SifA is a virulence protein required for assembly and tubulation of a modified phagosome that promotes Salmonella replication. We show that SifA expressed in yeast induces membrane invagination during peroxisome proliferation and requires functional Rho1p. This is consistent with SifA ability to interact with RhoA and the fact that it is a GEF structural homologue.
Abstract
The Salmonella typhimurium effector protein SifA regulates the assembly and tubulation of the Salmonella phagosome. SifA localizes to the phagosome and interacts with the membrane via its prenylated tail. SifA is a structural homologue of another bacterial effector that acts as a GTP-exchange factor for Rho family GTPases and can bind GDP-RhoA. When coexpressed with a bacterial lipase that is activated by RhoA, SifA can induce tubulation of mammalian endosomes. In an effort to develop a genetic system to study SifA function, we expressed SifA and characterized its activity in yeast. GFP-SifA predominantly localized to yeast peroxisomal membranes. Under peroxisome-inducing conditions, GFP-SifA reduced the number of free peroxisomes and promoted the formation of large peroxisomes with membrane invaginations. GFP-SifA activity depended on the recruitment to peroxisomes of wild-type Rho1p and Pex25p, a receptor for Rho1p. GFP-SifA could also rescue the actin organization defects in pex25Δ and rho1 mutants, suggesting that SifA may recruit and potentiate Rho1p activity. We reexamined the distribution of GFP-SifA in mammalian cells and found the majority colocalizing with LAMP1-positive compartment and not with the peroxisomal marker PMP70. Together, these data suggest that SifA may use a similar mode of action via Rho proteins to alter yeast peroxisomal and mammalian endosomal membranes. Further definition of SifA activity on yeast peroxisomes could provide more insight into its role in regulating host membrane dynamics and small GTPases.
INTRODUCTION
Salmonella are Gram-negative intracellular bacteria that cause diseases ranging from gastroenteritis to typhoid fever (Haraga et al., 2008). The well-studied Salmonella serovar Typhimurium uses type III secretion systems (TTSS), which are essential for pathogenesis and tissue colonization, to translocate SPI (Salmonella pathogenicity island)-1 and -2 effector proteins into the host cytosol. Although there appears to be some overlapping functions, SPI-1 effectors are mainly required during early steps of invading nonphagocytic cells, and SPI-2 effectors are translocated once bacteria have been internalized. SPI-2 effectors are important for the development of a modified phagosome called the Salmonella-containing vacuole (SCV) that is required for bacterial survival and replication (Haraga et al., 2008; Steele-Mortimer, 2008). In epithelial cells and macrophages, morphological alterations termed Salmonella-induced filaments (Sifs) are observed as microtubule-dependent growth extensions of the SCV (Portillo et al., 1993).
SCV biogenesis is a dynamic process that results in the progressive accumulation of host endosomal and lysosomal markers. Inside cells, SPI-2 effectors are targeted to the SCV, the Golgi, and the actin cytoskeleton, ultimately promoting alterations of the SCV (Ramsden et al., 2007a; Haraga et al., 2008; Steele-Mortimer, 2008). Microtubule and actin cytoskeletal filaments that are rearranged near the SCV and Sifs require specific SPI-2 effectors (Meresse et al., 2001; Brumell et al., 2002; Miao et al., 2003; Kuhle et al., 2004; Poh et al., 2008). Some SPI-2 effectors manipulate microtubules to modulate the recruitment of motors to the SCV (Guignot et al., 2004; Boucrot et al., 2005), to redirect vesicle trafficking (Kuhle et al., 2006) or to position the SCV in the vicinity of the Golgi (Abrahams et al., 2006; Ramsden et al., 2007b). However, biochemical activities and functions for many SPI-2 effectors are undefined and the molecular mechanism for how remodeling the cytoskeleton contributes to bacterial replication within the SCV is unknown.
Among the SPI-2 effectors, SifA plays an essential role in bacterial replication, virulence in inbred susceptible mice, the regulation of SCV formation, and phagosome tubulation (i.e., Sifs; Stein et al., 1996; Beuzon et al., 2000). SifA translocated from bacteria localizes to the SCV and Sifs (Brumell et al., 2002), whereas green fluorescent protein (GFP)-SifA transfected into epithelial cells show a mixed distribution in the cytoplasm, plasma membrane, and endosomal/lysosomal compartments (Boucrot et al., 2003; Ohlson et al., 2008). The C-terminal tail of SifA is prenylated and acylated at the CaaX sequence and is important for its function and membrane association (Boucrot et al., 2003; Reinicke et al., 2005). Structural studies indicate that SifA has two domains (Ohlson et al., 2008). The N-terminal domain binds mammalian SKIP, which binds kinesin and links microtubules to the SCV, contributing to phagosome tubulation (Boucrot et al., 2005). SifA also competes with the late endosomal GTPase Rab9 for binding to SKIP and alters lysosomal protein trafficking (Jackson et al., 2008). The C-terminal domain of SifA is similar to the Salmonella SPI-1 effector SopE, which functions as a GTPase-exchange factor (GEF) for Cdc42, Rac1, and RhoG (Ohlson et al., 2008). SifA can bind the GDP-bound form of RhoA, although no direct GEF activity has been demonstrated. Within the region of SifA structure that is analogous to the catalytic domain of SopE, is the WxxxE motif that is conserved among the family of bacterial effectors that targets small GTPases (Alto et al., 2006; Huang et al., 2009; Bulgin et al., 2010). Interestingly, Sif-like structures are generated when the SPI-2 effector SseJ is ectopically coexpressed in HeLa cells with either SifA or activated RhoA, B, or C, suggesting that SifA might function as a GEF to activate Rho proteins (Ohlson et al., 2008). SseJ is a lipase that is activated by RhoA in vitro and can esterify cholesterol in mammalian cell membranes, possibly contributing to membrane tubulation (Christen et al., 2009).
As a complementary approach to the challenging task of correlating biochemical activities of individual SPI-2 effectors with the cellular phenotypes seen in mammalian cells, we characterized SifA activity in the model organism Saccharomyces cerevisiae. Budding yeast has proven very useful for studying the interactions of bacterial effectors with host targets and gaining insight into their enzymatic functions (Valdivia, 2004; Siggers and Lesser, 2008; Curak et al., 2009). The pathways of protein trafficking, cytoskeletal organization, and signal transduction are well studied in yeast, and bacteria recognize analogous components in many steps of these conserved cellular processes. For example, bacterial effectors that alter the actin cytoskeleton have been shown to target yeast Rho1p or other components of the Rho family pathways when they are expressed in yeast (Von Pawel-Rammingen et al., 2000; Alto et al., 2006). We report here that SifA is recruited to the membrane domain of yeast peroxisomes and most likely promotes peroxisome-specific Rho1p activities which include regulating actin organization. Thus, SifA may be used as a tool to study the biology of yeast peroxisomes, and in return, the yeast peroxisome provides a model for studying the biochemical and functional properties of SifA interactions with Rho GTPases at the membrane.
MATERIALS AND METHODS
Yeast Strains and Plasmids
All S. cerevisiae strains used in this study were derivatives of either BY4743 (MATa/α), BY4742 (MATα) or BY4741 (MATa) and the corresponding deletion strain library (Winzeler et al., 1999), unless otherwise indicated. Strain DVY119 (MATa ABP1-CFP::KanMX6 GAL10prGFP-SifA::LEU2 can1 his31 leu2 ura3 lys2) was created by integrating CFP (cyan fluorescent protein) at the C-terminus of ABP1 in MOY14 (gift of M. Ohl, Miller laboratory) according to Wach et al., (1997). Strain MOY14 contains an integrated copy of GAL10prGFP-SifA in the W303 background and was generated as described in Lesser and Miller (2001). Strains containing Snf7-monomeric red fluorescent protein (mRFP) and Chc1-mRFP are gifts of E. O'Shea (Harvard). The PEX11-pA haploid strain (YOL14MC) contains a genomically encoded Pex11p tagged at the C-terminus with Staphylococcus aureus protein-A and was created in the BY4743 background (Smith et al., 2002). Strain rho1-4D contains the Ts− allele rho1-104 that was backcrossed into the BY4742 background (Marelli et al., 2004). The following plasmids were used in this study: pFusGAL10prGFP-SifA (pMO32, gift of M. Ohl) with GFP-SifA expression induced by galactose, contains SifA open reading frame (ORF) that was PCR-amplified from S. typhimurium chromosomal DNA and cloned in frame with the C-terminus of GFP in pFUSGAL10prGFP (Lesser and Miller, 2001); p426TEFprGFP-SifA (pDV116) with GFP-SifA constitutively expressed, generated by PCR amplifying the GFP-SifA fragment from pMO32 and introducing it into p426TEFpr (Mumberg et al., 1995) via homologous recombination; pVPS1 (pCKR19; (Vater et al., 1992); pDsRed-PTS1 contains the Discosoma sp. RFP gene FP583 fused to the peroximal signal sequence PTS1 (Ser-Lys-Leu) at the C-terminus and flanked at both ends with FAA2 regulatory sequences (Smith et al., 2002).
Yeast Media
Synthetic minimal media (SM) contained the following: 0.17% yeast nitrogen base (YNB) without amino acids, 0.5% NH4SO4; Saccharomyces Cerevisiae induction medium (SCIM), 0.7% YNB plus NH4SO4, 0.5% yeast extract, 0.5% peptone, and 1/10 amount of amino acids.
Microscopy
Strains containing plasmids were routinely maintained in SM supplemented with the proper amino acids and carbon source. For expressing pFUS-based genes in trafficking mutants, an overnight culture grown in the same noninducing media was first diluted into fresh SM plus 2% raffinose and grown at 25°C until early log at OD600 ∼0.2. Then, cultures were induced by adding 2% galactose and grown for another 2–3 h until midlog OD600 = 0.5–0.6. Because the distribution of GFP-SifA on peroxisomes was found to be more robust in media containing raffinose than in glucose, cells were routinely cultured overnight in media containing 2% raffinose before oleic acid induction. We routinely use single colonies from fresh transformation of pDV116 to avoid possible revertants resulting from constitutive expression of GFP-SifA. All oleic acid induction studies were performed at 30°C, unless otherwise indicated. To induce peroxisomes, an overnight culture was first diluted into fresh SM plus raffinose and grown until midlog (2 × 107 cells/ml). Cells were then pelleted and resuspended into fresh SM or SCIM containing 0.1% raffinose, 0.15% oleic acid, and 0.2% Tween-40 and incubated at 30°C for 16–18 h (final OD600 = 0.5–0.7). For optimal viability, rho1TS cells were induced at 24°C for 20–22 h in SCIM plus 0.1% raffinose, 0.15% oleic acid, and 0.2% Tween-40. At the appropriate time points, cells were collected and visualized either live by direct fluorescence microscopy, or fixed first in 2% paraformaldehyde (Ted Pella, Irvine, CA) at room temperature (RT) for 10 min, followed by several washes in a 1.2 M sorbitol and 0.1 M potassium phosphate buffer (pH 7.5). To visualize actin, fixed cells were washed in PBS plus 1 mg/ml BSA, and stained with 3.3 μM of AlexaFluor 660-phalloidin (Invitrogen) for 30 min at RT. For either live or fixed-cell imaging, a small cell suspension was dropped onto a glass slide, and a coverslip was gently applied on top. All image acquisition were kept to under 30 min to prevent sample dehydration. Imaging was performed at RT on a Nikon Eclipse TE2000E inverted microscope (Melville, NY) with a Planapochromat 100×/1.4 NA oil objective. Microscope functions were controlled by the Metamorph software (Molecular Device, Sunnyvale, CA). For each sample, five z-axis planes spaced by 0.8 μm were acquired. For each plane, separate images at the GFP, rhodamine, or Cy5 channel were taken with the appropriate exposure times. The planes from each channel were then merged into a single plane containing the maximal signal intensity (Stack Arith Max, Metamorph) and processed using a two-dimensional deconvolution algorithm (Nearest Neighbor 2D Decon, Metamorph).
EM analyses were performed on cells induced for 18 h in SCIM plus 0.1% raffinose, 0.15% oleic acid, and 0.2% Tween-40 at 30°C. Sample preparations, observations, and analyses are as previously described in Vizeacoumar et al. (2003).
Subcellular Fractionation and Extraction of Peroxisomes
Peroxisome isolation was based on Marelli et al. (2004) with modifications. Briefly, overnight culture of yeast cells harboring PEX11-PA and p426TEFprGFP-SifA (pDV116) were induced in SCIM + 0.1% raffinose, 0.15% oleic acid, and 0.2% Tween-40 for 17 h at 30°C. Cells were harvested, spheroplasts were generated, and homogenized to produce a whole cell lysate in MES buffer (5 mM MES, pH 5.5; 1 mM EDTA, 1 mM KCl) plus 0.6 M sorbitol. The whole cell lysate was subjected to a 20,000 × g spin to yield a supernatant (20KgS) and a pellet (20KgP). The 20KgP material containing most of GFP-SifA, was resuspended in MES/sorbitol buffer and ∼0.3 mg from this suspension was applied to an 11-ml step gradient consisting of 17, 25, 35, and 50% Nycodenz (Axis-Shield, Norton, MA) in MES buffer. Organelles were separated by isopycnic centrifugation at 107,000 × g for 4.5 h at 4°C in a SW41 rotor. Fractions of 1 ml were collected from the top of the gradient, TCA-precipitated, and analyzed by SDS-PAGE and Western blotting. Anti-GFP was a gift from A. Merz (U. of Washington), and anti-PA (PAP) was purchased from Sigma (St. Louis, MO).
To extract peroxisomes, a portion of fraction 9, 10, or 11 enriched for peroxisomes from the Nycodenz gradient (Figure 2B) was first diluted in MES/sorbitol buffer, and the organelles were pelleted at 20,000 × g for 1 h in an SW40 rotor. The supernatant was removed, and the pellet was incubated on ice for 1 h in Ti8 buffer (10 mM Tris, pH 8; 1 mM EDTA) containing protease inhibitors. After a 200,000 × g spin for 1 h in a TLA100 rotor (Beckman, Fullerton, CA), the supernatant (Ti8S) was separated from the pellet (Ti8P). The Ti8P pellet was further extracted by suspending in 0.1 M Na2CO3, pH 11.2, and incubated for 1 h on ice, followed by centrifugation at 200,000 × g for 1 h to yield a supernatant (CO3S) and pellet (CO3P). Samples were TCA-precipitated, and equivalent portions were subjected to SDS-PAGE and Western blot.
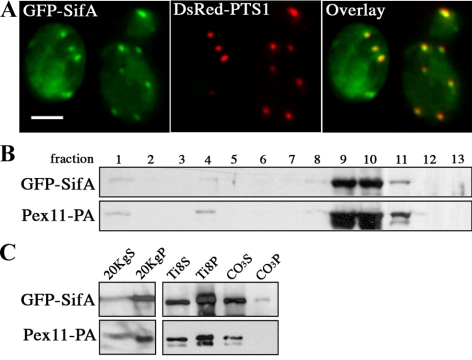
Figure 2.
SifA distributes to the peripheral membrane of peroxisomes in yeast. (A) GFP-SifA (pDV116) colocalizes with the peroxisomal reporter DsRed-PTS1 in wild-type yeast cells as indicated by yellow in the merged image. Cells containing two plasmids expressing the indicated fusion reporters were grown in media containing raffinose and then fixed and analyzed by direct fluorescence microscopy. Bar, 4 μm. (B) GFP-SifA cofractionates with the peroxisomal marker Pex11-PA by isopycnic centrifugation. Cells containing a chromosomal copy of Pex11-PA were transformed with pDV116 and induced in oleic acid for 17 h before biochemical fractionation. GFP-SifA–enriched material derived from the 20KgP crude organellar fraction (in C) was applied to a Nycodenz gradient and separated by centrifugation. Gradient fractions were collected from the top (fraction 1), and equal portions of each fraction were analyzed by Western blotting using anti-GFP and anti-pA antibodies. (C) Materials from fraction 10 were further extracted in a low-salt buffer to separate matrix (Ti8S) from membrane (Ti8P) peroxisomal proteins. The Ti8P fraction was again extracted with Na2CO3 to distinguish between peripheral membrane (CO3S) and integral membrane (CO3P) proteins. Equivalent portions of each fraction were analyzed by Western blot. Note that GFP-SifA and Pex11-PA behave mainly as peripheral membrane proteins.
Mammalian Cells Transfection and Immunofluorescence
Reagents.
Plasmid enhanced green fluorescent protein (pEGFP)-SifA (gift from S. Meresse Université de la Méditerranée, Marseille, France) contains the SifA ORF fused to the C-terminus of EGFP (Boucrot et al., 2003). UH1 LAMP1 mAb (1:20 dilution) was from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City); rabbit anti-PMP70 (1:1000) was from Invitrogen.
Immunofluorescence.
Chinese hamster ovary (CHO)-K1 fibroblasts (ATCC, Manassas, VA) were grown in F12 medium (Invitrogen) supplemented with 10% FBS, penicillin/streptomycin, at 37°C in 5% CO2. Cells were transfected with Fugene6 (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer's instructions in eight-well chamber slides (Nalge Nunc International, Rochester, NY). Cells were fixed in 3% paraformaldehyde for 15 min at 37°C and then permeabilized and blocked in 0.2% saponin and 5% normal goat serum in PBS for 30 min at RT. Primary antibodies diluted in blocking solution were added to cells and incubated for 2 h at RT or overnight 4°C. After three PBS washes, cells were incubated with appropriate Alexa Fluor–conjugated secondary antibodies (Invitrogen) for 30 min. Cells were again washed and mounted on coverslips with Fluoromount (Southern Biotechnology Associates, Birmingham, AL). Cells were observed under light fluorescence microscope with a Planapochromat 60×/1.4 NA oil objective lens.
RESULTS
SifA Distribution Is Not Associated with Yeast Endosomal Compartments
The intracellular distribution of SifA upon expression in yeast was first analyzed to determine whether it would be localized to endosomal compartments similar to its localization in mammalian cells (Brumell et al., 2002; Boucrot et al., 2003). A fusion protein construct containing SifA linked to GFP at the N-terminus was created similarly to the GFP-SifA construct previously characterized in mammalian cells (Boucrot et al., 2003). The expression of GFP-SifA was tested under either constitutive expression or under the control of an inducible promoter, but similar growth rates were observed in all media for either mode of expression. GFP-SifA localized to distinct punctate structures throughout the cell in a wild-type strain, in addition to the diffuse cytoplasmic and nuclear background distribution that is also observed in cells expressing only GFP (Figure 1A). The punctate staining of GFP-SifA is reminiscent of that found for yeast endosomal structures or the Golgi. Therefore, we sought to determine the identity of the GFP-SifA structures by looking for their codistribution relative to other yeast endocytic and Golgi markers: Abp1-CFP (early endocytic vesicle; Kaksonen et al., 2003), Snf7-RFP (late endosome; Huh et al., 2003), Chc1-RFP (late endosome/Golgi; Kaksonen et al., 2005; Newpher et al., 2005), and the vital dye FM4-64, which stains endocytic intermediates and the vacuole (yeast lysosome; Vida and Emr, 1995). Overexpression of GFP-SifA did not appear to affect the general distribution of Abp1-CFP, Snf7-RFP, or Chc1-RFP, or the uptake and trafficking of FM4-64 to the vacuolar membrane, suggesting normal endocytosis and protein sorting in these cells (data not shown). Although there was no evidence of colocalization in most cases, GFP-SifA dots transiently overlapped with Abp1-CFP (15% cells) and Snf7-RFP (10% cells) dots. However, GFP-SifA structures appeared to be substantially more static compared with Abp1-RFP or Snf7-RFP structures, which were highly dynamic and had a high turnover rate as they trafficked through the endocytic pathway (Kaksonen et al., 2005).
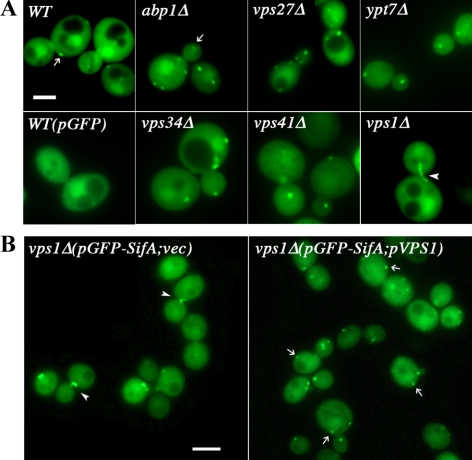
Figure 1.
Distribution of GFP-SifA in endocytic trafficking yeast mutants. (A) The distribution of GFP-SifA in fixed wild-type or endocytic trafficking deletion mutants was observed by direct fluorescence microscopy. Cells containing plasmids that expressed either the GFP-SifA fusion (pDV116) or GFP control were grown in media containing raffinose at 25°C. In all cases, except for vps1Δ cells, GFP-SifA localizes to distinct punctate structures (indicated by arrows). The diffuse cytoplasmic and nuclear fluorescence background was also observed in cells expressing only GFP. In vps1Δ mutants that lack the dynamin gene, GFP-SifA localizes to thin filamentous structures (arrowheads) anchored at the neck of large budded (G2) cells. Bar, 4 μm. (B) Introducing a plasmid expressing the wild-type dynamin VPS1 into vps1Δ cells was sufficient to redistribute GFP-SifA to punctate structures. Note that vps1Δ cells at other budding stages that harbor the control plasmid (vec) contain fewer (0–1 dot) and larger GFP-SifA punctate structures than those with the pVPS1 plasmid. Expression of GFP-SifA (pMO32) in vps1Δ cells was induced in galactose for 3 h, and fluorescent images were captured live. Bar, 4 μm.
The low frequency of association observed between GFP-SifA and yeast endocytic markers could be due either to the dynamic nature of certain endocytic compartments or that GFP-SifA might only traffic through part of the endocytic pathway. Further analyses in mutants blocked at specific endocytic trafficking steps might reveal organelle intermediates that accumulate GFP-SifA as they traffick through this step in the pathway (Bowers and Stevens, 2005). We studied the distribution of GFP-SifA in deletion mutants of genes regulating endosomal/vacuolar trafficking: early endocytosis (abp1, sla1, sla2, ark1, prk1; Kaksonen et al., 2003); to and from late endosomes (vps21: Rab5 homologue; vps27, vps34); and to and from the vacuole (ypt7: Rab7 homologue, vps1, vps41; Bowers and Stevens, 2005; Nickerson et al., 2009). The presence of overexpressed GFP-SifA did not affect the normal growth of any of these deletion mutants. As shown in Figure 1A, except for vps1Δ cells, the punctate distribution of GFP-SifA structures is unaffected in all trafficking mutants tested. In vps1Δ cells, there appears to be substantially fewer and larger GFP-SifA structures, and a single tubular GFP-SifA structure was found to be anchored at the dividing neck of most large budded/G2 cells (Figure 1, A and B). To eliminate the possibility that the novel phenotype was caused by nonspecific mutations outside the vps1 deletion, a plasmid containing a single copy of wild-type VPS1 was introduced together with plasmid GFP-SifA into vps1Δ cells. As shown in Figure 1B, plasmid-expressed Vps1p reverts the phenotype of GFP-SifA structures from tubules to patches in vps1Δ cells, suggesting that GFP-SifA resides in an organelle whose distribution is specifically affected by Vps1p function.
SifA Localizes to Yeast But Not Mammalian Peroxisome Membranes
Vps1p is a member of the dynamin-like protein family that facilitates the scission of vesicles after budding (Praefcke and McMahon, 2004). Although the role of dynamin in yeast endocytosis remains unclear, Vps1p participates in diverse pathways including vacuolar protein trafficking (Vater et al., 1992), actin organization (Yu and Cai, 2004) and peroxisome fission (Hoepfner et al., 2001). The striking GFP-SifA filament observed in vps1Δ cells is reminiscent of the elongated tubular peroxisomes previously seen in the same mutant (Hoepfner et al., 2001). To test whether GFP-SifA might be recruited to peroxisomes in wild-type cells, we cotransformed pGFP-SifA and pDsRed-PTS1, which contains the reporter protein RFP fused to the peroxisomal targeting sequence PTS1 at the C-terminus (Smith et al., 2002). As shown in Figure 2A, GFP-SifA punctate structures tightly colocalize with DsRed-PTS1–marked peroxisomes, suggesting that they reside in the same cellular compartment.
Subcellular fractionation was also used to confirm that GFP-SifA was stably associated with peroxisomes. GFP-SifA was expressed in a strain containing a chromosomal copy of the peroxisomal protein Pex11 fused to protein A, and cells were incubated with oleic acid to induce peroxisome proliferation. Whole cell lysate was centrifuged and separated into a cytosol-enriched supernatant (20KgS) and crude organellar pellet (20KgP). As shown in Figure 2C, most of GFP-SifA sedimented in the 20KgP fraction. This GFP-SifA enriched material was subjected to isopycnic gradient centrifugation, and nearly all GFP-SifA was observed to comigrate with fractions containing Pex11-PA (Figure 2B). We further extracted the peroxisome-enriched fractions from the gradient (no. 9 and 10, Figure 2B) to determine the subcellular distribution of GFP-SifA. After subjecting these fractions to hypotonic lysis, GFP-SifA was found predominantly in the membrane pellet (Ti8P) similar to the behavior of the membrane protein Pex11-PA (Figure 2C). The Ti8P fraction was further extracted with sodium carbonate to separate peripheral from integral membrane proteins of peroxisomes. Under this condition, virtually all GFP-SifA cofractionated with Pex11-PA in the supernatant (CO3S) enriched for peripheral membrane proteins (Figure 2C). Taken together, these data indicate that GFP-SifA behaves mainly as a peripheral membrane protein on yeast peroxisomes in a similar manner to its behavior on mammalian cell membranes (Boucrot et al., 2003).
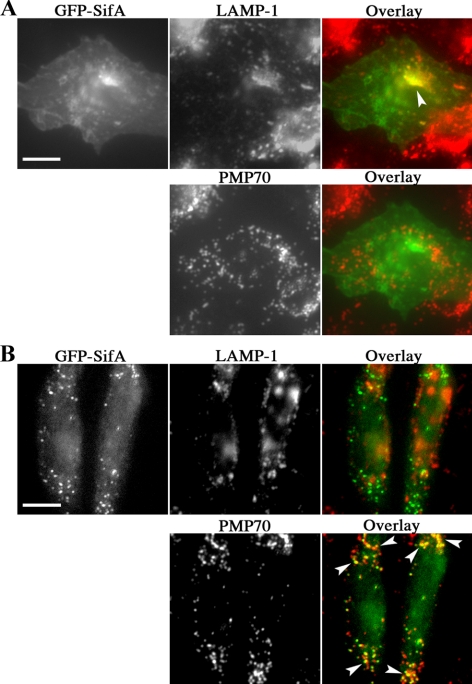
Localization of GFP-SifA to yeast peroxisomes was a surprising result given its localization in mammalian cells and raised the possibility that SifA might transiently associate with peroxisomes in mammalian cells before trafficking to the SCV. Therefore, SifA was analyzed for colocalization with the peroxisomal marker PMP70 in mammalian cells. On transfection of pEGFP-SifA (Boucrot et al., 2003) into epithelial CHO cells, the majority of EGFP-SifA localized to the plasma membrane and LAMP-1 positive compartment as has been reported in other cell types (Figure 3A). Interestingly, a small fraction of transfected CHO cells (<10%) showed EGFP-SifA colocalizing with PMP70 (Figure 3B). Colocalization was observed as early as on day 1 and again on day 5 after transfection. However, we were unable to increase this fraction of cells through induction of peroxisome proliferation with the drug 4-phenylbutyrate (Li et al., 2002). This pattern of colocalization was neither observed in transfection nor bacterial translocation studies of RAW mouse macrophages, HeLa human epithelial cells or primary human fibroblasts. We also found that Salmonella replication and phagosome tubulation appeared normal in primary human fibroblasts from Zellweger patients that completely lack peroxisomes (data not shown). Therefore, the localization of SifA to peroxisomes was unique to yeast and SifA does not usually associate specifically with mammalian peroxisomes.
Figure 3.
Distribution of GFP-SifA transfected in mammalian CHO cells. CHO cells were transfected with pEGFP-SifA for 24–48 h., followed by fixation, permeabilization, and immunostaining for the endosome/lysosome marker LAMP-1 and peroxisomal marker PMP70. An overlay of the corresponding images on each row is shown, with yellow color and arrowheads marking areas of overlap with GFP-SifA. Although the majority of cells show the typical codistribution of GFP-SifA and LAMP-1 in A, a small number of cells was found to have GFP-SifA distributed to PMP70 dots (B). Bar, 10 μm.
SifA Changes Yeast Peroxisome Abundance and Morphology
To further understand how GFP-SifA affects peroxisome activities, we characterized the abundance and morphology of peroxisomes labeled with DsRed-PTS1 when yeast cells were incubated in medium with or without the fatty acid oleic, a condition that requires peroxisome replication in order to maintain growth. As expected, the proliferation of peroxisomes was induced ∼40% in cells containing a control vector grown in medium with oleic acid compared with raffinose (Figures 4, A and B, and Table 1). In the absence of GFP-SifA, peroxisomes appeared punctate and distinct from one another in medium with raffinose or oleic acid. The introduction of GFP-SifA has no apparent effect on the growth rate of cells in raffinose and GFP-SifA colocalized tightly with peroxisomes. However, in oleic acid, GFP-SifA containing cells have fewer distinct peroxisomes than in vector containing cells (1.8 times less, p = 1.6 × 10−13; Table 1), and many of these peroxisomes were larger and occurred in clusters (Figure 4C). On closer examination, GFP-SifA appeared to localize to one side of the peroxisome clusters in many cells, and occasionally, was found between two adjacent clusters.
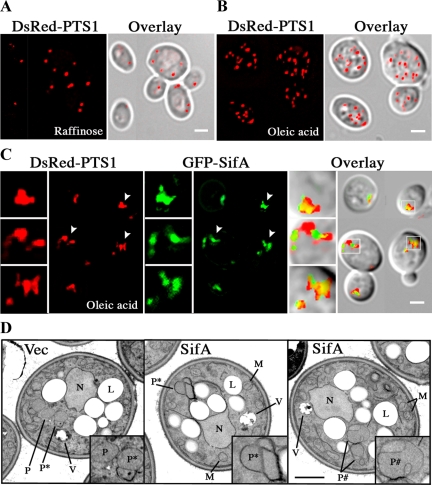
Figure 4.
GFP-SifA alters the morphology of peroxisomes. Distribution of peroxisomes (DsRed-PTS1 labeled) in fixed wild-type cells grown in media containing raffinose (A), or oleic acid (B and C). Cells were cotransformed with pDsRed-PTS1 and empty vector (A and B) or pGFP-SifA (C). Overlay indicates merged images of fluorescence and DIC; and yellow indicates the overlap of red and green signals. In C, boxed images to the left of each panel are 2.5× enlargements of smaller areas in each panel indicated by arrowheads or small white squares. Note that peroxisomes associated with SifA are less abundant, tend to clump, and are not always round in shape. The localization of SifA is to one side of peroxisomes and in many cases, appears to bridge nearby clusters of peroxisomes (middle enlarged box). All bars, 2 μm. (D) Cells containing either vector or pGFP-SifA were incubated in oleic acid for 18 h and then fixed and processed for EM. Boxed images at the lower right in each panel show 2× enlargement of specific areas containing peroxisomes that are depicted with letters. P, free and dispersed peroxisomes; P*, clusters of peroxisomes with adherent membranes; P#, enlarged peroxisomes with exaggerated membrane invaginations; N, nucleus; L, lipid body; M, mitochondria; V, vacuole. Bar, 1 μm.
Table 1.
Fluoresence microscopic analyses of peroxisomes in wild-type and mutant cells
| Strain background | Average no. of discrete dots/cella |
|
|---|---|---|
| Vector | pGFP-SifAWT | |
| Without oleic acid | ||
| WT | 5.4 ± 2.5 | 5.5 ± 1.9 |
| With oleic acid | ||
| WT | 7.2 ± 4.6 | 3.9 ± 3.7b |
| pex7Δ | 2.6 ± 2.6 | 1.2 ± 1.8 |
| pex11Δ | 3.2 ± 2.9 | 1.5 ± 1.6 |
| pex25Δ | 3.8 ± 3.6 | 4.0 ± 4.2b |
| rho1-4D | 3.5 ± 4.5 | 2.6 ± 2.2b |
Cells transformed with plasmids pDsRed-PTS1 and pGFP-SifAWT (pDV116) or empty vector (p426TEFpr) were grown in medium with or without oleic acid, then fixed and visualized by direct fluorescence microscopy. In experiments with pGFP-SifA, PTS1-labelled peroxisomes were scored in cells expressing GFP-SifA structures. In all cases, >95% cells have overlapping DsRed-PTS1 and GFP-SifA structures. Results were scored in an average of 100 cells per strain, and each experiment was repeated at least once. WT, wild-type strain.
a Average numbers of dots ± SD, are either dispersed or attached, but distinctly small round peroxisomes labelled with DsRed-PTS1.
b Statistically significant difference (p < 0.005) was observed for the average numbers of dots between vector and pGFP-SifA populations in all cases, except in pex25Δ (p = 0.56) and rho1 (p = 0.18) background.
Detailed EM analyses (Figure 4D and Table 2) showed that the peroxisome structures could be subdivided into two classes of morphology: 1) clusters of two or more peroxisomes with adherent, and in some cases, thickening membranes (P*) and 2) enlarged peroxisomes with exaggerated and invaginated membranes (P#). Consistent with the fluorescence data, fewer GFP-SifA containing cells were found to exhibit free and dispersed peroxisomes (category P in Figure 4D), and the average peroxisome count per cell area assayed was also substantially reduced in these cells (Table 2). In examining the EM data, although the frequency of cells showing adherent membranes (category 1 above) does not appear to be very different between vector and GFP-SifA populations, the percentage of cells exhibiting peroxisomes with invaginated membrane increased strikingly from 6.3 for control, to 63 for GFP-SifA plasmid (Table 2). Therefore, at least half of the peroxisome clusters observed by fluorescence microscopy in GFP-SifA–expressing cells contained enlarged peroxisomes with invaginated membranes.
Table 2.
EM analyses of peroxisomes in wild-type cells
| Plasmid | Free and distinct (%)a | Peroxisome countb | Clusters of ≥ two peroxisomes with adherent membranesc (%) | Invaginations and exaggerated membranesc (%) |
|---|---|---|---|---|
| Vector | 64.6 | 0.30 | 66.7 | 6.3 |
| pGFP-SifAWT | 38.2 | 0.07 | 63.2 | 62.5 |
Wild-type cells selected for either pGFP-SifAWT or vector were grown in oleic acid for 18 h before fixing and processing for EM.
a Percent of total cells analyzed (162 for vector, and 160 for SifA); values add up to more than 100% because cells may exhibit more than one phenotype.
b Number of free and distinct peroxisomes counted (437 for vector; 105 for pGFP-SifA) per μm2 of total cell area assayed (1454.1 μm2 for vector; 1496.7 μm2 for GFP-SifA).
c See Figure 4D for depiction of phenotypes. These peroxisomes are not included in the calculation of peroxisome count.
SifA Activities in Yeast Mutants Defective for Peroxisome Biogenesis
We sought further insight into how GFP-SifA might interact with peroxisomal proteins to affect the morphology of peroxisomes. Mutants deleted for gene products (the peroxins) that affect peroxisome assembly (pex3Δ, pex19Δ), size and number (pex11Δ, pex25Δ, vps1Δ), or matrix protein import and maturation (pex5Δ, pex7Δ; Fagarasanu et al., 2007; Platta and Erdmann, 2007; Smith and Aitchison, 2009), were transformed with pGFP-SifA and pDsRed-PTS1 and grown in oleic acid, and then cells were analyzed by fluorescence microscopy. Because Pex25p acts as a peroxisomal receptor for Rho1p, we also tested GFP-SifA effects on peroxisome distribution in a temperature-sensitive rho1 mutant (Marelli et al., 2004).
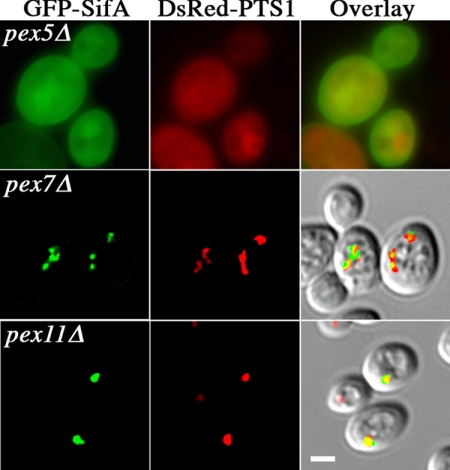
Overall, GFP-SifA did not improve the growth rate of any pexΔ mutants on oleic acid plates, but caused a slight delay in growth of wild-type, pex11Δ, pex25Δ, and vps1Δ mutants, but not of pex3Δ, pex5Δ, pex7Δ, or pex19Δ cells. Furthermore, GFP-SifA did not appear to improve any of the peroxisomal morphological defects found in these pexΔ mutants. Three peroxisome phenotypes were observed: 1) diffuse cytoplasmic DsRed-PTS1 staining as in pex5Δ (Figure 5), pex3Δ, and pex19Δ cells. In this category, functional mature peroxisomes were not formed, causing GFP-SifA distribution to be dispersed. Another phenotype (category 2)) included fewer, distinct peroxisomes as in pex7Δ (Figure 5 and Table 1). PEX5 and PEX7 encode receptors for peroxisomal matrix protein targeting sequences PTS1 and PTS2, respectively. Thus, DsRedPTS1-labeled peroxisomes would be observed in pex7Δ, but not in pex5Δ cells (Figure 5). Because GFP-SifA localization was virtually identical to DsRed-PTS1, GFP-SifA must be preferentially recruited to peroxisomes importing PTS1-specific matrix proteins. The final peroxisomal phenotype (category 3)) was found in vps1Δ (Figure 1), pex11Δ (Figure 5), and pex25Δ (Figure 6) mutants: the number of small and dispersed peroxisomes decreases and is replaced by a few large peroxisomes (Hoepfner et al., 2001; Rottensteiner et al., 2003; Tam et al., 2003). Similarly to its behavior in a wild-type background, GFP-SifA reduced the number of free PTS1-labeled peroxisomes in pex7Δ and pex11Δ cells (statistically significant difference between vector vs. GFP-SifA plasmid containing populations, p = 0.003 and p = 0.002, respectively; Table 1). Interestingly, GFP-SifA did not appear to reduce the number of free peroxisomes in a pex25Δ (p = 0.56) or a rho1 mutant (p = 0.18). Therefore, GFP-SifA activities on peroxisome morphology and abundance require the presence of functional Pex25p or Rho1p.
Figure 5.
Distribution of GFP-SifA in pexΔ mutants. pexΔ cells selected for expression of plasmids carrying GFP-SifA and DsRed-PTS1 were grown in oleic acid for 17 h and then fixed for direct fluorescence microscopy. PEX5 and PEX7 encode receptors for peroxisomal targeting-signal sequences PTS1 and PTS2, respectively; PEX11 regulates peroxisome proliferation. Note that the diffuse cytoplasmic staining in pex5Δ indicates that no functional peroxisomes were assembled. Overlay shows merged images of the GFP and DsRed channels, and in the bottom two cases, also overlaid with the corresponding DIC images. Bar, 2 μm.
Figure 6.
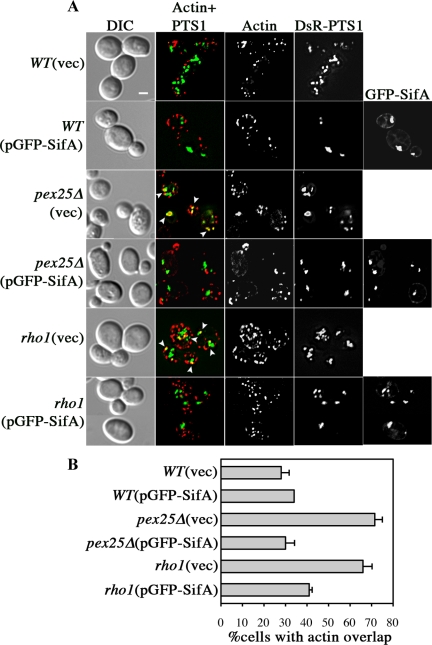
Effects of GFP-SifA on actin dynamics on yeast peroxisomes. (A) Distribution of actin relative to peroxisomes and GFP-SifA was analyzed by direct fluorescence microscopy. Various strains containing pDsRed-PTS1 and pGFP-SifA or vector alone were grown in oleic acid for either 18 h at 30°C or 22 h at 24°C (for rho1). Cells were fixed and stained with phalloidin-Alexa660 to visualize actin. [Actin+PTS1] panels show merged images with yellow areas (arrowheads), indicating colocalization of actin patches and peroxisomes in pex25Δ and rho1 cells. Note that in cases where GFP-SifA overlaps with peroxisomes, actin colocalization with peroxisomes is less prevalent. Bar, 2 μm. (B) The frequency of cells that have overlapping actin and peroxisomes are scored for each strain. Results are from examining more than 160 cells for each strain.
SifA Alters Actin Organization around Peroxisomes via Rho1p
SifA belongs to a family of bacterial effectors that promote Ras-like GTPase activities at mammalian cell membranes (Alto et al., 2006; Ohlson et al., 2008; Huang et al., 2009). The yeast Rho1p GTPase has previously been shown to be involved in regulating peroxisome maturation and abundance. On oleic acid induction, Rho1p is recruited to peroxisomes via Pex25p and regulates the organization of the surrounding actin cloud, thus affecting membrane dynamics during peroxisome division (Marelli et al., 2004). To further understand the role of SifA in peroxisome activities in yeast, we analyzed whether GFP-SifA affects the actin distribution around peroxisomes in pex25Δ or rho1 mutants. As shown in Figure 6, a higher percentage of cells in the pex25Δ and rho1 background compared with wild-type contains actin patches that mostly overlap with peroxisomes. These results are consistent with previous studies, suggesting that actin reorganization around peroxisomes depends on Rho1p and is altered in these mutants (Marelli et al., 2004). The expression of GFP-SifA in either pex25Δ or rho1 mutants suppressed the actin accumulating phenotype on peroxisomes (Figure 6B). GFP-SifA most likely compensates for a deficiency of Rho1p activity at peroxisomes in pex25Δ or rho1 mutants by recruiting Rho1p to peroxisomes in the absence of pex25 and/or potentiating its activity during rho1 depletion.
DISCUSSION
SifA is a type III effector protein required for Salmonella intracellular replication and virulence in mammals. In an effort to develop a genetic system to understand SifA activity and function in host cells, we expressed SifA in yeast cells. SifA was recruited to yeast peroxisomal membranes with a result of fewer, large peroxisomes that contain multiple membrane invaginations, suggesting hyperactive membrane constrictions without division. This phenotype may be analogous to the endosomal tubulation in mammalian cells caused by coexpressing SifA and another bacterial effector acting as a lipase. Furthermore, SifA appears to promote Rho1p activity on peroxisomes consistent with its ability to interact with mammalian GDP-RhoA and its structural similarity to bacterial GEFs. We propose that SifA mode of action is similar in yeast and mammalian cells, but on different membrane systems perhaps by binding to a protein conserved between mammalian endosomes and yeast peroxisomes.
SifA Effects on Peroxisome Division in Yeast
The effects of SifA on peroxisome morphology argue for it functioning to control peroxisome division. In yeast cells, peroxisome biogenesis occurs via two pathways: de novo formation from ER precursors or growth and division of preexisting peroxisomes (Fagarasanu et al., 2007; Motley and Hettema, 2007). Division of preexisting peroxisomes includes distinct steps of tubulation/elongation, constriction, and fission. Analyses of yeast peroxisome biogenesis mutants have separated them into gene products that affect assembly (de novo) or division (size and number). The expression of GFP-SifA slightly decreases the growth on oleic acid plates of wild-type and only the division mutants (pex11Δ, pex25Δ, vps1Δ), but not the assembly mutants (pex3Δ, pex5Δ, pex19Δ), implicating SifA as altering the division steps of peroxisome biogenesis.
By sequence similarity and overlapping functions, Pex11p, Pex25p, or Pex27p are grouped into the peroxin family (Pex11-protein family) involved in the division steps before fission, with Pex11p and Pex25p being the dominant regulators (Thoms and Erdmann, 2005; Yan et al., 2005). Overproducing Pex11p promotes peroxisome proliferation and elongation, and deletion in PEX11-family genes typically results in a few enlarged peroxisomes with rare occasion of membrane invaginations in the triple deletions (Tam et al., 2003; Rottensteiner et al., 2003; Koch et al., 2004). The peroxisome phenotype caused by SifA is unlikely to be a fission defect, which is often seen as beads-on-string peroxisomes in the dynamin-like vps1Δ mutant (Hoepfner et al., 2001). What is observed by EM might be large amorphous peroxisomes with multiple arms that look like invaginations at cross sections. One interpretation is that SifA promotes membrane tubulation and/or constrictions and because fission activities are limited, overly large peroxisomes with multiple extensions result.
Two lines of observation suggest that SifA exerts its effects on peroxisomes in conjunction with Pex25p and/or Rho1p activity. First, SifA reduced the number of free peroxisomes and increased the frequency of clustered peroxisomes in wild-type, pex7Δ, and pex11Δ cells, but not in pex25Δ or rho1 cells. Because wild-type Pex25p or Rho1p must be present for the morphological changes induced by SifA to be apparent, this is most likely an exaggeration of Pex25p/Rho1p activities. Other studies have reported that overproducing Pex25p can sometimes cause unusually overgrown membrane sheets resembling karmellae near the ER/nucleus, suggesting its role in membrane modification (Rottensteiner et al., 2003). This would be consistent with SifA working with Pex25p/Rho1p to cause membrane invaginations. Second, SifA is able to complement the ability of pex25Δ and rho1 mutants to regulate actin dynamics around peroxisomes. The fact that SifA does not cause a higher than wild-type frequency of cells with accumulating actin on peroxisomes suggests an already maximally measurable phenotype in the presence of Pex25p/Rho1p. The effects of SifA on actin is revealed only when Rho1p is deficient at the peroxisomes as in pex25Δ or rho1 cells, again suggesting that SifA largely acts by promoting Rho1p activity. SifA does not appear to improve all aspects of Rho1p function since it cannot complement the vegetative growth defects of rho1 mutants (Olson and Miller, unpublished data).
SifA Role in Membrane Dynamics
In yeast, peroxisome division under noninducing condition occurs mainly via replication of preexisting mature peroxisomes, but upon induction with oleic acid, more de novo assembly might be activated to accommodate rapid proliferation and turnover of the peroxisome membrane system (Schrader and Fahimi, 2006; Fagarasanu et al., 2007, Smith and Aitchison, 2009). This might involve fusion of ER-derived preperoxisome vesicles with each other to create new peroxisomes or with preexisting peroxisomes to promote growth and division. In general, peroxisomes in rho1 and pex25Δ cells are smaller than those in wild type, suggesting a defect in growth and maturation. Because both Rho1p and Pex25p are observed on peroxisomes only upon oleic acid induction, they may promote division involving fusion (Smith et al., 2002, Marelli et al., 2004). It is tempting to speculate that because SifA also only induces peroxisome morphological changes when cells are grown in oleic acid, it might act more effectively in association with Rho1p and Pex25p on certain peroxisome intermediates, such as those with membrane dynamics primed for fusion.
How might Rho1p and Pex25p affect peroxisomal membrane activity? Rho1-dependent actin dynamics are proposed to be important for peroxisomal budding and fusion (Marelli et al., 2004). According to the mesh hypothesis of membrane dynamics, local and transient assembly/disassembly of an actin mesh surrounding an organelle can provide active zones where vesicles can fuse or bud (Lorra and Huttner, 1999). It is plausible that Rho1p acts as a spatial landmark to start an actin remodeling event leading to membrane fusion (Eitzen, 2003). Recently, a systems approach to study regulators of yeast peroxisome biogenesis identified signaling proteins involved in actin organization and/or phosphatidylinositol metabolism (Saleem et al., 2008). Many of these proteins function through activation by Rho1p. To control actin dynamics, Rho GTPases either target regulators of actin nucleation, or change the abundance of membrane phosphoinositides to affect the recruitment of actin-binding proteins (Ridley, 2006). Interestingly, lipid raft-like membrane domains enriched with ergosterol and ceramide have been found on peroxisome precursors that act as signaling platforms to organize the fusion machinery including phosphoinositides and small GTPases (Boukh-Viner et al., 2005). Alternatively, Rho1p-dependent actin dynamics are used to change membrane tension and, in the presence of phosphoinositide-binding proteins that can assemble into a scaffold at discrete sites, can generate membrane curvature necessary for vesicle budding/fusion (McMahon and Gallop, 2005). All members of the Pex11-family proteins can interact with themselves, and some with each other, suggesting the ability to oligomerize (Thoms and Erdmann, 2005). Evidence also points to Pex-11 family proteins being able to bind phospholipids and playing a direct role in membrane modification (Fagarasanu et al., 2007). This would be consistent with a previous proposal that Rho1p might facilitate Pex11p activity based on the actin phenotype observed for a pex25Δ pex11Δ double mutant (Marelli et al., 2004).
The fact that SifA can affect Rho1p activity and change yeast peroxisome morphology and distribution is reminiscent of its role on the assembly and tubulation of the SCV during Salmonella infection. SifA effects on membrane dynamics can be drawn in parallel between yeast and mammalian cells. In both systems, SifA is involved in exaggerated membrane tubulation (Sif- and SifA-induced tubules in epithelial cells or invaginated peroxisomes in yeast); in conjunction with a lipid-modifying protein (Salmonella effector SseJ; or possibly Pex25p) and the small GTPase Rho-family proteins (Brumell et al., 2002; Ruiz-Albert et al., 2002; Ohlson et al., 2008). SseJ, like Pex25p, recruits RhoA (the mammalian equivalent of Rho1p) and in turn, is activated by RhoA to alter the membrane composition of cholesterol, a condition that can induce endosomal tubules (Christen et al., 2009). SifA can bind RhoA (Ohlson et al., 2008) and might recruit Rho proteins directly to both membrane systems. This can amplify RhoA effects on SseJ or other bacterial or host lipid-modifying proteins, resulting in SifA-induced phagosome tubulation, or can overpromote Rho1p effects on peroxisomes causing aberrant membrane invaginations. Consistent with this model, the ability of SifA to induce tubules in combination with SseJ was reduced in the presence of siRNA against Rho proteins (Ohlson et al., 2008), and SifA does not reduce the number of free peroxisomes in a rho1 or pex25Δ mutant. Interestingly, peroxisomal membrane invaginations are reminiscent of the phenotype in the lipase mutant lpx1Δ with defects in membrane lipid modification (Thoms et al., 2008). It is possible that SifA expression disrupts Rho1p normal regulation of a yeast lipase by promoting its membrane modifying activity at an inappropriate time during peroxisome biogenesis, causing an imbalance between membrane growth and fission and hence, the exaggerated invagination phenotype. A useful consequence of studying SifA activity on yeast peroxisome division is that this could be a means to increase Rho1p activity specifically on peroxisomes without having to overexpress Rho1p, a condition that could be lethal to vegetative growth.
Yeast Peroxisomes as Model System to Elucidate SifA Function
Inferring from the yeast studies, we reexamined the distribution of SifA in mammalian cells and found that GFP-SifA did not specifically colocalize with the peroxisomal protein PMP70 in most cells. There have been previous examples of bacterial virulence proteins that target common structural components of eukaryotic signaling pathways (e.g., MAP kinase) even though the outcomes of these pathways may not necessarily be conserved between mammalian and yeast systems (e.g., inflammation vs. mating; Valdivia 2004; Siggers and Lesser 2008). Because GFP-SifA still localizes to peroxisomes in pex25Δ mutants, it is plausible that a common protein exists on both yeast peroxisomes and mammalian endosomes that recruits SifA to the relevant membrane systems and link it to the local conserved Rho family members. This membrane recruiter would also have different cytoskeletal interactions. For example, there is a pronounced abundance of microtubules in mammalian cells compared with yeast cells, and there is also no homologue for SKIP in yeast cells that could link SifA to microtubules.
Because many aspects of endosomal fusion leading to SCV assembly are unknown, the studies described here could provide a model system to help define additional binding partners for SifA and its effects on Rho proteins and membrane dynamics. Specifically, the recruitment and activation of a GTPase at an inappropriate time in the functional development of the phagosome could result in aberrant membrane tubulation. How this activity promotes and maintains the bacteria replication niche remains to be defined. In conclusion, our data suggest that there are parallels between the effects of a bacterial virulence protein on yeast peroxisomes and the mammalian phagosome that may be exploited to better define activities of SifA.
ACKNOWLEDGMENTS
We thank T. Davis (University of Washington), D. Drubin (University of California, Berkeley), S. Fields (University of Washington), G. Eitzen (University of Alberta, Canada), T. Hazbun (Purdue University), S. Meresse, A. Merz, E. O'Shea, M. Ohl, T. Stevens (University of Oregon, Eugene), Y. Sun (University of California, Berkeley), and M. Vignali (University of Washington) for gifts of plasmids and antibodies; T. Davis for usage of the ultra microcentrifuge; C. Toret for work on GFP-SifA tracking; G. Martin from the Keck Center for advice with microscopy techniques; and M. Brittnacher for help on statistics. For discussions, we thank A. Merz; T. Davis, D. Drubin, M. Ohlson, Megha, and A. Haraga, and also thank other members of the Miller lab for their support. Finally, we are grateful to M. Welch and N. Salama for helpful comments on the manuscript. This work was supported by the National Institutes of Health Grant RO1-AI048683 to S.I.M.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0482) on August 25, 2010.
REFERENCES
- Abrahams G. L., Muller P., Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic. 2006;7:950–965. doi: 10.1111/j.1600-0854.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- Alto N. M., et al. Identification of a bacterial type III effectors family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Beuzon C. R., Meresse S., Unsworth K. E., Ruiz-Albert J., Garvis S., Waterman S. R., Ryder T. A., Boucrot E., Holden D. W. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Beuzon C. R., Holden D. W., Gorvel J. P., Meresse S. Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 2003;278(16):14196–14202. doi: 10.1074/jbc.M207901200. [DOI] [PubMed] [Google Scholar]
- Boucrot E., Henry T., Borg J. P., Gorvel J. P., Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308:1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- Boukh-Viner T., et al. Dynamic ergosterol- and ceramide-rich domains in the peroxisomal membrane serve as an organizing platform for peroxisome fusion. J. Cell Biol. 2005;168:761–773. doi: 10.1083/jcb.200409045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1774:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brumell J. H., Goosney D. L., Finlay B. B. SifA, a type III secreted effector of Salmonella typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic. 2002;3:407–415. doi: 10.1034/j.1600-0854.2002.30604.x. [DOI] [PubMed] [Google Scholar]
- Bulgin R., Raymond B., Garnett J. A., Frankel G., Crepin V. F., Berger C. N., Arbeloa A. Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect. Immun. 2010;78:1417–1425. doi: 10.1128/IAI.01250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M., Coye L. H., Hontz J. S., LaRock D. L., Pfuetzner R. A., Megha, Miller S. I. Activation of a bacterial virulence protein by the GTPase RhoA. ScienceSignal. 2009;95:1–6. doi: 10.1126/scisignal.2000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curak J., Rohde J., Stagljar I. Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 2009;12:18–23. doi: 10.1016/j.mib.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Eitzen G. Actin remodeling to facilitate membrane fusion. Biochim. Biophys. Acta. 2003;1641:175–181. doi: 10.1016/s0167-4889(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Rachubinski R. A. Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- Guignot J., Caron E., Beuzon C., Bucci C., Kagan J., Roy C., Holden D. W. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 2004;117:1033–1045. doi: 10.1242/jcs.00949. [DOI] [PubMed] [Google Scholar]
- Haraga A., Ohlson M. B., Miller S. I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H. F., Hettema E. H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2001;155:979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Sutton S. E., Wallenfang A. J., Orchard R. C., Wu X., Feng Y., Chai J., Alto N. M. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat. Struct. Mol. Biol. 2009;16:853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jackson L. K., Nawabi P., Hentea C., Roark E. A., Haldar K. The Salmonella virulence protein SifA is a G protein antagonist. Proc. Natl. Acad. Sci. USA. 2008;105:14141–14146. doi: 10.1073/pnas.0801872105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A Pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Koch A., Schneider G., Luers G. H., Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 2004;117:3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- Kuhle V., Jackel D., Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–370. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- Kuhle V., Abrahams G. L., Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2006;7:716–730. doi: 10.1111/j.1600-0854.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- Lesser C. F., Miller S. I. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 2001;20(8):1840–1841. doi: 10.1093/emboj/20.8.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Baumgart E., Dong G. X., Morrell J. C., Jimenez-Sanchez G., Valle D., Smith K. D., Gould S. J. PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 2002;22(23):8226–8240. doi: 10.1128/MCB.22.23.8226-8240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorra C., Huttner W. B. The mesh hypothesis of Golgi dynamics. Nat. Cell Biol. 1999;1:E113–115. doi: 10.1038/12939. [DOI] [PubMed] [Google Scholar]
- Marelli M., et al. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 2004;167(6):1099–1112. doi: 10.1083/jcb.200404119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T., Gallop J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Meresse S., Unsworth K. E., Habermann A., Griffiths G., Fang F., Martinez-Lorenzo M. J., Waterman S. R., Gorvel J.-P., Holden D. W. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell Microbiol. 2001;3:567–577. doi: 10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- Miao E. A., Brittnacher M., Haraga A., Jeng R. L., Welch M. D., Miller S. I. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 2003;48:401–415. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- Motley A. M., Hettema E. H. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., Brett C. L., Merz A. J. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson M. B., Huang Z., Alto N. M., Blanc M. P., Dixon J. E., Chai J., Miller S. I. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4:434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H. W., Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Poh J., Odendall C., Spanos A., Boyle C., Liu M., Freemont P., Holden D. W. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell Microbiol. 2008;10(1):20–30. doi: 10.1111/j.1462-5822.2007.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo G-D, F., Zwick M. B., Leung K. Y., Finlay B. B. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G.J.K., McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Ramsden A. E., Holden D. W., Mota L. J. Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 2007a;15:516–524. doi: 10.1016/j.tim.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Ramsden A. E., Mota L. J., Munter S., Shorte S. L., Holden D. W. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 2007b;9:2517–2529. doi: 10.1111/j.1462-5822.2007.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinicke A. T., Hutchinson J. L., Magee A. I., Mastroeni P., Trowsdale J., Kelly A. P. A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J. Biol. Chem. 2005;280:14620–14627. doi: 10.1074/jbc.M500076200. [DOI] [PubMed] [Google Scholar]
- Ridley A. J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Rottensteiner H., Stein K., Sonnenhol E., Erdmann R. Conserved function of Pex11p and the novel Pex25p and Pex27p in peroxisome biogenesis. Mol. Biol. Cell. 2003;14:4316–4328. doi: 10.1091/mbc.E03-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Albert J., Yu X. J., Beuzon C. R., Blakey A. N., Galyov E. E., Holden D. W. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microb. 2002;44:645–661. doi: 10.1046/j.1365-2958.2002.02912.x. [DOI] [PubMed] [Google Scholar]
- Saleem R. A., Knoblach B., Mast F. D., Smith J. J., Boyle J., Dobson C. M., Long-O'Donnell R., Rachubinski R. A., Aitchison J. D. Genome-wide analysis of signaling networks regulating fatty acid-induced gene expression and organelle biogenesis. J. Cell Biol. 2008;181:281–292. doi: 10.1083/jcb.200710009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M., Fahimi H. D. Growth and division of peroxisomes. Int. Rev. Cytol. 2006;255:237–290. doi: 10.1016/S0074-7696(06)55005-3. [DOI] [PubMed] [Google Scholar]
- Siggers K. A., Lesser C. F. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Marelli M., Christmas R. H., Vizeacoumar F. J., Dilworth D. J., Ideker T., Galitski T., Dimitrov K., Rachubinski R. A., Aitchison J. D. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 2002;158(2):259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Aitchison J. D. Regulation of peroxisome dynamics. Curr. Opin. Cell Biol. 2009;21:119–126. doi: 10.1016/j.ceb.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O. The Salmonella-containing vacuole-moving with the times. Curr. Opin. Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M. A., Leung K. Y., Zwick M., Garcia-del Portillo F., Finlay B. B. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 1996;20:151–164. doi: 10.1111/j.1365-2958.1996.tb02497.x. [DOI] [PubMed] [Google Scholar]
- Tam Y. Y., Torres-Guzman J. C., Vizeacoumar F. J., Smith J. J., Marelli M., Aitchison J. D., Rachubinski R. A. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4089–4102. doi: 10.1091/mbc.E03-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S., Erdmann R. Dynamin-related proteins an Pex11 proteins in peroxisome division and proliferation. FEBS J. 2005;272:5169–5181. doi: 10.1111/j.1742-4658.2005.04939.x. [DOI] [PubMed] [Google Scholar]
- Thoms S., Debelyy M. O., Nau K., Meyer H. E., Erdmann R. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 2008;275:504–514. doi: 10.1111/j.1742-4658.2007.06217.x. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H. ) Modeling the function of bacterial virulence factors in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:827–834. doi: 10.1128/EC.3.4.827-834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater C. A., Raymond C. K., Ekena K., Howald-Stevenson I., Stevens T. H. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J. Cell Biol. 1992;119(4):773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128(5):779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar F. J., Torres-Guzman J. C., Tam Y.Y.C., Aitchison J. D., Rachubinski R. A. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 2003;161:321–332. doi: 10.1083/jcb.200210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Pawel-Rammingen U., Telepnev M. V., Schmidt G., Aktories K., Wolf-Watz H., Rosqvist R. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 2000;36:737–748. doi: 10.1046/j.1365-2958.2000.01898.x. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Yan M., Rayapuram N., Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Yu X., Cai M. The yeast dynamin-related GTPase Vps1p functions in the organization of the actin cytoskeleton via interaction with Sla1p. J. Cell Sci. 2004;117:3839–3853. doi: 10.1242/jcs.01239. [DOI] [PubMed] [Google Scholar]