and nuclear retention of RNA (Shav-Tal and Zipori, 2002). It therefore is likely that in addition to influencing CD45 alternative splicing in T cells, the regulation of PSF by GSK3 and TRAP150 influences a wide spectrum of PSF-mediated gene expression events impinging on numerous cellular functions. In light of the many roles GSK3 has been shown to play in a variety of cell types, it will be interesting to elucidate the contribution of PSF in mediating these functions.
Experimental Procedures
Cell culture and reagents
Growth and stimulation of the JSL1 cell line was done as described previously (Lynch and Weiss, 2000). The GSK3 inhibitor SB216763 (Tocris, 10 uM), Cycloheximide (Sigma, 20 uM) or DMSO solvent controls were used for the times indicated. Purified human primary CD4+ and CD8+ T cells were obtained from the University of Pennsylvania Human Immunology Core (IRB protocol #811028). Cells were cultured in RPMI with 10% FBS at a density of 5×105 cells/ml. HEK293 cells were cultured in DMEM with Pen/Strep and 5% FBS.
For knock down experiments 10×106 JSL1 cells were transfected with 5–20 nmol of a morpholino-oligo (Gene Tools) blocking the translation start site of the respective target using electroporation. 24h post transfection fresh media was added and cells were grown for an additional 48h. Morpholino-sequences were
BTF: CTAGAATTGGAGCGACCCATTTCTT;
TRAP150: GTTTTTGACATCTCTGAAGAGAGGA;
GSK3beta: GGCCGCCCTGACATGATCACTCTCT.
PSF: GTCGAGGCAAAAGCGAAGAAGACGC.
Stable cell lines were produced as described (Rothrock et al., 2003). For minigene assays, cells were transfected as above with 1 ug of a CD45 exon 4 minigene with WT or mutated ESS1 sequence (Rothrock et. al. 2003) and 10 ug expression construct where indicated. In the case of testing expression of recombinant forms of PSF, a morpholino directed against the 5′-UTR of PSF was also cotransfected to reduce endogenous PSF expression (Melton et al., 2007).
For overexpression of proteins, HEK293 cells were transfected with Lipofectamine2000 using standard procedures. Cell lysates were prepared 48h post transfection.
Immunoprecipitation and protein purification
Whole cell lysates (WCE) were prepared in lysis buffer (25 mM Tris pH7.4, 150 mM NaCl, 1 mM CaCl2, 1% Triton X-100), nuclear extracts (NE) were prepared as described (Melton et al., 2007). For IPs from JSL1 cells, 100 ug NE was incubated with 5 ug PSF antibody (Sigma) or TRAP150 antibody (Abcam) for 1h and an additional 1h with Protein-G Sepharose (GE Healthcare) or 2h with M2-Flag affinity gel (Sigma) in 400 ul 1xRIPA buffer at 4°C under rotation. Beads were spun down, washed 6x with 200 ul 1xRIPA buffer and eluted with sample buffer. To compare bound and unbound protein fractions, the supernatant after the first spin was saved and analyzed by Western blot.
For small scale purification, up to 1 mg NE from JSL1 cells stably expressing Flag-tagged proteins or WCE from transiently transfected 293 cells was immunoprecipitated as above using M2-Flag affinity gel. Elution of bound proteins was performed for 1h on ice with 3xFlag pepide (Sigma) at a concentration of 500 ug/ml in GFB100 (20 mM Tris pH7.5, 100 mM KCl, 1 mM EDTA) with gentle shaking. For large scale protein purification, Flag-PSF was purified from JLS1 cells, or His-PSF from E. coli, using affinity chromatography as described in Melton et al, 2007.
Antibodies
The following antibodies were used throughout the manuscript as noted: PSF (Sigma P2860 for IP, abnova 269–362 for WB), Phospho-Threonine (42H4, Cell Signaling), TRAP150 (ab71985, abcam), SNIP1 (ab19611, abcam), TRAP220 (ab64965, abcam), GSK3beta (ab31826, abcam), GSK3alpha (ab28833, abcam), Flag (2368, Cell Signaling), BTF (ab51758, abcam), hnRNP L (4D11, abcam), hnRNP LL (ARP41102, Aviva Systems Biology), hnRNP U (3G6, abcam), Phospho-Tyrosine (P-Tyr-100, Cell Signaling).
RNA purification
RNA was isolated from JSL1 cells using RNA-Bee (Tel-Test) according to the manufacturer’s protocol. 32P-labeled ESS1 RNA for UV crosslinking was prepared using T7 polymerase as in Rothrock et al., 2005, cold competitor RNAs were chemically synthesized by Dharmacon.
RT-PCR
Radioactive CD45 RT-PCR was performed as previously described (Lynch and Weiss, 2000). Briefly, 1 ug of total RNA was reverse transcribed using a primer binding the junction of CD45 exons 9 and 10 (E9/10). Low-cycle PCR was then performed with a radio-labeled primer hybridizing to CD45 exon 3 and unlabeled E9/10 (sequences in (Topp et al., 2008)). PCR products were separated on denaturing 5% PAGE and visualized using autoradiography. Quantification was performed using a Phosphorimager (Typhoon 9200, GE Healthcare) and ImageQuant software.
RNA binding assays
RNA pulldown assays with biotinylated ESS1 RNA (Dharmacon) were performed as described in Melton et al., 2007. UV crosslink assays were carried out as described (Rothrock et al., 2005). Briefly, 1×105 cpm uniformly 32P-labeled ESS1 RNA was incubated with purified proteins and/or nuclear extracts or cold competitor RNAs. Reaction mixtures were crosslinked with 254 nm UV light for 20 min, RNase digested and analyzed by 8% SDS-PAGE. Purified proteins were mixed and preincubated at 30°C for 2 min prior to the addition of hot RNA probe. Similarly, mixtures including cold RNA competitor were incubated 2 min at 30°C before 32P-labeled RNA was added.
In vitro kinase assay
HEK293 cells expressing HA-tagged constitutively active GSK3beta mutant (S9A) were lysed in lysis buffer (as above but 0.1% Triton X-100) and 500 ug protein were used for HA immunoprecipitation in lysis buffer. The precipitate was washed 3 times in lysis buffer and 3 times in kinase buffer (25 mM HEPES pH7.5, 25 mM MgCl2, 1 mM DTT; 10 uM ATP; adopted from (Wu et al., 2009). The final pellet was resuspended in kinase buffer, mixed with purified PSF variants as indicated and incubated for 1h at 37°C in the presence of 5 mM ATP and/or 25 uCi 32P-gamma-ATP.
Supplementary Material
Acknowledgments
We thank Melanie Cobb for the kind gift of the GSK3-S9A expression construct and for helpful comments on the study. We thank Russ Carstens and members of the Lynch lab for critical reading of the manuscript. This work was supported by US National Institutes of Health grant R01 GM067719 to K.W.L. F.H. is funded by a fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biology. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfant CE, Mischak H, Watson JE, Winkler BC, Goodnight J, Farese RV, Cooper DR. Regulation of alternative splicing of protein kinase C beta by insulin. J Biol Chem. 1995;270:13326–13332. doi: 10.1074/jbc.270.22.13326. [DOI] [PubMed] [Google Scholar]
- Cohen P, Frame S. The renaissance of GSK3. Nat Revs. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends in Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D, Crabtree GR, Brown PO. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. PNAS (USA) 2002;99:11796–11801. doi: 10.1073/pnas.092284399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. PNAS (USA) 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C, Killick R, Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. J Neurochemistry. 2008;104:1433–1439. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Valcárcel J. Fas-activated Serine/Threonine Kinase (FAST K) Synergizes with TIA-1/TIAR Proteins to Regulate Fas Alternative Splicing. J Biol Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- Jacobsen M, Hoffmann S, Cepok S, Stei S, Ziegler A, Sommer N, Hemmer B. A novel mutation in PTPRC interferes with splicing and alters the structure of the human CD45 molecule. Immunogenetics. 2002;54:158–163. doi: 10.1007/s00251-002-0455-7. [DOI] [PubMed] [Google Scholar]
- Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biology. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hawkins IC, Harvey CD, Jennings JL, Link AJ, Patton JG. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol Cell Biol. 2003;23:7437–7447. doi: 10.1128/MCB.23.21.7437-7447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immuno. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;29:223–234. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohteki T, Parsons M, Zakarian A, Jones RG, Nguyen LT, Woodgett JR, Ohashi PS. Negative Regulation of T Cell Proliferation and Interleukin 2 Production by the Serine Threonine Kinase Gsk-3. J Exp Med. 2000;192:99–104. doi: 10.1084/jem.192.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner F, Angelo M, Giese KP. The Roles of Cyclin-dependent Kinase 5 and Glycogen Synthase Kinase 3 in Tau Hyperphosphorylation. J Biol Chem. 2006;281:25457–25465. doi: 10.1074/jbc.M603469200. [DOI] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Jayaraman P, Chilton PM, Casella CR, Mitchell TC. Unrestrained glycogen synthase kinase-3 beta activity leads to activated T cell death and can be inhibited by natural adjuvant. J Immunol. 2007;178:6083–6091. doi: 10.4049/jimmunol.178.10.6083. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Cohen M, Lapter S, Dye B, Patton JG, Vandekerckhove J, Zipori D. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol Biol Cell. 2001;12:2328–2340. doi: 10.1091/mbc.12.8.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS letters. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh GI, Miyamoto S, Price NT, Safer B, Proud CG. T-cell Activation Leads to Rapid Stimulation of Translation Initiation Factor eIF2B and Inactivation of Glycogen Synthase Kinase-3. J Biol Chem. 1996;271:11410–11413. doi: 10.1074/jbc.271.19.11410. [DOI] [PubMed] [Google Scholar]
- Wu G, Huang H, Garcia-Abreu J, He X. Inhibition of GSK3 phosphorylation of beta-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLoS One. 2009;4:e4926. doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Jia X, de la Cruz L, Su XC, Marzolf B, Troisch P, Zak D, Hamilton A, Whittle B, Yu D, et al. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity. 2008;29:863–875. doi: 10.1016/j.immuni.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Weiss A. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nature immunology. 2002;3:764–771. doi: 10.1038/ni822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.