Abstract
Telomeres protect the ends of eukaryotic chromosomes from being recognized and processed as double strand breaks. In most organisms, telomeric DNA is highly repetitive with a high GC-content. Moreover, the G residues are concentrated in the strand running 3′–5′ from the end of the chromosome towards its center. This G-rich strand is extended to form a 3′ single-stranded tail that can form unusual secondary structures such as T-loops and G-quadruplex DNA. Both the duplex repeats and the single-stranded G-tail are assembled into stable protein–DNA complexes. The unique architecture, high GC content, and multi-protein association create particularly stable protein–DNA complexes that are a challenge for replication, recombination, and transcription. Helicases utilize the energy of nucleotide hydrolysis to unwind base paired nucleic acids and, in some cases, to displace proteins from them. The telomeric functions of helicases from the RecQ, Pi., FANCJ, and DNA2 families are reviewed in this article. We summarize data showing that perturbation of their telomere activities can lead to telomere dysfunction and genome instability and in some cases human disease.
Keywords: Telomere, Telomerase, Helicase, G-quadruplex, Telomere replication
1. Helicases
Helicases are best known for their ability to harness the energy of nucleotide triphosphate (usually ATP) hydrolysis to catalyze the unwinding of duplex nucleic acids. Helicases can unwind a variety of different structures including DNA, RNA, and DNA/RNA duplexes as well as more exotic molecules, such as forked or bubbled replication intermediates, Holliday junctions, and non-Watson Crick base paired structures such as G-quadruplexes. In addition to unwinding base paired nucleic acids, some helicases can translocate along an RNA or DNA substrate and others can displace proteins from nucleic acids. Owing to this great versatility, helicases are required for virtually all biological processes involving nucleic acids including DNA replication, repair and recombination, and RNA transcription, splicing and translation. This wide variety of cellular functions may explain why organisms encode so many different helicases. For example, 134 (2%) of the open reading frames in the Saccharomyces cerevisiae genome encode predicted helicase proteins [1]. In this review we focus on DNA helicases with demonstrated functions at telomeres, the ends of eukaryotic chromosomes (see Fig. 1 for helicase families with roles at telomeres).
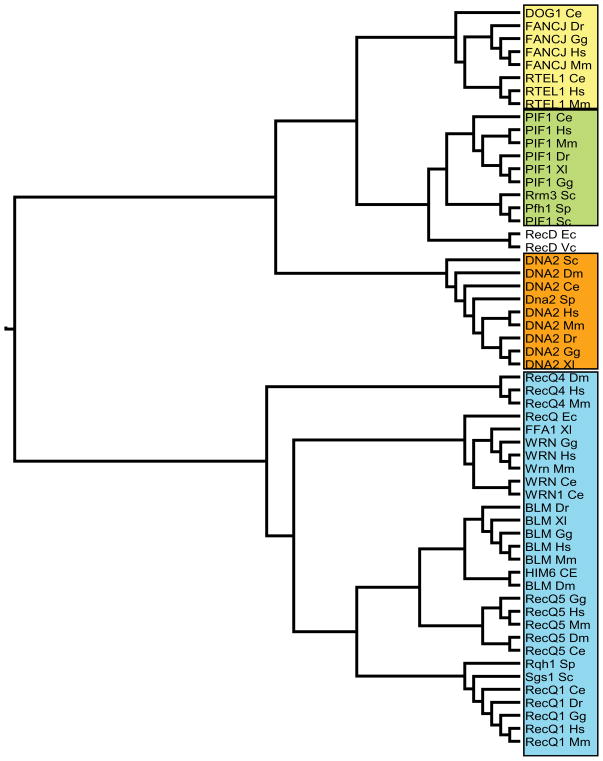
Fig. 1.
Sequence alignments of the RecQ, Pif1, FANC-J, and DNA2 family helicases. Sequences were aligned using ClustalX, and the phylogenetic relationship among them was drawn as a rooted tree using the unrelated human beta actin protein (NP_001092) as an outgroup (not shown) with TreeView v. 1.6.6. software (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). Tree lines are not scaled. Helicase family members from the following organisms were aligned: Caenorhabditis elegans (Ce), Danio rerio (Dr), Drosophila melanogaster (Dm), Gallus gallus (Gg), Escherichia coli (Ec), Homo sapiens (Hs), Mus musculus (Mm), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Vibrio cholera (Vc), Xenopus laevis (Xl). Drosophila melanogaster has four different annotated PIF1A isoforms (designated A–D; PIF1B is an unrelated protein), but none of the isoforms closely align to any known PIF1 helicase member. It is unclear if this is due to the misannotation of PIF1A or some other problem. Due to this discrepancy, PIF1 Dm is not shown in the tree. For each organism all related proteins are listed, if annotated as such in the database. The GenBank accession numbers are as follows: BLM Dm, AAD41441; BLM Dr, XP_701357; BLM Gg, NP_001007088; BLM Hs, NP_000048; BLM Mm, NP_001035992; BLM Xl, NP_001079095; DNA2 Ce, NP_496515; DNA2 Dm, ACS78060; DNA2 Dr, CAX13876; DNA2 Gg, NP_001006497; DNA2 Hs, AAI11741; DNA2 Mm, AAI15717; DNA2 Sc, AAB68010; DNA2 Sp, CAB38508; DNA2 Xl, NP_001079231; DOG1 Ce, NP_493618; FANCJ Dr, ABO27623; FANCJ Gg, Q3YK19; FANCJ Hs, NP_114432; FANCJ Mm, Q5SXJ3; HIM6 Ce, AAM26298; PIF1 Hs, NP_079325; PIF1 Ce, BAA28677; PIF1 Dr, NP_942102; PIF1 Gg, XP_426648; PIF1 Mm, AAH46611; PIF1 Sc, CAA86260; Rrm3 Sc, NP_011896; PIF1 Sp, NP_596488; PIF1 Xl, AAZ41379; RecD Bc, YP_085716; RecD Ec, CAQ33145; RecD Vc, NP_231950; RecQ Ec, YP_002331581; RecQ1 Ce, AAK21428; RecQ1 Dr, NP_001038561; RecQ1 Gg, NP_989724; RecQ1 Hs, NP_002898; RecQ1 Mm, NP_075529; RecQ4 Dm, AAF42939; RecQ4 Hs, NP_004251; RecQ4 Mm, BAD11131; RecQ5 ce, CAA86232; RecQ5 Dm, AAD43051; RecQ5 Gg, BAI79325; RecQ5 Hs, NP_004250; RecQ5 Mm, BAD11130; Rqh Sp, CAA91177.1; RTEL1 Hs, NP_116575; RTEL1 Ce, NP_492769; RTEL1 Mm, AAI44979; Sgs1 Sc, AAB60289.1; WRN Gg, BAI79323; WRN Hs, AAC63361; WRN Mm, AAH60700; WRN Xl, NP_001081838; WRN1 Ce, NP_495324.
The majority of helicases are classified into two superfamilies (SFI and SFII) defined by the presence of seven conserved amino acid motifs. The helicase motifs are found in a domain of approximately 300–500 amino acids and are observed by structural analysis to cluster in space (review [2]). Helicases are placed into families based on higher levels of sequence similarity, both within and outside of the helicase motifs. In this review we discuss helicases in the RecQ, Pif1, FANC-J, and DNA2 families.
Helicases are divided into DNA or RNA helicases depending on the chemical identity of the strand onto which they load (all of the helicases discussed in this review are DNA helicases). They are further divided by their direction of unwinding. Helicases typically unwind duplexes in a unidirectional manner, moving either 3′ to 5′ (RecQ family helicases) or 5′–3′ (Pif1, FANC-J, DNA2 families) along the strand onto which they load. The number of nucleic acid base pairs unwound before the helicase dissociates from its substrate defines its processivity. Helicases vary in their in vitro determined processivity, which may reflect the enzyme’s preferred substrate and/or necessary co-factors. For example, the S. cerevisiae Pif1 DNA helicase is poorly processive on conventional duplex DNA but is highly processive on forked RNA/DNA duplexes [3] and G-quadruplex structures (KP and VAZ, in preparation) (discussed in more detail in Section 4.1).
2. Telomeres
This section reviews the properties of telomeres with an emphasis on those aspects of telomere biology that are likely to involve DNA helicases. Telomeres are protein–DNA structures that distinguish natural chromosome ends from double strand breaks (DSBs). Because telomeres protect chromosome ends from processing events that result in degradation and/or end-to-end fusions, they are essential for chromosome integrity. Telomeric DNA is replenished by an unusual replication mechanism that involves a telomere-dedicated reverse transcriptase called telomerase that compensates for the inability of DNA polymerases to replicate the 5′ ends of linear chromosomes [4]. In addition, from yeasts to humans, telomeres are specialized sites for gene expression, as structural genes positioned near telomeres are transcriptionally repressed (reviewed in [5]).
Telomeric DNA consists of a tandem array of short repeated sequences in which the strand running 5′–3′ from the centromere towards the chromosome end is usually guanine-rich. The amount of duplex telomeric DNA per chromosome end varies enormously from organism to organism. For example, the yeasts S. cerevisiae and Schizosaccharomyces pombe have ~300 bps of telomeric repeats per end while many mammals have ~10 kb or more. Extension of the guanine-rich strand forms a 3′ single stranded overhang (G-tail). This G-tail is a conserved feature of telomere structure and is essential for telomere function [6–10]. Because duplex telomeric DNA is G–C rich, its thermal stability is high, which may explain why multiple helicases have roles in telomere dynamics. Because of the high concentration of guanines, telomeres are able to form stable G-quadruplex structures (discussed in detail below).
As part of their end-protection function, telomeric DNA is bound constitutively by a core group of proteins called shelterin. Shelterin components include both duplex (TRF proteins in mammals, Rap1 in S. cerevisiae, Taz1 in S. pombe) and single strand (Pot1 in mammals and S. pombe, Cdc13 in S. cerevisiae) binding proteins as well as proteins that associate via protein–protein interactions [8,11–13]. The overall composition of the shelterin complex varies between species in respect to protein content and higher order structure, but the general design is conserved [14]. The role of the shelterin complex in telomere function and regulation includes distinguishing natural ends from DNA breaks and recruiting telomerase to DNA ends [12].
In addition to the shelterin complex, higher-order DNA structures are thought to contribute to telomere functions. A well documented example of such structures is the T-loop that was first identified in human and mouse cells by electron microscopy [15] and later observed in a variety of other organisms, including trypanosomes, ciliates, and nematodes [15–23]. T-loops are duplex lariat structures formed when the single stranded telomeric G-overhang invades the double stranded telomeric region of the same chromosome [24]. Unwinding T-loops may require a specialized helicase. By sequestering chromosome ends, T-loops are proposed to protect telomeres against checkpoint recognition, DNA repair, and telomerase-mediated extension [15,25,26]. Since T-loops are structurally similar to Holliday junctions, they may also be important for telomere recombination [27,28]. It is not yet known if T-loops exist at each telomere, how they are regulated in the cell cycle, or how they are displaced to allow both semi-conservative and telomerase-mediated telomere replication.
G-quadruplex structures are another secondary DNA structure that can affect telomere function. G-quadruplexes involve the association of four guanines into a cyclic Hoogsteen hydrogen bonding arrangement in which each guanine shares a hydrogen bond with its neighbor (N1–O6 and N2–N7) (reviewed in [29,30]). The G-rich single stranded telomeric overhang can form intra- and intermolecular G-quadruplex structures in vitro. The in vivo occurrence of telomeric G-quadruplex structures has so far been demonstrated only in ciliates [31–33]. In ciliates, two telomere-binding proteins regulate and promote the formation of telomeric G-quadruplex DNA in vitro and in vivo [31,32,34]. Some DNA helicases with well documented effects on telomeres, such as WRN and Pif1, can unwind G-quadruplex structures in vitro [35,36]. G-quadruplexes have the potential to regulate telomerase activity: intramolecular G-quadruplexes block telomerase activity in vitro [37,38], and telomerase RNA exhibits a G-quadruplex motif, which might be regulated by an as yet unidentified RNA helicase [39]. The formation and regulation of secondary structures such as T-loops and G-quadruplexes may contribute to telomere function, but these structures have to be tightly regulated as they also present a problem for telomere maintenance. Secondary DNA structures are an obstacle for both semi-conservative and telomerase-mediated replication and must be resolved prior to these events.
Telomeres become shortened during every cell division due to incomplete replication of the lagging strand (the so called “end replication problem”). Additional loss of telomeric DNA occurs due to post-replicative degradation of the 5′ strand that generates long 3′ G-rich overhangs [40,41]. In most species, the loss of telomeric DNA is balanced by the action of telomerase that uses its RNA component to template extension of the 3′ G-tails [42–44]. The complementary C-strand is then synthesized by conventional RNA-primed DNA replication [4,45].
The importance of telomerase to genome integrity is best illustrated by situations where telomerase is not expressed, which occurs naturally in most human somatic cells or by mutation in genetically tractable organisms. Due to incomplete replication, telomeres progressively shorten in telomerase deficient cells: human telomeres lose ~100 bps of telomeric DNA in each cell division, while yeast telomeres lose ~4 bps. Although telomerase is not essential and its absence is tolerated for many cell divisions, extreme telomere shortening causes telomere dysfunction, which leads to chromosome instability, end-to-end fusions, and checkpoint-mediated cell cycle arrest and/or apoptosis (reviewed in [46,47]). The ability of human cells to divide in culture is finite. However, if the catalytic subunit of telomerase is introduced into these cells, its expression confers an unlimited division potential upon them [48]. Although telomerase is not expressed in most human somatic cells, inherited mutations in telomerase components or certain telomere structural proteins result in short telomeres and reduced life expectancy, probably as a result of stem cell failure [49,50]. Finally, telomerase is up regulated in the vast majority of human cancers [50–52], and this heightened activity contributes to the increased division potential of malignant cells. Thus, both down and up regulation of telomerase are associated with human disease.
Semi-conservative replication of duplex telomeric DNA is usually unidirectional, moving from an internal origin of replication towards the chromosome end. Because of this unidirectional replication, if replication forks stall within telomeric DNA they cannot be rescued by a converging replication fork. Moreover, because telomeric DNA is always replicated in the same direction, the telomeric G-rich strand is always the template for lagging strand synthesis [53–55]. During the unwinding of the parental duplex to allow the start of a new Okazaki fragment, the lagging strand is transiently single stranded, which at least theoretically provides an opportunity for it to form stable secondary structures, such as G-quadruplex DNA. The occurrence of such structures could impede replication fork progression through telomeric regions. Indeed, replication fork stalling within telomeres has been seen in diverse organisms using a variety of methods, although it is not known if G-quadruplex structures cause these replication problems.
Replication forks move slowly through S. cerevisiae telomeres as seen from both two-dimensional (2D) gel and genome-wide DNA Polymerase 2 association analyses [56,57] (see Section 4.2 for a discussion of the role of the Rrm3 helicase in semi-conservative replication of S. cerevisiae telomeres). 2D gels also reveal that semi-conservative replication of S. pombe telomeres is slowed when cells lack Taz1, the duplex telomere binding protein, in a manner that is independent of the direction of replication through the repeats [58] (see Section 3.2 for role of the RecQ helicase Rqh1 in S. pombe telomere replication). Moreover, in S. pombe, the leading strand polymerase arrives at the telomere before the lagging strand DNA polymerases, suggesting that the telomeric template for lagging strand synthesis may have a longer half-life as single stranded DNA than lagging strands elsewhere in the genome, a situation that is expected to exacerbate replication problems [59]. A situation seemingly similar to S. pombe Taz1 deficient cells exists in mammalian cells depleted for the Taz1 ortholog, TRF1. Using DNA combing, which allows the examination of replication at the single molecule level, replication forks move about half as fast through mouse telomeric DNA when it lacks TRF1 [60]. Moreover, when examined by FISH (fluorescence in situ hybridization), telomeres of metaphase chromosomes in TRF1 depleted cells have an aberrant structure in which telomeres are no longer discrete entities but often have a bipartite appearance that is reminiscent of structures at chromosomal fragile sites upon replication stress [60]. Both RecQ (BLM, WRN) and FANCJ (RTEL) family helicases are implicated in replication of mammalian telomeres (see Sections 3.3 and 5).
Given that telomeres from yeasts to humans are regions where transcription of structural genes is repressed [5], the finding that telomeric repeats and subtelomeric regions are transcribed was quite surprising [61–63]. This telomeric transcribed region, called TERRA, produces a non-coding G-rich RNA transcribed from the C-rich strand that is often telomere-associated. Although TERRA was only recently discovered, it may be a wide spread feature of telomeres, as it has been detected in S. cerevisiae [61] and mammals [62,63]. In yeast increased TERRA results in telomere shortening in cis, suggesting that TERRA interferes with telomerase-mediated telomere lengthening [64,65]. So far no helicases are identified that function during TERRA biogenesis, but it would not be surprising if helicases are required for TERRA synthesis, regulation, and/or removal from telomeric DNA.
Although telomerase is the major mechanism for telomere maintenance, homologous recombination (HR) provides an alternative (ALT) method for maintenance of telomeric DNA, especially in telomerase deficient cells. In S. cerevisiae, most cells lacking telomerase ultimately die, although a subset of cells emerge as survivors if the strain is recombination proficient. Yeast has two distinct ALT pathways, called type I and type II recombination [66]. The two pathways use different telomeric sequences as substrates for recombination and depend on different recombination proteins [66–68]. In type I recombination, the sub-telomeric Y′ elements are expanded into large tandem arrays at individual chromosome ends. In contrast, in type II recombination, the simple G-rich repeats at chromosome ends are expanded with some telomeres bearing 10–100 times more telomeric DNA than at wild type telomeres, whereas other telomeres are very short [67,68]. The heterogenous lengths and telomere dynamics in yeast type II survivors are very similar to what is seen in human tumors that maintain telomeres by ALT [69].
Because telomeric DNA is G-rich, it is particularly stable. This stability is true for standard B-form duplex telomeric DNA as well as unusual secondary structures, such as T-loops, G-quadruplex DNA, and RNA/telomeric DNA hybrids that form during TERRA and telomerase extension. Given this high inherent thermal stability, as well as the many shelterin components with which telomeric DNA is associated, it is probably not surprising that helicases play a large role in telomere biology. In the remainder of this review we focus on helicases with demonstrated roles in telomere metabolism. The telomere functions of helicases discussed in this review are summarized in Table 1.
Table 1.
Summary of helicase functions at telomeres.
| Function | RecQ | Pif1 | FancJ | Dog-l | RTEL | Dna2 |
|---|---|---|---|---|---|---|
| Telomere binding | (✔) hWRN | ✔ (ScPifl, ScRrm3, hPif) | ✔ (SpDna2) | |||
| Telomerase regulation/interaction | ✔ (scPifl, hPif, mPif) | |||||
| Homologous recombination at telomeres | ✔ (hBLM, hWRN, mWRN, SpTlh, Rqhl?, Sgsl) | ✔ (hFANCJ?) | ✔ (all) | |||
| Telomere length regulation by ALT | ✔ (hBLM, hWRN, mWRN, SpTlh, Rqhl?, Sgsl) | ✔ (scDNA) | ||||
| Telomere semi-conservative replication | ✔ (WRN?, hBLM?, Rqhl?) | ✔ (scRrm3) | (Dog-l?) | ✔ (mRTEL?) | ✔ (scDna2) | |
| G-quadruplex unwinding/regulation | ✔ (hWRN, hBLM, Sgsl) | ✔ (scPifl) | ✔ (hFANCJ) | ✔ (Dogl) | ✔ (hDNA2, scDna2) | |
| Telomere length regulation | ✔ (hBLM, hWRN) | ✔ (scPifl, ScRrm3, Pfhl?, hPifl?) | (Dog-l?) | ✔ (mRTEL) | ✔ (scDNA2) | |
| Telomeric overhang | ✔ (hWRN, Sgsl) | (scPifl?) | ✔ (hDna2, SpDna2, ScDna2) | |||
| Preventing telomere fusion | ✔ (Rqhl?) | ✔ (mRTEL) |
Columns indicate helicase families. Rows indicate predicted function at telomeres. Checkmark (✔) indicates a direct role for this helicase family at this telomere event. In parenthesis are the names of the specific helicase shown to have this functional activity. ? indicates a modest or indirectly demonstrated effect on this telomere event.
3. RecQ family helicases
The SFII RecQ family of 3′–5′ DNA helicases, named for its prototypical member from Escherichia coli, is conserved from bacteria through humans. As described in more detail below, RecQ helicases have important roles in various aspects of DNA metabolism and seem to be particularly important in preventing illegitimate recombination, repairing stalled replication forks, and processing DNA breaks to generate the 3′ single strand tails that initiate HR.
Most unicellular organisms including E. coli (RecQ), S. cerevisiae (Sgs1), and S. pombe (Rqh1) encode a single RecQ family member (Fig. 1). The null phenotypes of cells lacking their single RecQ homolog is complex, suggesting that these proteins act at multiple steps in DNA replication, recombination, and repair. Multicellular organisms including Caenorhabditis elegans, Xenopus laevis, Drosophila melanogaster, and Homo sapiens encode multiple RecQ family members. The human genome encodes five RecQ homologs called RECQ1, BLM (Bloom’s syndrome), WRN (Werner’s syndrome, WS), RECQ4 (Rothmund–Thomson syndrome), and RECQ5. Mutations in three of the human RecQ helicases result in inherited diseases (disease names are in parentheses) that affect genome integrity and cause an increased propensity to cancer (Bloom’s, Rothmund–Thomson) and/or accelerated aging (WS, Rothmund–Thomson). Although it is not clear if the five human RecQ proteins provide complementary or independent cellular roles, their different substrate specificities as defined in vitro allow for the possibility of their having specialized functions as does their distinguishable disease phenotypes [70–75].
Other metazoans also encode multiple RecQ proteins, and in most cases they are clearly homologs of one of the human proteins (Fig. 1). For example, the four C. elegans RecQ proteins can be identified as homologs of the human RECQ1, BLM (called HIM-6), WRN (WRN-1), and RECQ5 [76–80]. However, the number of distinct RecQ family proteins does not correlate directly with evolutionary position. For example, X. laevis encodes only two RecQ family members, Xl BLM (BLM) and FFA-1 (WRN) (Fig. 1).
RecQ family members range in size from 610 amino acids in E. coli to 1447 amino acids in S. cerevisiae. Although the N-termini are not well conserved [81,82], the middle and C-terminal regions have high similarity in three conserved regions: the helicase domain, the RecQ carboxy-terminal (RQC) domain, and the Helicase and RNase D C-terminal (HRDC) domain. Although different RecQ helicases have different substrate specificities, most act preferentially on structured substrates that resemble replication and recombination intermediates such as replication forks, Holliday junctions, D-loops, and 5′ flaps [71,83]. Additionally, several RecQ helicases are able to unwind G-quadruplexes, including the S. cerevisiae Sgs1 and the human WRN and BLM proteins [35,72,75]. Consistent with their more general roles in recombination, Sgs1, mouse WRN, and human BLM function in ALT, where telomeres are maintained by homologous recombination (Table 1) [69,84–86].
3.1. Telomere functions of the S. cerevisiae RecQ homolog Sgsl
Sgs1 has roles in the two major pathways for telomere maintenance, telomerase and ALT. Sgs1 participates in degradation of the C-strand of newly replicated telomeres to generate long 3′ overhangs, which are the presumed DNA substrate for telomerase [87]. This function is not specific to telomeres, as Sgs1 plays a similar role in resection of DSBs, which generates the 3′ single strand tails that initiate HR [88–91]. At telomeres Sgs1 may also help in promoting the formation of telomeric G-tails by unwinding G-quadruplex structures, as Sgs1 is able to efficiently unwind these structures in vitro [75,87].
In telomerase deficient cells, telomeres slowly shorten until most cells die. The rate of death in these senescing cultures is heightened by the absence of Sgs1. In cells lacking both telomerase and Sgs1, recombination-dependent X-shaped structures accumulate [92]. These structures are interpreted as late intermediates during telomere recombination that cannot be resolved in the absence of Sgs1. The interpretation that Sgs1-mediated recombination in telomerase deficient cells prevents cell death is supported by sequencing analysis that reveals reduced telomere recombination in sgs1 telomerase deficient cells [92].
A small fraction of the cells in senescing telomerase deficient cultures give rise to type I or II survivors in which telomeres are maintained by recombination. Sgs1 helicase activity, its interaction with topoisomerase III, and its modification by sumoylation at the C-terminus are required for generation and maintenance of type II survivors [84,85,92]. Remarkably, expression of mouse WRN helicase in yeast suppresses the slow growth and G2/M arrest observed in telomerase negative sgs1 yeast cells, while expression of human BLM allows telomerase deficient sgs1 cells to form type II survivors [85,93]. These findings suggest that Sgs1, mouse WRN, and human BLM have conserved functions in telomere recombination. Finally, in cells deficient for both telomerase and recombination, Sgs1 inhibits the generation of rare survivors that arise by recombination-independent mechanisms [94].
3.2. Telomeric functions of the S. pombe RecQ homolog Rqh1
Rqh1 is important to help cells recover from impaired DNA replication by stabilization and restart of stalled replication forks and activation of checkpoints in response to replication stress [95–97]. Several lines of evidence indicate that Rqh1 also has telomere functions, although these may be extensions of its more general role in DNA replication. Telomere length is at best modestly shorter in cells lacking Rqh1 [98,99]. However, mutations in rqh1+ affect telomere maintenance in certain mutant backgrounds where telomere length is already compromised. For example, cells lacking the telomeric double strand DNA binding protein Taz1 have exceptionally long telomeres that render them cold sensitive [100,101]. If taz1-cells are deficient in telomerase [98] or if they express a partial loss of function allele of an RPA subunit (rad11-D223Y; RPA, replication factor A, is a sequence non-specific single strand DNA binding protein that is essential for DNA replication, repair, and recombination) [102], they lose telomeric DNA extremely rapidly. In both taz1-cell types, telomere loss is suppressed if mutant cells also lack Rqh1.
As noted in Section 2, semi-conservative replication of S. pombe telomeric DNA is impaired in cells lacking Taz1 [58]. When telomeres are replicated in Taz1 defective cells, telomeres do not separate properly in mitosis, especially at low temperatures, a process called telomere entanglements. These entanglements result in DNA breaks, chromosome missegregation, and loss of viability [98,101]. This telomere dysfunction is promoted by sumolyated Rqh1 [98]. Rqh1 sumolyation appears to be fairly specific for telomeres as non-sumolyated Rqh1 is proficient for non-telomeric functions. Rqh1 sumoylation is proposed to affect its localization to telomeres or to allow unwinding of telomere-specific structures such as stalled replication forks or G-quadruplexes [98].
S. pombe encodes multiple telomere-linked helicase (tlh) genes that are reported to have significant sequence homology between residues ~1180 and ~1820 with RecQ helicases [103]. Tlh genes, evolutionarily conserved in fungi, are normally transcriptionally repressed by telomeric silencing. However, in telomerase deficient cells, these genes are transcriptionally activated once telomeres become very short, and cell division is slowed. Moreover, this activation is important for the emergence of cells able to maintain telomeres by the ALT pathway [104,105]. Indeed, if a wild type domain of one of the tlh1 genes is over-expressed in telomerase deficient cells, these cells exit from the growth crisis faster than controls [103]. The helicase activity of the Tlh1 proteins may be important for exit from crisis, as over-expression of a presumed helicase dead version of this domain does not hasten this process. Similarly, S. cerevisiae subtelomeric Y′ elements encode several helicases called Y′-helicase protein 1 (Y′Help1). The expression level of Y′-help is higher in telomerase deficient survivors cells [106]. Our analysis reveals no significant similarity of these full-length helicases to other helicase families discussed in this paper (KP and KRM, unpublished results).
3.3. Telomere functions of mammalian RecQ helicases
Two human RecQ helicases WRN and BLM are also implicated in telomere maintenance. Primary fibroblasts from WS patients recapitulate a tissue culture version of the most dramatic phenotype of WS patients, premature aging, as these cells have a dramatically reduced division potential in vitro (reviewed in [107]). In addition, cultured WS cells grow slowly and exhibit accelerated telomere shortening and genome instability [108,109]. These deleterious phenotypes are likely associated with telomere dysfunction as over-expression of the catalytic subunit of telomerase rescues the reduced division potential, slow growth, and chromosome instability phenotypes of WS cells [110,111]. Moreover, wrn knockout mice show premature aging only in a telomerase minus background [112]. As shown by live cell imaging, chromatin immunoprecipitation, and immunostaining, WRN associates with telomeres during S phase [109,113] and thus likely affects telomeres and their replication directly.
In primary fibroblasts of WS patients, WRN helicase activity is necessary during replication of the G-rich telomeric lagging strand [109]. Using FISH, lack of WRN results in a preferential loss of telomeres from the sister chromatid of the lagging strand; a phenotype referred to as sister telomere loss (STL) [109]. STL is observed in cells expressing helicase-deficient WRN but not in cells expressing wild-type WRN or nuclease-deficient WRN, suggesting that WRN helicase but not exonuclease activity is necessary for its function in telomere replication [109]. Furthermore, expression of telomerase rescues STL [109]. WRN could act by promoting semi-conservative replication through duplex telomeric DNA, perhaps by unwinding G-quadruplex structures [72] that form on the lagging strand. Alternatively or in addition, WRN could resolve telomeric D-loops to allow passage of the replication fork and/or telomerase access [109].
Although the WRN nuclease activity may not be important for telomere replication, there are suggestions that it plays a role in processing telomeric DNA to activate a DNA damage response. In support of this idea, if telomeric oligonucleotides with free 3′ ends are introduced into cells, they elicit a WRN exonuclease-mediated DNA damage response [114]. Moreover, this response does not occur if the 3′ ends are nuclease resistant, providing additional support for an important function of the WRN exonuclease activity in checkpoint activation in response to telomere perturbations.
In Section 2 we summarized data showing that replication of mammalian telomeres is impaired in cells lacking TRF1, as demonstrated by a reduced rate of fork movement through telomeric DNA and by the occurrence of fragile telomeres [60]. Depletion of the RecQ BLM helicase also generates fragile telomeres, while surprisingly depletion of WRN does not. Thus, while both BLM and WRN are implicated in telomere maintenance, their exact role at telomeres are likely different. However, WRN and BLM both unwind G-quadruplex structures in vitro [72], and it is possible that both promote telomere replication via this activity.
A variety of studies indicate that BLM and WRN interact biochemically and functionally with several shelterin components, providing further evidence that the two helicases affect telomeres. For example, both helicases interact with TRF1 and TRF2, the two sequence specific duplex telomere binding proteins, as well as with POT1, the sequence specific single strand telomere binding protein. TRF2 stimulates WRN and BLM helicase activity in vitro, especially during the unwinding of long telomeric substrates [115,116] while POT1 stimulates WRN and BLM activity on long telomeric forked duplexes and D-loop structures [117].
As with the S. cerevisiae Sgs1, WRN and BLM are also implicated in the recombination dependent ALT pathway. Both proteins localize to ALT telomeres and to ALT-associated promyelocytic leukemia bodies (PML), a nuclear structure whose exact function is not known but that is associated with DNA repair and recombination [83,118–120]. In the presence of TRF1 and TRF2, WRN unwinds artificial telomeric D-loops, which are thought to be intermediates in ALT [113]. Perhaps the most direct evidence for a role for RecQ helicases in ALT is that RNAi-mediated reduction in BLM expression results in shortening of ALT telomeres [121].
4. Pif1 family helicases
The SFI PIF1 family of 5′–3′ DNA helicases, named for its prototypical member from S. cerevisiae [122], is found in almost all eukaryotes (Fig. 1). Eukaryotic PIF1 helicase family proteins share sequence similarity in all pair-wise combinations over the 400–500 amino acid helicase domain (reviewed in [123]). In contrast, the N- and C-termini of Pif1 family helicases differ in size and sequence. S. cerevisiae Pif1 has low sequence similarity (~16%) to the bacterial RecD helicases with which it shares three additional motifs [124]. Phylogenetic comparisons show that a small subset of the prokaryotic RecD-like proteins cluster with the most divergent eukaryotic Pif1 family proteins, rather than with prokaryotic RecD proteins and that organisms with a Pif1/RecD-like gene also encode a more canonical RecD protein. Thus, this analysis suggests that the similarity between RecD and Pif1 proteins reflects a common evolutionary origin [123]. Eukaryotic Pif1 family proteins contain a highly conserved 21-residue signature motif between helicase motifs II and III that is not found in the prokaryotic RecD proteins [123].
S. cerevisiae, in which Pif1 family helicases were first discovered and where they have been the most extensively studied, encodes two Pif1 family members, Pif1 itself and a second protein called Rrm3, which is 40% identical to Pif1 over the helicase domain (Fig. 1). As described below, the two S. cerevisiae family members have quite different functions. Several other fungi also encode two distinct Pif1 family proteins, one that is clearly a homolog of the S. cerevisiae Pif1 and one that is clearly Rrm3-like. In contrast, all metazoans encode a single Pif1 family helicase that has roughly equal similarity to the two S. cerevisiae proteins (Fig. 1). At this time, Pif1 family helicases have been studied in detail only in S. cerevisiae (Pif1, Rrm3), S. pombe (Pfh1), mouse (mPIF), humans (hPIF), and parasites (which can encode up to seven family members, the majority of which reside in mitochondria). In all of these organisms except mouse, where the question has not been addressed, Pif1 family helicases are expressed as both nuclear and mitochondrial isoforms [125–129]. S. cerevisiae Pif1, S. pombe Pfh1, and several of the Trypanasoma Pif1 helicases are critical for maintenance of mitochondrial DNA. Clear telomere functions have been demonstrated for the two S. cerevisiae proteins and suggested for the S. pombe, human, and mouse proteins.
4.1. S. cerevisiae Pif1: general biology and telomere roles
Pif1 is the only family member that is relatively easy to purify and thus is the Pif1 family helicase for which there is the most extensive biochemical analyses [130]. On conventional linear DNA substrates, Pif1 is poorly processive, although its activity is higher on forked DNA substrates [3]. Although Pif1 cannot load onto RNA, it has higher activity removing RNA from an RNA/DNA hybrid than DNA from an equivalent DNA/DNA hybrid [3]. Moreover, Pif1 is even more active on forked RNA/DNA molecules than on linear RNA/DNA hybrids or forked DNA/DNA substrates. In fact, Pif1 can unwind these structures in a processive manner, that is, under single cycle conditions. Like many of the RecQ helicases, Pif1 also unwinds G-quadruplex structures [36]. In fact, in side by side comparisons, Pif1 unwinds G-quadruplex structures ~100-times faster than the human WRN protein and again is able to do so under single cycle conditions (KP and VAZ, in preparation).
Pif1 was first identified because of its role in promoting recombination in mitochondrial DNA [131]. It was rediscovered in a mutant screen for genes affecting telomeres [125]. Depletion of Pif1 results in long telomeres, and this lengthening is telomerase-dependent [125,132]. Over-expression of Pif1 leads to modest telomere shortening [132]. Thus, telomere length is inversely proportional to Pif1 levels. The effects of Pif1 on telomere length require its ATPase/helicase activity, and because Pif1 is telomere-associated in vivo, it likely acts directly to affect telomerase [132].
The most dramatic effect of Pif1 on telomerase is its inhibition of telomere addition to DSBs, which is increased ~600-fold in pif1Δ cells [125,133]. Pif1 is phosphorylated in response to DSB formation [134], and this phosphorylation activates Pif1 activity at DSBs. In contrast, Pif1 function at telomeres does not require phosphorylation [134]. Gross chromosomal rearrangements (GCRs), which are complex chromosome changes similar to those observed in human tumor cells, increase ~1000-fold in pif1Δ cells. Most of the GCR events recovered in pif1 cells are a result of telomerase dependent telomere addition to DSBs [135]. Thus, Pif1 inhibits both telomere lengthening and telomere addition by negatively regulating telomerase.
Pif1 also affects the specificity of telomere addition. Unlike wild type cells where the rare telomere addition events almost always occur near telomere-like sequences, telomere addition in pif1 cells occurs at many sites which often have virtually no telomere-like DNA near the break site [125,133,136]. By inhibiting telomere addition to DSBs, Pif1 promotes genome integrity by preventing the generation of terminally deleted chromosomes.
In vivo and in vitro experiments indicate that Pif1 inhibits telomerase not by altering telomerase activity but rather by removing it from DNA. In vivo, Pif1 over-expression reduces telomerase levels at telomeres, while Pif1 depletion increases these levels at both telomeres [137] and DSBs (J. Phillips, KP and VAZ, in preparation). The ability of Pif1 to inhibit telomerase depends on its interaction with Est2, the catalytic subunit of the telomerase holoenzyme [138].
Pif1 inhibits telomerase directly by removing it from its DNA substrate, as shown by in vitro studies using purified Pif1 and partially purified telomerase. Surprisingly, in this in vitro system, total synthesis of telomeric DNA is actually increased in the presence of Pif1. However, telomerase processivity is reduced and Est2, is released from DNA in the presence of catalytically active Pif1. As shown by competition experiments, the displaced telomerase is still active and therefore able to lengthen another DNA oligonucleotide, explaining why total synthesis is higher even though telomerase processivity is reduced. Presumably, when Pif1 releases telomerase from telomeres in vivo, the concentration of telomeres is not high enough for the released enzyme to find another telomere in the narrow window of the cell cycle in which yeast telomerase is active. Given its facility at unwinding forked RNA/DNA hybrids in vitro [3], Pif1 might displace telomerase from DNA ends by unwinding the hybrid between telomerase RNA and single strand telomeric DNA. Since there is no Est2 detectable at telomeres in the absence of telomerase RNA [139,140], dissociating this hybrid is likely to release the protein subunits of telomerase. Alternatively, like certain other helicases that can remove proteins from DNA, Pif1 could displace the holoenzyme directly [138]. Another possibility is that the effects of Pif1 on telomerase are linked to its ability to unwind G-quadruplexes [36]. However, in vitro assays show that telomerase activity at telomeres is blocked by intramolecular G-quadruplexes [37,38,141]. Thus, it is difficult to imagine how Pif1’s ability to unwind G-quadruplexes could explain its inhibitory effects on telomerase.
In addition to its telomeric and mitochondrial functions, Pif1 has more general roles in chromosomal DNA replication. Genetic [142] and biochemical [143] studies indicate that Pif1 has roles in Okazaki fragment maturation. Together with DNA polymerase δ, Pif1 contributes to the generation of long 5′ flaps on Okazaki fragments that are then cleaved by Dna2 [142–144]. Pif1 might also remove the last RNA primer during Okazaki fragment maturation at telomeres [142].
4.2. S. cerevisiae Rrm3: telomere functions
The second S. cerevisiae Pif1 family member (Fig. 1), RRM3 (rDNA recombination mutant), was identified by two groups, one finding that mutation in Rrm3 results in increased rDNA recombination [145] and the other noting its sequence similarity to Pif1 [146]. Like Pif1, Rrm3 is found in both nuclei and mitochondria. However unlike Pif1, Rrm3 is not important for the maintenance of mitochondrial DNA [147,148]. Furthermore unlike Pif1, which is recruited to its sites of action (KP and VAZ, unpublished results), Rrm3 travels with the replication fork [149]. 2D gel analyses and genome-wide microarray studies demonstrate that efficient replication fork progression at specific sites is facilitated by Rrm3 helicase activity [56,146,149,150]. Replication forks slow at over 1000 genomic loci in rrm3Δ cells including tRNA genes, inactive replication origins, centromeres, multiple sites within each rDNA repeat, and, as discussed below, telomeres. All Rrm3 sensitive sites are incorporated into stable protein complexes. Disruption of these complexes eliminates Rrm3 dependence [150,151]. These findings suggest that Rrm3 uses its helicase activity to dissociate proteins that are bound to these sites.
At first glance, Rrm3 seems to have only modest effects on telomeres as telomeres are only slightly longer, and telomeric silencing is only modestly reduced in its absence [56]. However, 2D gel analysis of replication intermediates [56] and genome-wide studies of replication fork movement by DNA polymerase 2 occupancy [57] demonstrate that in wild type cells replication forks slow as they move through telomeric DNA. Pausing within telomeric DNA is exacerbated about 10-fold in rrm3Δ cells. Slow replication through telomeres is not due to the terminal position of these sequences as replication through internal tracts of telomeric sequence is also slow in wild type cells, and this slowing is heightened in the absence of Rrm3. These studies were the first to demonstrate that semi-conservative replication of telomeric DNA is a problem for the conventional replication apparatus.
4.3. S. pombe Pfh1: telomere functions
Unlike S. cerevisiae and certain other fungi, S. pombe encodes only one PIF1 family helicase (Pfh1) (Fig. 1). Like its S. cerevisiae counterparts, Pfh1 is a 5′–3′ DNA helicase [132] that can unwind RNA/DNA as well as DNA/DNA duplexes and is more active on forked substrates [152]. Although neither of the S. cerevisiae Pif1 proteins is essential and even a pif1Δ rrm3Δ strain is viable [146], Pfh1 is essential [132,153]. Like the two S. cerevisiae Pif1 family proteins, Pfh1 is found in both nuclei and mitochondria, and its ATPase/helicase activity is essential in both compartments [126]. Although nuclear Pfh1 has roles in both repair and replication, only its replication function is essential. The best clue as to the nature of the essential nuclear function of Pfh1 is that it can be supplied by S. cerevisiae Rrm3, suggesting that Pfh1 might promote fork progression through hard to replicate sites [126]. However, Pfh1 may have a more general role in DNA replication as genetic assays suggest that Pfh1, like the S. cerevisiae Pif1, functions during Okazaki fragment maturation [152–154].
So far, the only evidence that Pfh1 functions at telomeres comes from telomere length analyses in Pfh1-depleted cells. When heterozygous pfh1+/pfh1Δ cells go through meiosis, the resulting pfh1Δ spore clones divide zero to four times before arresting, and telomeres in these pfh1Δ spore clones are modestly shorter than in wild type cells [128]. In a second study where Pfh1 expression was reduced to very low levels using a repressible promoter, there was no change in telomere length [126]. However, in more recent analyses, a more complete repression of Pfh1 results in telomere shortening (K.M., N. Sabouri, and VAZ, unpublished results). The question of whether or not Pfh1 affects telomeres is still not resolved, but given that its depletion does not result in telomere lengthening, it does not appear to be an inhibitor of telomerase like the S. cerevisiae Pif1.
4.4. Mammalian Pif1 family proteins
Like S. pombe, mouse (mPif) and humans (hPif) encode a single Pif1 family helicase (Fig. 1) [155]. An N-terminally truncated version as well as full length hPif1 protein have been purified and shown to unwind both DNA/DNA and DNA/RNA substrates with 5′–3′ directionality. However, hPIF is more active on forked structures that resemble replication intermediates [156–158]. In vitro hPif reduces telomerase processivity and binds preferentially to telomeric TTAGGG repeats [158]. mPIF is not essential because mice that are homozygously deleted for mPif1 are viable and show no differences in telomere length, DNA damage response, cell cycle progression, or chromosome integrity [159]. Thus, if mPIF is important in vivo, its activity is likely redundant with that of other helicases/translocases.
In cultured human cells, hPIF is a low abundance protein that shows dramatic cell cycle regulation with significant expression limited to late in the cell cycle [155]. This expression pattern is due to ubiquitin-mediated degradation via the anaphase promoting complex, a pattern of regulation that hPIF shares with the S. cerevisiae Pif1 [155,160]. However, not all Pif1 family proteins are cell cycle regulated, as levels of S. cerevisiae Rrm3 are constant throughout the cell cycle [149].
There are conflicting in vivo data for hPif function at telomeres as in one study [158] but not in another [155], over-expression of hPIF in tissue culture cells results in telomere shortening. An additional indication of mammalian PIF function at telomeres comes from co-immunoprecipitation experiments in which both mouse [159] and human [155] PIF proteins are associated with the catalytic subunit of telomerase.
5. FANCJ helicases
The SFI FANCJ family of 5′–3′ DNA helicases is the most recently identified helicase family with telomere effects. The human FANCJ helicase, which was originally called BACH1/BRIP1, was first identified as a DNA helicase that interacts with BRCA1, the product of a human gene whose mutation is associated with a high incidence of early onset breast cancer [161]. Like BRCA1 mutations, FANCJ mutations can also lead to an inherited predisposition to early onset breast cancer [162]. However, FANCJ is probably best known for being one of 13 genes whose mutation leads to the human genetic disorder Fanconi anemia (FA) [163–165]. FA is characterized by genome instability, especially hyper-sensitivity to inter-strand DNA cross-linking agents, such as mitomycin C. FA patients suffer bone marrow failure and increased cancer rates.
BLAST analysis and alignment to FANCJ and RTEL family members show that FANCJ family helicases are also found in some prokaryotes and single-celled eukaryotes. Widespread in metazoans, they are best studied in humans, but also in mouse, chicken and C. elegans. The family includes the FANCJ proteins themselves as well as the more distantly related Dog1 protein (C. elegans) and the RTEL helicases (Fig. 1). C. elegans, as well as some vertebrates, encodes several FANCJ family proteins, FANCJ itself (dog-1 in C. elegans) and RTEL. In general, FANCJ-like helicases function in response to DNA damage, DNA repair, and maintenance of genomic stability (review [166,167]).
In almost all studied FANCJ helicases, a Fe–S cluster is present in the ATP binding domain that is essential for helicase activity and recognition of damaged DNA [168,169]. The BRCA1 binding site, which is important for DNA repair, is found at the C-terminus of the human and mouse FANCJ proteins (reviewed in [167]).
5.1. C. elegans DOG-1 helicase
Although the C. elegans Dog-1 protein is 32% identical to human FANCJ over the helicase domain, they differ in size with Dog-1 being 983 amino acids and FANCJ being 1249 amino acids in length. This size difference is due to the lack of the BRCA1 interaction domain at the C-terminus of Dog-1 [170,171]. The Dog-1 gene (deletions of guanine-rich DNA) was studied initially because of its sequence similarity to mammalian RTEL (see Section 5.3.2). Its mutation yields a mutator phenotype and reduced brood size [172]. Closer examination of the dog-1 associated mutations led to the realization that they are largely due to deletions that initiate throughout the genome in tracts of G-rich sequences with the potential to form G-quadruplex structures [172,173]. In some loci, mutation frequencies are as high as 4% per animal generation [173], and the mutation frequency correlates positively with the length of the G-rich tract that initiates the deletion [172]. In addition, large chromosomal rearrangements are detected genome-wide in dog-1 mutants [174]. Similar to what is seen in humans defective for FANCJ, dog-1 mutants are hypersensitive to DNA cross linking agents [170]. Dog-1 mutations do not have dramatic effects on telomere length, perhaps because the C. elegans telomeric repeat (TTAGGC) has low G-quadruplex forming potential [172].
5.2. Human FANCJ
In vitro, human FANCJ preferentially binds and unwinds forked duplex substrates. Given its anticipated roles in recombination and repair, it is perhaps surprising that it cannot unwind Holliday junctions. However, it does unwind D-loop structures, another potential recombination intermediate [175]. Purified FANCJ also unwinds G-quadruplexes [176–178]. Cell lines from human FA patients with mutations in FANCJ show accumulation of genomic deletions that overlap predicted G-quadruplex motifs [178]. Moreover, telomestatin, a compound that stabilizes G-quadruplexes in vitro [179–181], causes impaired proliferation and elevated levels of apoptosis and DNA damage in FANCJ-deficient cells [177]. Although these data are consistent with a telomeric role for FANCJ, there are as yet no experiments that link the helicase directly to telomeres.
5.3. RTEL helicases
Mouse RTEL (regulator of telomere length) is the founding member of this helicase family [182]. This gene was first identified as a locus that affects telomere length in crosses between mouse species with different starting telomere lengths [183]. As described below, human RTEL is the only family member that has been subjected to biochemical analysis. Human RTEL has ATPase activity [184], but no helicase assays have been reported.
5.3.1. C. elegans RTEL
Like mammals, C. elegans encodes a second FANCJ family protein called RTEL [184]. This protein is 32% identical (62% similar) over the entire ORF to the C. elegans FANCJ homolog Dog-1 and 31% identical to the human RTEL1 ORF. It was identified in a genetic strategy to find C. elegans helicases with an anti-recombination activity similar to that of the S. cerevisiae Srs2 helicase. Since S. cerevisiae srs2 sgs1 double mutants are not viable, the authors searched for helicases whose mutation confers a synthetic lethal phenotype on him-6 deficient worms (him-6 encodes the C. elegans BLM, a RecQ helicase). This analysis identified rtel-1 as a gene that is essential in a him-6 deficient background [184].
Although the RTEL1 protein has no sequence similarity to the S. cerevisiae Srs2 helicase, genetic and biochemical studies indicate that it too inhibits homologous recombination. For example, in meiosis, rtel-1 deficient worms have large numbers of RAD51 foci, which are diagnostic for recombination events as well as elevated rates of recombination. Despite its similarity to Dog-1, the functions of the two FANCJ family helicases are different as G-rich tracts are not unstable in rtel-1 deficient cells. So far, the C. elegans rtel-1 has not been linked to telomeres.
5.3.2. Mammalian RTEL
Telomeres in laboratory mice (M. musculus) are much longer than in wild type Mus spretus mice. Because the two species are inter-fertile, they were crossed to identify genes involved in telomere length regulation. From linkage analysis, a region on chromosome 2 was identified that when derived from the M. musculus parent acts in a dominant manner to elongate telomeres. Although the interval had no previously identified telomere factor, it encodes a helicase-like gene that is highly conserved among mammals, which was named RTEL (regulator of telomere length) [182,183]. In humans, the RTEL locus encodes a protein that is 27% identical (55.5% similar) over its entire ORF to human FANCJ. The amino terminal 750 amino acids of the predicted ~1200 amino acid protein contain the seven helicase motifs.
Subsequent genetic analyses showed that the mouse RTEL is an essential gene, with homozygous null animals dying early in embryogenesis with abnormalities in multiple organs [182]. Homozygous null ES cells are viable and have variable length telomeres that are ~70% shorter than in wild type ES cells. If the null ES cells are allowed to differentiate, the differentiating cells display massive genome instability, including telomere signal free ends, chromosome fusions, broken chromosomes, and multi-chromosome fusions. Some of the chromosomal abnormalities suggest that RTEL functions at non-telomere sites as well as at telomeres. Because mammalian RTEL shares sequence similarity to C. elegans Dog-1, which is needed to maintain G-rich DNA (see Section 5.1), the authors asked if non-telomeric G-rich internal tracts are unstable in RTEL null ES cells. However, none of the more than 30 G-rich sites examined in this study had deletions or insertions [182].
One suggestion to explain the telomeric role of RTEL is that it resolves G-quadruplex structures formed during DNA replication. In support of this idea, mouse cells depleted for RTEL using siRNA have a fragile telomere phenotype similar to that seen in TRF1 or BLM depleted cells [60]. Another possibility is that RTEL regulates telomere length by inhibiting telomere recombination. This possibility is supported by biochemical studies using purified human RTEL that were inspired by genetic studies that suggested that the C. elegans rtel-1 inhibits recombination in vivo [184]. Depletion of human RTEL1 with siRNA in cultured cells results in a four-fold increase in DSB repair via homologous recombination. In vitro, purified human RTEL1 has ATPase activity and can prevent D-loop formation as well as disrupting preformed D-loops. However, unlike the S. cerevisiae Srs2, human RTEL1 does not disrupt Rad51-DNA filaments vitro. Human Rtel is amplified in certain gastric tumors [185], but RTEL function during tumor development is not known.
6. Dna2 helicases
The SFI Dna2 family of 5′–3′ DNA helicases was first discovered in S. cerevisiae as an essential gene required for complete synthesis of chromosomal DNA due its role in Okazaki fragment processing (reviewed in [186]). Dna2 is conserved throughout eukaryotes, and it has been well studied in S. cerevisiae, S. pombe, C. elegans, and humans [154,186–191]. The Dna2 helicase motifs are located at the C-terminus, while the N-terminal region is not conserved in size or sequence [192,193]. Although Dna2 helicase motifs are well defined, the detection of helicase activity in human Dna2 is disputed, and it has not been possible to demonstrate helicase activity for purified S. pombe or Xenopus Dna2 [153,186,194,195]. Moreover, the helicase activity of S. cerevisiae Dna2 is not essential in some growth conditions, suggesting it may not be necessary for Okazaki fragment maturation [196]. A single stranded DNA specific endonuclease activity is an essential feature of Dna2 in S. cerevisiae and S. pombe [197,198].
Genetic and biochemical studies show that in S. cerevisiae and S. pombe, the Dna2 endonuclease has a genome-wide role in processing the 5′ ends of “long flap” Okazaki fragments. These flaps are generated by DNA pol δ extending the 3′ end of an Okazaki fragment and thereby displacing the 5′ end of the adjacent Okazaki fragment. Genetic and biochemical data indicate that the S. cerevisiae Pif1 helicase, described in Section 4.1, participates in generating these long flaps [142], and its deletion can suppress the lethality of dna2Δ cells [142]. Human Dna2 is found in both nuclei and mitochondria and has key roles in replication of both genomes [199,200].
6.1. Telomeric roles of S. cerevisiae and S. pombe Dna2 proteins
So far, telomere effects of Dna2 proteins have been reported only in S. cerevisiae and S. pombe. The N-terminus of S. cerevisiae Dna2 was identified as one of ten genes whose over-expression reduces telomeric silencing (DOT gene, disruptor of telomeric silencing) [201]. Over-expression of Dna2 also results in telomere shortening and increased levels of single stranded G-overhangs [196,202]. Levels of telomere silencing correlate positively with telomere length in wild type cells [5]. Thus, the telomere shortening that results from over-expressing Dna2 may explain why overexpression decreases silencing. Multiple different mutations in dna2 result in telomere lengthening [196]. Reduced Dna2 levels also result in faster senescence of telomerase deficient cells and faster appearance of type II survivors [203].
Although more complicated models are also possible, it is not unreasonable that all of the effects of Dna2 on telomere structure are due to its role in Okazaki fragment processing [204], as impaired lagging strand replication is expected to inhibit C-strand resynthesis and thereby affect G-tails. Changes in G-tails could in turn affect telomerase access. Dna2 likely affects telomeres directly as it localizes to telomeres by both one-hybrid and ChIP analyses, and this association is cell cycle regulated [203]. Finally, in vitro S. cerevisiae and human Dna2 can unwind G-quadruplex DNA structures [205], providing the only suggestion that the helicase activity, rather than the Dna2 endonuclease activity, has a role in telomere biology.
In S. pombe, like S. cerevisiae, Dna2 is a flap endonuclease that is important for Okazaki fragment maturation and hence viability [198]. That this activity is important for telomeric G-tails comes from their loss in dna2-C2 mutant cells growing at semi-permissive temperatures [206]. Since wild type Dna2 binds telomeres as shown by ChIP, and this binding is reduced in dna2-2 mutants at high temperatures, S. pombe Dna2 likely affects telomeres directly. Telomeres progressively shorten, and telomerase telomere binding is reduced in dna2-2 cells growing at semi-permissive temperatures. Thus, the data from both S. cerevisiae and S. pombe indicate that Dna2 is important for maintaining G-tails and telomerase-mediated telomere replication. It is possible in both organisms that the effects of dna2 mutants on telomeres are due to faulty Okazaki fragment maturation.
7. Future directions
Helicases are critically involved in all processes involving nucleic acids. This dependence is true throughout the genome but is likely to be particularly important for G-rich and shelterin bound telomeric DNAs. To date, members of four helicase families – RecQ, Pif1, FANCJ, and DNA2 – are known to have roles in telomere biology. Mutations in RecQ and FANCJ helicases cause inherited human diseases, characterized by genome instability, increased cancer susceptibility, and premature aging. Given the relatively young age of telomere-helicase research and the large number of eukaryotic helicases, there are likely to be additional helicases with telomeric roles that are important for human health and longevity.
One critical area of helicase-telomere research over the next years is to elucidate the role of G-quadruplex structures in telomere biology. Do G-quadruplex structures form during semi-conservative telomere replication? If so, does their formation affect replication fork progression? Does the demonstrated ability of certain helicases to unwind G-quadruplex structures in vitro explain their in vivo effects on telomeres? Another interesting question is whether the unusual chromatin structure of telomeres impacts their replication in organisms other than S. cerevisiae, and if so does their replication involve helicases, such as the S. cerevisiae Rrm3, with the specialized ability to bypass stable protein–DNA structures. Identifying helicases that regulate telomerase is another important area. So far only the S. cerevisiae Pif1 is known to have a direct effect on telomerase. Do other Pif1 family helicases affect telomerase? The case is not yet made, but there are intriguing links between mammalian Pif proteins and telomerase. The exact roles of human RecQ helicases in ALT may clarify the roles of these proteins in cancer. Another intriguing question is to determine if (and if so, how) the S. cerevisiae and S. pombe subtelomeric helicases contribute to the recombination that maintains telomeres when telomerase is not active as well as to determine if subtelomeric helicases are a general feature of telomeric DNA. The involvement of helicases in TERRA is totally unexplored and a likely area for exciting findings. Future experiments will continue to elucidate the diverse and important roles that helicases perform in telomere metabolism and will lead to the identification of additional helicases with telomere functions.
Acknowledgments
We thank M. Bochman for help with computer analysis of helicase families and for careful reading of the manuscript. Work in the Zakian lab is supported by grants from the NIH. K.P. and K.R.M. were supported in part by grants from the NJCCR.
References
- 1.Shiratori A, Shibata T, Arisawa M, Hanaoka F, Murakami Y, Eki T. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast. 1999;15:219–253. doi: 10.1002/(SICI)1097-0061(199902)15:3<219::AID-YEA349>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 3.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 5.Mondoux M, Zakian V. Telomere position effect: silencing near the end. In: de Lange T, Lundblad V, Blackburn EH, editors. Telomeres. 2. CSHL Press; Cold Spring Harbor, New York: 2005. pp. 261–316. [Google Scholar]
- 6.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 7.Chan SWL, Blackburn EH. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9.Dionne I, Wellinger RJ. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 11.Rhodes D, Fairall L, Simonsson T, Court R, Chapman L. Telomere architecture. EMBO Rep. 2002;3:1139–1145. doi: 10.1093/embo-reports/kvf246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega L, Mateyak M, Zakian V. Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 14.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 16.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD. Telomere looping in P. sativum (common garden pea) Plant J. 2003;36:271–279. doi: 10.1046/j.1365-313x.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 19.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 20.Munoz-Jordan JL, Cross GA, de Lange T, Griffith JD. t-loops at trypanosome telomeres. EMBO J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murti KG, Prescott DM. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc Natl Acad Sci USA. 1999;96:14436–14439. doi: 10.1073/pnas.96.25.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomaska L, Willcox S, Slezakova J, Nosek J, Griffith JD. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J Biol Chem. 2004;279:50764–50772. doi: 10.1074/jbc.M409790200. [DOI] [PubMed] [Google Scholar]
- 24.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 25.Poulet A, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Steensel B, Smogorzewska A, de lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 27.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 28.Stansel RM, Subramanian D, Griffith JD. p53 binds telomeric single strand overhangs and t-loop junctions in vitro. J Biol Chem. 2002;277:11625–11628. doi: 10.1074/jbc.C100764200. [DOI] [PubMed] [Google Scholar]
- 29.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 30.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 31.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 32.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 33.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang G, Cech TR. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 35.Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum Mol Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- 36.Ribeyre C, Lopes J, Boulé J, Piazza P, Guédin A, Zakian V, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 38.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gros J, Guedin A, Mergny JL, Lacroix L. G-quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem. 2008;9:2075–2079. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- 40.Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 41.Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 42.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 43.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 44.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 45.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 46.Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 47.Shore D, Bianchi A. Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shay JW, Wright WE. Mutant dyskerin ends relationship with telomerase. Science. 1999;286:2284–2285. doi: 10.1126/science.286.5448.2284. [DOI] [PubMed] [Google Scholar]
- 49.Carroll KA, Ly H. Telomere dysfunction in human diseases: the long and short of it! Int. J Clin Exp Pathol. 2009;2:528–543. [PMC free article] [PubMed] [Google Scholar]
- 50.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 53.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wellinger RJ, Wolf AJ, Zakian VA. Structural and temporal analysis of telomere replication in yeast. Cold Spring Harb Symp Quant Biol. 1993;58:725–732. doi: 10.1101/sqb.1993.058.01.080. [DOI] [PubMed] [Google Scholar]
- 55.Wellinger RJ, Zakian VA. Lack of positional requirements for autonomously replicating sequence elements on artificial yeast chromosomes. Proc Natl Acad Sci USA. 1989;86:973–977. doi: 10.1073/pnas.86.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivessa AS, Zhou JQ, Schulz VP, Monson EM, Zakian VA. Saccharomyces Rrm3p, a 5′–3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azvolinsky A, Giresi P, Lieb J, Zakian V. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 59.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′–3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 63.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 64.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci USA. 1994;91:12061–12065. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 67.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 68.Teng SC, Zakian VA. Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashton TM, Hickson ID. Yeast as a model system to study RecQ helicase function. DNA Repair (Amst) 2010;9:303–314. doi: 10.1016/j.dnarep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Cejka P, Kowalczykowski SC. The full-length S. cerevisiae SGS1 protein is a vigorous DNA helicase that preferentially unwinds Holliday junctions. J Biol Chem. 2010 doi: 10.1074/jbc.M109.083196. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 75.Sun H, Bennett R, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G–G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyun M, Bohr VA, Ahn B. Biochemical characterization of the WRN-1 RecQ helicase of Caenorhabditis elegans. Biochemistry. 2008;47:7583–7593. doi: 10.1021/bi800197m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–4114. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grabowski MM, Svrzikapa N, Tissenbaum HA. Bloom syndrome ortholog HIM-6 maintains genomic stability in C. elegans. Mech Ageing Dev. 2005;126:1314–1321. doi: 10.1016/j.mad.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 79.Kim YM, Yang I, Lee J, Koo HS. Deficiency of Bloom’s syndrome protein causes hypersensitivity of C. elegans to ionizing radiation but not to UV radiation, and induces p53-dependent physiological apoptosis. Mol Cell. 2005;20:228–234. [PubMed] [Google Scholar]
- 80.Lee SJ, Yook JS, Han SM, Koo HS. A Werner syndrome protein homolog affects C. elegans development, growth rate, life span and sensitivity to DNA damage by acting at a DNA damage checkpoint. Development. 2004;131:2565–2575. doi: 10.1242/dev.01136. [DOI] [PubMed] [Google Scholar]
- 81.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 82.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 83.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J Biol Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 84.Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 2006;34:506–516. doi: 10.1093/nar/gkj452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohen H, Sinclair D. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc Natl Acad Sci USA. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bhattacharyya S, Sandy A, Groden J. Unwinding protein complexes in ALTernative telomere maintenance. J Cell Biochem. 2010;109:7–15. doi: 10.1002/jcb.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 88.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 2007;5:e160. doi: 10.1371/journal.pbio.0050160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lillard-Wetherell K, Combs KA, Groden J. BLM helicase complements disrupted type II telomere lengthening in telomerase-negative sgs1 yeast. Cancer Res. 2005;65:5520–5522. doi: 10.1158/0008-5472.CAN-05-0632. [DOI] [PubMed] [Google Scholar]
- 94.Lee JY, Mogen JL, Chavez A, Johnson FB. Sgs1 RecQ helicase inhibits survival of Saccharomyces cerevisiae cells lacking telomerase and homologous recombination. J Biol Chem. 2008;283:29847–29858. doi: 10.1074/jbc.M804760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 96.Willis N, Rhind N. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Mol Biol Cell. 2009;20:819–833. doi: 10.1091/mbc.E08-08-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 99.Wilson S, Warr N, Taylor DL, Watts FZ. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 101.Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 102.Kibe T, Ono Y, Sato K, Ueno M. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol Biol Cell. 2007;18:2378–2387. doi: 10.1091/mbc.E06-12-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mandell JG, Goodrich KJ, Bahler J, Cech TR. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J Biol Chem. 2005;280:5249–5257. doi: 10.1074/jbc.M412756200. [DOI] [PubMed] [Google Scholar]
- 104.Hansen KR, Ibarra PT, Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 2006;34:78–88. doi: 10.1093/nar/gkj415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mandell JG, Bahler J, Volpe TA, Martienssen RA, Cech TR. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 2005;6:R1. doi: 10.1186/gb-2004-6-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamada M, Hayatsu N, Matsuura A, Ishikawa F. Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem. 1998;273:33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- 107.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schulz VP, Zakian VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 109.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 110.Ouellette MM, McDaniel LD, Wright WE, Shay JW, Schultz RA. The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum Mol Genet. 2000;9:403–411. doi: 10.1093/hmg/9.3.403. [DOI] [PubMed] [Google Scholar]
- 111.Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 112.Chang S, et al. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 113.Opresko PL, et al. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 114.Eller MS, Liao X, Liu S, Hanna K, Backvall H, Opresko PL, Bohr VA, Gilchrest BA. A role for WRN in telomere-based DNA damage responses. Proc Natl Acad Sci. 2006;103:15073–15078. doi: 10.1073/pnas.0607332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 116.Lillard-Wetherell K, et al. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 117.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 118.Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 120.Johnson F, Marciniak R, McVey M, Stewart S, Hahn W, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase Ilalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmidt KH, Kolodner RD. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 126.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol Cell. 2009;35:490–501. doi: 10.1016/j.molcel.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou JQ, Qi H, Schulz V, Mateyak M, Monson E, Zakian V. Schizosaccharomyces pombe pfh1+ encodes an essential 5′–3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol Biol Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 130.Boule JB, Zakian VA. Methods in Molecular Biology Series. Humana Press; 2009. Characterization of the helicase activity and anti-telomerase properties of yeast Pif1p in vitro Helicases Methods and Protocols; pp. 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou JQ, Monson EM, Teng SC, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 133.Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 136.Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 137.Boule J, Vega L, Zakian V. The yeast Pif1p helicase removes telomerase from DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 138.Eugster A, et al. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat Struct Mol Biol. 2006;13:734–739. doi: 10.1038/nsmb1126. [DOI] [PubMed] [Google Scholar]
- 139.Chan A, Boule JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:3509–3519. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taggart AKP, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 141.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Keil RL, McWilliams AD. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 147.O’Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene. 2005;354:86–92. doi: 10.1016/j.gene.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 148.Prokisch H, et al. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004;2:e160. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Azvolinsky A, Dunaway S, Torres J, Bessler J, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 151.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ryu GH, et al. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill SA, Yuasa Y. The fission yeast pfh1+ gene encodes an essential 5′–3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002;30:4728–4739. doi: 10.1093/nar/gkf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gould K, Burns C, Feoktistova A, Hu C, Pasion S, Forsburg S. Fission yeast cdc24(+) encodes a novel replication factor required for chromosome integrity. Genetics. 1998;149:1221–1233. doi: 10.1093/genetics/149.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mateyak M, Zakian V. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;23:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 156.George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM. Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res. 2009;37:6491–6502. doi: 10.1093/nar/gkp671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–6308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Snow B, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vega L, Phillips J, BRT, DPT, Onigbanjo M, Zakian V. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 2007;3:1065–1075. doi: 10.1371/journal.pgen.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cantor SB, et al. Bach1, a novel helicase-like protein, interacts directly with brca1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 162.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 164.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 165.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 166.Hiom K. FANCJ: solving problems in DNA replication. DNA Repair (Amst) 2010;9:250–256. doi: 10.1016/j.dnarep.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 167.Wu Y, Brosh RM., Jr FANCJ helicase operates in the Fanconi Anemia DNA repair pathway and the response to replicational stress. Curr Mol Med. 2009;9:470–482. doi: 10.2174/156652409788167159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.White MF. Structure, function and evolution of the XPD family of iron-sulfur-containing 5′→3′ DNA helicases. Biochem Soc Trans. 2009;37:547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 169.Boal AK, Yavin E, Lukianova OA, O’Shea VL, David SS, Barton JK. DNA-bound redox activity of DNA repair glycosylases containing [4Fe–4S] clusters. Biochemistry. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 170.Youds JL, Barber LJ, Ward JD, Collis SJ, O’Neil NJ, Boulton SJ, Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 172.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 173.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 174.Zhao Y, Tarailo-Graovac M, O’Neil NJ, Rose AM. Spectrum of mutational events in the absence of DOG-1/FANCJ in Caenorhabditis elegans. DNA Repair (Amst) 2008;7:1846–1854. doi: 10.1016/j.dnarep.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 175.Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM., Jr Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 176.Maizels N. Genomic stability: FANCJ-dependent G4 DNA repair. Curr Biol. 2008;18:R613–R614. doi: 10.1016/j.cub.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arola A, Vilar R. Stabilisation of G-quadruplex DNA by small molecules. Curr Top Med Chem. 2008;8:1405–1415. doi: 10.2174/156802608786141106. [DOI] [PubMed] [Google Scholar]
- 180.Gomez D, Paterski R, Lemarteleur T, Shin-Ya K, Mergny JL, Riou JF. Interaction of telomestatin with the telomeric single-strand overhang. J Biol Chem. 2004;279:41487–41494. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- 181.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 182.Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 183.Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. Telomere length regulation in mice is linked to a novel chromosome locus. Proc Natl Acad Sci. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bai C, et al. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a fourgene cluster. Proc Natl Acad Sci. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit Rev Biochem Mol Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 187.Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Eki T, et al. Assignment of the closest human homologue (DNA2L:KIAA0083) of the yeast Dna2 helicase gene to chromosome band 10q21.3-q22.1. Genomics. 1996;37:408–410. doi: 10.1006/geno.1996.0581. [DOI] [PubMed] [Google Scholar]
- 190.Kim DH, et al. Enzymatic properties of the Caenorhabditis elegans Dna2 endonuclease/helicase and a species-specific interaction between RPA and Dna2. Nucleic Acids Res. 2005;33:1372–1383. doi: 10.1093/nar/gki255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu Q, Choe W, Campbell JL. Identification of the Xenopus laevis homolog of Saccharomyces cerevisiae DNA2 and its role in DNA replication. J Biol Chem. 2000;275:1615–1624. doi: 10.1074/jbc.275.3.1615. [DOI] [PubMed] [Google Scholar]
- 192.Bae SH, Kim DW, Kim J, Kim JH, Kim DH, Kim HD, Kang HY, Seo YS. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J Biol Chem. 2002;277:26632–26641. doi: 10.1074/jbc.M111026200. [DOI] [PubMed] [Google Scholar]
- 193.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J Biol Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 194.Bae SH, et al. Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 2001;29:3069–3079. doi: 10.1093/nar/29.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Budd ME, Campbell JL. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci USA. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kang HY, et al. Genetic analyses of Schizosaccharomyces pombe dna2(+) reveal that dna2 plays an essential role in Okazaki fragment metabolism. Genetics. 2000;155:1055–1067. doi: 10.1093/genetics/155.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zheng L, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Parenteau J, Wellinger RJ. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol Cell Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Choe W, Budd M, Imamura O, Hoopes L, Campbell JL. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol Cell Biol. 2002;22:4202–4217. doi: 10.1128/MCB.22.12.4202-4217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J Biol Chem. 2006;281:38555–38564. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 205.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tomita K, Kibe T, Kang HY, Seo YS, Uritani M, Ushimaru T, Ueno M. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol. 2004;24:9557–9567. doi: 10.1128/MCB.24.21.9557-9567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]