Slx and Slxl1 are genes present in multiple copies on the mouse X chromosome. Using transgenically-delivered small interfering RNAs to disrupt their function, we show that Slx and Slxl1 are important for normal sperm differentiation and male fertility.
Abstract
The human and mouse sex chromosomes are enriched in multicopy genes required for postmeiotic differentiation of round spermatids into sperm. The gene Sly is present in multiple copies on the mouse Y chromosome and encodes a protein that is required for the epigenetic regulation of postmeiotic sex chromosome expression. The X chromosome carries two multicopy genes related to Sly: Slx and Slxl1. Here we investigate the role of Slx/Slxl1 using transgenically-delivered small interfering RNAs to disrupt their function. We show that Slx and Slxl1 are important for normal sperm differentiation and male fertility. Slx/Slxl1 deficiency leads to delay in spermatid elongation and sperm release. A high proportion of delayed spermatids are eliminated via apoptosis, with a consequent reduced sperm count. The remaining spermatozoa are abnormal with impaired motility and fertilizing abilities. Microarray analyses reveal that Slx/Slxl1 deficiency affects the metabolic processes occurring in the spermatid cytoplasm but does not lead to a global perturbation of sex chromosome expression; this is in contrast with the effect of Sly deficiency which leads to an up-regulation of X and Y chromosome genes. This difference may be due to the fact that SLX/SLXL1 are cytoplasmic while SLY is found in the nucleus and cytoplasm of spermatids.
INTRODUCTION
Spermatogenesis is the process during which spermatogonial stem cells multiply and generate spermatocytes, which through two meiotic divisions form haploid spermatids that differentiate into spermatozoa. The differentiation of haploid round spermatids into spermatozoa (spermiogenesis) involves major alteration of cell structure and function as the nucleus is restructured via chromatin remodeling and compaction to form the sperm head, and sperm-specific structures, such as the acrosome and the flagellum, are formed (Russell et al., 1990).
The X and Y chromosomes are enriched in genes presumed to be important for sperm differentiation (Burgoyne and Mitchell, 2007; Mueller et al., 2008) but functional studies remain rare (for review, see Stouffs et al., 2009). The sex chromosomal complement of spermatid-expressed genes in man and mouse is enriched for multicopy genes (Skaletsky et al., 2003; Toure et al., 2005; Mueller et al., 2008), and this has hindered gene function studies as a classical targeting strategy is not applicable (Burgoyne and Mitchell, 2007). Using an RNA interference–based strategy to disrupt the function of the multiple copies (>100) of the Y-encoded Sly (Sycp3 like Y-linked) gene, we found that its protein is crucial for the epigenetic regulation of sex chromosome expression after meiosis and for sperm differentiation (Cocquet et al., 2009). The X chromosome carries two multicopy genes related to Sly: Slx (formerly known as Xmr; ∼43 copies) and Slx-like1 (Slxl1, formerly known as AK015913 or 4930527E24Rik; ∼16 copies) (Reynard et al., 2007; Scavetta and Tautz, 2010). Like Sly, Slx and Slxl1 are specifically expressed in male postmeiotic germ cells. Slx encodes a cytoplasmic protein of unknown function, while for Slxl1 it has been unclear whether transcripts are translated in the testis (Reynard et al., 2007). In the case of Sly deficiency, Slx and Slxl1 genes are up-regulated, along with other sex-chromosome genes, and thus are candidates to explain the aberrant sperm differentiation phenotypes and the near sterility of Sly-deficient males (Ellis et al., 2005; Cocquet et al., 2009). Intriguingly, genomic and genetic evidence suggests the existence of an ongoing postmeiotic intragenomic conflict between multicopy X and Y genes (Partridge and Hurst, 1998; Ellis and Affara, 2006), and Slx and Sly genes have been hypothesized to be key mediators of this ‘competition’ between X-bearing and Y-bearing gametes (Ellis et al., 2005).
In the present study, we sought to determine the function of Slx and Slxl1 and to see whether, like their Y-encoded counterpart Sly, they have a critical role in the control of sex chromosome expression during spermiogenesis. For this, we produced Slx/Slxl1-deficient mice using transgenically-delivered small interfering RNAs (siRNA). We show that the Slx gene family is important for normal mouse sperm differentiation (and thus for male fertility), but in contrast to Sly this is not a consequence of a global perturbation of sex chromosome expression.
MATERIALS AND METHODS
Plasmid Construction, Generation, and Breeding of Transgenic Mice
To generate the U6shSLX constructs, we used a PCR-based approach similar to that described in Harper et al., 2005, using primers designed to generate the short hairpin SLX sequences (Harper et al., 2005) (Supplementary Table 1). The PCR products were cloned into the pCR2.1 vector (TOPO TA Cloning, Invitrogen, Life technologies, Carlsbad, CA). An insulator element from the chicken β-globin domain (Chung et al., 1993) and a genotype tag (gTag) were then added to each construct between SalI and BamHI restriction enzyme sites. These genotyping tags were inserted to enable discrimination of the different shSLX constructs by PCR (see Supplementary Table 1 for primer sequences). All constructs were sequenced before testing their specificity and efficiency.
Before injection, the plasmids were linearized at PvuI and BamHI sites and on-column purified from agarose gels (Gel Extract II kit, Macherey Nagel, Germany). Fertilized eggs from CBA/Ca × C57BL/10 matings were microinjected with the construct, using standard protocols. Transgenic founders carrying an shSLX construct were identified by PCR and crossed with random-bred MF1 (National Institute for Medical Research colony) mice. Two founders (one male shSLX1 and one female shSLX2) transmitted the transgene to their offspring. The lines have been maintained by backcrossing shSLX transgenic females to MF1 XYRIII males (i.e., with a Y chromosome originating from the RIII strain maintained on the random-bred MF1-NIMR background). Two-month-old MF1 XYRIII males with (tsgic) and without (neg sib) the transgene were processed for all the analyses presented here except IVF, sperm motility, and electron microscopy for which males with a mixed B6D2F1 (C57BL/6 x DBA/2)/MF1 background were used. Animal procedures were in accordance with the United Kingdom Animal Scientific Procedures Act 1986 and were subject to local ethical review. For IVF, sperm motility, and electron microscopy, animal procedures were in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and guidelines presented in the National Research Council's Guide for Care and Use of Laboratory Animals published by the Institute for Laboratory Animal Research of the National Academy of Science, Bethesda, MD, 1996.
Elutriation of Spermatids
Fractions enriched in round spermatids (>90%) were obtained using an adapted protocol of Meistrich (Meistrich, 1977), as described previously (Cocquet et al., 2009).
Western Blot and Immunofluorescence
Western blot analyses were performed as described previously (Reynard et al., 2007). Briefly, 10–15 micrograms of testis or spermatid fraction protein extracts were run on a 12% SDS/polyacrylamide gel or a 4–12% gradient Bis/Tris gel (NuPage, Invitrogen). After transfer and blocking, membranes were incubated overnight with either anti-SLX/SLXL1 antibody (Reynard et al., 2007) diluted at 1/1000, anti–β-actin (SIGMA-Aldrich, St. Louis, MO) at 1/50000, or anti-GFP (Invitrogen) at 1/1000. Incubation with the corresponding secondary antibody coupled to peroxidase and detection by chemiluminescence were carried out as described by the manufacturer (SuperSignal West Pico, Pierce, Rockford, IL).
Immunofluorescence experiments were performed on testis material fixed in 4% buffered paraformaldehyde as described before (Cocquet et al., 2009). Anti-SLX/SLXL1 antibody (Reynard et al., 2007) and anti-H4K12Ac (Millipore, Bedford, MA) were used overnight at 1/100. Testis sections were stained using an in situ cell death detection kit as described by the manufacturer (Roche Diagnostics, Indianapolis, IN). This detects apoptotic cells by fluorescently labeling DNA strand breaks (terminal deoxynucleotidyltransferase dUTP nick end labeling [TUNEL]). Approximately 100 tubules were counted per male (four individuals per genotype) to determine the percentage of tubules containing TUNEL-positive elongating/condensing spermatids. Alexa Fluor 594-conjugated peanut agglutinin lectin (Invitrogen), which stains the developing acrosome of spermatids, was used to stage the testis tubules.
Histology and Analysis of Sperm Head Morphology
For the analysis of spermatid development and sperm shedding delay, testes were fixed in Bouin (Sigma) and wax-embedded; five micron sections were stained with periodic acid–Schiff (PAS). Sperm smears obtained from the initial caput epididymis were silver stained and analyzed as described previously (Touré et al., 2004). Sperm count was done on cauda epididymis. Sperm count, assessment of sperm morphology, and timing of sperm shedding were performed on two-month-old animals (at least four mice per genotype). Transmission electron microscopy was done on sperm from the cauda epididymis as previously described (Yamauchi et al., 2010).
Fertility Testing and In Vitro Fertilization
To assess fertility, six MF1 XYRIII males of each genotype were mated with MF1 females over a period of six and a half months. Mating was confirmed by the presence of copulatory plugs. In vitro fertilization (IVF) analysis was performed with sperm from four shSLX1 males, six shSLX2 males, and six nontransgenic siblings (WT) from a mixed B6D2F1/MF1 background, using B6D2F1 oocytes as described before (Cocquet et al., 2009). Sperm from the same males were subjected to motility analysis.
Real-Time PCR and Microarray Analyses
For real-time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and microarray analyses, RNA was extracted from total testis or from fractions of 1 × 106 round spermatids using Trizol (Invitrogen). Reverse transcription of polyadenylated RNA and real-time PCR were performed as described previously (Cocquet et al., 2009). Briefly, real-time PCR was performed using Absolute qPCR SYBR Green ROX mix (ThermoFisher Scientific, Waltham, MA) on an ABI PRISM 7500 machine (Applied Biosystems, Life technologies, Carlsbad, CA). Testis samples from four to five transgenic mice and six nontransgenic siblings (negative controls), all at two months of age, were analyzed. All reactions were carried out in triplicate per assay and Acrv1 (an autosomal spermatid specific gene) was included on every plate as a loading control. The difference in PCR cycles with respect to Acrv1 (ΔCt) for a given experimental sample was subtracted from the mean ΔCt of the reference samples (negative siblings) (ΔΔCt). For quantification on purified round spermatid samples, β-actin was used as the loading control. Primer sequences are available in Supplementary Table 1.
Microarray analyses were performed on RNA from purified round spermatid fractions as previously described (Cocquet et al., 2009) using two groups (a pool of 2–4 individuals) of shSLX1 transgenic mice and four groups of WT mice. Results were compared with those obtained with purified round spermatid fractions from Sly-deficient males (Cocquet et al., 2009). Differentially expressed genes were classified according to their likely biological function in Onto-Express.
Statistical Analysis
For comparisons of the incidence of sperm head abnormalities, differences between genotypes were assessed by ANOVA after angular transformation of percentages, using the Generalized Linear Model provided by NCSS statistical data analysis software. The same test was applied to the frequency of abnormal head-tail connections, the proportion of oocytes fertilized in vitro, tubules with sperm shedding delay, TUNEL+ elongating spermatids, abnormal H4K12Ac staining, or sperm motility. Student's t test was used to compare the data obtained for fecundity, sperm number, testis weight, Western blot quantification, and real-time PCR (performed on the ΔΔCt values). For microarray analysis, quantile normalization of all expression data was performed using BeadStudio (Illumina Inc., San Diego, CA). Data for the shSLX1/control spermatids was compared in BeadStudio, using the Illumina custom error model. Significant up- or down-regulation was defined as a false discovery rate-corrected p value of ≤0.05 and a fold change of ≥1.5.
RESULTS
Specific Knockdown of Slx and Slxl1 Transcripts by Transgenically-Delivered siRNAs
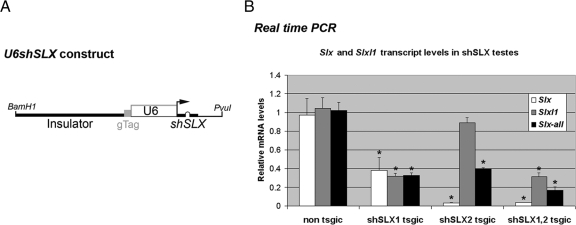
To study the function of the Slx multicopy gene family, we used a strategy similar to the one previously developed for Sly (Cocquet et al., 2009), i.e., the transgenic delivery of short hairpin RNAs (shRNAs) that generate small interfering RNAs (siRNAs). These siRNAs mediate gene-specific knockdown via RNA interference. In our strategy, the shRNAs are expressed under the control of the strong and ubiquitous U6 promoter (Figure 1).
Figure 1.
Transgenically delivered shSLX constructs knockdown Slx and Slx-like1 at the transcript level. (A) Structure of the shSLX construct. The U6 promoter drives the expression of a short hairpin RNA targeting Slx/Slxl1 (shSLX). An insulator sequence is located upstream of the U6 promoter. A ‘genotyping’ tag (gTag) that is specific to each shSLX construct has been inserted to facilitate genotyping in double transgenic mice. (B) Quantification of the knockdown of Slx and Slxl1 transcripts in shSLX testes by real-time RT-PCR. Values were normalized to Acrv1 (an autosomal spermatid-specific gene). ‘Slx-all’ represents amplification with primers designed to amplify both Slx and Slxl1 transcripts. One star indicates significant difference from nontransgenic (WT) value (p < 0.005; t test).
Four shRNA sequences expected to target Slx and Slx-like 1 (Slxl1) transcripts were carefully selected (to avoid cross-reaction with related sequences such as Sly and Xlr) and used to design shRNA constructs. The efficiency and specificity of these shRNA constructs were tested in cell culture by cotransfection (Supplementary Figure 1). Based on these results, two efficient and specific shRNA constructs (shSLX1 and shSLX2) were used to produce transgenic mice via pronuclear injection. Each transgene was stably inserted in the genome and was transmitted to the progeny. The resulting transgenic lines (designated shSLX1 and shSLX2) showed a dramatic knockdown of Slx transcripts: ∼62% decrease in shSLX1 testes and ∼97% decrease in shSLX2 testes, as quantified by real-time PCR (Figure 1). The shSLX2 transgene did not have a significant effect on Slxl1 expression; however, shSLX1 testes displayed a ∼68% decrease in Slxl1 transcript level. These results are in agreement with predictions from cotransfection experiments (cf. Supplementary Figure 1). Males carrying both shSLX1 and shSLX2 transgenes (shSLX1,2 transgenic mice) had decreased levels in both Slx and Slxl1 transcripts (∼97% and ∼69% decrease, respectively). When using primers designed to amplify together Slx and Slxl1 transcripts (Slx-all), global knockdown was estimated as ∼68% for shSLX1, ∼59% for shSLX2, and ∼83% for shSLX1,2.
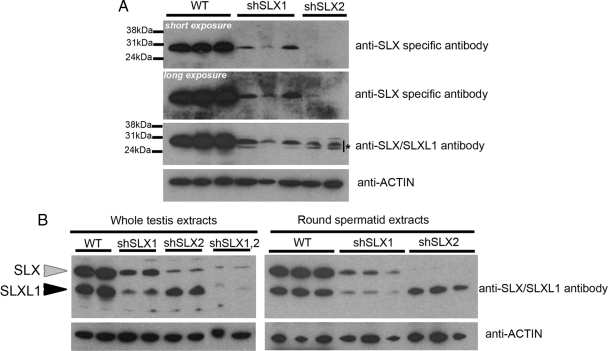
SLX and SLXL1 Protein Levels Are Reduced in shSLX Testes
Two antibodies had been developed for the study of SLX (Reynard et al., 2007). In cell lines transiently expressing an Slx or Slxl1 transgene, one antibody (anti-SLX aka anti-SLX69-81) detected specifically SLX protein, while the other (anti-SLX/SLXL1 aka anti-SLX96-106) detected both SLX and SLXL1 protein. In testes, SLX protein was detected by both antibodies but it remained unclear whether Slxl1 transcripts were translated (Reynard et al., 2007). Making use of our two shSLX mouse lines (shSLX1 targeting Slx and Slxl1 transcripts, and shSLX2 only targeting Slx) we decided to reinvestigate Slxl1 translation. SLX protein is predicted to be 25 kDa, while SLXL1 is expected at 18 kDa. After separation on a 12% polyacrylamide gel, Western blot detection with anti-SLX specific antibody showed a fainter band in shSLX1 testis protein samples compared with wild-type testes (i.e., nontransgenic siblings). In shSLX2 testes, the band was barely visible (Figure 2A) in agreement with transcript quantifications. Detection with the anti-SLX/SLXL1 antibody showed a double band of reduced intensity in shSLX1 and shSLX2 testes (Figure 2A). After separation on a 4–12% gradient gel, it became clear that the upper band (indicated by the gray arrowhead) corresponds to SLX protein (decreased in shSLX1 and in shSLX2 testes) while the bottom band (indicated by the black arrowhead) corresponds to SLXL1 protein (reduced in shSLX1 but almost unaffected in shSLX2 testes). As expected, double transgenic shSLX1,2 mice showed a deficiency in both types of proteins. Similar results were obtained on round spermatid protein extracts (Figure 2B). Immunofluorescence on testis section using anti-SLX/SLXL1 antibody confirmed a dramatic decrease in SLX/SLXL1 protein levels in round spermatids (Supplementary Figure 2).
Figure 2.
Transgenically delivered shSLX constructs lead to a dramatic decrease of SLX and SLXL1 proteins. (A) Western blot detection of SLX/SLXL1 proteins on WT (i.e., nontransgenic siblings), shSLX1 and shSLX2 adult testicular extracts, after separation by 12% PAGE. A double band (indicated by a star) can be observed when detecting with the anti-SLX/SLXL1 antibody. Actin antibody was used for normalization. (B) Left panel, Western-blot detection of SLX/SLXL1 proteins on WT, shSLX1, shSLX2, and shSLX1,2 adult testicular extracts, after separation by 4–12% gradient PAGE. The upper band (gray arrowhead) corresponds to SLX protein. The bottom band (black arrowhead) is SLXL1 protein. Similar results were obtained on purified round spermatid extracts from WT, shSLX1, and shSLX2 mice (right panel, same conditions). Actin antibody was used for normalization.
All in all, we have produced mouse models deficient for Slx/Slxl1 genes. We proceeded to analyze their phenotypes to elucidate the roles of these genes in spermiogenesis and the extent to which Slx/Slxl1 depletion mimics Sly deficiency.
Slx/Slxl1 Deficiency Leads to Impaired Male Fertility
The first phenotype of Slx/Slxl1-deficient males we observed was poor fertility. Over a breeding period of 6½ months with young WT (MF1) females, shSLX1 and shSLX2 males had a considerable reduction in the number of litters and in litter sizes compared with WT males (nontransgenic siblings) (Table 1). Double transgenic shSLX1,2 males were sterile. Transgenic females did not have obvious breeding problems (data not shown). In vitro, the fertilizing ability of Slx/Slxl1-deficient epididymal sperm was also severely impaired relative to controls (Table 1). Testis weight for shSLX1 and shSLX2 males did not differ from controls but shSLX double transgenic testes were slightly smaller (90 mg vs. 102 mg in WT) (Table 1).
Table 1.
Analysis of the reproductive parameters of Slx/Slxl1-deficient males and controls
| Parameters | Characteristics from males with following genotypes |
|||
|---|---|---|---|---|
| WT | shSLX1 | shSLX2 | shSLX1,2 | |
| Number of litters per malea | 6 | 1.83* | 3.33* | 0* |
| Total number of offspring per malea | 70.17 | 9.83* | 18.83* | 0* |
| Average litter sizea | 11.8 | 3.92* | 4.26* | 0* |
| Oocytes fertilized in vitro (%) | 54.5 | 4.6** | 1.0** | N/D |
| Testis weight (mean value in mg ± SEs) | 102.3 ± 2.5 | 99.8 ± 3.0 | 104.1 ± 3.1 | 90.1* ± 3.4 |
| Total motile sperm (%) | 46.3 | 25.4** | 22.9** | N/D |
| Sperm number/cauda (mean value × 10−6 ± SEs) | ||||
| MF1 | 13.6 ± 0.8 | 2.0* ± 0.8 | 6.6* ± 1.2 | 0.2* ± 0.1 |
| B6D2F1 | 7.0 ± 1.8 | 5.9 ± 1.9 | 5.2 ± 2.0 | N/D |
a Six males of each genotype were mated with MF1 females over a period of six and a half months.
* Significantly different from WT (p < 0.05; t test).
** Significantly different from WT (p < 0.05; ANOVA).
Slx/Slxl1 Deficiency Leads to Abnormal Spermatid Elongation
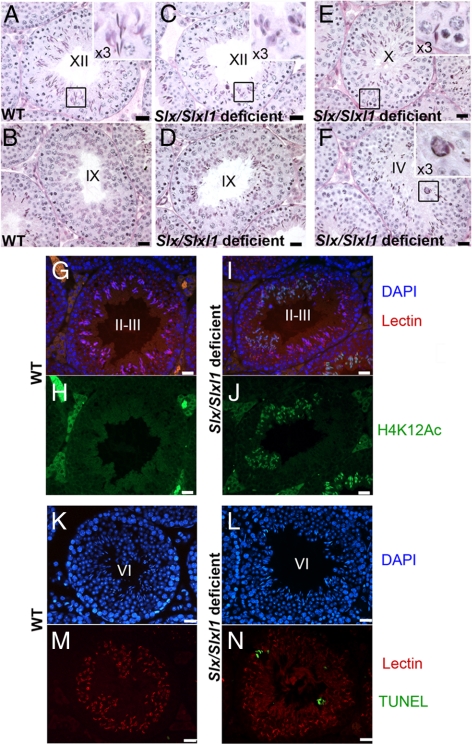
We sought to determine the origin of the fertility defect observed in shSLX males. Histology of all types of Slx/Slxl1-deficient males did not differ from WT until post-meiotic stages. This is in agreement with the observation that Slx and Slxl1 expression is restricted to the postmeiotic stage of male germ cells (i.e., spermatids) (Reynard et al., 2007). There was no detectable delay of round spermatid development; however, Slx/Slxl1-deficient testes had a defect in spermatid elongation (from stage IX and onwards). For instance, in PAS-stained stage XII tubules of Slx/Slxl1-deficient mice, elongating spermatids are visibly less developed (with the presence of step10 spermatids instead of step12) than in controls (Figure 3, A and C). Histone H4 hyperacetylation is a good marker of the spermatid elongation stages, as it occurs in early elongating spermatids before histone replacement by transition proteins (Hazzouri et al., 2000; Gaucher et al., 2009). Using antibodies directed against acetylated lysine 8 and 12 of histone H4 (H4K8Ac and H4K12Ac) as markers of stage 9–12 elongating spermatids, we observed a major delay in spermatid elongation (Figure 3, G–J): H4K12Ac positive spermatids were retained in ∼70% of stage I to IV tubules in shSLX1 testes, 11% in shSLX2 and 93% in shSLX1,2, versus 4% in WT (shSLX1 and shSLX1,2 values significantly different from WT; ANOVA, p < 0.0001).
Figure 3.
Impaired spermiogenesis and increased apoptosis of elongating spermatids in Slx/Slxl1-deficient mice. (A–D) In PAS-stained stage XII testis tubules of Slx/Slxl1-deficient (shSLX1 transgenic) mice, elongating spermatids are less developed than in controls (A). Typically, elongating spermatids with morphology of step 10 spermatids were observed in step XII tubules (C). In PAS-stained stage IX testis tubules of Slx/Slxl1-deficient mice (D) mature sperm are retained in stage IX tubules, whereas in WT testes (B) mature sperm have already been released into the lumen. (E) PAS-stained stage X Slx/Slxl1-deficient testis tubule. Several mature sperm are retained near the basal lamina (enlargement, right top corner). (F) PAS-stained stage IV Slx/Slxl1-deficient testis tubule. A typical group of dying sperm organized in circle can be observed (enlargement, right top corner). (G–J) stage II-III testis tubules of Slx/Slxl1-deficient mice present a great number of delayed elongating spermatids (marked with H4K12Ac antibody in green). Same stage tubules of control mice do not show any remaining H4K12Ac signal in spermatids. (K–M) Detection of apoptosis (TUNEL) in WT and Slx/Slxl1-deficient testes. Representative pictures show the presence of several apoptotic elongating spermatids (in green) in a stage VI testis tubule of a Slx/Slxl1-deficient male. No or very few apoptotic cells can be observed in a control testis (shSLX nontransgenic sibling). DAPI (in blue) was used to stain nuclei, and Lectin (in red) was used to stage the seminiferous tubules. Scale bar, 20 μm.
Slx/Slxl1 Deficiency Leads to Delay in Sperm Shedding and Reduction of Sperm Count
We then observed that mature (step 16) sperm were retained in most stage IX to XI tubules, instead of being normally shed at stage VIII (Figure 3, B and D). The occurrence of sperm shedding delay was significantly higher in Slx/Slxl1-deficient males compared with WT (95 and 77% of stage IX to XI tubules with retained sperm in shSLX1 and shSLX2 males respectively vs. 12% in WT; ANOVA p < 0.0005). Interestingly, a delay in sperm shedding has previously been observed in Sly-deficient mice and MSYq-deficient mice (Touré et al., 2004; Cocquet et al., 2009), but in these cases the unshed sperm were located adjacent to, and were eventually released into, the lumen; in Slx/Slxl1-deficient tubules, unshed sperm were often located near the basal lamina or form a group of dying cells (Figure 3, E and F). Apoptotic (TUNEL-positive) elongating spermatids were found in all types of tubule stages of the seminiferous epithelium cycle (Figure 3, K–N, and Supplementary Figure 4). The proportion of testicular tubules containing TUNEL-positive elongating/condensing spermatids was determined and found to be significantly higher in Slx/Slxl1-deficient testes compared with controls (∼70% in shSLX1 and shSLX1,2 testes, ∼25% in shSLX2 testes vs. ∼3% in WT; ANOVA, p < 0.0005). The substantial loss of these spermatids was associated with reduced sperm counts in cauda epididymis from Slx/Slxl1-deficient males (Table 1).
Slx/Slxl1-Deficient Sperm Are Abnormally Formed and Exhibit Reduced Motility
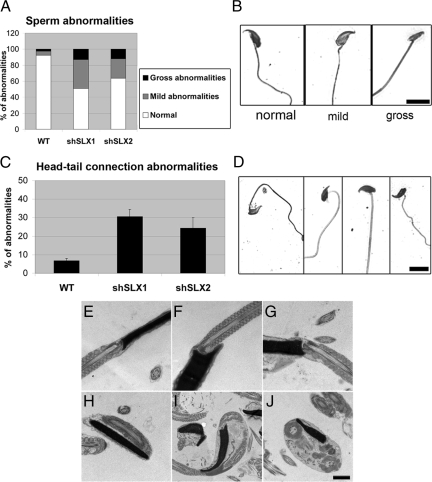
Analysis of Slx/Slxl1-deficient epididymis revealed a significant increase in the incidence of abnormal sperm with ∼49% of spermhead abnormalities in shSLX1 and ∼36% in shSLX2 (vs. ∼8% in WT; ANOVA, p < 0.0005) (Figure 4, A and B; Supplementary Figure 3). In addition to spermhead malformations, shSLX1 and shSLX2 spermatozoa displayed abnormal head to tail connections (values significantly different from WT; ANOVA, p < 0.005) (Figure 4, C and D). Using electron microscopy, we observed epididymal sperm with partially detached heads or abnormally-oriented tails: either aligned parallel to the head, curved or looped (Figure 4, E–J). Abnormal connection between the spermhead and its tail could have an effect on sperm motility, and, indeed, sperm motility was impaired as shown by the decrease in the proportion of motile sperm (Table 1).
Figure 4.
Sperm abnormalities in Slx/Slxl1-deficient mice. (A) Bar graph representing the percentage of sperm with sperm head abnormalities in Slx/Slxl1-deficient mice and control. (B) Examples of sperm head abnormalities observed in Slx/Slxl1-deficient mice (silver-stained epididymal sperm). Scale bar, 10 μm. (C) Bar graph representing the percentage of sperm with abnormal head-tail connections in epididymal sperm from Slx/Slxl1-deficient mice and control. (D) Examples of head-tail connection anomalies in Slx/Slxl1-deficient spermatozoa. Scale bar, 10 μm. (E–J) Electron microscopy pictures of WT (E and F) and Slx/Slxl1-deficient epididymal sperm with partial detachment of the tail (G), parallel alignment of the head and tail (H), curved tail (I), or looped tail (J). Pictures were taken using the same parameters. Scale bar, 1 μm.
Transcriptome Analyses Show that Slx/Slxl1 Deficiency Alters Metabolic Processes Occurring in the Spermatids
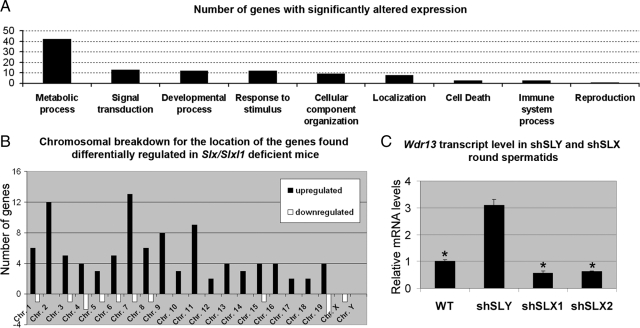
To better understand the consequences of Slx/Slxl1 deficiency, we performed microarray analyses on purified round spermatids (the site of Slx and Slxl1 expression) from shSLX1 mice and controls. First, this analysis confirmed the knockdown of Slx and Slxl1 transcripts. Second, in Slx/Slxl1-deficient round spermatids, 101 genes were found to be significantly up-regulated compared with controls. Of these 101 genes, 77 are annotated and, of these, 54% (42/77) encode proteins involved in various metabolic processes, among which are the serine proteases belonging to Kallikrein 1 family (klk1, klk1b26, klk1b5, klk1b9, klk1b4, klk1b27) and Ela2a; several enzymes implicated in lipid metabolism (Acsl5, Cyp2a12, St8sia5, Agpat2, Ppap2b) and energy metabolism (Nqo2, Atp6v1h, 4933437F05Rik); and proteins of the ubiquitin pathway (Usp3, Ube2g2). Several genes coding for proteins of the cytoskeleton and the extracellular matrix (classified in category ‘cellular component organization’) were also affected by Slx/Slxl1 deficiency such as Tmod4 (Tropomodulin 4), Myl2 (myosin light chain 2), Col20a1 (collagen, type XX, alpha 1), Pstpip1 (proline-serine-threonine phosphatase interacting protein 1), which is a member of the actin-associated protein family (Spencer et al., 1997), and LOC100045019 (similar to Tubulin, gamma 2) (Figure 5A). Interestingly, several genes involved in positive or negative regulation of apoptosis were also found affected (such as Hipk2, Tnfsf13b, Jund1). Complete results can be found in Supplementary Figure 5.
Figure 5.
Transcriptome analysis of Slx/Slxl1-deficient round spermatids versus WT round spermatids. (A) Bar graph representing the number of genes found differentially expressed in round spermatids from Slx/Slxl1-deficient males compared with those of WT males by microarray. Differentially expressed genes are shown by classification according to their likely biological function in Onto-Express. (B) Chromosome location of the genes found up- or down-regulated in case of Slx/Slxl1 deficiency. Note the enrichment of up-regulated genes on chromosome 7. This is due to the presence of the Klk1 genes that are affected by the loss of Slx/Slxl1. (C) Quantification of the transcript level of the X-encoded Wdr13 gene in purified round spermatids of shSLY, SLX1, shSLX2, and WT males by real-time RT-PCR. Values were normalized to β-actin. One star indicates significant difference from WT value (p < 0.05; t test). Wdr13 transcript level is differentially affected by Slx/Slxl1 or Sly deficiency.
Slx/Slxl1 Deficiency Does Not Lead to Global Perturbation of Sex Chromosome Gene Expression
Slx and Slxl1 genes are related to the Y-encoded Sly multicopy gene. SLY protein has been shown to repress sex chromosome genes in spermatids and, as a result, X- and Y-encoded transcripts are up-regulated in the case of Sly deficiency (Cocquet et al., 2009). Here, in round spermatids from Slx/Slxl1-deficient males, none of the genes found up-regulated were located on the sex chromosomes (Figure 5B). Only one X-encoded transcript (apart from Slx and Slxl1 transcripts and cross-hybridization with an ‘Sly’ probe), Wdr13, was found significantly down-regulated in Slx/Slxl1-deficient round spermatids. Real-time PCR analysis on Slx/Slxl1- and Sly-deficient round spermatid fractions confirmed that Wdr13 is differentially regulated by SLY and SLX/SLXL1 (Figure 5C).
DISCUSSION
Using an siRNA approach to produce mice with markedly reduced transcript levels for the multicopy genes Slx and Slxl1, we have demonstrated that the Slx gene family is important for normal mouse sperm development. Slx/Slxl1 deficiency considerably delays spermatid elongation and sperm release in the lumen. These perturbations of sperm differentiation are associated with the elimination of many elongating/condensing spermatids via apoptosis, with a consequent reduced sperm count. The remaining spermatozoa are abnormally shaped and have decreased motility and fertilizing abilities. All these factors are likely to contribute together to the near sterility observed in Slx/Slxl1-deficient males.
Microarray analyses revealed that Slx/Slxl1 deficiency does not lead to global up-regulation of X- and Y-linked genes after meiosis; this is in striking contrast with what we had observed in the case of Sly deficiency, where the vast majority of the genes found up-regulated were encoded either by the X or the Y chromosome (Cocquet et al., 2009). This difference may stem from the fact that i) SLX/SLXL1 proteins are restricted to the cytoplasm while SLY is found in both the nucleus and cytoplasm of spermatids (Reynard et al., 2007; Cocquet et al., 2009); ii) SLX/SLXL1 only share ∼43% overall homology with SLY (while SLX and SLXL1 share ∼66% identity).
Only a few genes (mainly kallikrein 1-related genes) were found up-regulated in both models (Slx/Slxl1- and Sly-deficient males). Kallikrein-related peptidases are serine proteases with a variety of physiological roles; they have notably been reported to be involved in semen liquefaction and consequently in sperm motility but through their production by the prostate (Emami et al., 2009). Their biological role in the testis remains to be defined, but they could act (as they do in other contexts) as signaling molecules (for review, see Sotiropoulou et al., 2009). Interestingly, kallikrein 1–related transcripts are also found up-regulated in mouse models with deletions of the Y long arm (which represents a non siRNA model of Sly deficiency) (Cocquet et al., 2009).
Among the few genes that were found differentially affected by Slx/Slxl1 and Sly deficiency (i.e., up-regulated in one model and down-regulated in the other) are Fndc3 and Wdr13. Fndc3 has been implicated in spermatid–Sertoli cell adhesion (Obholz et al., 2006). Wdr13 is predominantly expressed in male germ cells, but its function is yet unknown (Singh et al., 2003; Suresh et al., 2005). Future work (such as gain or loss of function approaches) will be required to determine the contribution of these candidate genes to the spermiogenic phenotype(s) observed in Slx/Slxl1- and Sly-deficient males.
In the case of Sly deficiency, ∼100 X and Y-linked spermiogenic genes are up-regulated, among which are Slx and Slxl1 (Cocquet et al., 2009). Both Slx/Slxl1-deficient males and Sly-deficient males present sperm differentiation defects, though some of these defects appear to be specific to Slx/Slxl1 deficient males, such as spermatid elongation delay and abnormal head-tail connections. The increase of Slx and Slxl1 transcript levels are likely to contribute to the spermiogenic defects observed in Sly-deficient males. Slx and Sly genes have been hypothesized to be involved in an intragenomic conflict occurring between X-bearing and Y-bearing gametes (Ellis et al., 2005). The production of double transgenic mice deficient for both Slx/Slxl1 and Sly would address this question.
The structural changes, such as acrosome and flagellum formation, that lead to the transformation of a round cell into a highly specialized sperm cell start soon after meiosis in the round spermatid (Russell et al., 1990). Many of the genes found altered in our microarray analysis of Slx/Slxl1-deficient round spermatids encode enzymes associated with specific cellular organelles and involved in various metabolic processes (such as energy production, ubiquitin-mediated degradation, etc.). Several proteins of the cytoskeleton were also affected. These results suggest that a ‘cytoplasmic’ defect underlies the above-mentioned sperm differentiation abnormalities. The mechanism by which SLX/SLXL1 proteins regulate these processes remains mysterious as no functional domain (apart from the Cor1 domain) is known and previous attempts to identify potential protein partners have failed (L Reynard and P Burgoyne, unpublished data). Because SLX/SLXL1 proteins are restricted to the spermatid cytoplasm, it is unlikely that the genes found altered in our microarray analysis are direct transcriptional targets of SLX/SLXL1. One attractive hypothesis is that a yet unidentified partner of SLX/SLXL1 is at the basis of the transcriptional changes that are necessary for normal spermatid elongation.
Finally, before the elimination of elongating spermatids by apoptosis, several ‘cell death’ genes were found altered in round spermatids deficient for Slx/Slxl1. Hipk2, a potentiator of p53-mediated transcriptional activation of proapoptotic genes (Hofmann et al., 2002), was up-regulated while the proto-oncogene Jund1 was down-regulated. SLX/SLXL1 may consequently be important for the balance between cell proliferation/differentiation and cell death during sperm development. This relates to a recent study in which expression in tumor cells of SYCP3, SLX, or the related protein XLR was shown to induce activation of AKT and up-regulation of antiapoptotic proteins (Kang et al., 2010). Ectopic expression of SYCP3 was found in some human cancers (such as cervical cancer and acute leukemia) (Niemeyer et al., 2003; Kang et al., 2010). AKT proteins are known to have a crucial role in cell proliferation, survival, and differentiation in many tissues (Manning and Cantley, 2007). In the testis, the phosphatidylinositol 3-kinase (Pi3k)/AKT pathway has been shown to be required for spermatogonial stem cell survival and proliferation (Blume-Jensen et al., 2000; Chen et al., 2001). To our knowledge, the impact of the Pi3k/AKT pathway on spermatid development and differentiation remains unclear, but AKT1 protein has been described to be present and activated (phosphorylated) at different stages of the rat germ cell development, including spermatids (Hixon and Boekelheide, 2003). Interestingly, several genes involved in the AKT regulation cascade were found up-regulated in round spermatids deficient for Slx/Slxl1: Eif4ebp1 is a gene of the Pi3K signaling pathway and acts downstream of AKT/PKB; while Tnfsf13b (aka Baff) and Aatf encode two positive regulators of AKT (Otipoby et al., 2008; Ishigaki et al., 2010). The mechanism by which SLX/SLXL1 acts upon the Pi3k/AKT pathway in postmeiotic germ cells or in case of ectopic activation remains to be determined.
In conclusion, our work provides insight into the function of multicopy X-linked genes. With the transgenic-delivery of siRNA we have targeted both Slx and Slxl1 transcripts and characterized their biological role in spermatogenesis. To the best of our knowledge, our study is the first to describe the targeted disruption of the function of X chromosome-encoded multicopy genes in mammals. A similar strategy could be applied to the study of other multicopy genes such as the numerous ‘cancer/testis antigens’ found on the X chromosome for which a role in germ cells remains largely unknown (for review, see Simpson et al.).
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Wood for pronuclear injections; R. Mahmood and E. Grigorieva for histology; the Department of Pathology Centre for Microarray Resources for array hybridization; N. Vernet for her expertise in testicular histology and helpful discussions; and S. Mahadevaiah and J.M.A. Turner for helpful discussions. This work was supported by the Medical Research Council (MRC, U117532009 to P.S.B.), U.S. National Institutes of Health grants HD058059 and RR024206-Project 2 (to M.A.W.), and Biotechnology and Biological Sciences Research Council grant BB/F007434/1 (to N.A.A. and P.E.). J.C. was a recipient of a MRC Career Development Fellowship.
Abbreviation used:
- shRNAs
short hairpin RNAs.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0601) on August 25, 2010.
REFERENCES
- Blume-Jensen P., Jiang G., Hyman R., Lee K. F., O'Gorman S., Hunter T. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nature Genet. 2000;24:157–162. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- Burgoyne P. S., Mitchell M. J. The roles of mouse Y chromosome genes in spermatogenesis. In: Lau Y.-F.C., Chan W.Y., editors. Y Chromosome and Male Germ Cell Biology. Hackensack, NJ: World Scientific Publishers; 2007. pp. 27–45. [Google Scholar]
- Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J. H., Whiteley M., Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Cocquet J., Ellis P. J., Yamauchi Y., Mahadevaiah S. K., Affara N. A., Ward M. A., Burgoyne P. S. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P. J., Affara N. A. Spermatogenesis and sex chromosome gene content: an evolutionary perspective. Hum. Fertil. (Camb) 2006;9:1–7. doi: 10.1080/14647270500230114. [DOI] [PubMed] [Google Scholar]
- Ellis P.J.I., Clemente E. J., Ball P., Toure A., Ferguson L., Turner J.M.A., Loveland K. L., Affara N. A., Burgoyne P. S. Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Hum. Mol. Genet. 2005;14:2705–2715. doi: 10.1093/hmg/ddi304. [DOI] [PubMed] [Google Scholar]
- Emami N., Scorilas A., Soosaipillai A., Earle T., Mullen B., Diamandis E. P. Association between kallikrein-related peptidases (KLKs) and macroscopic indicators of semen analysis: their relation to sperm motility. Biol. Chem. 2009;390:921–929. doi: 10.1515/BC.2009.094. [DOI] [PubMed] [Google Scholar]
- Gaucher J., Reynoird N., Montellier E., Boussouar F., Rousseaux S., Khochbin S. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J. 2009;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Harper S. Q., Staber P. D., He X., Eliason S. L., Martins I. H., Mao Q., Yang L., Kotin R. M., Paulson H. L., Davidson B. L. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M., Pivot-Pajot C., Faure A. K., Usson Y., Pelletier R., Sele B., Khochbin S., Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur. J. Cell Biol. 2000;79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- Hixon M. L., Boekelheide K. Expression and localization of total Akt1 and phosphorylated Akt1 in the rat seminiferous epithelium. J. Androl. 2003;24:891–898. doi: 10.1002/j.1939-4640.2003.tb03141.x. [DOI] [PubMed] [Google Scholar]
- Hofmann T. G., Moller A., Sirma H., Zentgraf H., Taya Y., Droge W., Will H., Schmitz M. L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell. Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Ishigaki S., Fonseca S. G., Oslowski C. M., Jurczyk A., Shearstone J. R., Zhu L. J., Permutt M. A., Greiner D. L., Bortell R., Urano F. AATF mediates an antiapoptotic effect of the unfolded protein response through transcriptional regulation of AKT1. Cell Death Differ. 2010;17:774–786. doi: 10.1038/cdd.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. H., Noh K. H., Kim J. H., Bae H. C., Lin K. Y., Monie A., Pai S. I., Hung C. F., Wu T. C., Kim T. W. Ectopic expression of X-linked lymphocyte-regulated protein pM1 renders tumor cells resistant to antitumor immunity. Cancer Res. 2010;70:3062–3070. doi: 10.1158/0008-5472.CAN-09-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B. D., Cantley L. C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M. L. Separation of spermatogenic cells and nuclei from rodent testes. In: Prescott D.M., editor. Methods in Cell Biology. New York, San Francisco, London: Academic Press; 1977. pp. 15–54. [DOI] [PubMed] [Google Scholar]
- Mueller J. L., Mahadevaiah S. K., Park P. J., Warburton P. E., Page D. C., Turner J. M. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer P., Tureci O., Eberle T., Graf N., Pfreundschuh M., Sahin U. Expression of serologically identified tumor antigens in acute leukemias. Leukemia Res. 2003;27:655–660. doi: 10.1016/s0145-2126(02)00230-8. [DOI] [PubMed] [Google Scholar]
- Obholz K. L., Akopyan A., Waymire K. G., MacGregor G. R. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev. Biol. 2006;298:498–513. doi: 10.1016/j.ydbio.2006.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otipoby K. L., Sasaki Y., Schmidt-Supprian M., Patke A., Gareus R., Pasparakis M., Tarakhovsky A., Rajewsky K. BAFF activates Akt and Erk through BAFF-R in an IKK1-dependent manner in primary mouse B cells. Proc. Natl. Acad. Sci. USA. 2008;105:12435–12438. doi: 10.1073/pnas.0805460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Hurst L. D. Sex and conflict. Science. 1998;281:2003–2008. doi: 10.1126/science.281.5385.2003. [DOI] [PubMed] [Google Scholar]
- Reynard L. N., Turner J. M., Cocquet J., Mahadevaiah S. K., Toure A., Hoog C., Burgoyne P. S. Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR. Biol. Reprod. 2007;77:329–335. doi: 10.1095/biolreprod.107.061101. [DOI] [PubMed] [Google Scholar]
- Russell L. D., Hikim A.P.S., Ettlin R. A., Clegg E. D. Cache River Press; 1990. Histological and Histopathological Evaluation of the Testis. [Google Scholar]
- Scavetta R. J., Tautz D. Copy number changes of CNV regions in intersubspecific crosses of the house mouse. Mol. Biol. Evol. 2010;27:1845–1856. doi: 10.1093/molbev/msq064. [DOI] [PubMed] [Google Scholar]
- Simpson A. J., Caballero O. L., Jungbluth A., Chen Y. T., Old L. J. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Singh B. N., Suresh A., UmaPrasad G., Subramanian S., Sultana M., Goel S., Kumar S., Singh L. A highly conserved human gene encoding a novel member of WD-repeat family of proteins (WDR13) Genomics. 2003;81:315–328. doi: 10.1016/s0888-7543(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P. J., Cordum H. S., Hillier L., Brown L. G., Repping S., Pyntikova T., Ali J., Bieri T., Chinwalla A., Delehaunty A., Delehaunty K., Du H., Fewell G., Fulton L., Fulton R., Graves T., Hou S. F., Latrielle P., Leonard S., Mardis E., Maupin R., McPherson J., Miner T., Nash W., Nguyen C., Ozersky P., Pepin K., Rock S., Rohlfing T., Scott K., Schultz B., Strong C., Tin-Wollam A., Yang S. P., Waterston R. H., Wilson R. K., Rozen S., Page D. C. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G., Pampalakis G., Diamandis E. P. Functional roles of human kallikrein-related peptidases. J. Biol. Chem. 2009;284:32989–32994. doi: 10.1074/jbc.R109.027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S., Dowbenko D., Cheng J., Li W., Brush J., Utzig S., Simanis V., Lasky L. A. PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J. Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffs K., Tournaye H., Liebaers I., Lissens W. Male infertility and the involvement of the X chromosome. Hum. Reprod. Update. 2009;15:623–637. doi: 10.1093/humupd/dmp023. [DOI] [PubMed] [Google Scholar]
- Suresh A., Shah V., Rani D. S., Singh B. N., Prasad G. U., Subramanian S., Kumar S., Singh L. A mouse gene encoding a novel member of the WD family of proteins is highly conserved and predominantly expressed in the testis (Wdr13) Mol. Reprod. Dev. 2005;72:299–310. doi: 10.1002/mrd.20362. [DOI] [PubMed] [Google Scholar]
- Toure A., Clemente E. J., Ellis P., Mahadevaiah S. K., Ojarikre O. A., Ball P. A., Reynard L., Loveland K. L., Burgoyne P. S., Affara N. A. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 2005;6:R102. doi: 10.1186/gb-2005-6-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A., Szot M., Mahadevaiah S. K., Rattigan A., Ojarikre O. A., Burgoyne P. S. A new deletion of the mouse Y chromosome long arm associated with loss of Ssty expression, abnormal sperm development and sterility. Genetics. 2004;166:901–912. doi: 10.1534/genetics.166.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Riel J. M., Stoytcheva Z., Burgoyne P. S., Ward M. A. Deficiency in mouse Y chromosome long arm gene complement is associated with sperm DNA damage. Genome Biol. 2010;11:R66. doi: 10.1186/gb-2010-11-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.