Abstract
COX-2 catalyzes the rate-limiting step in the synthesis of prostaglandins. Its overexpression induces numerous tumor-promoting phenotypes and is associated with cancer metastasis and poor clinical outcome. Although COX-2 inhibitors are promising chemotherapeutic and chemopreventative agents for cancer, the risk of significant cardiovascular and gastrointestinal complications currently outweighs their potential benefits. Systemic complications of COX-2 inhibition could be avoided by specifically decreasing COX-2 expression in epithelial cells. To that end, we have investigated the signal transduction pathway regulating COX-2 expression in response to DNA damage in breast epithelial cells. In variant human mammary epithelial cells that have silenced p16 (vHMEC), double strand DNA damage or telomere malfunction results in a p53-and activin A-dependent induction of COX-2 and continued proliferation. In contrast, telomere malfunction in HMEC with an intact p16/Rb pathway induces cell cycle arrest. Importantly, in ductal carcinoma in situ (DCIS) lesions, high COX-2 expression is associated with high γH2AX, TRF2, activin A and telomere malfunction. These data demonstrate that DNA damage and telomere malfunction can have both cell autonomous and cell non-autonomous consequences and provides a novel mechanism for the propagation of tumorigenesis.
Keywords: DNA damage, telomere, COX-2, Activin A, DCIS
INTRODUCTION
Cyclooxygenase II (COX-2) is one of two enzymes that catalyze the rate-limiting step in the synthesis of prostaglandins. In contrast to the widespread constitutive expression of COX-1, COX-2 is strongly inducible by a variety of agents in a limited number of cell types, including neoplastic tissues (1, 2). Through the production of prostaglandins, COX-2 plays a significant role in cancer by increasing proliferation, motility, invasion, angiogenesis and resistance to apoptosis, while also repressing host immune response to tumors (3-6). COX-2 has a profound impact on every aspect of breast cancer including initiation, progression to invasive disease and finally metastasis. For example, high COX-2 in benign atypical hyperplasia lesions is associated with increased risk of developing breast cancer (7). COX-2 is associated with mammographic density, a significant risk factor for breast cancer (8). In combination with p16/Rb pathway malfunction, high COX-2 expression predicts progression of pre-invasive ductal carcinoma in situ (DCIS) lesions to invasive ductal carcinoma (IDC) (9) . COX-2 enhances breast cancer metastasis to bone in mouse models (10) and is associated with bone marrow and brain metastasis (11, 12). Consequently, high COX-2 expression is associated with poor prognosis in both DCIS and IDC (9, 13, 14).
COX-2 is an ideal target for chemopreventatives and chemotherapeutics. Non-selective COX-2 inhibitors (NSAIDs), such as aspirin, and COX-2 specific inhibitors (coxibs) decrease cancer mortality, recurrence and incidence (15-18). However, the use of these drugs is associated with gastrointestinal and cardiovascular complications (1, 19). One plausible explanation for these side effects is the eicosanid imbalance theory, i.e. the inhibition of COX-2 in the vasculature decreases the vasodilator PGI2, without reducing COX-1-dependent vasoconstrictive thromboxanes (1, 19). Decreasing COX-2 expression in a single cell type might mitigate these side effects; for instance, loss of COX-2 in breast epithelial cells could reduce breast cancer incidence and progression without inducing the complications associated with NSAIDS or coxibs. Such an approach requires a detailed understanding of the cell-type specific mechanisms that govern COX-2 expression. We have identified a subpopulation of epithelial cells within the human breast that up-regulate COX-2, an event that coincides with increased genomic instability, telomere malfunction (3, 20) and accumulation of the telomere binding protein, TRF2. Here we identify the upstream pathways responsible for this COX-2 induction and unmask novel biology that has cell-autonomous as well as cell non-autonomous repercussions for tumor progression.
MATERIALS AND METHODS
Cell Culture
HMEC were isolated from disease-free breast tissue of nine individuals (CM7, RM9, RM15, RM16, RM18, RM40, RM45, RM46, RM146). Epithelial cells were propagated as described (20). Activin A and the p38 inhibitor, SB203580, (Sigma) were added to culture media for 48h and 24h, respectively, prior to harvest at the doses shown in Figures 4 and 2. Etoposide and NU7026 (Sigma) were added to the culture media for the times indicated in Figures 1 and 3. Media was removed prior to exposure to UVC and immediately replaced.
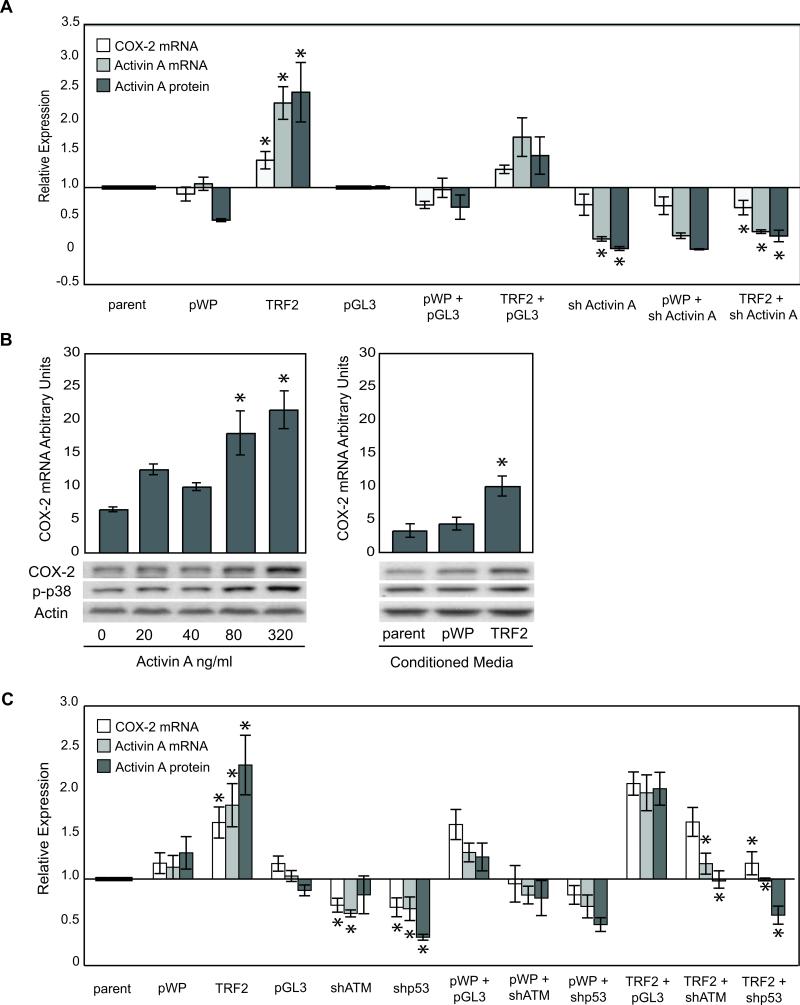
Figure 4. Activin A and p53 are Necessary for COX-2 Induction.
A) Activin A levels were reduced using shRNA. Mean relative COX-2 and activin A mRNA and activin A protein levels measured as described in Figure 3B in RM9, RM15, RM16 and expressed relative to parent (parent, pWP1 or TRF2) or pGL3 (all others). B) COX-2 mRNA levels in vHMEC incubated with exogenous activin A (left panel). Immunoblot showing COX-2, phospho-p38 (p-p38) and actin (loading control) protein levels for each treatment. Conditioned media from parent, vector or TRF2-vHMEC obtained from 2 donors (RM9, 15) was diluted 1:1 and added to vHMEC obtained from 3 donors (RM9, 15, 16). Corresponding immunoblots of treated cells showing COX-2, p-p38 and actin (loading control) expression (right panel). C) ATM and p53 levels were reduced using shRNA. Mean COX-2 and activin A mRNA and activin A protein levels in RM9, RM15 and RM16 expressed relative to parent (parent, pWP1 or TRF2) or pGL3 (all others). For A and C, asterisk (*) indicates statistically significant changes compared to either parent, pGL3 or TRF2+pGL3.
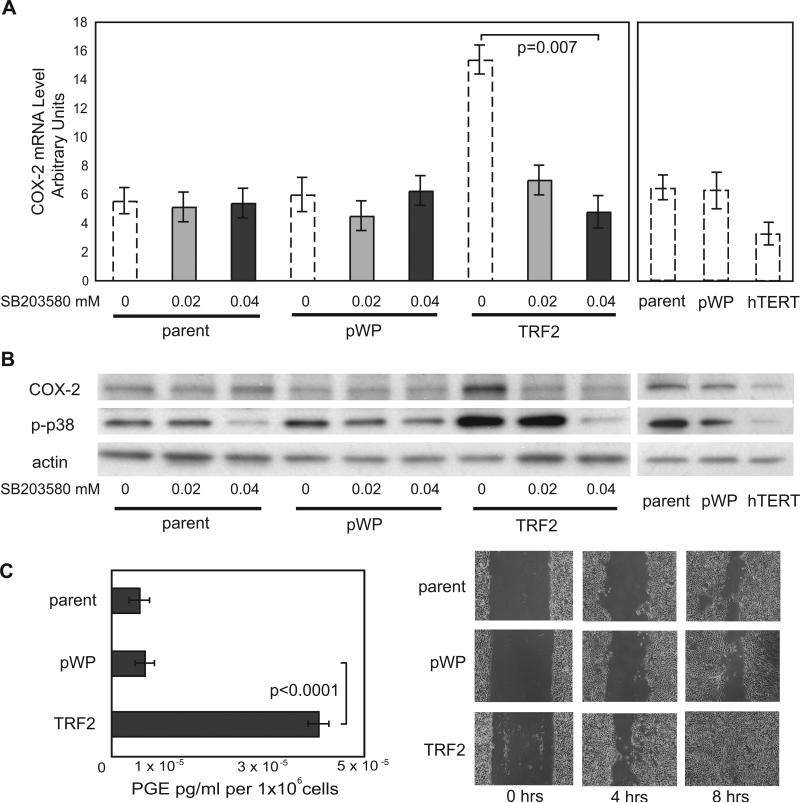
Figure 2. TRF2 Induces COX-2 via a Phospho-p38-Dependent Mechanism.
A) Mean COX-2 mRNA levels in untreated parent vHMEC, pWP1, TRF2-vHMEC or hTERT-vHMEC (hatched bars, shown in Figure 1C) or treated with the phospho-p38 inhibitor, SB203580 for 24h (grey bars). B) Representative immunoblot showing COX-2, phospho-p38 (p-p38) and actin (loading control) protein levels for the conditions described in A. C) Mean levels of prostaglandins in conditioned media (left panel). Wounding assay for parent, pWP or TRF2-vHMEC (right panel).
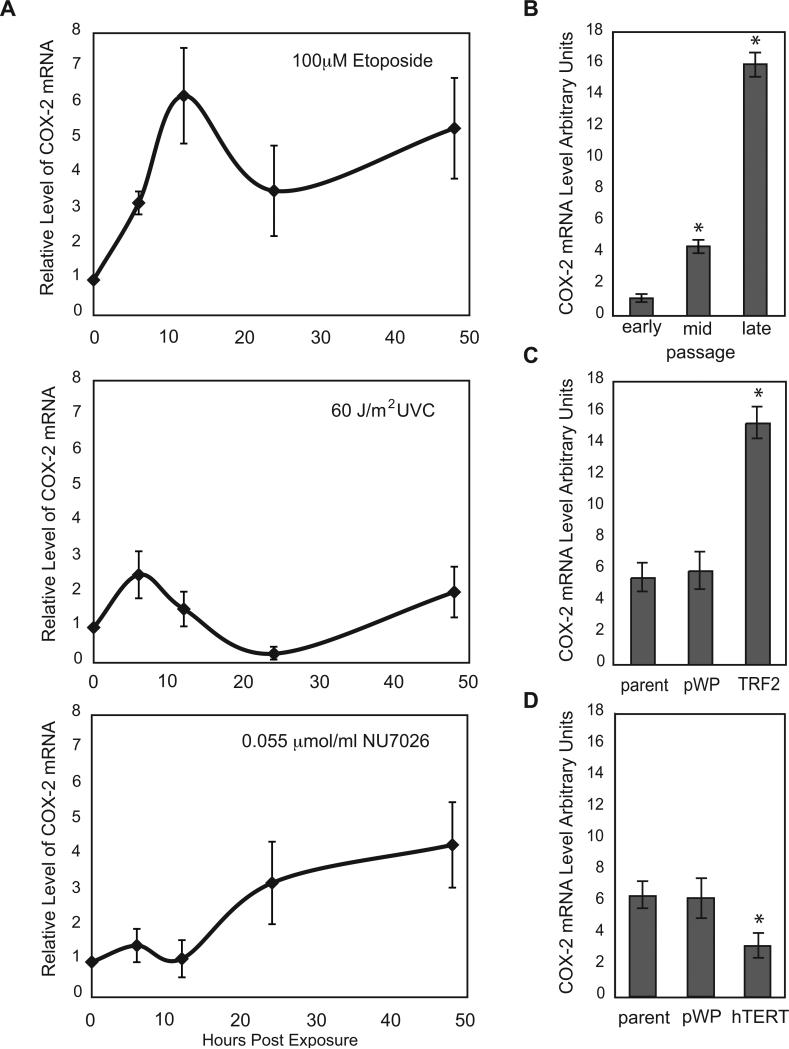
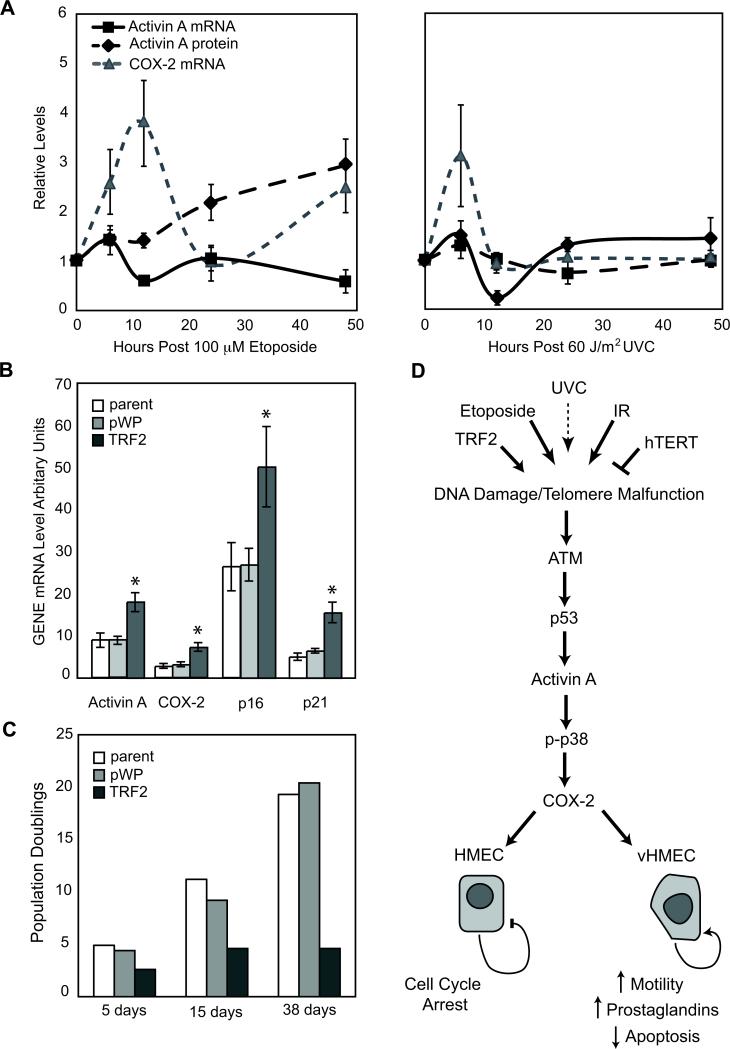
Figure 1. DNA Damage Induces COX-2 in vHMEC.
RM9, RM15 and RM16 were used in panels A-C. A) vHMEC were exposed to 100μM Etoposide (top), 60J/m2 UVC (middle) and 0.055μmol/ml NU7026 (bottom) and mean COX-2 mRNA levels were measured at the indicated times. Error bars represent the standard error of the mean. B) COX-2 mRNA levels in early, mid or late passage vHMEC. C) COX-2 mRNA levels in vHMEC infected with lentivirus containing either TRF2 or empty vector (pWP1). D) COX-2 mRNA levels in vHMEC (RM15, RM16, RM18) infected with lentivirus containing either hTERT or empty vector (pWP1). Asterisks indicate statistically significant (p<0.05) changes in expression compared to vector or untreated control.
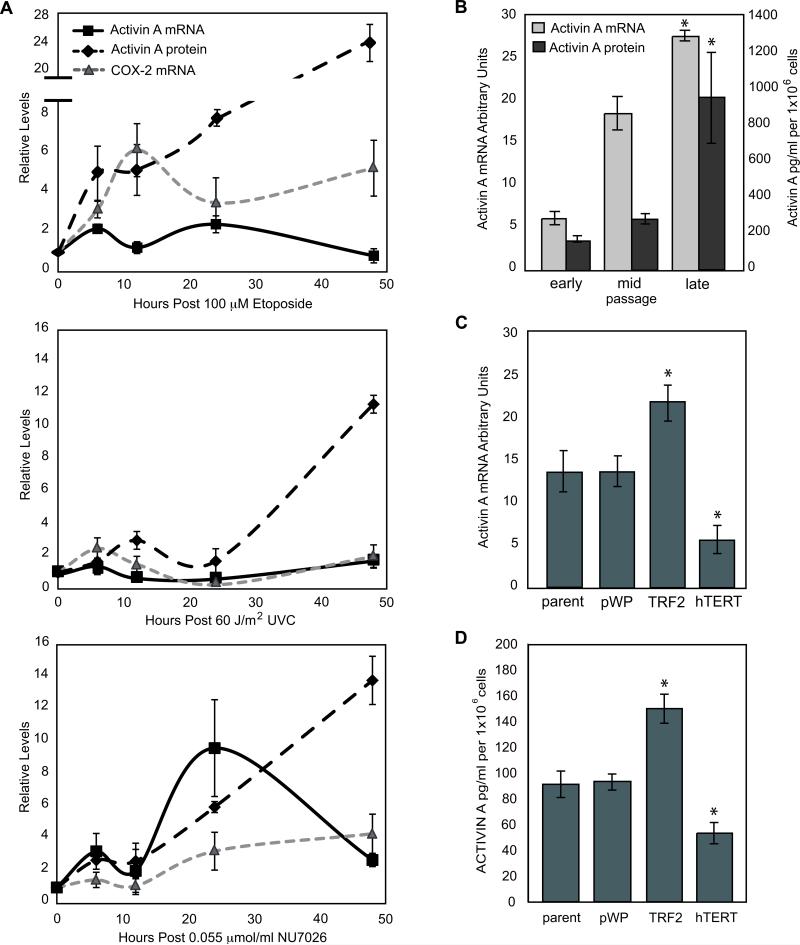
Figure 3. Activin A is Induced by DNA Damage and Coincides with COX-2 Expression.
A) vHMEC (RM9, RM15, RM16) were treated as described in Figure 1A. Line graphs indicate the relative level of COX-2 and activin A mRNAs and activin A protein in conditioned media compared to the untreated control. COX-2 mRNA levels (shown in Figure 1A) are indicated for comparison (light grey dashed line). B) Mean activin A mRNA levels (light grey bars) in vHMEC (RM9, RM15, RM16) and protein levels (dark grey bars) in early, mid and late passage RM9 vHMEC in duplicate experiments. Activin A mRNA (panel C) and protein (panel D) levels in RM9, RM15, RM16 and RM18 vHMEC overexpressing either TRF2, hTERT or vector (pWP) or mock infected (parent).
Expression of TRF2, hTERT and short hairpins
The TRF2 gene, provided by Dr. T. De Lange (Rockefeller University, NY), was inserted into the pWP1 lentiviral expression vector obtained from Dr. D. Trono (EPFL, Switzerland). The pWP1-hTERT construct was provided by Dr. E. Blackburn (UCSF, CA). The pWP1-TRF2 and pWP1-hTERT constructs were packaged in 293T cells. Infection efficiency was monitored using GFP expression driven by an IRES sequence. The control luciferase short hairpin (pGL3) and the pLKO vector were provided by Dr. M. McManus (UCSF, CA). The activin A short hairpin (5’ GAACTGTTGCTCTCTGAAA 3’) was designed using PSICO Oligo Maker. ATM and p53 short hairpins were published previously (21, 22). Infected cells were maintained in 2μg/ml puromycin (Sigma).
Wound Closure Assay
Wound closure assays were performed as described (23).
Quantitative PCR
Q-PCR was performed using standard methods. Primer-probe sets for COX-2 (Hs00153133), TRF2 (Hs00194619), hTERT (Hs00162669), activin A/inhibin A (Hs00170103), ATM (HS00175892), p53 (Hs99999147) and p21 (Hs00355782) were obtained from ABI. Glucoronidase B (GUSB; IDT) expression was used to normalize for variances in input cDNA.
Microarray Analysis
RNA was purified (RNeasy, Qiagen) and amplified using the MessageAmp II-Biotin Kit (Ambion). Amplified RNA was labeled using the enzo BioArray High Yield RNA transcript labeling kit (Affymetrix). Microarray hybridization and analysis were performed at the J. David Gladstone Institutes Core Genomics Laboratory.
Western Blotting and ELISA
Western blots were performed as described (4). Activin A protein and prostaglandin levels were measured using the Duo-Set Activin ELISA kit (R&D Systems) and the Prostaglandin E2 E1A Elisa Kit (Cayman).
Tissue Samples
High (n=7) and low (n=8) grade non-recurrent DCIS specimens were obtained with institutional review board approval from the Helen Diller Comprehensive Cancer Tissue Core (University of California, San Francisco). Patients were identified through anonymous reference numbers in accordance with federal guidelines.
Telomere PNA Hybridization
Chromosome spreads were prepared, processed and analyzed as described (20). An average of 75 nuclei each were evaluated for parent, pWP, and TRF2- and hTERT-vHMEC.
Tissue Preparation and Immunohistochemistry
5μm sections were cut from paraffin-embedded tissue blocks adjacent to sections used for telomere DNA content determination. Immunohistochemistry was performed using standard protocols. Microwave antigen retrieval was accomplished using 0.001M EDTA, pH=8 for COX-2, Antigen Unmasking Solution (Vector Laboratories) for TRF2 and 0.01 M citrate buffer, pH=6.0 for activin A and γH2AX. Antibodies against COX-2 (1:200, Dako), TRF2 (1:20, Imgenex), γH2AX (1:150, Upstate) and activin A (1:100, AbD Serotech) were used. A blocking step (0.01% Triton-X 100 in PBS for 1h) was performed prior to addition of the TRF2 antibody. Evaluation of γH2AX TRF2, COX-2 or activin A staining intensity was performed in a blinded fashion. For COX-2 and activin A, staining intensity was scored as low to absent (low) or moderate to strong (high). The number of γH2AX or TRF2 positive nuclei were counted in at least 500 cells and expressed as a percentage of the total number of nuclei per DCIS lesion. The mean number of γH2AX (28%) or TRF2 (29%) positive nuclei was used to divide the lesions into high or low expression groups.
Telomere DNA Content Determination
For each case, a 5μm tissue section was stained with hematoxylin and eosin (H&E) and cellular morphology was evaluated and used to guide microdissection of 6 × 25 μm serial sections. Telomere content was determined as described previously (24).
Statistical Methods
Two-sided t-tests assuming unequal variance were used to test the relationships between gene expression or activin A protein or prostaglandin levels in cells overexpressing vector, TRF2 or hTERT, or exposed to drugs and the relationship between telomere DNA content and γH2AX, TRF2, activin A and COX-2. Chi-Square test was used to evaluate the relationship between staining intensity (high or low) for γH2AX, TRF2 and activin A or COX-2. The Jmp 7.0 statistical package (SAS Institute) was used for all analyses.
RESULTS
Double-Strand, in contrast to Single-Strand DNA Damage, is a Potent Inducer of COX-2 in Mammary Epithelial Cells with a Compromised p16/Rb Pathway
Several types of DNA damage induce COX-2 (3, 4) in variant human mammary epithelial cells with a compromised p16/Rb pathway (vHMEC). To more fully elucidate these observations, we exposed vHMEC to agents that induce double- (etoposide) or single-strand (UVC) DNA breaks. Treatment of vHMEC with etoposide (1, 10 and 100 μmol) and UVC (0, 30, 60 and 120 J/m2) induced a cell death of approximately 5.6% (100 μmol etoposide) and 3.5% (60 J/m2 of UVC), respectively, after 24h. COX-2 mRNA levels, measured by quantitative PCR (Q-PCR), were greater in cells treated with 10 and 100 μmol etoposide (3.1- and 4.6-fold; p=0.009 and p=0.0005, respectively) compared to controls and exhibited a sustained induction. In contrast, exposure to 60 or 120 J/m2 UVC decreased COX-2 mRNA levels 3.7- and 3.6-fold compared to controls (p=0.0006 and p=0.0006, respectively) after 24h. We then compared the kinetics of COX-2 induction in vHMEC exposed to 100 μmol etoposide or 60 J/m2 of UVC for 6, 12, 24 and 48h (selected treatment conditions were based on the similar cytotoxicity and significant changes in COX-2 mRNA levels described above). Exposure to both treatments resulted in a biphasic induction of COX-2 mRNA; expression peaked at 6-12h and 48h (Figure 1A). However, the magnitude of induction was lower and of shorter duration following exposure to UVC compared to etoposide, suggesting that double-strand DNA (dsDNA) breaks are more potent and sustained inducers of COX-2 in vHMEC than single-strand DNA (ssDNA) breaks.
Endogenously Induced Double-Strand DNA Damage, via Telomere Malfunction, Induces COX-2
As vHMEC proliferate, they lose telomeric DNA, accumulate genomic instability and up-regulate COX-2 (3, 20). We hypothesized that telomere malfunction, a type of dsDNA damage, which occurs early and nearly universally in epithelial cancers, might also induce COX-2. Based on a report showing that inhibition of DNA-PK kinase activity by NU7026 resulted in telomere malfunction (25), we tested the effect of this treatment (0.055umol/ml NU7026) on COX-2 expression in vHMEC. We observed a 10.8% cell death and a 3.2-fold increase in COX-2 mRNA 24h after exposure (Figure 1A). Subsequent kinetic experiments demonstrated that treatment with NU7026 also resulted in a biphasic induction of COX-2 mRNA with peak expression at 6 (1.5-fold) and 48 hours (4.32-fold).
To directly assess if telomere malfunction induced COX-2 in vHMEC, we modulated telomere function by overexpressing two telomere binding proteins: TRF2 and hTERT (Supplemental Figure 1). TRF2 is a negative regulator of telomere length (26-29) while hTERT is the reverse transcriptase that lengthens telomeres (Supplemental Figure 1). Consistent with previous reports, TRF2 overexpression, to levels observed in cancer cell lines and tissues, induced telomere malfunction and a > 6-fold increase in abnormal metaphase spreads including duplications, deletions, translocations and chromosome breaks (p<0.0001). Cells exhibited a concomitant accumulation of the DNA damage protein γH2AX. In stark contrast, hTERT, when expressed in these cells, maintained telomere function (Supplemental Figure 2) and failed to induce significant chromosomal abnormalities. Mean COX-2 mRNA and protein levels increased ~3-fold in vHMEC overexpressing TRF2 (TRF2-vHMEC) compared to vector (p=0.01, Figure 1C, 2B). In contrast, COX-2 mRNA and protein levels were significantly down-regulated (3.2 units, p=0.02) in vHMEC overexpressing hTERT (hTERT-vHMEC) (Figure 1D, 2B). Thus, proteins that affect telomere function modulate COX-2 expression.
COX-2 Overexpression Resulting from TRF2-Induced Telomere Malfunction Alters Cellular Phenotypes
Next, we determined if TRF2-vHMEC displayed alterations of known COX-2 dependent phenotypes, i.e. production of prostaglandins and cell motility. Mean levels of prostaglandins increased ~6-fold in TRF2-vHMEC compared to vector (p<0.0001, Figure 2C), illustrating that even a moderate COX-2 overexpression can result in dramatic downstream events. Cell-wounding assays revealed that TRF2-vHMEC were more motile, filling a “wound” in 8h, while parent and vector required 12h (Figure 2C). Thus, increased COX-2 expression in TRF2-vHMEC alters cellular phenotypes.
Up-Regulation of COX-2 Induced by TRF2 is Phospho-p38 Dependent
Phosphorylation of p38 is necessary for COX-2 mRNA stabilization and, consequently, for increased protein levels in late-passage vHMEC (3, 4). Phospho-p38 was up-regulated in TRF2-vHMEC and repressed in hTERT-vHMEC (Figure 2B). To determine if phospho-p38 was necessary for COX-2 induction in response to TRF2, we exposed early passage parent, vector and TRF2-vHMEC to the phospho-p38 inhibitor SB203580. Exposure to 0.02 or 0.04 μM SB203580 reduced mean COX-2 mRNA levels by 49% and 71% (p=0.007), respectively, in TRF2-vHMEC. Correspondingly, COX-2 and phospho-p38 protein levels were decreased following exposure to SB203580, demonstrating that phospho-p38 was indeed necessary for TRF2 to induce COX-2 in vHMEC.
Telomere Malfunction Induces Activin A
Gene expression profiling analysis for genes showing at least a 1.4-fold expression change in TRF2-vHMEC compared to vector (Supplemental Table 1) enabled us to identify candidate molecules upstream of p38 in TRF2-vHMEC. Among the up-regulated genes was an attractive candidate, activin A. Activin A is a secreted member of the TGF-β superfamily and can promote p38 phosphorylation (30, 31). Q-PCR validated up-regulation of activin A mRNA in TRF2-vHMEC and its down-regulation in hTERT-vHMEC compared to vector control (Figure 3C). Consistent with this, activin A protein is elevated in conditioned media from TRF2-vHMEC and reduced in conditioned media from hTERT-vHMEC compared to control (p=0,007 and p=0.01, respectively).
Since telomere function can be impaired by the progressive loss of telomere DNA during cellular replication, we predicted that activin A would increase during continued propagation of vHMEC. We found that activin A protein and mRNA levels increased 4-to 5-fold in late vs. early passage vHMEC (Figure 3B). Similarly, treatment with NU7026, which induces telomere malfunction, increased activin A mRNA and protein levels (Figure 3A).
We then asked if DNA damage caused by etoposide or UVC could also induce activin A. Activin A mRNA and protein were significantly up-regulated in vHMEC exposed to either agent (Figure 3A). However, activin A levels were greater in vHMEC treated with etoposide or NU7026 than with UVC. As observed for COX-2, activin A mRNA and protein induction was biphasic and peaked at similar intervals. Collectively these data demonstrated that activin A mRNA and protein were preferentially induced by dsDNA damage and coincided with the expression of COX-2.
Activin A is Necessary and Sufficient for COX-2 induction
Next we assessed whether activin A was necessary for COX-2 induction. TRF2-vHMEC, parent and vector were infected with lentivirus expressing either a control short hairpin RNA (shRNA) against luciferase (pGL3) or a shRNA against activin A to reduce its expression. Basal activin A mRNA and protein levels were reduced by 79% and 75%, (p<0.001 and p<0.0001), respectively, compared to control shRNA (Figure 4A). Activin A shRNA reduced basal COX-2 mRNA in vHMEC by 27%, but was not statistically significant (Figure 4A). In contrast, mean COX-2 mRNA levels decreased 46% in TRF2-vHMEC expressing activin A shRNA compared to TRF2-vHMEC expressing pGL3 (p=0.002). We then investigated whether activin A was sufficient to induce COX-2 by exposing vHMEC to exogenous activin A or to conditioned media from either TRF2-vHMEC or controls. At the two highest doses of activin A (30, 32), mean levels of COX-2 mRNA were increased ~3-fold (p=0.04 and p=0.01, respectively, Figure 4B). Likewise, vHMEC propagated in conditioned media from TRF2-vHMEC had increased COX-2 mRNA (p=0.008) and protein (Figure 4B) compared to vHMEC propagated in conditioned media from vector. COX-2 and phospho-p38 protein levels were also increased by both treatments (Figure 3B). Thus, activin A is both necessary and sufficient for COX-2 expression, demonstrating that secreted activin A can act in a cell non-autonomous fashion to induce COX-2 in absence of DNA damage.
The dsDNA Damage Response Effectors ATM and p53 are Necessary for COX-2 Induction
ATM and p53 are two essential members of the dsDNA damage response. Importantly, p53 is necessary for COX-2 induction (33). Using shRNAs, we repressed ATM and p53 expression by 68% and 93%, respectively, in control and TRF2-vHMEC (Supplemental Figure 3). ATM or p53 silencing decreased basal levels of COX-2 mRNA, activin A mRNA and activin protein in vHMEC by 40%, 41%, 53% and 42%, 36%, 61%, respectively, compared to control shRNA (Figure 4C). Similarly, repression of ATM and p53 in TRF2-vHMEC, decreased COX-2 mRNA, activin A mRNA and activin protein by 16.5%, 41% and 48%, and 43%, 51%, 71%, respectively, compared to TRF2-vHMEC expressing a control shRNA (pGL3).
COX-2 is Induced by DNA Damage in p16-Competent HMEC
Previously, we reported that exogenous expression of COX-2 in HMEC, with an intact p16/pRb pathway, results in cell cycle arrest and induction of p16 and p21 (9), implying that HMEC and vHMEC might respond differently to DNA damage. Upon treatment of HMEC with etoposide or UVC, COX-2 mRNA, activin mRNA and activin A protein were induced, although at a lower level than in vHMEC. Consistent with our observations in vHMEC, HMEC treated with etoposide exhibited a biphasic induction of COX-2 and activin A with peaks at 6-12h and 48h. Exposure to UVC induced COX-2 and activin A, both of whose expression peaked at 6h, but in contrast to treatment with etoposide, then returned to basal levels after 24h (Figure 5A). Thus, COX-2 is induced by DNA damage in HMEC with an intact p16/Rb pathway.
Figure 5. DNA Damage Induces Activin A, COX-2 and Growth Arrest in p16-Competent HMEC.
A) HMEC were exposed to 100μM Etoposide (RM40, RM45, RM46) or to 60J/m2 of UVC (RM45) as described in Figure 1A. Line graphs indicate relative COX-2 and activin A mRNA levels and activin A protein levels in conditioned media compared to the untreated control. B) Activin A, COX-2, p16 and p21 mRNA levels in parent, vector (pWP1) or TRF2-HMEC (CM7, RM146). C) Population doublings in HMEC (CM7) infected with lentivirus containing TRF2 or empty vector (pWP1) or parent HMEC. D) Overview of the DNA damage-dependent COX-2 induction and its consequences in HMEC (intact p16/Rb) or vHMEC (silenced p16).
COX-2 Overexpression in p16-Competent HMEC is Growth Suppressive
We then examined how HMEC (p16-competent) would respond to sustained DNA damage induced by TRF2 overexpression. When compared to vector, TRF2-HMEC had a statistically significant increase in COX-2 (p=0.006) and activin A (p=0.005) mRNAs (Figure 5B). However, TRF2-HMEC also up-regulated p16 and p21 (Figure 5B), displayed flattened vacuolated morphology, entered growth arrest (Figure 5C) and exhibited a decreased fraction of cells in S-phase (Supplemental Figure 4), all phenotypes consistent with senescence. Thus, in the context of an intact p16/Rb pathway, DNA damage results in increased activin A and COX-2, but also induction of growth arrest. These data show that, in contrast to vHMEC, the cellular response to COX-2 in HMEC is both self-limiting and growth suppressive.
COX-2 Expression in Ductal Carcinoma In Situ is Associated with Manifestations of a Double-Strand DNA Damage Response and Increased TRF2 and Activin A Expression
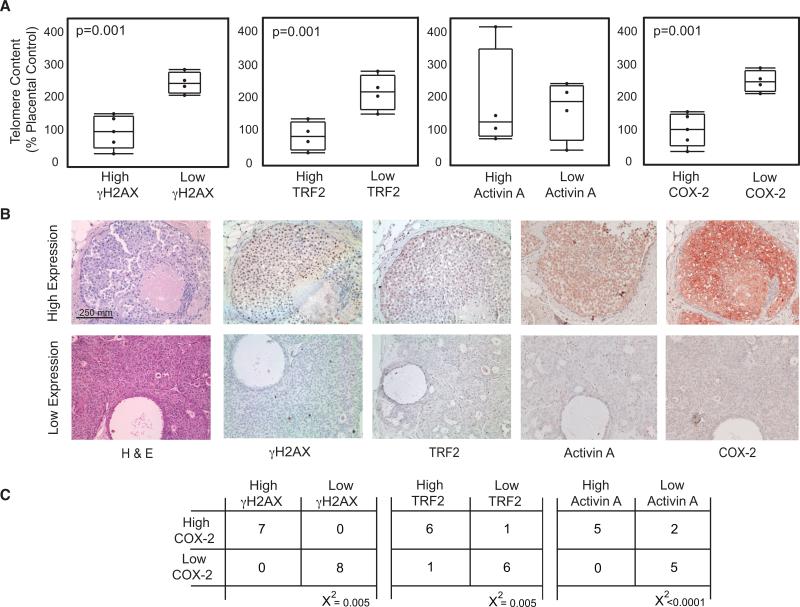
COX-2 is often up-regulated in DCIS (34) where it has prognostic significance (9, 13). Additionally, DCIS is the earliest stage in breast cancer at which loss of telomere DNA can be widely detected (35). We used a pilot cohort of fifteen DCIS cases to determine whether changes in telomere content, a proxy for telomere length (24), were associated with alterations in γH2AX, TRF2, activin A and COX-2 expression. DCIS lesions were microdissected to produce enriched populations of tumor cells that were used for DNA purification. Telomere content was measured in 9 of the 15 lesions from which sufficient DNA was obtained.
Expression levels of γH2AX, TRF2, activin A and COX-2 were evaluated by immunohistochemistry in serial sections of the 15 DCIS cases (Supplemental Table 2) and were used to divide lesions into two groups (see Materials and Methods). DCIS lesions with high γH2AX, TRF2 or COX-2 expression had lower telomere content than those with low γH2AX, TRF2 or COX-2 expression (p=0.001, p=0.001, p=0.001, respectively). Although not statistically significant, mean telomere content was reduced in 3 of the 4 lesions with high activin A (Figure 6). Moreover, COX-2 expression levels were correlated with those of γH2AX (x2=0.005), TRF2 (x2=0.005) and activin A (x2<0.0001). Thus, our findings in vitro are recapitulated in vivo: DNA damage induced by telomere malfunction and characterized by low telomere content, high γH2AX and high TRF2 expression, are associated with increased activin A and COX-2 expression.
Figure 6. Telomere Content is Inversely Associated with TRF2, Activin A and COX-2 Expression in Ductal Carcinoma in Situ (DCIS).
A) γH2AX, TRF2, activin A and COX-2 protein levels were evaluated by immunohistochemistry. Lesions were divided into two groups based on staining intensity for each protein. Telomere content in DCIS lesions was expressed as a percentage of placental DNA. Box plots show the relationship between γH2AX (far left), TRF2 (left), activin (right) and COX-2 (far right) and telomere content in high and low expression lesions. p-values for each comparison are shown in the inset. B) Examples of serial sections for high and low expression DCIS lesions (20x) stained with H&E, and γH2AX, TRF2, activin A and COX-2 antibodies. Nuclei counterstained with hematoxylin (blue) and primary antibodies against each antigen detected using AEC (red). C) Contingency tables showing the number of cases with high or low COX-2 levels versus the number of cases with high or low γH2AX, TRF2 and activin A.
DISCUSSION
Here we show for the first time that dsDNA damage and telomere malfunction in human breast epithelial cells results in a p53- and activin A-dependent COX-2 induction. By identifying signaling events leading to COX-2 induction, this study complements our previous work establishing a direct link between COX-2 and malignant phenotypes (3). Strikingly, COX-2 expression, and its associated phenotypes, are not confined to the initial cell with telomere malfunction, but are also induced in cells in absence of DNA damage through the cell non-autonomous action of activin A (Figure 5D). While induction of this pathway is self-limiting, i.e. leading to cell cycle arrest, in HMEC (intact p16/Rb pathway), it is not in vHMEC (silenced p16), where cells continue to proliferate. Finally, we demonstrate in vivo that high COX-2 expression is associated with high levels of γH2AX, TRF2 and activin A in a pilot cohort of DCIS lesions.
Our study highlights the coordinated action of p53, activin A and p38 in inducing COX-2 following dsDNA damage. To our knowledge this is the first report of an activin A-dependent COX-2 induction in human cells. A similar observation has been reported in rat macrophages (36). Multiple stimuli can induce COX-2 including mitogens, cytokines, hormones, irradiation, oncogene activation and inflammation (33, 37). Many of these factors induce p53, which can in turn increase COX-2 expression. Interestingly, COX-2 induction decreases p53-dependent apoptosis in vHMEC, suggesting that COX-2 represses p53 function (38). This feed-back loop may explain the biphasic activin A and COX-2 induction in response to DNA damage described here.
Activin A is a TGF-β superfamily member whose signaling requires receptor dimer formation and downstream effectors including Smads, MAPKs, ERK, and p38 among others. The effects of activin A signaling are cell context-dependent. For example, through interaction with p53, activin A can induce either dorsal or ventral mesoderm formation during Xenopus embryogenesis (39), erythroid differentiation (40) or cell cycle arrest via p21 induction and CDK4 down-regulation (41).
In breast tumor cell lines, activin A induces growth arrest and inhibits HGF-induced tubule formation in primary mammary epithelial cells grown in a collagen matrix (32, 42). However, the role of activin A in breast tumorigenesis remains ambiguous. Activin A mRNA and protein are frequently up-regulated in DCIS and in IDC compared to normal breast. Moreover, breast tumors with local recurrence or metastasis to lymph nodes have the highest levels of activin A expression (43, 44) highlighting the potential predictive value of activin A as biomarker. Gains in chromosomes 3p, 7p, 12q and 15q, which contain the genes encoding the type II receptor, activin A, two other activin family members and Smad 3, are often observed in breast cancer (45-48). Last, activin A overexpression increases tumor volume by inhibiting apoptosis in mouse xenographs (49). This paradox is reminiscent of that described for TGF-β. The induction of growth arrest described here in HMEC (intact p16) and elsewhere (32, 50) supports that activin A acts as a barrier to tumor initiation. In contrast, the observation that activin A is frequently up-regulated in IDC and metastatic breast lesions suggests that activin A, like TGF-β, may facilitate tumorigenesis in the context of impaired growth inhibitory response (for example due to loss of p16 in vHMEC) by decreasing immune response and altering the tumor microenvironment. Despite the similarity between activin A and TGF-β, these proteins are not synonymous; since vHMEC arrest in response to TGF-β (4). Inhibiting activin A, either through induction of physiological regulators (follistatin, inhibin A or follistatin-related protein FLRG) or use of inhibitors (SB432542 or type I and II receptor antibodies), is an attractive therapeutic approach to ablate the COX-2 overexpression triggered by DNA damage, although side effects of such therapies remain to be investigated.
DNA damage is an early and nearly universal event in epithelial cancers. It is well appreciated that in the absence of cell cycle checkpoints (e.g. p53 or p16/Rb), DNA damage generates genomic instability and, consequently, may result in random loss of tumor suppressors, gain of oncogenes and clonal expansion. Thus, DNA damage indirectly contributes to tumorigenesis through generation of genomic instability. Here we show that DNA damage may also directly contribute to tumorigenesis through a separate mechanism, the specific induction of activin A and COX-2. Since activin A and the prostaglandins are secreted, DNA damage in one cell could drive tumorigenic phenotypes in an adjacent p16-compromised precursor cell or lead to proliferative arrest in an adjacent p16-intact cell. We postulate that these cell non-autonomous effects might provide a proliferative advantage to precursor lesions and facilitate the expression of pre-malignant phenotypes. Understanding how DNA damage contributes to cell autonomous and cell non-autonomous events will further elucidate the tumorigenic process. For example, it may provide novel insights into the ecology of breast tissues and cell-cell interactions that modulate early events in malignancy, identify people with an increased propensity to develop aggressive tumors, or finally, provide an opportunity to prevent the progression of a precursor lesion to a fully tumorigenic state.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Tlsty laboratory for thoughtful discussions. We thank Drs. Judy Tjoe and James Marx at the Comprehensive Breast Health Center Aurora Sinai Medical Center (Milwaukee, WI), Dr. David Baer at Kaiser Foundation Research Institute (Oakland, CA), Dr. Sara Sukumar at the Johns Hopkins University School of Medicine (Baltimore, MD), Dr. Michael Idowu at Virginia Commonwealth University (Richmond, VA) and Ms. Kerry Wiles at the Cooperative Human Tissue Network (Nashville, TN) for providing us with breast tissue samples.
Financial Support: This work was supported by NCI/NIH R01 CA122024, R01 CA097214, P01 CA107584, CIRM grant RS1-00444-1, CBCRP grant A110281 and Avon Foundation grant 07-2007-074 to TDT. CAF was supported by postdoctoral fellowship DAMD17-03-1-0424 from the DOD and NIH RO1 Research Supplement for Underrepresented Minorities
REFERENCES
- 1.Marnett LJ. The COXIB experience: a look in the rearview mirror. Annu Rev Pharmacol Toxicol. 2009;49:265–90. doi: 10.1146/annurev.pharmtox.011008.145638. [DOI] [PubMed] [Google Scholar]
- 2.Stack E, DuBois RN. Role of cyclooxygenase inhibitors for the prevention of colorectal cancer. Gastroenterol Clin North Am. 2001;30:1001–10. doi: 10.1016/s0889-8553(05)70225-9. [DOI] [PubMed] [Google Scholar]
- 3.Crawford YG, Gauthier ML, Joubel A, et al. Histologically normal human mammary epithelia with silenced p16(INK4a) overexpress COX-2, promoting a premalignant program. Cancer Cell. 2004;5:263–73. doi: 10.1016/s1535-6108(04)00023-6. [DOI] [PubMed] [Google Scholar]
- 4.Gauthier ML, Pickering CR, Miller CJ, et al. p38 regulates cyclooxygenase-2 in human mammary epithelial cells and is activated in premalignant tissue. Cancer Res. 2005;65:1792–9. doi: 10.1158/0008-5472.CAN-04-3507. [DOI] [PubMed] [Google Scholar]
- 5.Greenhough A, Smartt HJ, Moore AE, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55:115–22. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher DW, Pankratz VS, Santisteban M, et al. Association between cyclooxygenase-2 expression in atypical hyperplasia and risk of breast cancer. J Natl Cancer Inst. 2008;100:421–7. doi: 10.1093/jnci/djn036. [DOI] [PubMed] [Google Scholar]
- 8.Yang WT, Lewis MT, Hess K, et al. Decreased TGFbeta signaling and increased COX2 expression in high risk women with increased mammographic breast density. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–91. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Schem C, Shi YH, Medina D, Zhang M. Increased COX2 expression enhances tumor-induced osteoclastic lesions in breast cancer bone metastasis. Clin Exp Metastasis. 2008;25:389–400. doi: 10.1007/s10585-007-9117-3. [DOI] [PubMed] [Google Scholar]
- 11.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucci A, Krishnamurthy S, Singh B, et al. Cyclooxygenase-2 expression in primary breast cancers predicts dissemination of cancer cells to the bone marrow. Breast Cancer Res Treat. 2009;117:61–8. doi: 10.1007/s10549-008-0135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boland GP, Butt IS, Prasad R, Knox WF, Bundred NJ. COX-2 expression is associated with an aggressive phenotype in ductal carcinoma in situ. Br J Cancer. 2004;90:423–9. doi: 10.1038/sj.bjc.6601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ristimaki A, Sivula A, Lundin J, et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–5. [PubMed] [Google Scholar]
- 15.Baron JA, Sandler RS, Bresalier RS, et al. A randomized trial of rofecoxib for the chemoprevention of colorectal adenomas. Gastroenterology. 2006;131:1674–82. doi: 10.1053/j.gastro.2006.08.079. [DOI] [PubMed] [Google Scholar]
- 16.Bertagnolli MM, Eagle CJ, Zauber AG, et al. Celecoxib for the prevention of sporadic colorectal adenomas. N Engl J Med. 2006;355:873–84. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- 17.Kwan ML, Habel LA, Slattery ML, Caan B. NSAIDs and breast cancer recurrence in a prospective cohort study. Cancer Causes Control. 2007;18:613–20. doi: 10.1007/s10552-007-9003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thun MJ, Namboodiri MM, Heath CW., Jr. Aspirin use and reduced risk of fatal colon cancer. N Engl J Med. 1991;325:1593–6. doi: 10.1056/NEJM199112053252301. [DOI] [PubMed] [Google Scholar]
- 19.Dajani EZ, Islam K. Cardiovascular and gastrointestinal toxicity of selective cyclooxygenase-2 inhibitors in man. J Physiol Pharmacol. 2008;59(Suppl 2):117–33. [PubMed] [Google Scholar]
- 20.Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–7. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- 21.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–55. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- 22.Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J Cell Sci. 2006;119:3128–40. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- 23.Dumont N, Bakin AV, Arteaga CL. Autocrine transforming growth factor-beta signaling mediates Smad-independent motility in human cancer cells. J Biol Chem. 2003;278:3275–85. doi: 10.1074/jbc.M204623200. [DOI] [PubMed] [Google Scholar]
- 24.Fordyce CA, Heaphy CM, Griffith JK. Chemiluminescent Measurement of Telomere DNA Content in Biopsies. Biotechniques. 2002;33:144–8. doi: 10.2144/02331md02. [DOI] [PubMed] [Google Scholar]
- 25.Bailey SM, Brenneman MA, Halbrook J, Nickoloff JA, Ullrich RL, Goodwin EH. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair (Amst) 2004;3:225–33. doi: 10.1016/j.dnarep.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–5. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 27.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–71. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 28.Oh BK, Kim YJ, Park C, Park YN. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol. 2005;166:73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter T, Saretzki G, Nelson G, Melcher M, Olijslagers S, von Zglinicki T. TRF2 overexpression diminishes repair of telomeric single-strand breaks and accelerates telomere shortening in human fibroblasts. Mech Ageing Dev. 2007;128:340–5. doi: 10.1016/j.mad.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Cocolakis E, Lemay S, Ali S, Lebrun JJ. The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. J Biol Chem. 2001;276:18430–6. doi: 10.1074/jbc.M010768200. [DOI] [PubMed] [Google Scholar]
- 31.de Guise C, Lacerte A, Rafiei S, et al. Activin inhibits the human Pit-1 gene promoter through the p38 kinase pathway in a Smad-independent manner. Endocrinology. 2006 doi: 10.1210/en.2006-0444. [DOI] [PubMed] [Google Scholar]
- 32.Burdette JE, Jeruss JS, Kurley SJ, Lee EJ, Woodruff TK. Activin A mediates growth inhibition and cell cycle arrest through Smads in human breast cancer cells. Cancer Res. 2005;65:7968–75. doi: 10.1158/0008-5472.CAN-04-3553. [DOI] [PubMed] [Google Scholar]
- 33.de Moraes E, Dar NA, de Moura Gallo CV, Hainaut P. Cross-talks between cyclooxygenase-2 and tumor suppressor protein p53: Balancing life and death during inflammatory stress and carcinogenesis. Int J Cancer. 2007;121:929–37. doi: 10.1002/ijc.22899. [DOI] [PubMed] [Google Scholar]
- 34.Shim V, Gauthier ML, Sudilovsky D, et al. Cyclooxygenase-2 expression is related to nuclear grade in ductal carcinoma in situ and is increased in its normal adjacent epithelium. Cancer Res. 2003;63:2347–50. [PubMed] [Google Scholar]
- 35.Meeker AK, Argani P. Telomere shortening occurs early during breast tumorigenesis: a cause of chromosome destabilization underlying malignant transformation? J Mammary Gland Biol Neoplasia. 2004;9:285–96. doi: 10.1023/B:JOMG.0000048775.04140.92. [DOI] [PubMed] [Google Scholar]
- 36.Nusing RM, Barsig J. Induction of prostanoid, nitric oxide, and cytokine formation in rat bone marrow derived macrophages by activin A. Br J Pharmacol. 1999;127:919–26. doi: 10.1038/sj.bjp.0702626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Signalling networks regulating cyclooxygenase-2. Int J Biochem Cell Biol. 2006;38:1654–61. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 38.Han JA, Kim JI, Ongusaha PP, et al. P53-mediated induction of Cox-2 counteracts p53- or genotoxic stress-induced apoptosis. EMBO J. 2002;21:5635–44. doi: 10.1093/emboj/cdf591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takebayashi-Suzuki K, Funami J, Tokumori D, et al. Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development. 2003;130:3929–39. doi: 10.1242/dev.00615. [DOI] [PubMed] [Google Scholar]
- 40.Sehy DW, Shao LE, Yu AL, Tsai WM, Yu J. Activin A-induced differentiation in K562 cells is associated with a transient hypophosphorylation of RB protein and the concomitant block of cell cycle at G1 phase. J Cell Biochem. 1992;50:255–65. doi: 10.1002/jcb.240500306. [DOI] [PubMed] [Google Scholar]
- 41.Zauberman A, Oren M, Zipori D. Involvement of p21(WAF1/Cip1), CDK4 and Rb in activin A mediated signaling leading to hepatoma cell growth inhibition. Oncogene. 1997;15:1705–11. doi: 10.1038/sj.onc.1201348. [DOI] [PubMed] [Google Scholar]
- 42.Liu QY, Niranjan B, Gomes P, et al. Inhibitory effects of activin on the growth and morpholgenesis of primary and transformed mammary epithelial cells. Cancer Res. 1996;56:1155–63. [PubMed] [Google Scholar]
- 43.Mylonas I, Jeschke U, Shabani N, Kuhn C, Friese K, Gerber B. Inhibin/activin subunits (inhibin-alpha, -betaA and -betaB) are differentially expressed in human breast cancer and their metastasis. Oncol Rep. 2005;13:81–8. [PubMed] [Google Scholar]
- 44.Reis FM, Luisi S, Carneiro MM, et al. Activin, inhibin and the human breast. Mol Cell Endocrinol. 2004;225:77–82. doi: 10.1016/j.mce.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 46.Bieche I, Khodja A, Driouch K, Lidereau R. Genetic alteration mapping on chromosome 7 in primary breast cancer. Clin Cancer Res. 1997;3:1009–16. [PubMed] [Google Scholar]
- 47.Bieche I, Lidereau R. Genetic alterations in breast cancer. Genes Chromosomes Cancer. 1995;14:227–51. doi: 10.1002/gcc.2870140402. [DOI] [PubMed] [Google Scholar]
- 48.Aubele MM, Cummings MC, Mattis AE, et al. Accumulation of chromosomal imbalances from intraductal proliferative lesions to adjacent in situ and invasive ductal breast cancer. Diagn Mol Pathol. 2000;9:14–9. doi: 10.1097/00019606-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 49.Krneta J, Kroll J, Alves F, et al. Dissociation of angiogenesis and tumorigenesis in follistatin- and activin-expressing tumors. Cancer Res. 2006;66:5686–95. doi: 10.1158/0008-5472.CAN-05-3821. [DOI] [PubMed] [Google Scholar]
- 50.Kalkhoven E, Roelen BA, de Winter JP, et al. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6:1151–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.