The pH and lumenal environment of intracellular organelles is considered essential for protein sorting and trafficking through the cell. We provide the first evidence that a mammalian sodium (potassium)/proton exchanger, NHE8, plays a key role in the control of protein trafficking and endosome morphology.
Abstract
The pH and lumenal environment of intracellular organelles is considered essential for protein sorting and trafficking through the cell. We provide the first evidence that a mammalian NHE sodium (potassium)/proton exchanger, NHE8, plays a key role in the control of protein trafficking and endosome morphology. At steady state, the majority of epitope-tagged NHE8 was found in the trans-Golgi network of HeLa M-cells, but a proportion was also localized to multivesicular bodies (MVBs). Depletion of NHE8 in HeLa M-cells with siRNA resulted in the perturbation of MVB protein sorting, as shown by an increase in epidermal growth factor degradation. Additionally, NHE8-depleted cells displayed striking perinuclear clustering of endosomes and lysosomes, and there was a ninefold increase in the cellular volume taken up by LAMP1/LBPA-positive, dense MVBs. Our data points to a role for the ion exchange activity of NHE8 being required to maintain endosome morphology, as overexpression of a nonfunctional point mutant protein (NHE8 E225Q) resulted in phenotypes similar to those seen after siRNA depletion of endogenous NHE8. Interestingly, we found that depletion of NHE8, despite its function as a sodium (potassium)/proton antiporter, did not affect the overall pH inside dense MVBs.
INTRODUCTION
The eukaryotic NHE family of sodium/proton exchangers is a large group of transmembrane proteins involved in the electroneutral exchange of sodium (or, in some cases, potassium) ions for protons across membranes. These exchangers catalyze ion transport down concentration gradients and are involved in many diverse cellular processes including the regulation of intracellular pH, cell volume, absorption of sodium into epithelia, salt tolerance, cell adhesion, cell proliferation, organelle biogenesis, and protein trafficking (Orlowski and Grinstein, 2004). The NHE proteins can be broadly divided into two subfamilies, based on cellular localization and protein sequence alignments: a cell surface subfamily, including mammalian NHEs 1–5; and an intracellular subfamily localized to the membranes of organelles, including mammalian NHEs 6–9, and yeast (Saccharomyces cerevisiae) Nhx1p (Orlowski and Grinstein, 2004, 2007; Brett et al., 2005). Further phylogenetic analysis of the intracellular subfamily NHE genes results in their subdivision into three groups (Brett et al., 2005):
The plant vacuolar NHE group, including rice (Oryza sativa) OsNHX1 and OsNHX2, and Japanese morning glory (Ipomoea nil) InNHX1. This group of exchangers is proposed to be involved in salt tolerance and pH regulation of the plant vacuole (Serrano and Rodriguez-Navarro, 2001);
The endosome/trans-Golgi network (TGN)-localized NHE group, including S. cerevisiae NHX1 and mammalian NHE6, NHE7, and NHE9. These exchangers are proposed to be involved in pH regulation inside endosomal and Golgi compartments within the cell (Nass and Rao, 1998, 1999; Bowers et al., 2000; Nakamura et al., 2005);
A group of NHE genes designated “NHE8-like,” including mammalian NHE8 and slime mold (Dictyostelium discoideum) DdNHE. NHE8 has been proposed to regulate medial/trans-Golgi pH, as well as having a role in sodium uptake in the kidney and intestine (Goyal et al., 2005; Nakamura et al., 2005; Xu et al., 2005, 2008; Bobulescu and Moe, 2009).
NHX1 is the only NHE gene in the S. cerevisiae genome, and we have previously shown that its deletion or mutation resulted in cells that fail to sort proteins correctly to the yeast vacuole (Bowers et al., 2000). Our results showed that Nhx1p was localized to the yeast endosome and suggested that the ion exchange activity of Nhx1p was essential for its role in protein trafficking (Bowers et al., 2000). Furthermore, our data suggested that NHX1 belongs to the E class of VPS genes that, when mutated, result in a distinctive aberrant endosome compartment (often referred to as the “class E compartment”; Raymond et al., 1992). Interestingly, Nhx1p is the only transmembrane protein in this class of Vps proteins: the other class E proteins are peripheral membrane proteins and are components of, or associate with, the four endosomal sorting complexes required for transport (ESCRT-0, I, II, and III; Bowers and Stevens, 2005; Babst, 2006; Raiborg and Stenmark, 2009). In the endocytic pathway, ESCRTs are essential for the sorting of protein cargoes into the intralumenal vesicles of multivesicular endosomes/multivesicular bodies (MVBs; Hurley and Emr, 2006; Hurley, 2008; Raiborg and Stenmark, 2009). Much recent work has concentrated on finding and studying the functions of the mammalian ESCRT proteins. However, it remains unclear whether one or more of the mammalian NHEs have a functional role in endosomal protein sorting.
In this study, we have investigated the possible involvement of NHE proteins in endosomal protein sorting and trafficking in mammalian cells. We chose to study the ubiquitously expressed, intracellular NHEs 6–9, as they are most closely related to yeast Nhx1p and the most likely to be found on endosomal organelles.
MATERIALS AND METHODS
Plasmid Construction
Human NHE8 was amplified from clone DKFZp686C03237 from the RZPD Deutsches Ressourcenzentrum fuer Genomforschung GmbH (Berlin, Germany). PCR was used to amplify the NHE8 sequence with a Kozak sequence before the ATG and a single hemagglutinin (HA) epitope tag immediately before the stop codon. The resulting NHE8-HA fragment was subcloned into the pIRESneo2 vector (Clontech, Palo Alto, CA) for expression in mammalian cells (pKEB316). To generate the siRNA-resistant version of NHE8-HA (pKEB366), 22 silent mutations were introduced into pKEB316 by PCR, changing the sites targeted by the four siRNA oligos to no. 1: CAATACTGCAGAGGGATTA; no. 2: GACGTTATTTCAAAGCTGA; no. 3: GGACCGCTATTTCTGCCTT; and no. 4: GAAGAAAGATTTCCGAACA (silent mutations are shown in bold type). The numbers correspond to the oligos for knockdown (see below). To generate the siRNA-resistant NHE8 E225Q HA mutant (pKEB367), site-directed mutagenesis by PCR using pKEB366 as a template was used to change the GAA (E) codon to CAA (Q).
Primary Antibodies
Mouse monoclonal antibodies against transferrin receptor (TfR; H68.4), EEA1 (14), lysosome-associated membrane protein 1 [LAMP1 (H4A3)], and HA tag (HA.11), were from Invitrogen (Carlsbad, CA), BD Biosciences (San Jose, CA), the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City) and Covance (Madison, WI), respectively. The rabbit anti-mouse IgG was from Dako (Carpinteria, CA). The rabbit polyclonal antibodies against calreticulin (PA3–900), green fluorescent protein (GFP; ab290) and dinitrophenol (DNP) were from Affinity BioReagents (Golden, CO), Abcam (Cambridge, MA), and Molecular Probes (Invitrogen), respectively. The sheep anti-TGN46 (AHP500G) was from Serotec (Oxford, United Kingdom). The anti-LBPA antibody was a gift from Prof. Jean Gruenberg, University of Geneva, Switzerland, the rabbit anti-TGN46 (for electron microscopy [EM]) was a gift from Dr. Vas Ponnambalam (University of Leeds, United Kingdom).
Small Interfering RNA Knockdowns
The small interfering RNA (siRNA) oligonucleotides against NHE6, 7, 8, or 9 coding sequences were SMARTpools of four oligonucleotides purchased from the siGENOME collection of Dharmacon (Boulder, CO), The target sequences of the NHE8 oligonucleotides were as follows: no. 1: CAACACAGCTGAAGGTTTA; no. 2: GATGTAATCTCTAAACTCA; no. 3: GGACGGCAATCTCCGCTTT; and no. 4: GAGGAGAGGTTCCCCAATA. The TSG101 and Alix siRNA duplexes were as described (Garrus et al., 2001; Matsuo et al., 2004). All oligonucleotides had 3′-UU overhangs. HeLa M-cells (n = 4 × 105; Tiwari et al., 1987) were seeded per 6-cm dish and transfected the following day with siRNA oligonucleotides as described (Motley et al., 2003). This was a double-knockdown protocol with siRNA transfections on days 0 and 2. For TSG101 siRNA transfections, the cells were transfected on days 1 and 2 to avoid the cell death seen when using the longer protocol.
Western Blotting
Western blotting was performed as previously described (Bowers et al., 2006).
Immunofluorescence
On day 3 of the siRNA transfection protocol, HeLa M-cells were seeded onto glass coverslips for processing on day 4. Cells were fixed in 3% (wt/vol) paraformaldehyde for 30 min and permeablized with 0.05% (wt/vol) saponin. All antibody incubations were carried out in PBS with 0.2% (wt/vol) bovine serum albumin and 3% (wt/vol) goat serum (Sigma). Secondary antibodies were Alexa Fluor conjugates used at a final concentration of 4 μg/ml (Molecular Probes, Invitrogen). To label acidic organelles, cells were incubated on day 4 of the siRNA transfection protocol for 30 min at 37°C in culture medium containing 30 μM 3-(2,4-dinitroanilino)-3′-amino-N-methyldipropylamine (DAMP; Molecular Probes, Invitrogen) before fixation and staining with anti-DNP antibodies and secondary antibodies as described above.
Images of stained cells were collected with either a Zeiss Axioskop 2 (plus) upright microscope (Thornwood, NY) with 63× Plan-APOCHROMAT/100× Plan-NEOFLUOR objectives (NA 1.4 and 1.3, respectively) connected to a Hamamatsu ORCA-ER camera (Bridgewater, NJ) and processed using Openlab 4.04 (Improvision, Lexington, MA) and Adobe Photoshop 10.0.1 (San Jose, CA) or a Leica confocal upright microscope (TCS SPE; Milton Keynes, United Kingdom) with a 63× Apochromat objective (numerical aperture 1.3). Leica optimized confocal Z-sections were taken and displayed as an extended focus projection using Volocity LE software 5.1 (Improvision).
Epidermal Growth Factor Degradation Assays
Epidermal growth factor (EGF) degradation assays were performed 2 d after the second siRNA transfection (day 4), exactly as described previously (Bowers et al., 2006). 125I-EGF (Perkin Elmer-Cetus Life Sciences, Waltham, MA) was used at 2.5 ng/ml.
Immunogold Electron Microscopy
HeLa M-cells stably expressing HA-tagged NHE8 were washed with PBS, fixed with 4% (wt/vol) paraformaldehyde/0.1% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2), and pelleted in an Eppendorf tube (13,000 rpm for 5 min). The fixative was aspirated, and the cell pellet was resuspended in warm 10% (wt/vol) gelatin in PBS. The cells were then pelleted (13,000 rpm for 5 min) and the gelatin-enrobed cells were set on ice, trimmed into 1-mm3 blocks, and infused with 1.7 M sucrose/15% (wt/vol) polyvinylpyrolidone for 24 h at 4°C. The blocks were subsequently mounted on cryostubs and snap-frozen in liquid nitrogen. Frozen ultrathin sections were cut using a diamond knife in an ultramicrotome with a cryochamber attachment (Leica) at −120°C, collected from the knife-edge with 50:50 2% (wt/vol) methyl cellulose: 2.3 M sucrose (Liou et al., 1996) and mounted on formvar-carbon–coated EM grids.
Sequential immunolabeling of TGN46 and HA (using rabbit anti-TGN46 and mouse anti-HA) was performed using the protein A-gold technique at room temperature (Slot et al., 1991). The sections were contrasted by embedding in 1.8% (wt/vol) methyl cellulose/0.3% (wt/vol) uranyl acetate and air-dried before observation in a Philips CM100 transmission electron microscope (Eindhoven, The Netherlands) at an operating voltage of 80 kV.
Transmission Electron Microscopy
After treatment with NHE8 siRNA HeLa M-cells were fixed with 2% (wt/vol) paraformaldehyde/2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate, pH 7.2, at room temperature, processed for transmission electron microscopy as previously described (Bright et al., 1997), and embedded in Agar 100 resin (Agar Scientific, Stansted, United Kingdom). Ultrathin sections (50 nm) were cut and mounted on formvar/carbon-coated EM grids. The sections were stained with uranyl acetate and lead citrate and observed in a Philips CM100 transmission electron microscope at an operating voltage of 80 kV.
EM Quantitation
MVB Volume Fraction.
Quantitation of the cytosolic volume occupied by MVBs was performed by point counting with a 500-nm lattice overlay (Griffiths, 1993) using iTEM software (Olympus Soft Imaging Solutions, Melville, NY). EM blocks, grids, and sections were selected randomly, sections were scanned systematically and a MegaView III CCD camera was used to record 200 images for each experiment. MVBs containing ≤10 internal vesicles (electron lucent appearance by EM) were defined as electron lucent MVBs and MVBs containing >10 internal vesicles (electron-dense appearance by EM) were defined as electron-dense MVBs.
Distribution of HA Immunolabeling.
Quantitation of the labeling density of HAs associated with the TGN, MVBs, or vesicular/tubular clusters (VTCs) greater than 500 nm from a Golgi stack (including particles that were at or near the plasma membrane [PM]) was determined by counting gold particles associated with these morphologically identified compartments from three independent labeling experiments.
Estimation of MVB Lumenal pH Using DAMP Accumulation and Anti-DNP Immunolabeling.
The pH of MVBs was estimated using the formula below, as previously described (Orci et al., 1986). Electron-dense MVB were defined as those with >10 internal vesicles. Forty images per condition were analyzed, as follows: pH = 7.0 − log D/N, where 7.0 = pH at neutrality, N = density of gold particles in a pH 7.0 compartment (the nucleus), and D = density of DAMP-specific gold particles in the endosomes.
Vesicular Stomatitis Virus Glycoprotein–Enhanced Green Fluorescent Protein Transport Assay
HeLa M-cells stably expressing vesicular stomatitis virus glycoprotein (VSVG)–enhanced green fluorescent protein (EGFP) were transfected with NHE8 siRNA in a double-knockdown protocol with transfections on days 0 and 2. On day 3, the cells were transferred into six-well plates and incubated at 40°C for 24 h to accumulate VSVG–EGFP in the ER. Cells were shifted to 32°C for 0, 30, 60, and 180 min in the presence of cycloheximide (50 μg/ml). The cells were then cooled and surface-labeled with 0.5 mg/ml of the cell impermeable EZ-Link Sulfo-NHS-SS-Biotin for 40 min. Excess label was quenched, cells were lysed, and equal amounts of protein were incubated with neutravidin agarose beads for 2 h. The precipitated, biotinylated proteins were Western-blotted using anti-GFP and anti-rabbit IRDye680 (Li-Cor Biosciences, Lincoln, NE), and the resultant images were quantified using the Li-Cor Odyssey Infrared Imaging System.
Secretion Assay Using Metabolic Labeling
HeLa M-cells were transfected with NHE8 siRNA using a double-knockdown protocol. On day 4 of the siRNA transfection protocol HeLa M-cells were incubated in methionine-, cysteine-, and serum-free DMEM for 1 h, pulsed for 15 min with 50 μCi/ml EasyTag EXPRESS35S protein-labeling mix (Perkin Elmer-Cetus), washed in PBS, and chased in DMEM with 5 mM methionine and 1 mM cysteine for 0, 30, 60, and 180 min. The culture medium was removed, cells and debris were pelleted, and secreted proteins were precipitated with TCA onto glass microfiber discs that were then mixed with scintillant and counted in a liquid scintillation analyzer (TRI-CARB 2900TR, Packard Instruments, Downers Grove, IL).
Total protein levels were determined following lysis of the attached cells for each sample, and secreted protein was normalized as cpm per μg total protein.
RESULTS
NHE8 Regulates EGF Degradation and Endosomal Morphology
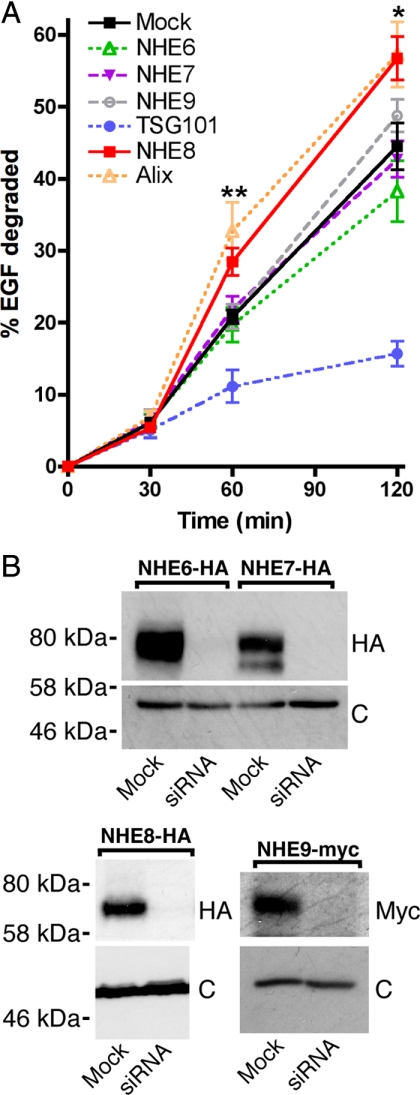
Our previous data demonstrated that yeast Nhx1p is essential for protein sorting at the MVB in yeast (Bowers et al., 2000). To establish whether the mammalian intracellular NHE proteins were also required for endocytic trafficking, we investigated the effect of depletion of NHE6, NHE7, NHE8, or NHE9 on MVB protein sorting in HeLa M-cells. 125I-EGF degradation was chosen as a readout, as the lysosomal degradation of EGF is preceded by the sorting of the growth factor/receptor complex into the intralumenal vesicles of MVBs. Indeed, perturbation of EGF sorting into intralumenal MVB vesicles resulting from the depletion of TSG101 (a subunit of ESCRT-1) significantly inhibits 125I-EGF degradation (Figure 1A; Doyotte et al., 2005; Bowers et al., 2006; Razi and Futter, 2006; Raiborg et al., 2008). To investigate the effect of NHE depletion on EGF degradation, we used pools of siRNA to deplete endogenous NHE6, 7, 8, or 9 from HeLa M-cells and followed a double-transfection protocol. To show efficient depletion of the NHE proteins (and in the absence of reliable anti-NHE antibodies), we generated stable cell lines expressing epitope-tagged versions of NHE6, 7, 8, or 9. As shown in Figure 1B, transfection of siRNA against NHE6, -7, -8, or -9 depleted NHE6-HA, NHE7-HA, NHE8-HA, or NHE9-Myc, respectively. As our siRNA transfection procedure depleted overexpressed, tagged versions of the proteins, we concluded that it was also effective toward endogenous NHE6, 7, 8, or 9 (as in Figure 1A).
Figure 1.
Depletion of human NHE8 affects EGF degradation. (A) HeLa M-cells were transfected with either water (mock) or siRNA oligos directed against TSG101, Alix, or pools of 4 oligos against NHE6, 7, 8, or 9 at 100 nM using a double-transfection protocol. 125I-EGF degradation over time was assayed as described in Materials and Methods. Error bars, ±SE over 10, 3, 4, 6, or 6 separate experiments for mock, NHE6-, TSG101-, NHE8-, or Alix-depleted cells, respectively; and the deviation from the mean of two separate experiments for NHE7 or 9-depleted cells. Unpaired t tests showed that mock- and NHE8-depleted cells had significant differences in EGF degradation at 60 and 120 min. *p = 0.024, **p = 0.0012. There were no significant differences between EGF degradation in the mock cells compared with cells depleted for NHE6, 7, or 9. (B) Depletion of NHE6, 7 or 9. HeLa M-cells stably expressing NHE6-HA, NHE7-HA, NHE8-HA or NHE9-myc were transfected with water (mock) or pools of siRNA oligos against NHE6, 7, 8, or 9 at 100 nM using a double-transfection protocol. Western blots were developed using either anti-HA or anti-myc antibodies (top panels). Levels of calreticulin were assessed in the same samples as loading controls (c, bottom panels).
As shown in Figure 1A, depletion of endogenous NHE6, 7, or 9 had no significant effect on 125I-EGF degradation. However, depletion of NHE8 resulted in a modest but significant increase in EGF degradation at both the 60- (p < 0.005) and 120-min (p < 0.05) time points compared with mock-transfected cells. Whereas depletion of most ESCRT proteins inhibits MVB protein sorting, depletion of the ESCRT-III- associated protein Alix does not, and in our hands Alix depletion actually led to an increase in EGF degradation (Figure 1A and Bowers et al., 2006).
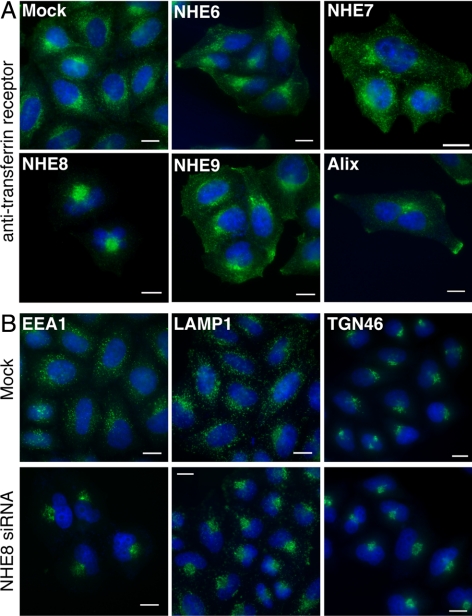
Mutation of ESCRT genes or NHX1 in yeast results in a clear morphological defect, the presence of the aberrant “class E” compartment. The class E compartment appears as a multilamellar structure by EM (Rieder et al., 1996; Odorizzi et al., 1998). Depletion of ESCRT proteins in mammalian cells gives more varied morphological phenotypes, e.g., TSG101-depleted cells have swollen endosomes visible by light microscopy and large multicisternal structures and tubular clusters by EM, but Alix-depleted cells show clustered endosomes in the perinuclear region that do not appear significantly enlarged (Cabezas et al., 2005; Doyotte et al., 2005; Bowers et al., 2006; Razi and Futter, 2006). To examine whether depletion of the mammalian NHEs caused a change in endosomal morphology, we investigated the localization of an endosomal marker protein after endogenous NHE6, 7, 8, or 9 depletion. The TfR at steady state in mock-transfected HeLa M-cells was found on the cell surface, in perinuclear recycling endosomes and occasionally at the cell tips (Figure 2A, Figure S3). Cell surface and perinuclear staining of the TfR was also seen in NHE6, 7, 8, and 9-depleted cells, but recycling endosomes were clustered into a tighter perinuclear spot in NHE8-depleted cells. This clustering phenotype is similar to, though more pronounced than, the phenotype seen upon Alix depletion (Figure 2A; Cabezas et al., 2005). Alix-depleted cells were frequently binucleate, consistent with the role of Alix in cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007). In Alix-depleted cells, we also observed that the peripheral TfR staining was almost always localized to the cell tips, unlike in the mock or NHE-depleted cells (Figure 2A and data not shown). Alix is proposed to be a negative regulator of the inward vesiculation of MVBs in vitro (Falguieres et al., 2008), and the similarity of the phenotypes of Alix-depletion versus NHE8-depletion led us to hypothesize that NHE8 might also negatively regulate this process. Therefore, we chose NHE8 for further study.
Figure 2.
Depletion of NHE8 affects endosome morphology (A) HeLa M-cells were transfected with different siRNA oligos as in Figure 1 and fixed and stained using an antibody against the TfR (green). Nuclei were stained with DAPI (blue). (B) Cells were transfected with either water (mock) or the pool of four NHE8 siRNA oligos at 100 nM (NHE8 siRNA) using a double-transfection protocol and were stained using antibodies to EEA1, LAMP1, or TGN46 (green). Nuclei were stained with DAPI (blue). Scale bars, 15 μm.
To analyze further the endosomal clustering phenotype in the NHE8-depleted cells, we transfected HeLa M-cells with NHE8 siRNA and stained the cells with antibodies against a variety of markers. In mock-transfected cells, early endosomes (stained with antibodies against early endosome autoantigen 1 [EEA1]) and late endosomes/lysosomes (stained with antibodies against LAMP1) showed punctate staining patterns in the perinuclear region of the cells (Figure 2B). After NHE8 depletion, both early endosome and late endosome/lysosome distribution was altered, with a tight cluster of staining observed in the perinuclear region (Figure 2B). Costaining experiments suggested that although the early and late endosomes and lysosomes clustered in the same region of the NHE8-depleted cells, the compartments were still distinct (Figure S1). The perinuclear pattern of TGN staining, as labeled by TGN46, seemed largely unaffected by NHE8 depletion (Figure 2B).
At Steady State, Epitope-tagged NHE8 Is Localized to the TGN, MVBs, and VTCs
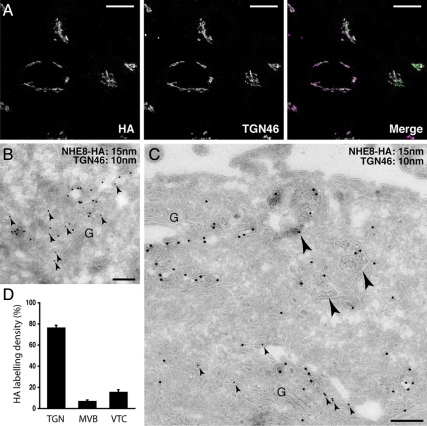
A previous study suggested that endogenous or transiently expressed, tagged NHE8 was localized to medial and/or trans-Golgi compartments in COS7 cells, due to fact that NHE8 staining was adjacent to that of the cis-Golgi marker GM130 (Nakamura et al., 2005). Although a commercial antibody against NHE8 exists, we were unable to detect specific staining by immunofluorescence or Western blot for endogenous NHE8 in HeLa M-cells using this reagent. Therefore, to study the subcellular distribution of NHE8 in HeLa M-cells, we made a stable cell line expressing human NHE8 with a C-terminal HA tag (NHE8-HA cells) and localized the HA tag in these cells by confocal microscopy. We found that NHE8 was predominantly in the TGN, where it colocalized with the trans-Golgi marker protein TGN46 (Figure 3A). To investigate the localization of NHE8 in more detail, we used electron microscopy. As shown in Figure 3, B–D, and in agreement with our immunofluorescence experiments, 77% of total NHE8-HA was found in the TGN at steady state, where it colocalized with TGN46. However, 7% of NHE8-HA was found on MVBs, and 16% on VTCs (Figure 3, C and D). The localization of NHE8-HA to structures other than the TGN was consistent with the punctate staining seen throughout the cytosol when the brightness and contrast of immunofluorescence images was increased (Figure S2, bottom left image).
Figure 3.
NHE8-HA is localized to the TGN and MVB. (A) HeLa M-cells were stably transfected with NHE8-HA and immunolabeled with anti-HA antibodies or anti-TGN46 antibodies. A series of confocal Z-stacks (Leica optimized) were taken and are displayed as an extended focus view with the merged image in the right panel. Colocalization of TGN46 (magenta) and NHE8-HA (green) appears white in the merged image. Scale bar, 17 μm. (B and C) HeLa M-cells stably expressing NHE8-HA were prepared for immuno-EM and labeled with antibodies to TGN46 (small arrowheads; 10-nm gold) or the HA tag (15-nm gold). (B) TGN46 and NHE8-HA coimmunolabeled vesicles and cisternae of the TGN adjacent to the Golgi stack (G). (C) NHE8-HA immunolabeling could also be identified within MVB (large arrowheads) on ultrathin cryosections. (D) Quantitation of the labeling density of HA associated with the TGN, MVBs, or vesicular/tubular clusters (VTCs) greater than 500 nm from the Golgi stack from three independent immunolabeling experiments ± SEM. Scale bars, 200 nm.
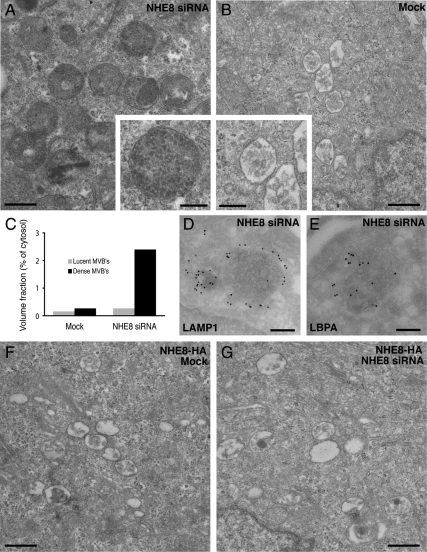
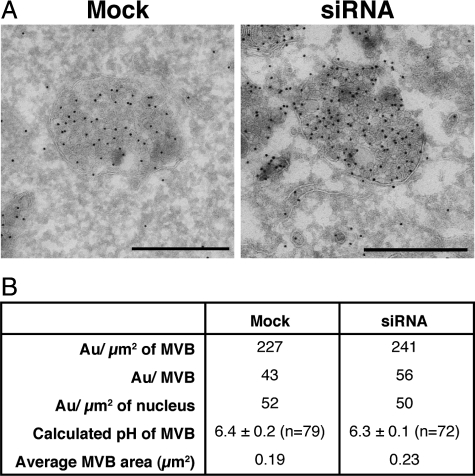
NHE8-depleted Cells Show an Increase in Electron-Dense MVBs
To examine the phenotype of NHE8-depleted HeLa M-cells further, we turned to EM. By EM, it was striking that although the mock-transfected cells contained both MVBs with <10 internal vesicles (lucent MVBs) and MVBs with >10 internal vesicles (dense MVBs), the proportion of the cell taken up by dense MVBs in the NHE8-depleted cells was increased (Figure 4, A and B). This difference was quantified by calculating the volume fraction taken up by lucent and dense MVBs in mock versus NHE8-depleted cells. As depicted in Figure 4C, there was a ninefold increase in the percentage of cytosol taken up by the dense MVBs in NHE8-depleted cells. Immunolabeling of sections from the NHE8-depleted cells showed that the dense MVBs in these cells were positive for both lysosome-associated membrane protein 1 (LAMP1) and lysobisphosphatidic acid (LBPA) (Figure 4, D and E), suggesting that these compartments are late endocytic organelles.
Figure 4.
Down-regulation of NHE8 results in an accumulation of enlarged electron-dense MVBs. (A) HeLa M-cells were treated with NHE8 siRNA using a double-knockdown protocol and processed for transmission EM. The resultant phenotype was characterized by an accumulation of enlarged electron-dense MVBs compared with mock-treated cells (B). C) Quantitation of the proportion of cytosol occupied by electron-lucent or electron-dense MVBs in mock or NHE8 siRNA-treated cells. Electron-dense MVBs in NHE8 siRNA-treated cells were immunolabeled with antibodies to LAMP1 (D) or LBPA (E) followed by protein A–15 nm gold on ultrathin cryosections. HeLa M-cells expressing NHE8-HA possess MVBs with morphology indistinguishable from mock-transfected cells (F), and expression of NHE8-HA rescues the dense MVB phenotype in cells depleted of NHE8 by treatment with siRNA (G). Scale bars, (A and B, F and G) 500 nm; (D and E) 200 nm.
To test whether the phenotype we saw after NHE8-depletion was a specific effect of the siRNA, we treated cells overexpressing NHE8 (the stable cell line overexpressing NHE8-HA in addition to the endogenous NHE8) with the pool of siRNA oligos against NHE8. Although a double-transfection procedure with the pool of siRNA oligos gave the most effective depletion, a small amount of NHE8-HA was still detectable after the double siRNA transfection protocol (Figure S2), and we saw no increase in dense MVB compared with mock-transfected cells (Figure 4, F and G). This suggests that overexpression of NHE8-HA is enough to “rescue” the morphological phenotype of NHE8 depletion and also suggests that a very low level of NHE8 is required for its endosomal function. This latter observation is consistent with our results from yeast, where we found that yeast Nhx1p was expressed at very low levels endogenously (Bowers et al., 2000) and also with our immunofluorescence microscopy studies of NHE8-HA, where we found that a double-transfection protocol, and high concentrations (100 nM) of siRNA were required to see the endosome-clustering phenotype in HeLa M-cells (data not shown).
To test further whether the altered endosome morphology phenotype was specific to NHE8-depletion rather than a result of an off-target effect, we analyzed HeLa M-cells that had been transfected with individual siRNA oligos against NHE8. As shown in Figure S3, transfection of the individual NHE8 siRNA oligos effectively depleted NHE8 and also resulted in the endosomal clustering and increased dense MVB typical of our NHE8 depletions using the oligo pool. In addition, as shown in Figure 5D, stable expression of siRNA-resistant NHE8-HA rescued the endosomal clustering phenotype seen in NHE8-depleted cells. Taken together (and with our data for the nonfunctional mutant of NHE8; see below), our data suggest that the endosomal phenotypes we observe are specific to depletion of endogenous NHE8.
Figure 5.
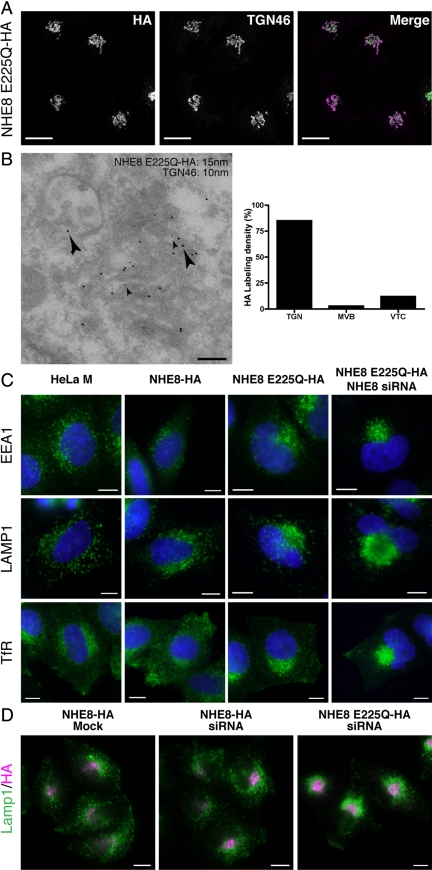
A nonfunctional mutant of NHE8 is localized to the TGN and causes endosomal clustering. (A) HeLa M-cells stably expressing NHE8 E225Q-HA were fixed and stained using anti-HA (green) and anti-TGN46 (magenta) antibodies. A series of confocal Z-stacks (Leica optimized) were taken and are displayed as an extended focus view. Scale bar, 17 μm. (B) Left, HeLa M-cells stably expressing NHE8 E225Q-HA were prepared for immuno-EM and labeled with antibodies to TGN46 (small arrowheads; 10-nm gold) or the HA tag (large arrowheads; 15-nm gold). Right, quantitation of the labeling density of HA associated with the TGN, MVBs or vesicular/tubular clusters (VTCs) greater than 500 nm from the Golgi stack from two independent immunolabeling experiments ± range. Scale bar, 200 nm. (C) HeLa M-cells, HeLa M-cells stably expressing NHE8-HA or NHE8 E225Q-HA, or HeLa M-cells stably expressing NHE8 E225Q-HA but with endogenous NHE8 depleted were fixed and stained with antibodies to label early or recycling endosomes (EEA1 or TfR, respectively) or late endosomes/lysosomes (LAMP1). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. (D) HeLa M-cells stably expressing siRNA-resistant NHE8-HA or siRNA resistant NHE8 E225Q-HA were transfected with siRNA to deplete endogenous NHE8. Cells were fixed and stained with antibodies to the HA tag (magenta) or to LAMP1 (green). Scale bar, 15 μm.
A Nonfunctional NHE8 Mutant Causes Endosome/Lysosome Clustering
To define further the functional role of NHE8 in the cell, we created a catalytically inactive form of NHE8. All NHE proteins have at least three highly conserved acidic residues in transmembrane domains that are essential for ion transport. For example, E262 in human NHE1 is essential for ion transport (Fafournoux et al., 1994), and mutation of D201, E225, or D230 in yeast Nhx1p renders the exchanger nonfunctional for protein trafficking (Bowers et al., 2000). E225 in NHE8 is the equivalent (by multiple amino acid sequence alignment of the NHE family) of E262 in NHE1 and E225 in Nhx1p and has been shown, in combination with D230, to be essential for NHE8 catalyzed ion transport in reconstituted liposomes (Nakamura et al., 2005; the amino acid numbering in the reference differs from ours by 4 amino acids at the N-terminus). Therefore, we made a nonfunctional version of NHE8-HA by mutating E225 (in predicted transmembrane domain 7) to Q. In addition, we rendered this construct resistant to siRNA by introducing 22 silent mutations in the regions targeted by the four siRNA oligos and generated a stable HeLa M-cell line expressing this mutant protein (NHE8 E225Q-HA cells). By immunofluorescence using anti-HA antibodies, we found that NHE8 E225Q-HA colocalized with TGN46 (Figure 5A), and by EM we saw NHE8 E225Q-HA in the TGN, MVBs, and VTCs (Figure 5B). However, although NHE8 E225Q-HA was localized similarly to NHE8-HA, the fraction found in MVBs at steady state was lower (3 compared with 7%) and the fraction found in the TGN higher (85 compared with 77%) than NHE8-HA. This difference may be a reflection of the nonfunctional state of the NHE8 protein, but may also be due to differences in expression levels in the stable cell lines.
To determine the consequence of overexpressing NHE8 E225Q-HA, we studied endosome morphology in the NHE8 E225Q-HA cells. As shown above, HeLa M-cells depleted of NHE8 by siRNA transfection had clustered early/recycling endosomes and late endosomes/lysosomes in the perinuclear region of the cell (Figure 2, Figure S1). The expression of NHE8 E225Q-HA in the presence of wild-type, endogenous NHE8 also resulted in a similar, although less dramatic, endosomal clustering phenotype (Figure 5C). These data suggest that the NHE8 E225Q mutant may act as a “dominant negative” mutant, inhibiting the function of wild-type NHE8. One possible explanation of the dominant negative effect is that NHE8, like NHE1, may function as a dimer (Hisamitsu et al., 2006; Moncoq et al., 2008). In this case, expression of NHE8 E225Q might inhibit the function of endogenous NHE8. Alternatively, we may be replacing functional NHE8 with nonfunctional NHE8 in MVBs due to overexpression of the NHE8 E225Q-HA mutant. The most severe endosome-clustering phenotype was consistently observed when endogenous NHE8 was depleted by siRNA transfection in NHE8 E225Q-HA cells (Figure 5, C and D). This severe endosome-clustering phenotype was not seen when endogenous NHE8 was depleted in cells stably expressing siRNA-resistant NHE8-HA (Figure 5D). Thus, cells depleted of wild-type NHE8 and expressing only a nonfunctional mutant protein show the strongest endosome morphology phenotype. This suggests that the ion exchange function of NHE8 is required for its role in maintaining endosome morphology.
At Steady State NHE8 Depletion Does Not Alter the pH of MVBs
As shown above, the depletion of NHE8 resulted in an increase in EGF degradation and dramatic morphological changes to endosomes. Intracellular NHEs have been proposed to regulate intracellular organellar pH (Nakamura et al., 2005); thus we decided to analyze the acidification of endosomal compartments in NHE8-depleted cells. To visualize acidic compartments, we utilized the properties of the fixable weak-base DAMP (Anderson et al., 1984) that readily diffuses and accumulates inside compartments due to their acidic pH. DAMP localizes to endocytic structures and TGN elements (Anderson et al., 1984; Anderson and Pathak, 1985).
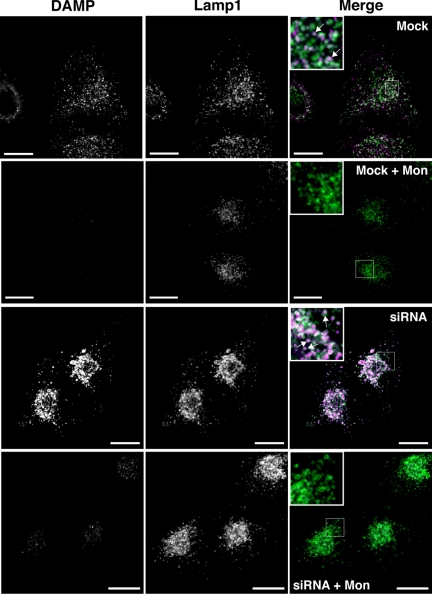
In mock-transfected HeLa M-cells, DAMP- and LAMP1-labeled structures colocalized and DAMP puncta were often encircled by LAMP1 staining, suggesting that these compartments were acidic late endosomes/lysosomes (Figure 6). After NHE8 depletion, the perinuclear LAMP1/DAMP-positive structures were enlarged and had visibly more fluorescent staining. DAMP accumulation is known to be dependent on the presence of a proton gradient (Anderson et al., 1984), and we found that incubation of HeLa M-cells with DAMP for 20 min followed by treatment with the artificial sodium/proton exchanger monensin for a further 10 min (still in the presence of DAMP) dramatically reduced the accumulation of DAMP (Figure 6). Similarly, preincubation of the cells in 24 mM NH4Cl for 10 min followed by a 30-min incubation in 24 mM NH4Cl with DAMP almost completely prevented DAMP accumulation (not shown). To study further the accumulation of DAMP and to see whether there was any pH change in these compartments, we labeled sections using anti-DNP antibodies (which recognize DAMP) and analyzed these by EM (Figure 7A). Although the electron-dense MVBs were enlarged with multiple intralumenal vesicles in NHE8-depleted cells, analysis of DAMP labeling showed that the pH of these compartments was almost identical to that of dense MVBs in mock-transfected cells (Figure 7B).
Figure 6.
NHE8 depletion causes an enlargement of acidic compartments. HeLa M-cells were treated with NHE8 siRNA using a double-knockdown protocol. Cells were treated with 30 μM DAMP for 20 min and then treated with or without 25 μM monensin (in the presence of DAMP) for a further 10 min before fixation. Cells treated with monensin are labeled “+ Mon” in the figure. Cells were immunolabeled with anti-LAMP1 antibodies (green) to label late endosomes/lysosomes and anti-DNP antibodies (magenta) to label the compartments containing DAMP. A series of confocal Z-stacks were taken with identical image settings and are displayed as an extended focus view. Enlarged areas are presented as inset boxes, and the arrows represent areas where the LAMP1 staining encircles the DNP staining. Scale bars, 17 μm.
Figure 7.
NHE8-depletion does not affect the overall lumenal pH of dense MVB. (A) HeLa M-cells were treated with NHE8 siRNA using a double-knockdown protocol. Cells were treated with 30 μM DAMP for 30 min before processing for EM. Immunolabeling was carried out with anti-DNP antibodies followed by protein A–15 nm gold on ultrathin cryosections to label the compartments containing DAMP. (B) Quantitation of the data in A. Bar, 500 nm.
NHE8 Depletion Does Not Affect the Rate of Constitutive Secretion
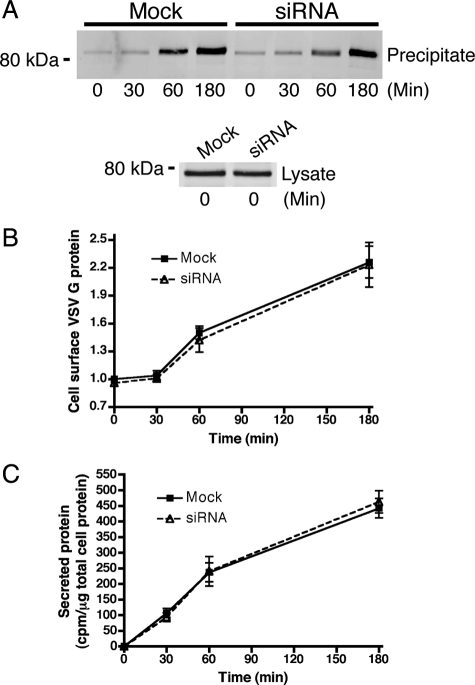
Although the overall aim of our study was to investigate the role of NHE proteins in endosomal protein trafficking and morphology, our discovery that the majority of NHE8 was in the TGN prompted us to investigate the effect of NHE8-depletion on constitutive secretion. As a reporter, we used the ts045 temperature-sensitive mutant of the VSVG tagged with EGFP (ts045 VSVG–GFP). The mutant protein is misfolded and retained in the ER at 40°C, but a pulse of protein can be chased into the secretory pathway and its transport through the secretory pathway followed by inhibiting protein synthesis and shifting the temperature to 32°C (Bergmann, 1989; Keller et al., 2001).
To quantify secretion of ts045 VSVG–GFP, we generated a stable HeLa M-cell line expressing the protein. After a 24-h incubation at 40°C and 0, 30, 60, or 180 min at 32°C, the cell surface was labeled with biotin, and the biotinylated surface proteins were precipitated with neutravidin beads. The amount of ts045 VSVG–GFP at the cell surface at each time point was assessed by Western blot (Figure 8A). Quantitation of the Western blot (Figure 8B) showed no significant difference in secretion of ts045 VSVG–GFP between mock and NHE8-depleted cells. A similar, qualitative experiment where we followed ts045 VSVG–GFP using microscopy to follow the fluorescent GFP tag gives a similar result and is shown in Figure S4. As controls, the chase was performed in the presence of the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or monensin. Both CCCP and monensin are known to inhibit the transport of ts045 VSVG–GFP from the TGN to the PM (Alonso-Caplen and Compans, 1983; Strous et al., 1983; Burkhardt and Argon, 1989). After 180 min of chase with CCCP, the arrival of ts045 VSVG–GFP on the cell membrane was dramatically slowed, whereas in the presence of monensin, ts045 VSVG–GFP transport out of the TGN was blocked (Figure S4).
Figure 8.
NHE8 depletion does not affect the rate of secretion. (A) HeLa M-cells stably expressing ts045 VSVG–GFP were transfected with NHE8 siRNA using a double-knockdown protocol. After incubation at the nonpermissive temperature to accumulate ts045 VSVG–GFP in the ER, cells were shifted to the permissive temperature for 0, 30, 60, or 180 min and surface-labeled with biotin. After cell lysis, cell surface, biotinylated proteins were precipitated with neutravidin agarose beads and levels of ts045 VSVG–GFP were determined at each time point using anti-GFP antibodies (top). Equal starting levels of ts045 VSVG–GFP were assessed in whole cell lysates (bottom). (B) Quantitation of the blot in A. Levels of cell surface ts045 VSVG–GFP are expressed relative to the level in mock-treated cells at t = 0 min (set to 1). Error bars, SE of the mean for each time point; n = 6. (C) Hela M-cells were treated with NHE8 siRNA using a double-knockdown protocol. The secretion of total protein was followed with a pulse with EasyTag EXPRESS35S protein-labeling mix and chase for 0, 30, 60, and 180 min. Error bars, SE of the mean, n = 3.
In addition to studying ts045 VSVG–GFP secretion, we also measured bulk constitutive secretion in metabolically labeled cells. As shown in Figure 8C and consistent with our ts045 VSVG–GFP experiments, no significant difference was seen between secretion of radiolabeled proteins in mock and NHE8-depleted cells.
In summary, our data shows endosomal phenotypes resulting from the depletion of endogenous NHE8: perinuclear clustering of early and late endosomes and lysosomes and an accumulation of dense MVBs. We saw a modest but significant increase in EGF degradation in NHE8-depleted cells, but no significant difference in the rate of protein secretion.
DISCUSSION
Our previous work identified a member of the NHE-family, Nhx1p, as essential for endosomal protein trafficking in the yeast S. cerevisiae. Here, we investigated the role of the mammalian intracellular NHEs and identified NHE8 as an intracellular NHE whose function is required for maintaining endosomal morphology and for the regulation of protein sorting.
NHE8 Is Found in the Golgi and Endosomes in HeLa M-Cells
Previous reports have localized NHE8 to the medial and trans-Golgi in COS7 cells (Nakamura et al., 2005), to the apical brush border membrane of kidney proximal tubule cells (Goyal et al., 2005), and to the apical membrane of rat intestinal epithelial cells (Xu et al., 2005). Our data suggest that in HeLa M-cells at steady state, the majority of NHE8-HA is localized to the TGN where it colocalizes with TGN46. However, we also find a population of NHE8-HA molecules in MVBs and VTCs (Figure 3). From our EM pictures using immunolabeling of the HA epitope tag on the extreme C-terminus of the protein, it would appear that there is a significant proportion of NHE8 on the intralumenal membranes as well as the limiting membrane of the MVBs. Although we cannot rule out possible mislocalization of NHE8-HA in our stable cell line due to over expression or the effect of the C-terminal epitope tag, the TGN/endosomal localization of NHE8-HA suggests that NHE8, like other TGN proteins such as furin (reviewed in Molloy et al., 1999), cycles in the cell. This is also consistent with a cell surface localization for the protein in some cell types (Goyal et al., 2003; Becker et al., 2007; Zhang et al., 2007), as it is conceivable that NHE8, like furin, may traffic over the PM during this cycle. Interestingly, NHE8 has putative targeting signals in its C-terminal domain that closely resemble those of furin: a tyrosine-based motif followed by an acidic cluster. Further work will be required to determine the trafficking route that endogenous NHE8 follows in the cell and the signals that direct it.
NHE8 Depletion Affects Endosomal Trafficking
The aim of this study was to investigate the role of intracellular NHEs in MVB protein sorting. To this end, the first assay we used measured EGF degradation, an indicator of MVB sorting. We discovered that depletion of NHE8 (but not NHE6, 7, or 9) resulted in a significant increase in the rate of EGF degradation (Figure 1). EGF remains associated with epidermal growth factor receptor (EGFR) until the pH drops to 5 (i.e., when the ligand/receptor complex reaches the lysosome; Sorkin et al., 1988). If NHE8 depletion were to reduce the pH of the MVBs, then it is possible that EGF could dissociate from its receptor and degradation of EGF would become independent of sorting into the intralumenal vesicles. However, we found no change in the overall pH of dense MVBs (i.e., MVBs with more than 10 intralumenal vesicles) after NHE8-depletion. We therefore conclude that EGF remained bound to EGFRs at the MVBs in the NHE8-depleted HeLa M-cells, and that the increase in EGF degradation that we observed was due to a perturbation in MVB protein sorting. It remains possible that the MVB sorting defect we see is an indirect result of NHE8-depletion (i.e., that NHE8 is required at a location other than the MVB for the trafficking or function of a factor essential for sorting of EGFR at MVBs). As the majority of NHE8 is found in the TGN, it is plausible that NHE8-depletion causes indirect effects on MVB sorting due to a direct effect on TGN function. However, we saw no change in the rate of secretion in NHE8-depleted cells (Figure 8) and no gross changes in the compartmentalization of any of the marker proteins that we studied (EEA1, TfR, LAMP1, and TGN46: Figure S2; cathepsin D, CD63, mannose 6-phosphate receptor: data not shown), although endosomes and lysosomes were clustered in the perinuclear region. In addition, we studied transport of a chimeric construct with the lumenal and transmembrane domains of CD8 and cytosolic domain of mannose 6-phosphate chimera and found that there was no major disruption of its transport from MVBs to the TGN (Figure S5). Therefore, all the evidence together suggests that the MVB sorting and morphology defects we observe after NHE8 depletion are direct effects on endosomes, because we were unable to identify any affect of NHE8 depletion on the functioning of the TGN.
Previous studies have shown that as early endosomes mature into late endosomes they have increasing numbers of intralumenal vesicles (Woodman and Futter, 2008). Therefore, we propose that the “lucent” MVBs we see by EM are earlier endosomes that will mature over time into the “dense” MVBs that label with LBPA and LAMP1 and will eventually fuse with lysosomes. Depletion of NHE8 results in an accumulation of dense MVBs in vivo (Figure 4) and so perhaps the larger numbers of intralumenal vesicles can sequester more EGF/EGFR per MVB for lysosomal degradation after MVB/lysosome fusion. One possibility to explain the increased dense MVBs in NHE8-depleted cells is the inhibition of fusion of these compartments with lysosomes. However, this is in contradiction to the increased EGF degradation that we observe. There are two remaining possibilities: 1) NHE8 is a negative regulator of inward vesiculation, or 2) NHE8 promotes fusion of the intralumenal vesicles with the limiting MVB membrane (“back fusion”). Interestingly, Alix has been proposed to be a negative regulator of inward MVB vesiculation and a positive factor in back fusion (Matsuo et al., 2004; Le Blanc et al., 2005; Falguieres et al., 2008; Luyet et al., 2008). Our results for NHE8 are also consistent with this idea, but currently, we cannot distinguish between these two possibilities or rule out an involvement for NHE8 in both.
How would a sodium/proton exchanger control inward vesiculation and/or back fusion? Matsuo et al. (2004) showed that liposomes with a lipid composition similar to that of late endosomes were capable of spontaneous inward vesiculation if the lumen was acidic. Endosomes are acidic mainly due to the action of the vacuolar ATPase (V-ATPase), which pumps protons into the lumen. NHE8 on the limiting membrane of an acidic MVB is predicted to contribute to “proton leak” (i.e., release of protons into the cytosol in exchange for sodium or potassium ions into the MVB lumen). If NHE8 were depleted, one would predict that concentration of protons inside the organelle would increase, and thus internal pH would decrease. It follows that if the acidic internal pH of MVBs promotes inward vesiculation then the action of NHE8 would oppose this. However, we failed to detect any change in lumenal pH in dense MVBs at steady state after NHE8 depletion (Figure 7). This implies that it may be the lumenal sequestration of the counterion (sodium or potassium) that is important for inward vesiculation or back fusion. Our previous results from yeast support this idea because we found that Nhx1p function is still required for trafficking even when the yeast endosomal system is inefficiently acidified (in the absence of the V-ATPase; Plant et al., 1999; Bowers et al., 2000).
NHE8 and the Regulation of Endosomal Ion Balance
Why does NHE8 depletion not affect overall MVB pH? Nakamura et al. showed that overexpression of NHE8 or NHE9 resulted in an alkalinization of the Golgi or recycling endosomes, respectively (Nakamura et al., 2005). However, Roxrud et al. found no change in pH after depletion of NHE6 or NHE9 (Roxrud et al., 2009). Our data suggest that NHE8 is required in mammalian cells at very low levels as overexpression of an HA-tagged copy of NHE8 was enough to rescue the dense MVB accumulation phenotype of the siRNA-depleted cells (Figure 4). In addition, Nhx1p is expressed at very low levels in yeast (Bowers et al., 2000). Therefore, an exchanger present at such low cellular levels may be unlikely to contribute to gross MVB pH regulation. An alternative view is that perhaps NHE8 depletion affects pH in particular areas of the MVB, specifically the lumenal, aqueous area not taken up by intralumenal vesicles (which we here term the “matrix”). A previous in vitro study suggests that the intralumenal vesicles remain at near neutral pH (Falguieres et al., 2008). In NHE8-depleted cells there was increased DAMP labeling per MVB but, since the MVBs were larger in these cells, the labeling density per unit volume was the same as in mock-treated cells. However, because the lumen of MVBs in the NHE8-depleted cells contained very densely packed intralumenal vesicles (assumed to have near neutral pH), this implies a reduced matrix volume and hence a reduced matrix pH. It was not possible to accurately quantify the change in matrix volume with respect to intralumenal vesicles in the MVB of NHE8-depleted cells because the intralumenal vesicles were packed so tightly. The analysis of differences in matrix pH versus intralumenal vesicle pH will be an area for future research.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vas Ponnambalam for the anti-TGN46 antibody and Jean Gruenberg for the anti-LBPA antibody. We are grateful to Matthew Seaman for help and advice, for the anti-Vps26 and anti-Vps35 antibodies and for the HeLa M CD8-CIMPR cell line. We thank Mark Turmaine for technical assistance. This work was supported by a British Heart Foundation Intermediate Fellowship (K.B.), a Wellcome Trust project grant (K.B.), Medical Research Council UK program grant G0900113 (J.P.L.), and Wellcome Trust Strategic Award 079895 to C.I.M.R.
Abbreviations used:
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- DAMP
3-(2,4-dinitroanilino)-3′-amino-N-methyldipropylamine
- DNP
dinitrophenol
- EEA1
early endosome autoantigen 1
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EM
electron microscopy
- LAMP1
lysosome-associated membrane protein 1
- LBPA
lysobisphosphatidic acid
- MVB
multivesicular body
- NHE
sodium proton exchanger
- oligo
oligonucleotide
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- TfR
transferrin receptor
- TGN
trans-Golgi network
- V-ATPase
vacuolar ATPase
- VTC
vesicular/tubular cluster.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1053) on August 18, 2010.
REFERENCES
- Alonso-Caplen F. V., Compans R. W. Modulation of glycosylation and transport of viral membrane glycoproteins by a sodium ionophore. J. Cell Biol. 1983;97:659–668. doi: 10.1083/jcb.97.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Falck J. R., Goldstein J. L., Brown M. S. Visualization of acidic organelles in intact cells by electron microscopy. Proc. Natl. Acad. Sci. USA. 1984;81:4838–4842. doi: 10.1073/pnas.81.15.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Pathak R. K. Vesicles and cisternae in the trans Golgi apparatus of human fibroblasts are acidic compartments. Cell. 1985;40:635–643. doi: 10.1016/0092-8674(85)90212-0. [DOI] [PubMed] [Google Scholar]
- Babst M. A close-up of the ESCRTs. Dev. Cell. 2006;10:547–548. doi: 10.1016/j.devcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Becker A. M., Zhang J., Goyal S., Dwarakanath V., Aronson P. S., Moe O. W., Baum M. Ontogeny of NHE8 in the rat proximal tubule. Am. J. Physiol. Renal Physiol. 2007;293:F255–F261. doi: 10.1152/ajprenal.00400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J. E. Using temperature-sensitive mutants of VSV to study membrane protein biogenesis. Methods Cell Biol. 1989;32:85–110. doi: 10.1016/s0091-679x(08)61168-1. [DOI] [PubMed] [Google Scholar]
- Bobulescu I. A., Moe O. W. Lumenal Na(+)/H(+) exchange in the proximal tubule. Pfluegers Arch. 2009;458:5–21. doi: 10.1007/s00424-008-0595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Levi B. P., Patel F. I., Stevens T. H. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Piper S. C., Edeling M. A., Gray S. R., Owen D. J., Lehner P. J., Luzio J. P. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brett C. L., Donowitz M., Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005;288:C223–C239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- Bright N. A., Reaves B. J., Mullock B. M., Luzio J. P. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J. Cell Sci. 1997;110(Pt 17):2027–2040. doi: 10.1242/jcs.110.17.2027. [DOI] [PubMed] [Google Scholar]
- Burkhardt J. K., Argon Y. Intracellular transport of the glycoprotein of VSV is inhibited by CCCP at a late stage of post-translational processing. J. Cell Sci. 1989;92(Pt 4):633–642. doi: 10.1242/jcs.92.4.633. [DOI] [PubMed] [Google Scholar]
- Cabezas A., Bache K. G., Brech A., Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J. Cell Sci. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Doyotte A., Russell M. R., Hopkins C. R., Woodman P. G. Depletion of TSG101 forms a mammalian “Class E” compartment: a multicisternal early endosome with multiple sorting defects. J. Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- Fafournoux P., Noel J., Pouyssegur J. Evidence that Na+/H+ exchanger isoforms NHE1 and NHE3 exist as stable dimers in membranes with a high degree of specificity for homodimers. J. Biol. Chem. 1994;269:2589–2596. [PubMed] [Google Scholar]
- Falguieres T., Luyet P. P., Bissig C., Scott C. C., Velluz M. C., Gruenberg J. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol. Biol. Cell. 2008;19:4942–4955. doi: 10.1091/mbc.E08-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Goyal S., Mentone S., Aronson P. S. Immunolocalization of NHE8 in rat kidney. Am. J. Physiol. Renal Physiol. 2005;288:F530–F538. doi: 10.1152/ajprenal.00229.2004. [DOI] [PubMed] [Google Scholar]
- Goyal S., Vanden Heuvel G., Aronson P. S. Renal expression of novel Na+/H+ exchanger isoform NHE8. Am. J. Physiol. Renal Physiol. 2003;284:F467–F473. doi: 10.1152/ajprenal.00352.2002. [DOI] [PubMed] [Google Scholar]
- Griffiths G. Berlin: Springer-Verlag; 1993. Fine Structure Immunocytochemistry. [Google Scholar]
- Hisamitsu T., Ben Ammar Y., Nakamura T. Y., Wakabayashi S. Dimerization is crucial for the function of the Na+/H+ exchanger NHE1. Biochemistry. 2006;45:13346–13355. doi: 10.1021/bi0608616. [DOI] [PubMed] [Google Scholar]
- Hurley J. H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P., Toomre D., Diaz E., White J., Simons K. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 2001;3:140–149. doi: 10.1038/35055042. [DOI] [PubMed] [Google Scholar]
- Le Blanc I., et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 2005;7:653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W., Geuze H. J., Slot J. W. Improving structural integrity of cryosections for immunogold labelling. Histochem. Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Luyet P. P., Falguieres T., Pons V., Pattnaik A. K., Gruenberg J. The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic. 2008;9:2279–2290. doi: 10.1111/j.1600-0854.2008.00820.x. [DOI] [PubMed] [Google Scholar]
- Matsuo H., et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Molloy S. S., Anderson E. D., Jean F., Thomas G. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/s0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- Moncoq K., Kemp G., Li X., Fliegel L., Young H. S. Dimeric structure of human Na+/H+ exchanger isoform 1 overproduced in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:4145–4154. doi: 10.1074/jbc.M704844200. [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Bright N. A., Seaman M. N., Robinson M. S. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Tanaka S., Teko Y., Mitsui K., Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem. 2005;280:1561–1572. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- Nass R., Rao R. Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast. Implications for vacuole biogenesis. J. Biol. Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- Nass R., Rao R. The yeast endosomal Na+/H+ exchanger, Nhx1, confers osmotolerance following acute hypertonic shock. Microbiology. 1999;145(Pt 11):3221–3228. doi: 10.1099/00221287-145-11-3221. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Amherdt M., Madsen O., Perrelet A., Vassalli J. D., Anderson R. G. Conversion of proinsulin to insulin occurs coordinately with acidification of maturing secretory vesicles. J. Cell Biol. 1986;103:2273–2281. doi: 10.1083/jcb.103.6.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J., Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pfluegers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- Orlowski J., Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 2007;19:483–492. doi: 10.1016/j.ceb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant P. J., Manolson M. F., Grinstein S., Demaurex N. Alternative mechanisms of vacuolar acidification in H(+)-ATPase-deficient yeast. J. Biol. Chem. 1999;274:37270–37279. doi: 10.1074/jbc.274.52.37270. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Malerod L., Pedersen N. M., Stenmark H. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 2008;314:801–813. doi: 10.1016/j.yexcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M., Futter C. E. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. E., Banta L. M., Kohrer K., McCaffery J. M., Emr S. D. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxrud I., Raiborg C., Gilfillan G. D., Stromme P., Stenmark H. Dual degradation mechanisms ensure disposal of NHE6 mutant protein associated with neurological disease. Exp. Cell Res. 2009;315:3014–3027. doi: 10.1016/j.yexcr.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Seaman M. N. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J. Cell Sci. 2007;120:2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- Serrano R., Rodriguez-Navarro A. Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 2001;13:399–404. doi: 10.1016/s0955-0674(00)00227-1. [DOI] [PubMed] [Google Scholar]
- Slot J. W., Geuze H. J., Gigengack S., Lienhard G. E., James D. E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A. D., Teslenko L. V., Nikolsky N. N. The endocytosis of epidermal growth factor in A431 cells: a pH of microenvironment and the dynamics of receptor complex dissociation. Exp. Cell Res. 1988;175:192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Strous G. J., Willemsen R., van Kerkhof P., Slot J. W., Geuze H. J., Lodish H. F. Vesicular stomatitis virus glycoprotein, albumin, and transferrin are transported to the cell surface via the same Golgi vesicles. J. Cell Biol. 1983;97:1815–1822. doi: 10.1083/jcb.97.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari R. K., Kusari J., Sen G. C. Functional equivalents of interferon-mediated signals needed for induction of an mRNA can be generated by double-stranded RNA and growth factors. EMBO J. 1987;6:3373–3378. doi: 10.1002/j.1460-2075.1987.tb02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman P. G., Futter C. E. Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 2008;20:408–414. doi: 10.1016/j.ceb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chen H., Dong J., Lynch R., Ghishan F. K. Gastrointestinal distribution and kinetic characterization of the sodium-hydrogen exchanger isoform 8 (NHE8) Cell Physiol. Biochem. 2008;21:109–116. doi: 10.1159/000113752. [DOI] [PubMed] [Google Scholar]
- Xu H., Chen R., Ghishan F. K. Subcloning, localization, and expression of the rat intestinal sodium-hydrogen exchanger isoform 8. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G36–G41. doi: 10.1152/ajpgi.00552.2004. [DOI] [PubMed] [Google Scholar]
- Zhang J., Bobulescu I. A., Goyal S., Aronson P. S., Baum M. G., Moe O. W. Characterization of Na+/H+ exchanger NHE8 in cultured renal epithelial cells. Am. J. Physiol. Renal Physiol. 2007;293:F761–F766. doi: 10.1152/ajprenal.00117.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.