Insulin increases GLUT4 at the muscle cell surface, and this process requires actin remodeling. We show that a dynamic cycle of actin polymerization and severing is induced by insulin, governed by Arp2/3 and dephosphorylation of cofilin, respectively. The cycle is self-perpetuating and is essential for GLUT4 translocation.
Abstract
GLUT4 vesicles are actively recruited to the muscle cell surface upon insulin stimulation. Key to this process is Rac-dependent reorganization of filamentous actin beneath the plasma membrane, but the underlying molecular mechanisms have yet to be elucidated. Using L6 rat skeletal myoblasts stably expressing myc-tagged GLUT4, we found that Arp2/3, acting downstream of Rac GTPase, is responsible for the cortical actin polymerization evoked by insulin. siRNA-mediated silencing of either Arp3 or p34 subunits of the Arp2/3 complex abrogated actin remodeling and impaired GLUT4 translocation. Insulin also led to dephosphorylation of the actin-severing protein cofilin on Ser-3, mediated by the phosphatase slingshot. Cofilin dephosphorylation was prevented by strategies depolymerizing remodeled actin (latrunculin B or p34 silencing), suggesting that accumulation of polymerized actin drives severing to enact a dynamic actin cycling. Cofilin knockdown via siRNA caused overwhelming actin polymerization that subsequently inhibited GLUT4 translocation. This inhibition was relieved by reexpressing Xenopus wild-type cofilin-GFP but not the S3E-cofilin-GFP mutant that emulates permanent phosphorylation. Transferrin recycling was not affected by depleting Arp2/3 or cofilin. These results suggest that cofilin dephosphorylation is required for GLUT4 translocation. We propose that Arp2/3 and cofilin coordinate a dynamic cycle of actin branching and severing at the cell cortex, essential for insulin-mediated GLUT4 translocation in muscle cells.
INTRODUCTION
A major function of insulin is to regulate glucose uptake by muscle and fat tissues. This is achieved through a rapid and dynamic gain in glucose transporter-4 (GLUT4) at the cell surface (Huang and Czech, 2007; Larance et al., 2008; Zaid et al., 2008). Notably, this process becomes defective in insulin resistance states and type 2 diabetes (Klip et al., 1990; Zierath et al., 1996; Garvey et al., 1998; Mora and Pessin, 2002). To date, defects in insulin signaling and GLUT4 traffic per se have been invoked to underlie such defects (Krook et al., 2004; Patel et al., 2006).
Skeletal muscle is the primary site of insulin-dependent glucose disposal in vivo, and muscle cells in culture are useful to scrutinize principles of this response. L6 myoblasts, like further differentiated myotubes and skeletal muscle, mount robust responses of insulin signaling via insulin receptors, the insulin receptor substrate-1 (IRS-1), phosphatidylinositol 3-kinase (PI3K), and Akt (Ruderman et al., 1990; Tsakiridis et al., 1995; Thong et al., 2007). In muscle cells, signaling bifurcates downstream of PI3K into two independent arms characterized by phosphorylation of Akt (Wang et al., 1999) and GTP activation of the small GTPase Rac leading to actin remodeling (Khayat et al., 2000; JeBailey et al., 2004). The two pathways are independent of one another because neither Akt dominant-negative mutants (Wang et al., 1999) nor the Akt inhibitor Akti (A. Koshkina and A. Klip, unpublished data) prevent insulin-induced Rac activation or its consequent actin remodeling, and disruption of Rac via small inhibitory RNA (siRNA) fails to reduce Akt phosphorylation by insulin (JeBailey et al., 2007). Both signaling arms are required to elicit proper insulin-mediated GLUT4 translocation as perturbation of either one significantly reduces the GLUT4 response to insulin in muscle cells (Wang et al., 1999; JeBailey et al., 2007; Ishikura and Klip, 2008; Zaid et al., 2008).
Although much emphasis has been placed on the effectors downstream of Akt such as Akt substrate of 160 (AS160) and Rab GTPases (Sano et al., 2003; Miinea et al., 2005; Gonzalez and McGraw, 2006; Ishikura et al., 2007; Sano et al., 2007; Thong et al., 2007; Ishikura and Klip, 2008), the function of Rac remains less explored. Rac belongs to the small Rho GTPase family whose activity is regulated by GTP loading (Bernards and Settleman, 2004; Rossman et al., 2005). Insulin promotes GTP loading of Rac within the first 1–5 min of stimulation (JeBailey et al., 2004; Ishikura et al., 2008). Once activated, Rac induces the reorganization of cortical actin filaments (Khayat et al., 2000; JeBailey et al., 2007). This actin remodeling is a critical component in insulin-stimulated GLUT4 translocation because overexpression of a dominant negative Rac mutant (Khayat et al., 2000) or siRNA-mediated Rac knockdown (JeBailey et al., 2007) not only prevent actin remodeling but also markedly diminish the insulin-mediated recruitment of GLUT4 to the surface. A similar reduction in insulin response is observed upon preventing actin remodeling with inhibitors of actin polymerization such as latrunculin B (LB) and cytochalasin D (Tsakiridis et al., 1994; Khayat et al., 2000) or by precluding actin depolymerization with jasplakinolide (Tong et al., 2001). Altogether, these findings reveal the importance of peripheral actin reorganization in insulin-dependent GLUT4 translocation in muscle cells. However, the precise regulation of this dynamic actin change and the elements acting downstream of Rac are undefined. Moreover, a model that incorporates both actin polymerization and depolymerization, as required for dynamic remodeling, has not been proposed.
Here we test the hypothesis that insulin produces a dynamic regulation of actin remodeling involving cycles of branching and depolymerization. Using a well-established muscle cell model of L6GLUT4myc myoblasts displaying insulin-induced GLUT4myc exocytosis, we identify the Arp2/3 complex as a downstream effector of Rac that governs cortical actin polymerization and also identify cofilin as a regulator promoting actin filament depolymerization. Cofilin dephosphorylation requires prior buildup of polymerized actin driven by Arp2/3. Therefore, an integrated mode for active actin cycling is proposed that enables insulin-mediated GLUT4 translocation/insertion into the muscle cell membrane.
MATERIALS AND METHODS
Reagents, siRNA, and Constructs
Polyclonal anti-myc antibody (A-14) and anti-p34-ARC were from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal anti-phosphorylated cofilin-1, anti-Akt, anti-phosphorylated Akt (Ser473), and anti-phosphorylated LIMK1 antibodies were from Cell Signaling Technology (Beverly, MA). Polyclonal anti-Arp3 was from BD Biosciences (San Jose, CA). Polyclonal anti-slingshot-1 (anti-SSH1) was from ECM Biosciences (Versailles, KY). Affinity-purified antibodies against P-AC and cofilin-1 were as described (Meberg et al., 1998; Shaw et al., 2004). Monoclonal anti-ACTN1 (mouse IgM isotype, clone BM-75.2), anti-β-actin, polyclonal anti-LIMK1, LB, and DMSO (high-pressure liquid chromatography grade) were from Sigma-Aldrich (St. Louis, MO). Indocarbocyanine (Cy3) or horseradish peroxidase (HRP)-conjugated goat-anti-mouse, goat-anti-rabbit, and rabbit-anti-goat IgG antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Rhodamine-bound phalloidin, Alexa 488–conjugated goat anti-mouse and anti-rabbit IgG antibodies were from Invitrogen (Carlsbad, CA). Human insulin was purchased form Eli Lilly (Indianapolis, IN). siRNAs targeted against p34 (AAGGAACTTCAGGCACACGGA), Arp3 (GAAAGGCGTGGATGACCTATT; Korobova and Svitkina, 2008), cofilin-1 (AAGGTGTTCAATGACATGAAA), LIMK1 (AAGGAATGTGCCGCTGGCAGATT), slingshot-1 (AAGAACTGAGCGTCTCATTAA), and nonrelated control (AATAAGGCTATGAAGAGATAC) were purchased from Qiagen (Chatsworth, CA) and are here named, respectively, sip34, siArp3, siCofilin, siLIMK, and siSSH1. Green fluorescent protein (GFP)-tagged Dictyostelium discoideum Arp3 was kindly provided by Dr. Sergio Grinstein (University of Toronto, Toronto, ON, Canada). GFP-tagged wild-type (WT) and S3E Xenopus cofilin were generated in the laboratory of Dr. J. Bamburg (Colorado State University; Ashworth et al., 2003).
Cell Culture and Transfection
Rat L6 muscle cells expressing GLUT4 with an exofacial myc epitope (L6 GLUT4myc) were cultured as described previously (Ueyama et al., 1999). GLUT4 transfected into these cells segregates away from the recycling pathway and responds to insulin to a similar extent in myoblasts and myotubes (Ueyama et al., 1999; Niu et al., 2003). Myoblasts, however, offer the advantage of being easier to transfect and to scrutinize by confocal fluorescence microscopy. Transfection of siRNAs used the calcium phosphate-based CellPhect Transfection Kit (GE Healthcare Bio-Sciences, Piscataway, NJ). siRNA-calcium phosphate precipitates, 200 nM, were removed 12–16 h after addition, and cells were maintained for 72 h until experimentation. Transfection of cDNA used Lipofectamine 2000 (Invitrogen). The cDNA-Lipofectamine mixture was removed after 4–6-h incubation, and cells were allowed to recover for 24 h before experimentation.
Cell-Surface GLUT4myc detection by Immunofluorescence Microscopy
Immunofluorescent detection of surface GLUT4myc in adhered L6 myoblasts was performed as previously described (Torok et al., 2004). After 3-h serum starvation, cells were stimulated with and without 100 nM insulin for 10 min at 37°C. Cells were quickly washed twice with cold PBS, fixed with 3% (vol/vol) paraformaldehyde, and blocked with 5% (vol/vol) milk. Surface GLUT4myc was stained by incubating anti-myc primary antibody followed by Cy2- or Cy3-coupled secondary antibody. For costaining of surface GLUT4myc and intracellular filamentous (F)-actin, surface GLUT4myc was labeled first before membrane permeabilization for subsequent F-actin staining. Fluorescent images were obtained with a Zeiss LSM 510 laser-scanning confocal microscope (Thornwood, NY). Whole cells were scanned along the z-axis, and a single composite image (collapsed xy projection) of the optical cuts per cell was generated using LSM5 Image software. Pixel intensity of single cells (>30 cells per condition) was quantified by ImageJ software (http://rsb.info.nih.gov/ij/). Surface GLUT4myc was also detected by immunofluorescence in intact rounded-up myoblasts prepared as described previously (Randhawa et al., 2008; Talior-Volodarsky et al., 2008).
Detection and Quantification of Actin Remodeling/Polymerized Dorsal Actin
After treatment with or without insulin for 10 min at 37°C, adhered or rounded-up L6 myoblasts were fixed, permeabilized with 0.1% Triton X-100 in PBS for 3 min, blocked with 5% milk, and stained with rhodamine-phalloidin for F-actin. Cells were imaged by Zeiss LSM 510 laser scanning confocal microscope. Acquisition parameters were adjusted to minimize signal saturation. To quantify actin remodeling, the F-actin pixel intensity from compiled dorsal optical slices of basal and insulin-stimulated cells were analyzed with ImageJ software (>30 cells per condition). The optical slices quantified began from the outermost confocal fluorescent signal detected and continued toward the interior of the cell but skipped the last 2–3 optical slices dominated by actin stress fibers. By eliminating from the quantification the focal planes enriched in parallel arrays of stress fibers, which are still devoid of crisscrossed arrays of branched actin (vastly present in the top half of the cells toward the dorsal surface), we quantify cortical remodeling. The method may potentially underestimate this response, but not magnify it.
Cell Lysates and Immunoblotting
After treatments, cells were washed quickly with cold phosphate-buffered saline (PBS) and lysed with 1% Triton X-100 in PBS. Lysates were passed through a 29-gauge syringe needle five times, and supernatants were collected by a 10-min spin at 12,000 × g. Equal protein samples were resolved by 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Bio-Rad, Richmond, CA), and immunoblotted with respective antibodies. Primary antibodies were detected with the appropriate HRP-conjugated species-specific IgG antibodies. Detection was completed with Western Lightning Chemiluminescence Reagent Plus and HyBlot CL autoradiography film from Denville Scientific (Denville, NJ).
125I-Transferrin Recycling
The recycling of transferrin (Tf) was measured essentially as previously described (Yan et al., 2005). After siRNA treatment and 3-h serum starvation, myoblasts were labeled with 125I-Tf (1 μg/ml) for 30 min at 37°C. Cells were placed on ice, washed once with cold medium (α-MEM, 1% BSA, 20 mM HEPES), once with cold acid solution (0.15 M NaCl, 0.1 M glycine, pH 3.0), and again with cold medium. Cells were then chased with medium containing 200 μg/ml holo-Tf with/without insulin at 37°C, and the medium was collected at the indicated time points. After another wash with cold medium, cells were scraped and lysed in 1 M NaOH. Radioactivity in cell lysates (internalized Tf) and medium collected (externalized Tf) was measured in a gamma counter. Data for each time point were measured in triplicate. Tf recycling was expressed as the ratio of externalized Tf to the internalized Tf. Data were corrected for nonspecific binding of 125I-Tf by the addition of 200 μg/ml holo-Tf during the initial labeling.
Statistical Analysis
Statistical analyses were carried out using Prism 4.0 software (GraphPad Software, San Diego, CA). Groups were compared using one-way ANOVA analysis. p < 0.05 was considered statistically significant.
RESULTS
Arp2/3 Is Required for Insulin-mediated Actin Remodeling
We have previously reported that insulin causes marked actin polymerization at the cortical periphery of L6 myoblasts and myotubes that has a branched appearance when visualized by fluorescence (Khayat et al., 2000; Patel et al., 2003) and electron (Randhawa et al., 2008) microscopy and that manifests even in suspended myoblasts lacking stress fibers (JeBailey et al., 2004; Randhawa et al., 2008). The small GTPase Rac is a major regulator of such cortical actin remodeling (Khayat et al., 2000; JeBailey et al., 2004; JeBailey et al., 2007).
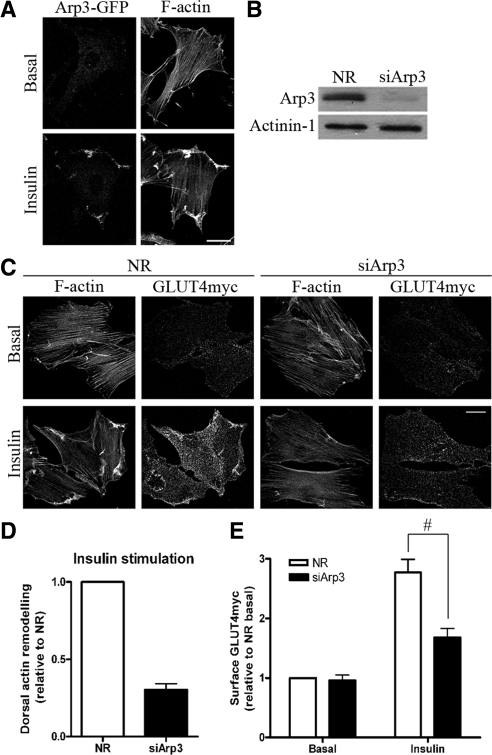
Known effectors of the Rho-family of small G proteins that modify actin filament organization in general include the branching complex Arp2/3 (Goley and Welch, 2006), capping/severing proteins such as gelsolin, and the severing protein cofilin (Ono, 2007). Here we hypothesize that Arp2/3 acts directly downstream of Rac to initiate the insulin-dependent actin polymerization process. Arp2/3 is a seven-subunit complex consisting of Arp2, Arp3, p16, p20, p21, p34, and p40 (Goley and Welch, 2006). By transiently expressing the Arp3-GFP subunit into L6-GLUT4myc myoblasts, we sought to examine changes in the localization of Arp2/3 after insulin stimulation. In the unstimulated state, myoblasts displayed normal cellular stress fibers and Arp3-GFP was mainly cytosolic (Figure 1A). When challenged with insulin, an actin meshwork formed at the periphery of the cells, and Arp3-GFP redistributed to the zone of remodeled actin, suggesting its involvement at this region (Figure 1A).
Figure 1.
Down-regulation of Arp3 prevents insulin-induced actin remodeling and reduces GLUT4 translocation. (A) L6GLUT4myc myoblasts were transiently transfected with Arp3-GFP. After 3-h serum starvation, cells were stimulated with 100 nM insulin for 10 min. Subsequent staining of F-actin by rhodamine-phalloidin was performed to observe the changes in the localization of Arp3-GFP with respect to the remodeled actin. Representative images of four independent experiments are shown. Bars, 20 μm. (B) Myoblasts were transfected with 200 nM of nonrelated (NR) siRNA control or Arp3 siRNA for 72 h. Total cell lysates were prepared, and 10 μg protein was loaded and immunoblotted for Arp3 and actinin-1 (as loading control). Representative blots of five independent experiments are shown. (C) Myoblasts transfected with NR or Arp3 siRNA were treated with/without insulin for 10 min followed by staining surface GLUT4myc in nonpermeabilized cells and then permeabilized to label actin filaments with rhodamine-phalloidin. Dorsal actin remodeling was calculated from the pixel quantification in fluorescence optical cuts of the dorsal surface of adhered myoblasts (see Materials and Methods). Representative images of three independent experiments are shown. Bars, 20 μm. (D) Quantification of changes in insulin-stimulated dorsal actin remodeling relative to NR control (mean ± SE). (E) Quantification of fold increases in surface GLUT4myc relative to NR basal in NR and Arp3 knockdown conditions (mean ± SE, #p < 0.05).
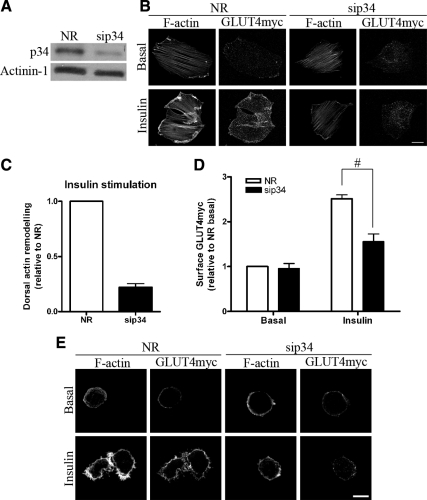
To functionally establish the role of Arp2/3 in insulin-responsive actin remodeling, we interfered with the complex by silencing expression of its Arp3 subunit via siRNA-mediated knockdown. Significant down-regulation of Arp3 (by 77%) was achieved in siArp3-treated cells compared with cells treated with a nonrelated siRNA (siNR) sequence (Figure 1B). In Arp3 knockdown cells, the aspect and density of basal-state stress fibers remained unchanged. However, the insulin-induced actin remodeling was lost (Figure 1C). Quantitative analysis of remodeled actin revealed a 70% decrease in dorsal actin remodeling upon Arp3 down-regulation compared with control siNR-treated cells (Figure 1D). This deleterious effect was further validated by inhibiting the function of Arp2/3 via down-regulation of another Arp2/3 subunit, p34. As with siArp3, siRNA-mediated knockdown of p34 (sip34) by 60% also prevented the formation of remodeled actin structures at the cell periphery upon insulin stimulation (Figure 2, A and B).
Figure 2.
Down-regulation of p34 inhibits the formation of remodeled actin and decreases GLUT4 translocation. (A) L6GLUT4myc myoblasts were transfected with 200 nM of nonrelated (NR) siRNA control or p34 siRNA for 72 h. Total cell lysates were prepared, and 10 μg protein was loaded and immunoblotted for p34 and actinin-1 (as loading control). Representative blots of five independent experiments are shown. (B) Myoblasts transfected with NR or p34 siRNA were treated with/without insulin for 10 min followed by costaining of surface GLUT4myc in the nonpermeabilized state and then permeabilized to label actin filaments with rhodamine-phalloidin. Representative images of five independent experiments are shown. Bars, 20 μm. (C) Quantification of changes in insulin-stimulated dorsal actin remodeling relative to NR control (mean ± SE). (D) Quantification of fold increases in surface GLUT4myc relative to NR basal in NR and p34 knockdown conditions (mean ± SE, #p < 0.05). (E) p34 knockdown abolishes cortical actin remodeling in rounded-up myoblasts and prevents gain in surface GLUT4myc in response to insulin. Bars, 10 μm.
To illustrate that Arp2/3 functions downstream of Rac to generate the dorsal actin meshwork, we took advantage of a constitutively active Rac-GFP mutant (CA-Rac-GFP) that remodels actin without insulin stimulation when transfected into myoblasts. In the control of siNR-transfected cells expressing CA-Rac-GFP, distinctive actin remodeling was observed, compared with neighboring nontransfected cells. However, CA-Rac-GFP failed to elicit actin remodeling after Arp3 knockdown (Figure S1A). This result suggests that Arp2/3 is a major effector downstream of Rac governing actin remodeling in muscle cells.
Depletion of Arp2/3 Reduces Insulin-mediated GLUT4 Gain on the Cell Surface
Because inhibition of Arp2/3 function averted actin remodeling, we explored its effect on insulin-responsive GLUT4 translocation. After siRNA-mediated disruption of Arp2/3 function, costaining of surface GLUT4myc and intracellular F-actin was applied to identify cells lacking the insulin-induced actin rearrangement and their corresponding GLUT4myc levels on the plasma membrane. Surface GLUT4myc showed the typical speckled distribution (Patel et al., 2003; Thong et al., 2007) that may suggest insertion into hot spots. These may represent specific plasma membrane domains or may correspond to ruffled areas supported by the remodeled actin, consistent with the predominant localization of GLUT4myc in ruffles determined by immunogold labeling and scanning electron microscopy (Tong et al., 2001). Down-regulation of Arp3 did not alter the surface level of GLUT4myc in the basal state, compared with control cells treated with siNR (Figure 1, C and E). In contrast, siRNA to Arp3 not only prevented insulin-mediated peripheral actin remodeling but also significantly decreased the amount of GLUT4myc at the plasma membrane (Figure 1C). Quantification of surface GLUT4myc indicated a 44% inhibition in GLUT4 translocation after the down-regulation of Arp3 (Figure 1E). Furthermore, a strong reduction (60%) was achieved by knocking down the p34 subunit of Arp2/3 (Figure 2, B and D). Neither Arp3 nor p34 knockdown altered the overall insulin-stimulated Akt phosphorylation (Figure S1B), suggesting that the Akt signaling arm of insulin action remained intact.
To demonstrate the specificity of Arp2/3 in GLUT4 translocation, we rescued Arp2/3 expression by transfecting D. discoideum Arp3-GFP into siArp3-treated cells. Under this setting, insulin-stimulated actin remodeling was restored (Figure S2, A and B), and the deleterious effect of Arp3 down-regulation on insulin-mediated GLUT4 translocation was alleviated (Figure S2C). More importantly, expression of Dictyostelium Arp3-GFP alone did not change the basal actin filament morphology or surface GLUT4 in unstimulated cells, indicating that Arp2/3 only exerts its functional action upon insulin stimulation.
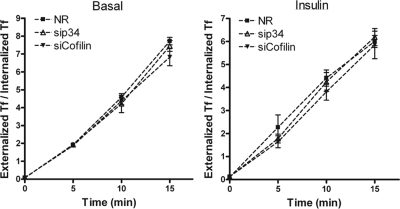
The fact that depletion of Arp2/3 components only altered the insulin-dependent component of surface GLUT4 but did not change the basal levels of the transporters at the cell membrane suggests that the constitutive recycling of GLUT4 does not require Arp2/3 input. To establish if the effect of Arp2/3 interference is selective to GLUT4 traffic, we examined the effect of depletion of p34 of the Arp2/3 complex on Tf recycling, which depends on endosome recycling. As shown later in Figure 6, Tf recycling in either basal or insulin stimulated state was similar in cells treated with siRNA to p34 as in siNR-treated cells, implying that the inhibition of Arp2/3 did not affect the traffic of Tf.
Figure 6.
Knockdown of either p34 or cofilin does not alter Tf recycling. L6GLUT4myc myoblasts were treated with NR, p34, or cofilin siRNA. After 3-h serum starvation, to 125I-Tf recycling was determined as described in Materials and Methods, in basal (left)and insulin-stimulated (right) conditions. Tf recycling is displayed as the ratio of externalized Tf to internalized Tf (mean ± SD).
The above results indicate that Arp2/3 is required for cortical actin remodeling and GLUT4 translocation including full exposure at the cell surface. To ascertain that the effect of Arp2/3 knockdown is independent of possible changes in actin stress fibers, we analyzed suspended myoblasts devoid of stress fibers. In this rounded-up configuration, myoblasts show actin filaments exclusively at the cell periphery (JeBailey et al., 2004; Randhawa et al., 2008; Talior-Volodarsky et al., 2008). Myoblasts treated with siRNA to p34 of the Arp2/3 complex or with siNR were suspended, stimulated with insulin, and analyzed for actin remodeling and surface GLUT4 levels. Although siNR-treated cells showed the habitual cortical actin arborizations and gain in surface GLUT4 in response to insulin, sip34 treatment markedly abolished both responses to insulin (Figure 2E). These results suggest that it is the cortical actin structures regulated by Arp2/3 that are required for GLUT4 translocation in response to the hormone.
Insulin Causes Cofilin Dephosphorylation That Depends on Slingshot
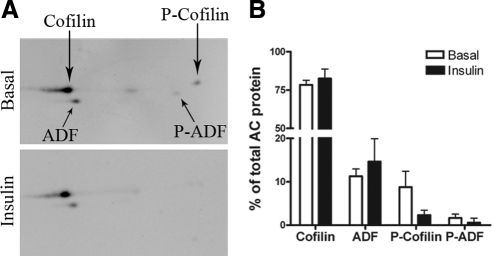
Actin dynamics is regulated by concerted actin polymerization and depolymerization. In fact, a spatial/temporal burst in actin polymerization and branching, as caused by Arp2/3, requires available sources of actin monomer, which are typically provided by the continuous depolymerization of actin filaments (Pollard, 2007; Insall and Machesky, 2009). Hence, we sought to identify molecules in the actin-depolymerization pathway that may respond to insulin, which would contribute to the balance of actin dynamics. ADF/cofilin are highly related, actin-severing, and monomer-sequestering proteins whose activity is mainly regulated by the phosphorylation status of Ser3 (Agnew et al., 1995; Moriyama et al., 1996). Phosphorylation and dephosphorylation of ADF/cofilin leads to its inactivation and activation, respectively. Insulin elicited ADF/cofilin dephosphorylation in cells that do not represent metabolically determining tissues and do not express GLUT4 (HT4 neuronal and 293T cells; Meberg et al., 1998; Nishita et al., 2004), but the function of cofilin in these cells and the metabolic consequences of its activation were not investigated. We therefore examined whether insulin stimulation in muscle cells alters the ADF/cofilin phosphorylation status and its possible contribution to GLUT4 traffic. We utilized a rabbit antibody that recognizes both ADF and cofilin-1 and their phosphorylated forms to an equal extent and cofilin-2 to a somewhat lesser extent (Shaw et al., 2004). By two-dimensional (2D) gel electrophoresis of L6 cell lysates, we calculated the molar ratio of cofilin to ADF in L6 myoblasts to be 7:1. The major cofilin isoform present is cofilin-1 with a faint immunoreactive spot probably representing cofilin-2 focusing at the more acidic position identified for mouse cofilin-2 (Ono et al., 1994). This 2D immunoblotting approach also revealed that ∼9% of the cellular content of cofilin is phosphorylated (inactive) in unstimulated cells, and insulin caused a fourfold decrease in the phosphorylation of this protein (Figure 3). By SDS-PAGE and immunoblotting with an antibody selective to cofilin (1 and 2) phosphorylated on p-Ser3, phosphorylation of cofilin in unstimulated cells was also ascertained (Figure 4A). On insulin stimulation, a reduction in the level of phosphorylated cofilin (P-cofilin) was observed without changes in total cofilin, indicating a shift to its active state (Figure 4A). This was evident as early as 3 min and was most significant 10 min after insulin treatment (Figure 4B). Inhibiting PI3K with wortmannin prevented cofilin dephosphorylation in response to insulin (not shown), paralleling the response in 293T cells (Nishita et al., 2004).
Figure 3.
2D Western blots of ADF/cofilin from which the relative percent of cofilin, ADF, phosphorylated-cofilin, and phosphorylated ADF can be calculated. Basal and insulin-stimulated L6GLUT4myc myoblasts were lysed and subjected to 2D gel electrophoresis and assessment of ratios of ADF, cofilin, and their phosphorylated versions, essentially as described (Shaw et al., 2004). (A) Representative gel. Faint spot midway between cofilin and phospho-cofilin probably represents the cofilin-2 isoform. (B) Quantification of three experiments.
Figure 4.
Insulin stimulation in L6GLUT4myc muscle cells causes dephosphorylation of cofilin that is dependent on SSH1. L6GLUT4myc myoblasts were serum-starved for 3 h before insulin stimulation (100 nM) for the indicated time periods. (A) Total lysates immunoblotted for P-cofilin, cofilin, P-Akt, and actin (as loading control). Representative blots of four experiments are shown. (B) Quantification of insulin-dependent cofilin dephosphorylation expressed as P-Cofilin/Cofilin ratio relative to time 0 (n = 4, mean ± SE, #p < 0.05). (C) Lysates from myoblasts treated with SSH1 siRNA (siSSH1) were prepared to determine the knockdown effect on SSH1 and its contribution toward insulin-induced cofilin dephosphorylation and Akt phosphorylation by immunoblotting for P-cofilin and p-Akt (Ser-473). Myoblasts were (D) transfected with p34 siRNA or (E) subjected to 250 nM LB treatment for 30 min, and the effect of insulin on P-Cofilin was assessed. Representative blots of more than three independent experiments are shown.
The balance of cofilin phosphorylation/dephosphorylation is primarily achieved by the action of kinases LIM kinase (LIMK) and testicular kinase and of the phosphatases slingshot (SSH), chronophin (Van Troys et al., 2008), and to some degree PP1/PP2A (Meberg et al., 1998). Because net dephosphorylation of cofilin was observed after insulin stimulation, we examined first the participation of the phosphatases in this response. Treatment of myoblasts with SSH1 siRNA reduced the expression of SSH1 by 73% (Figure 4C). Down-regulation of this phosphatase notably prevented the dephosphorylation of cofilin evoked by insulin compared with the degree observed in NR siRNA controls (Figure 4C). This revealed that insulin signals primarily via SSH1 to dephosphorylate cofilin, which would lead to cofilin activation. Regarding the kinases mediating cofilin phosphorylation, LIMK is attractive because it is typically activated by phosphorylation via Rac-dependent, p21-activated-kinase (Edwards et al., 1999). Although LIMK1 knockdown did not increase the steady-state phosphorylation of cofilin in the basal state, LIMK knockdown potentiated cofilin dephosphorylation in response to insulin (Figure S3). This observation suggests that, in response to insulin, LIMK1 may partially restore phosphorylation of cofilin, which is however more dominantly dephosphorylated by SSH1.
Arp2/3-mediated Actin Remodeling Signals to Insulin-stimulated Cofilin Dephosphorylation
The net effect of cofilin dephosphorylation induced by insulin suggests the activity of SSH1 outweighed that of LIMK1. One of the possible explanations could be a surge in the phosphatase activity of SSH1 after insulin stimulation. Indeed, adding F-actin to purified SSH1 markedly augments its phosphatase activity (Nagata-Ohashi et al., 2004; Soosairajah et al., 2005; Kurita et al., 2008). Analogously, the remodeled actin produced in muscle cells by insulin could serve as the stimulus that boosts the activity of SSH1 in vivo to cause a shift toward cofilin dephosphorylation because SSH1 accumulated at the remodeled actin after insulin stimulation (Figure S4). Knowing that the inhibition of Arp2/3 prevented actin remodeling, we examined this hypothesis by examining changes in P-cofilin levels upon p34 knockdown. In control cells treated with siNR, cofilin dephosphorylation occurred normally as evident from the decrease in P-cofilin level after 5 and 10 min of insulin stimulation (Figure 4D). In contrast, and notably, this insulin-dependent decrease in the level of P-cofilin was lacking in p34 knockdown cells (Figure 4D). Similar results were obtained when the actin filament–disrupting agent LB was used to stop the formation of remodeled actin by insulin. Under LB treatment, the dephosphorylation of cofilin by insulin was also prevented, so that the levels of P-cofilin at 5 and 10 min of stimulation were comparable to that of unstimulated cells (Figure 4E). In neither case was the ability of insulin to signal to Akt affected. These findings support the idea that the remodeled actin functions as a stimulus that augments the activity of SSH1 to generate the net dephosphorylation of cofilin after insulin stimulation in muscle cells.
Cofilin Knockdown Promotes F-Actin Accumulation and Decreases Insulin-mediated GLUT4 Translocation
Given that insulin activates cofilin via its dephosphorylation, it became intriguing to explore the functional implication that cofilin has on the dynamics of insulin-responsive actin remodeling. Immunofluorescence detection of endogenous cofilin in myoblasts revealed its redistribution from the cytosol to the peripheral zone where actin remodels upon insulin stimulation (Figure 5A). The localization of the phosphorylated form was determined using an antibody to phosphorylated ADF/cofilin that is compatible with immunofluorescence approaches. Costaining of total cofilin and phosphorylated ADF/cofilin (P-AC) revealed that insulin stimulation decreased the ratio of P-AC/cofilin at the cell periphery/remodeled actin (by 31%, p < 0.05), indicating that cofilin undergoes dephosphorylation in response to the hormone at this region of the cell (Figure S5, A and B), consistent with the localization of SSH1.
Figure 5.
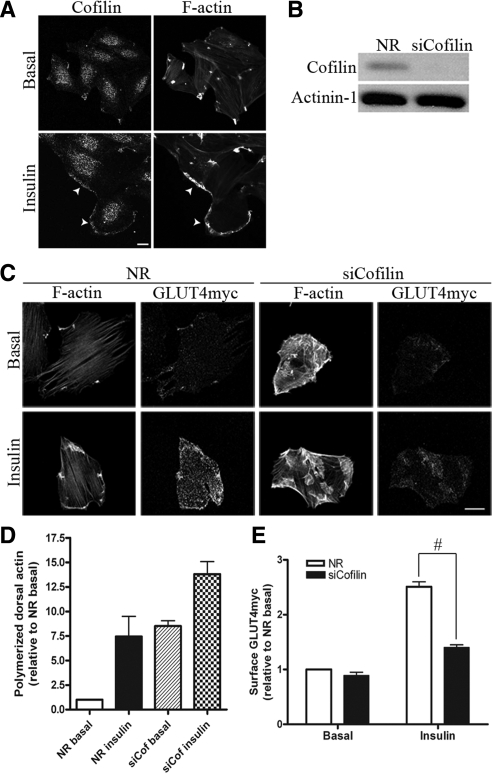
Cofilin localizes with insulin-induced remodeled actin and down-regulation of cofilin increases F-actin aggregates and reduces insulin-induced GLUT4 translocation. (A) L6GLUT4myc myoblasts were serum-starved for 3 h and stimulated with 100 nM insulin for 10 min, followed by labeling with rhodamine-phalloidin for F-actin and cofilin-specific antibody for endogenous cofilin. Representative images of three independent experiments are shown. Bars, 10 μm. (B) Total lysates from myoblasts transfected with NR or cofilin siRNA were prepared and immunoblotted for cofilin and actinin-1 (loading control). Representative blots of five independent experiments are shown. (C) Myoblasts with siNR or siCofilin were treated with/without insulin followed by costaining of surface GLUT4myc and F-actin. Representative images of four independent experiments are shown. Bars, 20 μm. (D) Quantification of dorsal polymerized actin relative to NR basal after cofilin knockdown (mean ± SE). (E) Quantification of fold increases in surface GLUT4myc relative to NR basal in NR and siCofilin conditions (mean ± SE, #p < 0.05).
To explore if cofilin dephosphorylation is required for insulin action on actin dynamics and GLUT4 translocation, siRNA against cofilin-1 was applied to achieve an 82% knockdown compared with the siNR control (Figure 5B). The siRNA sequence used to target cofilin-1 is unlikely to target cofilin-2, given a four-base difference (rat sequences). Under these conditions, the signal of phospho-cofilin was proportionately reduced (results not shown), confirming the 2D immunoblot data showing that cofilin 1 is the predominant isoform expressed and phosphorylated in L6 myoblasts. As expected from existing literatures (Nishita et al., 2005; Sidani et al., 2007), down-regulation of cofilin caused a massive increase in the level of random F-actin aggregates in the basal state, and this morphological change was scored via polymerized dorsal actin assay (Figure 5, C and D). When challenged with insulin, these cells displayed a further elevation in polymerized actin (Figure 5D), suggesting that potentially there were still some available actin monomers to respond to the actin polymerization cues evoked by insulin. However, this remodeled actin was morphologically more diverse and was not confined to the cell periphery compared with that in NR control cells stimulated by insulin (Figure 5C).
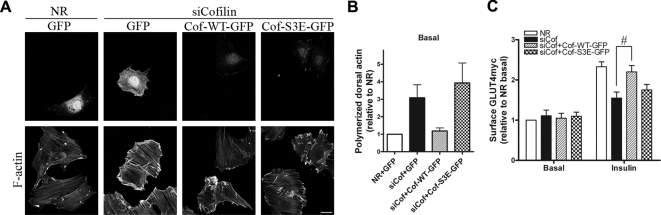
Although insulin-dependent actin polymerization continued upon down-regulation of cofilin, the visually excessive/abnormal remodeling suggests that the dynamics of F-actin at the zone of remodeling may have been impeded. We therefore explored whether under these conditions the insulin-dependent GLUT4 translocation was affected. As shown in Figure 5C and E, cofilin knockdown caused a major reduction (66%) in insulin-dependent gain in surface GLUT4, yet Tf recycling, in either absence or presence of insulin, was not significantly disrupted (Figure 6). This observation allowed us to test whether cofilin reexpression restores normal GLUT4 translocation and if such restoration would be dependent on the phosphorylation status of cofilin. Transfecting Xenopus cofilin-WT-GFP into myoblasts with down-regulated cofilin expression restored the normal F-actin pattern in the basal state by eliminating the excessive F-actin accumulation (Figure 7, A and B). Moreover, such reexpression also alleviated the defect in insulin-dependent GLUT4 translocation (Figure 7C). Strikingly, neither the recovery of actin dynamics nor that of GLUT4 translocation was achieved when the inactive cofilin-S3E-GFP mutant was transfected into myoblasts with down-regulated cofilin. This mutant is unable to sever actin filaments. Overall, these results argue that the insulin-induced increase in the severing function of cofilin is critical for the dynamics of actin remodeling, which in turn allows proper insulin-stimulated GLUT4 translocation to proceed.
Figure 7.
Expression of Xenopus cofilin-WT-GFP, but not cofilin-S3E-GFP mutant, restores normal F-actin morphology and GLUT4 translocation. (A) L6GLUT4myc myoblasts transfected with NR or cofilin siRNA were further transfected with cDNA to either GFP as control, cofilin-WT-GFP, or cofilin-S3E-GFP. After serum starvation, F-actin was labeled with rhodamine-phalloidin in the basal state to detect changes in actin morphology. Representative images of six independent experiments are shown. Bars, 20 μm. (B) Quantification of F-actin aggregates relative to NR+GFP basal (mean ± SE). (C) Surface GLUT4myc levels in siCofilin-treated myoblasts cotransfected with cofilin-WT or S3E mutant expression in siCofilin myoblasts were measured by fluorescent detection of single cells and are presented as fold increases in surface GLUT4myc relative to NR basal are also shown (mean ± SE, n = 6, #p < 0.05).
DISCUSSION
Because muscle is the major insulin-regulated storage of blood glucose, it is imperative to define the signaling events that promote the gain of GLUT4 at the surface to increase glucose uptake into this tissue. L6GLUT4myc myoblasts have been a bona fide cell culture model to study GLUT4 traffic in a muscle cell context (Ueyama et al., 1999; Wang et al., 1999; Khayat et al., 2000; Tong et al., 2001; JeBailey et al., 2004, 2007; Thong et al., 2007; Ishikura and Klip, 2008). In these cells, we found that in parallel to the well-established Akt-AS160 pathway (Thong et al., 2007), insulin induces a rapid activation of the small GTPase Rac that is PI3K-dependent and leads to cortical actin remodeling (Khayat et al., 2000; Tong et al., 2001; JeBailey et al., 2004, 2007). However, as with any cell culture system, lessons learned from L6 muscle cells should eventually be tested in the mature tissue. In this regard, insulin-dependent reorganization of nonsarcomeric actin and Rac activation were recently confirmed in rodent skeletal muscles (Brozinick et al., 2004; Ueda et al., 2010; Hansen et al., 2010). Rac activation and actin remodeling fail in several models of insulin resistance, without detectable defects in upstream IRS-1 phosphorylation or PI3K activation (Tong et al., 2001; McCarthy et al., 2006; JeBailey et al., 2007). Here we identify Arp2/3 as the effector downstream of Rac that promotes formation of remodeled actin, whereas the activity of cofilin maintains the active turnover of actin in the remodeling zone. Disruption of either protein yielded abnormal actin remodeling and subsequent inhibition in insulin-mediated GLUT4 translocation (Figures 1C, 2B, and 5C).
Insulin Induces Concerted Actin Branching and Severing
F-actin contributes to maintaining structural integrity, promoting cellular migration, and aiding in vesicle traffic. To be functional, these cellular processes depend on the dynamic nature of actin regulation. Cytoplasmic actin exists as monomers and oligomerized F-actin. Uncapped F-actin undergoes constant polymerization at the barbed ends, whereas depolymerization occurs at the point ends to regenerate a steady pool of monomeric actin for further rounds of polymerization (Pollard, 2007). Such dynamic turnover is tightly controlled by actin-modifying proteins including actin-nucleating and -severing proteins (Pollard, 2007). Arp2/3 initiates branching at 70° on existing actin filaments and is firmly established in the Rac-dependent formation of lamellipodia in migrating cells (Ridley et al., 1992; Miki et al., 2000). Here we report that Arp2/3 contributes to the Rac-mediated, cortical actin remodeling after insulin stimulation. Down-regulation of Arp2/3 through siRNA against its Arp3 or p34 subunits abrogated insulin-induced actin remodeling, suggesting that Arp2/3-initiated actin polymerization is responsible for the genesis of dorsally remodeled actin in muscle cells (Figures 1C and 2B). Overexpressing Arp3-GFP did not promote actin polymerization in the basal state (Figure 1A), suggesting that Arp2/3 activation is tightly controlled by insulin stimulation. Conceivably, this occurs in response to insulin-activated Rac and its downstream effectors. Indeed, constitutively active Rac caused Arp2/3-mediated actin branching (Figure S1A).
In both L6 muscle cells (this study) and 3T3-L1 adipocytes (Jiang et al., 2002) insulin caused Arp2/3 colocalization with the peripheral remodeled actin, and cortical actin remodeling is required for GLUT4 translocation (Kanzaki and Pessin, 2001; Tong et al., 2001; Torok et al., 2004). However, this phenomenon is differentially regulated in both cell types, because only in the muscle cells is the insulin-dependent actin remodeling downstream of PI3K (Patel et al., 2003; JeBailey et al., 2004). Moreover, the Rho-family G proteins engaged by each cell type also differ, with Rac determining actin remodeling in the muscle cells and TC10 (related to Cdc42) in adipocytes (Kanzaki et al., 2002; JeBailey et al., 2004). Arp2/3 is regulated by the nucleation-promoting factors N-WASP, WAVE, and cortactin (Machesky et al., 1999; Suetsugu et al., 1999; Weed et al., 2000), and a dominant-negative mutant of N-WASP prevented the TC10-mediated insulin-induced actin reorganization in adipocytes (Jiang et al., 2002). In contrast, WAVE, rather than N-WASP, is the nucleating factor downstream of Rac that activates Arp2/3 and its consequent actin-filament branching (Suetsugu et al., 1999). Thus, WAVE2 may be the Arp2/3 activator in insulin-stimulated muscle cells, a possibility worthy of future exploration.
Insulin also activated the actin-severing protein cofilin by inducing its dephosphorylation on Ser3 (Figure 3A). This may be required to generate a dynamic, rapid turnover of the branching network and to provide a continuous supply of monomeric actin.
Because cofilin phosphorylation and dephosphorylation are governed by LIMK1 and SSH1, respectively (Yang et al., 1998; Niwa et al., 2002), we propose that insulin must activate the phosphatase and that this enzyme has a predominant effect over the kinase to produce the net dephosphorylation (Soosairajah et al., 2005). In muscle cells, cofilin is already phosphorylated in the absence of stimuli (Figures 3 and 4), ostensibly due to a low, tonic activity of LIMK1 and a relatively inactive SSH1. On insulin stimulation, cofilin undergoes SSH1-dependent dephosphorylation (Figure 4C). The surge in SSH1 activity may arise in response to the formation of the cortical network of branched, polymerized actin, because F-actin binding is required for significant phosphatase activity of SSH1 (Nagata-Ohashi et al., 2004; Soosairajah et al., 2005; Kurita et al., 2008). This model is supported by our observations that LB and p34 knockdown, strategies that prevent insulin-induced actin polymerization, avert cofilin dephosphorylation (Figure 4, D and E). Consistent with this scenario, SSH1 concentrated along the peripherally remodeled actin upon insulin stimulation (Figure S4). Moreover, there is precedent for F-actin tilting the balance from LIMK1 to SSH1 dominance over cofilin (Jovceva et al., 2007). A dynamic interplay between LIMK1 and SSH1 likely enables spatial temporal control over cofilin activity especially at the zone of insulin-induced actin remodeling.
Arp2/3 Activation and Cofilin Dephosphorylation Are Required for Insulin-dependent GLUT4 Translocation
A key observation of this study is that silencing two components of the Arp2/3 complex prevents GLUT4 translocation in response to insulin. This result is reminiscent of the well-documented abrogation of GLUT4 translocation by LB in fat cells (Wang et al., 1998; Kanzaki and Pessin, 2001) and muscle cells (Khayat et al., 2000; Torok et al., 2004) in culture and in primary adipocytes (Omata et al., 2000) and muscle tissue (Brozinick et al., 2004). However, because LB inhibits actin remodeling by sequestering actin monomers, it impedes actin polymerization not only at the cell cortex but also at the level of stress fiber (Spector et al., 1983). Instead, disruption of Arp2/3 allowed us to assign the selective participation of branched actin remodeling in GLUT4 translocation, irrespective of stress fibers or other actin filament-containing structures. The inhibition of GLUT4 translocation by Arp2/3 silencing is here ascribed to inhibition of actin branching, but may also be due to the consequent loss of insulin-dependent cofilin dephosphorylation, consistent with an Arp2/3- and cofilin-controlled actin polymerization–depolymerization cycle.
When actin severing was impaired by siRNA-mediated knockdown of cofilin, actin polymerization was extensive and widely prevalent across the cell (Figure 5C), and this abnormality paralleled a reduction in insulin-dependent GLUT4 translocation (Figure 5E). This finding is reminiscent of the effect of jasplakinolide, an actin-stabilizing agent that causes excessive actin aggregates resembling those produced in cells with cofilin knockdown (Bubb et al., 1994). Likewise, jasplakinolide also interferes with GLUT4 vesicle traffic to the surface of muscle and adipose cells (Kanzaki and Pessin, 2001; Tong et al., 2001; Torok et al., 2004). All these considerations point to the need for a dynamic, concerted actin polymerization and severing induced by insulin and required for GLUT4 translocation. Perhaps the strongest support of this model is the fact that the defect in GLUT4 traffic caused by cofilin knockdown was rescued by expression of active cofilin-WT-GFP (which is amenable to phosphorylation and dephosphorylation) but could not be restored by the expression of inactive cofilin-S3E-GFP (Figure 7C). These results buttress the participation of insulin-induced, cofilin-mediated actin depolymerization to facilitate GLUT4 translocation. Further, although other interpretations may be possible, the results are consistent with cofilin dephosphorylation being required for insulin-dependent vesicle traffic.
Possible Mechanisms Whereby a Dynamic Actin Network Supports GLUT4 Traffic
The remodeled actin filaments may act as a tether for GLUT4 vesicles close to the plasma membrane, so that their subsequent docking/fusion can occur more readily (Khayat et al., 2000; Tong et al., 2001; Randhawa et al., 2008). Indeed, in insulin-stimulated muscle cells, GLUT4 vesicles accumulate within the cortical actin mesh visualized by electron microscopy (Tong et al., 2001; Randhawa et al., 2008). GLUT4 itself may tether to actin filaments via α-actinin-4 (Foster et al., 2006; Talior-Volodarsky et al., 2008), and indeed upon α-actinin-4 silencing, GLUT4 vesicles do not accumulate at the muscle cell periphery, and there is no insulin-dependent gain in surface GLUT4 (Talior-Volodarsky et al., 2008). Similarly, when actin filament remodeling is inhibited by expressing dominant negative Rac or by LB, the enrichment of GLUT4 beneath the plasma membrane is lost, leading to the collapse of GLUT4 vesicles back to perinuclear regions (Randhawa et al., 2008). It is also plausible that the remodeled actin clusters insulin-signaling molecules close to GLUT4 vesicles near the plasma membrane (Patel et al., 2003). Although preventing actin remodeling by Arp2/3 knockdown did not reduce the overall level of P-Akt (Figure S1B), failure to accumulate phosphorylated Akt near the membrane may be a factor in the loss of insulin-induced GLUT4 translocation (Peyrollier et al., 2000; Eyster et al., 2005; Ng et al., 2008; Gonzalez and McGraw, 2009).
On the other hand, actin polymerization is abundant in cells with down-regulated cofilin, yet the resulting polymerized actin filaments seem morphologically distinct and disseminated across the cell (Figure 5C). We speculate that, in this case, the impaired gain in surface GLUT4 is due to vesicle retention in a static mesh that no longer undergoes severing. By analogy, in neuronal cells, thick patches of submembrane F-actin present at rest act as a barrier to inhibit basal exocytosis of neurotransmitters (Trifaro et al., 2008). In cells with down-regulated cofilin, the imbalance in dynamic cycles of actin depolymerization and branching may lead to uncoordinated growth of “static” actin filaments that could similarly turn into a barrier for GLUT4 vesicles, preventing them from gaining access to interaction with membrane SNAREs required for fusion (Randhawa et al., 2008).
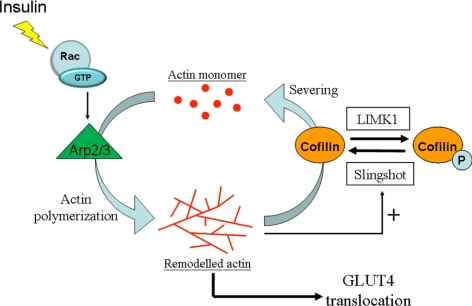
We propose a model (Figure 8) whereby insulin stimulation leads to Arp2/3 activation downstream of Rac that initiates dorsal actin polymerization. The increase in polymerized actin stimulates the phosphatase activity of SSH1, leading to cofilin dephosphorylation and consequential increase in its severing action. Active actin depolymerization by cofilin can then free up actin monomers for Arp2/3-dependent polymerization and a dynamic actin turnover of insulin-stimulated actin remodeling is achieved. This dynamic status must be sustained to allow GLUT4 vesicle positioning and proper interaction with elements of the vesicle fusion machinery. This framework will allow testing the fidelity of its individual steps during insulin resistance and may reveal so far unsuspected steps that may be altered in this condition.
Figure 8.
Proposed mechanism of insulin-regulated actin dynamics in muscle cells. Insulin stimulation in muscle cells promotes dynamic actin remodeling. The formation of the remodeled actin is achieved by the polymerization activity of Arp2/3 acting downstream of active Rac. The accumulation of polymerized F-actin poses a stimulatory factor in the phosphatase activity of SSH, which leads to net dephosphorylation and activation of cofilin. Hereon, the actin-severing function of cofilin maintains the flexibility of remodeled actin and enables regeneration of free monomeric actin for further polymerization. This active cycling of actin mediated by insulin keeps proper actin dynamics at the cortical zone in order to facilitate GLUT4 insertion onto the cellular surface.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sagar Dugani and Alex Wong for their participation in the early determination of cofilin dephosphorylation. This work was supported by Grant MOT 7307 from the Canadian Institutes of Health Research to A.K., and by National Institutes of Health Grant NS40371 to J.B. T.C. was supported by a studentship from the National Science and Engineering Research Council and the Research Training Centre from The Hospital for Sick Children. N.P. was supported by a doctoral research award from the Canadian Institutes of Health Research.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0316) on August 25, 2010.
REFERENCES
- Agnew B. J., Minamide L. S., Bamburg J. R. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Ashworth S. L., Southgate E. L., Sandoval R. M., Meberg P. J., Bamburg J. R., Molitoris B. A. ADF/cofilin mediates actin cytoskeletal alterations in LLC-PK cells during ATP depletion. Am. J. Physiol. Renal. Physiol. 2003;284:F852–F862. doi: 10.1152/ajprenal.00210.2002. [DOI] [PubMed] [Google Scholar]
- Bernards A., Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Brozinick J. T., Jr., Hawkins E. D., Strawbridge A. B., Elmendorf J. S. Disruption of cortical actin in skeletal muscle demonstrates an essential role of the cytoskeleton in glucose transporter 4 translocation in insulin-sensitive tissues. J. Biol. Chem. 2004;279:40699–40706. doi: 10.1074/jbc.M402697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L., Korn E. D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Edwards D. C., Sanders L. C., Bokoch G. M., Gill G. N. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Eyster C. A., Duggins Q. S., Olson A. L. Expression of constitutively active Akt/protein kinase B signals GLUT4 translocation in the absence of an intact actin cytoskeleton. J. Biol. Chem. 2005;280:17978–17985. doi: 10.1074/jbc.M409806200. [DOI] [PubMed] [Google Scholar]
- Foster L. J., Rudich A., Talior I., Patel N., Huang X., Furtado L. M., Bilan P. J., Mann M., Klip A. Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC) J. Proteome Res. 2006;5:64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- Garvey W. T., Maianu L., Zhu J. H., Brechtel-Hook G., Wallace P., Baron A. D. Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J. Clin. Invest. 1998;101:2377–2386. doi: 10.1172/JCI1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E. D., Welch M. D. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gonzalez E., McGraw T. E. Insulin signaling diverges into Akt-dependent and -independent signals to regulate the recruitment/docking and the fusion of GLUT4 vesicles to the plasma membrane. Mol. Biol. Cell. 2006;17:4484–4493. doi: 10.1091/mbc.E06-07-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E., McGraw T. E. Insulin-modulated Akt subcellular localization determines Akt isoform-specific signaling. Proc. Natl. Acad. Sci. USA. 2009;106:7004–7009. doi: 10.1073/pnas.0901933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L. S., Jensen T. E., Chiu T., Niu W., Bilan P., Klip A., Richter E. Rac1 activation is a major requirement for GLUT4 translocation and glucose uptake in both muscle cells and mouse skeletal muscle. 70th American Diabetes Association, Abstract 2463-PO. 2010:A645. [Google Scholar]
- Huang S., Czech M. P. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Insall R. H., Machesky L. M. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Ishikura S., Bilan P. J., Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem. Biophys. Res. Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- Ishikura S., Klip A. Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation. Am. J. Physiol. Cell Physiol. 2008;195:C1016–C1025. doi: 10.1152/ajpcell.00277.2008. [DOI] [PubMed] [Google Scholar]
- Ishikura S., Koshkina A., Klip A. Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol. 2008;192:61–74. doi: 10.1111/j.1748-1716.2007.01778.x. [DOI] [PubMed] [Google Scholar]
- JeBailey L., Rudich A., Huang X., Di Ciano-Oliveira C., Kapus A., Klip A. Skeletal muscle cells and adipocytes differ in their reliance on TC10 and Rac for insulin-induced actin remodeling. Mol. Endocrinol. 2004;18:359–372. doi: 10.1210/me.2003-0294. [DOI] [PubMed] [Google Scholar]
- JeBailey L., Wanono O., Niu W., Roessler J., Rudich A., Klip A. Ceramide- and oxidant-induced insulin resistance involve loss of insulin-dependent Rac-activation and actin remodeling in muscle cells. Diabetes. 2007;56:394–403. doi: 10.2337/db06-0823. [DOI] [PubMed] [Google Scholar]
- Jiang Z. Y., Chawla A., Bose A., Way M., Czech M. P. A phosphatidylinositol 3-kinase-independent insulin signaling pathway to N-WASP/Arp2/3/F-actin required for GLUT4 glucose transporter recycling. J. Biol. Chem. 2002;277:509–515. doi: 10.1074/jbc.M108280200. [DOI] [PubMed] [Google Scholar]
- Jovceva E., Larsen M. R., Waterfield M. D., Baum B., Timms J. F. Dynamic cofilin phosphorylation in the control of lamellipodial actin homeostasis. J. Cell Sci. 2007;120:1888–1897. doi: 10.1242/jcs.004366. [DOI] [PubMed] [Google Scholar]
- Kanzaki M., Pessin J. E. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J. Biol. Chem. 2001;276:42436–42444. doi: 10.1074/jbc.M108297200. [DOI] [PubMed] [Google Scholar]
- Kanzaki M., Watson R. T., Hou J. C., Stamnes M., Saltiel A. R., Pessin J. E. Small GTP-binding protein TC10 differentially regulates two distinct populations of filamentous actin in 3T3L1 adipocytes. Mol. Biol. Cell. 2002;13:2334–2346. doi: 10.1091/mbc.01-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat Z. A., Tong P., Yaworsky K., Bloch R. J., Klip A. Insulin-induced actin filament remodeling colocalizes actin with phosphatidylinositol 3-kinase and GLUT4 in L6 myotubes. J. Cell Sci. 2000;113(Pt 2):279–290. doi: 10.1242/jcs.113.2.279. [DOI] [PubMed] [Google Scholar]
- Klip A., Ramlal T., Bilan P. J., Cartee G. D., Gulve E. A., Holloszy J. O. Recruitment of GLUT-4 glucose transporters by insulin in diabetic rat skeletal muscle. Biochem. Biophys. Res. Commun. 1990;172:728–736. doi: 10.1016/0006-291x(90)90735-6. [DOI] [PubMed] [Google Scholar]
- Korobova F., Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook A., Wallberg-Henriksson H., Zierath J. R. Sending the signal: molecular mechanisms regulating glucose uptake. Med. Sci. Sports Exerc. 2004;36:1212–1217. doi: 10.1249/01.mss.0000132387.25853.3b. [DOI] [PubMed] [Google Scholar]
- Kurita S., Watanabe Y., Gunji E., Ohashi K., Mizuno K. Molecular dissection of the mechanisms of substrate recognition and F-actin-mediated activation of cofilin-phosphatase Slingshot-1. J. Biol. Chem. 2008;283:32542–32552. doi: 10.1074/jbc.M804627200. [DOI] [PubMed] [Google Scholar]
- Larance M., Ramm G., James D.E. The GLUT4 code. Mol. Endocrinol. 2008;22:226–233. doi: 10.1210/me.2007-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E., Pollard T. D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. M., Spisak K. O., Brozinick J. T., Elmendorf J. S. Loss of cortical actin filaments in insulin-resistant skeletal muscle cells impairs GLUT4 vesicle trafficking and glucose transport. Am. J. Physiol. Cell Physiol. 2006;291:C860–C868. doi: 10.1152/ajpcell.00107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meberg P. J., Ono S., Minamide L. S., Takahashi M., Bamburg J. R. Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell Motil. Cytoskelet. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Miinea C. P., Sano H., Kane S., Sano E., Fukuda M., Peranen J., Lane W. S., Lienhard G. E. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem. J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Yamaguchi H., Suetsugu S., Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- Mora S., Pessin J. E. An adipocentric view of signaling and intracellular trafficking. Diabetes Metab. Res. Rev. 2002;18:345–356. doi: 10.1002/dmrr.321. [DOI] [PubMed] [Google Scholar]
- Moriyama K., Iida K., Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- Nagata-Ohashi K., et al. A pathway of neuregulin-induced activation of cofilin-phosphatase Slingshot and cofilin in lamellipodia. J. Cell Biol. 2004;165:465–471. doi: 10.1083/jcb.200401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y., Ramm G., Lopez J. A., James D. E. Rapid activation of Akt2 is sufficient to stimulate GLUT4 translocation in 3T3–L1 adipocytes. Cell Metab. 2008;7:348–356. doi: 10.1016/j.cmet.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M., Wang Y., Tomizawa C., Suzuki A., Niwa R., Uemura T., Mizuno K. Phosphoinositide 3-kinase-mediated activation of cofilin phosphatase Slingshot and its role for insulin-induced membrane protrusion. J. Biol. Chem. 2004;279:7193–7198. doi: 10.1074/jbc.M312591200. [DOI] [PubMed] [Google Scholar]
- Niu W., Huang C., Nawaz Z., Levy M., Somwar R., Li D., Bilan P. J., Klip A. Maturation of the regulation of GLUT4 activity by p38 MAPK during L6 myogenesis. J. Biol. Chem. 2003;278:17953–17962. doi: 10.1074/jbc.M211136200. [DOI] [PubMed] [Google Scholar]
- Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Omata W., Shibata H., Li L., Takata K., Kojima I. Actin filaments play a critical role in insulin-induced exocytotic recruitment but not in endocytosis of GLUT4 in isolated rat adipocytes. Biochem. J. 2000;346(Pt 2):321–328. [PMC free article] [PubMed] [Google Scholar]
- Ono S., Minami N., Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Patel N., Huang C., Klip A. Cellular location of insulin-triggered signals and implications for glucose uptake. Pfluegers Arch. 2006;451:499–510. doi: 10.1007/s00424-005-1475-6. [DOI] [PubMed] [Google Scholar]
- Patel N., Rudich A., Khayat Z. A., Garg R., Klip A. Intracellular segregation of phosphatidylinositol-3,4,5-trisphosphate by insulin-dependent actin remodeling in L6 skeletal muscle cells. Mol. Cell. Biol. 2003;23:4611–4626. doi: 10.1128/MCB.23.13.4611-4626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrollier K., Hajduch E., Gray A., Litherland G. J., Prescott A. R., Leslie N. R., Hundal H. S. A role for the actin cytoskeleton in the hormonal and growth-factor-mediated activation of protein kinase B. Biochem. J. 2000;352(Pt 3):617–622. [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Randhawa V. K., Ishikura S., Talior-Volodarsky I., Cheng A. W., Patel N., Hartwig J. H., Klip A. GLUT4 vesicle recruitment and fusion are differentially regulated by Rac, AS160, and Rab8A in muscle cells. J. Biol. Chem. 2008;283:27208–27219. doi: 10.1074/jbc.M804282200. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Rossman K. L., Der C. J., Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Ruderman N. B., Kapeller R., White M. F., Cantley L. C. Activation of phosphatidylinositol 3-kinase by insulin. Proc. Natl. Acad. Sci. USA. 1990;87:1411–1415. doi: 10.1073/pnas.87.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Eguez L., Teruel M. N., Fukuda M., Chuang T. D., Chavez J. A., Lienhard G. E., McGraw T. E. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sano H., Kane S., Sano E., Miinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- Shaw A. E., Minamide L. S., Bill C. L., Funk J. D., Maiti S., Bamburg J. R. Cross-reactivity of antibodies to actin- depolymerizing factor/cofilin family proteins and identification of the major epitope recognized by a mammalian actin-depolymerizing factor/cofilin antibody. Electrophoresis. 2004;25:2611–2620. doi: 10.1002/elps.200406017. [DOI] [PubMed] [Google Scholar]
- Sidani M., et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosairajah J., Maiti S., Wiggan O., Sarmiere P., Moussi N., Sarcevic B., Sampath R., Bamburg J. R., Bernard O. Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 2005;24:473–486. doi: 10.1038/sj.emboj.7600543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector I., Shochet N. R., Kashman Y., Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Miki H., Takenawa T. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun. 1999;260:296–302. doi: 10.1006/bbrc.1999.0894. [DOI] [PubMed] [Google Scholar]
- Talior-Volodarsky I., Randhawa V. K., Zaid H., Klip A. Alpha-actinin-4 is selectively required for insulin-induced GLUT4 translocation. J. Biol. Chem. 2008;283:25115–25123. doi: 10.1074/jbc.M801750200. [DOI] [PubMed] [Google Scholar]
- Thong F. S., Bilan P. J., Klip A. The Rab GTPase-activating protein AS160 integrates Akt, protein kinase C, and AMP-activated protein kinase signals regulating GLUT4 traffic. Diabetes. 2007;56:414–423. doi: 10.2337/db06-0900. [DOI] [PubMed] [Google Scholar]
- Tong P., Khayat Z. A., Huang C., Patel N., Ueyama A., Klip A. Insulin-induced cortical actin remodeling promotes GLUT4 insertion at muscle cell membrane ruffles. J. Clin. Invest. 2001;108:371–381. doi: 10.1172/JCI12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok D., Patel N., Jebailey L., Thong F. S., Randhawa V. K., Klip A., Rudich A. Insulin but not PDGF relies on actin remodeling and on VAMP2 for GLUT4 translocation in myoblasts. J. Cell Sci. 2004;117:5447–5455. doi: 10.1242/jcs.01421. [DOI] [PubMed] [Google Scholar]
- Trifaro J. M., Gasman S., Gutierrez L. M. Cytoskeletal control of vesicle transport and exocytosis in chromaffin cells. Acta Physiol. 2008;192:165–172. doi: 10.1111/j.1748-1716.2007.01808.x. [DOI] [PubMed] [Google Scholar]
- Tsakiridis T., McDowell H. E., Walker T., Downes C. P., Hundal H. S., Vranic M., Klip A. Multiple roles of phosphatidylinositol 3-kinase in regulation of glucose transport, amino acid transport, and glucose transporters in L6 skeletal muscle cells. Endocrinology. 1995;136:4315–4322. doi: 10.1210/endo.136.10.7664650. [DOI] [PubMed] [Google Scholar]
- Tsakiridis T., Vranic M., Klip A. Disassembly of the actin network inhibits insulin-dependent stimulation of glucose transport and prevents recruitment of glucose transporters to the plasma membrane. J. Biol. Chem. 1994;269:29934–29942. [PubMed] [Google Scholar]
- Ueda S., et al. Crucial role of the small GTPase Rac1 in insulin-stimulated translocation of glucose transporter 4 to the mouse skeletal muscle sarcolemma. FASEB J. 2010;24:2254–2261. doi: 10.1096/fj.09-137380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama A., Yaworsky K. L., Wang Q., Ebina Y., Klip A. GLUT-4myc ectopic expression in L6 myoblasts generates a GLUT-4-specific pool conferring insulin sensitivity. Am. J. Physiol. 1999;277:E572–E578. doi: 10.1152/ajpendo.1999.277.3.E572. [DOI] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 2008;87:649–667. doi: 10.1016/j.ejcb.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Wang Q., Bilan P. J., Tsakiridis T., Hinek A., Klip A. Actin filaments participate in the relocalization of phosphatidylinositol3-kinase to glucose transporter-containing compartments and in the stimulation of glucose uptake in 3T3–L1 adipocytes. Biochem. J. 1998;331(Pt 3):917–928. doi: 10.1042/bj3310917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Somwar R., Bilan P. J., Liu Z., Jin J., Woodgett J. R., Klip A. Protein kinase B/Akt participates in GLUT4 translocation by insulin in L6 myoblasts. Mol. Cell. Biol. 1999;19:4008–4018. doi: 10.1128/mcb.19.6.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Sun W., Kujala P., Lotfi Y., Vida T. A., Bean A. J. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol. Biol. Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Higuchi O., Ohashi K., Nagata K., Wada A., Kangawa K., Nishida E., Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Zaid H., Antonescu C. N., Randhawa V. K., Klip A. Insulin action on glucose transporters through molecular switches, tracks and tethers. Biochem. J. 2008;413:201–215. doi: 10.1042/BJ20080723. [DOI] [PubMed] [Google Scholar]
- Zierath J. R., He L., Guma A., Odegoard Wahlstrom E., Klip A., Wallberg-Henriksson H. Insulin action on glucose transport and plasma membrane GLUT4 content in skeletal muscle from patients with NIDDM. Diabetologia. 1996;39:1180–1189. doi: 10.1007/BF02658504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.