This article shows that mitochondrial respiratory dysfunction activates a stress signaling that induces Akt1 activation. Akt1 activation occurs through calcineurin-mediated IGF1R/PI3-K pathway. Akt1-mediated phosphorylation of hnRNPA2 is a key requirement for the propagation of stress signaling and activation of nuclear target genes.
Abstract
Mitochondrial respiratory stress (also called mitochondrial retrograde signaling) activates a Ca2+/calcineurin-mediated signal that culminates in transcription activation/repression of a large number of nuclear genes. This signal is propagated through activation of the regulatory proteins NFκB c-Rel/p50, C/EBPδ, CREB, and NFAT. Additionally, the heterogeneous ribonucleoprotein A2 (hnRNPA2) functions as a coactivator in up-regulating the transcription of Cathepsin L, RyR1, and Glut-4, the target genes of stress signaling. Activation of IGF1R, which causes a metabolic switch to glycolysis, cell invasiveness, and resistance to apoptosis, is a phenotypic hallmark of C2C12 myoblasts subjected to mitochondrial stress. In this study, we report that mitochondrial stress leads to increased expression, activation, and nuclear localization of Akt1. Mitochondrial respiratory stress also activates Akt1-gene expression, which involves hnRNPA2 as a coactivator, indicating a complex interdependency of these two factors. Using Akt1−/− mouse embryonic fibroblasts and Akt1 mRNA-silenced C2C12 cells, we show that Akt1-mediated phosphorylation is crucial for the activation and recruitment of hnRNPA2 to the enhanceosome complex. Akt1 mRNA silencing in mtDNA-depleted cells resulted in reversal of the invasive phenotype, accompanied by sensitivity to apoptotic stimuli. These results show that Akt1 is an important regulator of the nuclear transcriptional response to mitochondrial stress.
INTRODUCTION
Mitochondria play critical roles in respiration-coupled energy production, amino acid and fatty acid metabolism, Ca2+ homeostasis, heme biosynthesis, Fe2+ homeostasis, and the integration of apoptotic signals (Babcock et al., 1997; Kroemer et al., 1998; Loeb et al., 2005; Taylor and Turnbull, 2005). Mitochondrial dysfunction is associated with various human diseases, from tissue-specific conditions to generalized whole-body disorders, including cancer (Taylor and Turnbull, 2005). Proliferating tumors in human patients and animal models have been shown to contain mutated or deleted mitochondrial DNA, and/or dysfunctional mitochondria. Mutations in mtDNA and reduced mtDNA copy number have been reported for numerous tumors (Horton et al., 1996; Parrella et al., 2001; Okochi et al., 2002; Mazurek and Eigenbrodt, 2003; Lee et al., 2004; Linnartz et al., 2004; Meierhofer et al., 2004; Biswas et al., 2005a; Lievre et al., 2005; Petros et al., 2005; Taylor and Turnbull, 2005; Shidara et al., 2005b; Kulawiec et al., 2009; Singh et al., 2009). A cybrid cell line carrying mutations in the mtDNA-encoded ATPase 6 gene exhibited increased tumorigenicity and reduced apoptosis, indicating a direct role for mtDNA mutations in tumor progression (Petros et al., 2005; Shidara et al., 2005a). Similarly, heteroplasmic but not homoplasmic mutations in mtDNA encoded ND5 gene in human 143B osteosarcoma cells were shown to be tumorigenic (Park et al., 2009). We reported earlier that partial mtDNA depletion in C2C12 skeletal myoblasts and A549 lung carcinoma cells results in highly invasive phenotypes that are resistant to chemically induced apoptosis (Amuthan et al., 2002; Biswas et al., 2005a). Induced proliferation and tumor growth promotion by mitochondrial stress signaling in immortalized C2C12 myoblasts is of special significance for cancer promotion (Amuthan et al., 2001; Biswas et al., 2005a; Guha et al., 2007) Others have shown that mtDNA depletion induces resistance to apoptosis in 143B osteosarcoma (Dey and Moraes, 2000), T47D breast carcinoma (Yu et al., 2009), SK-Hep1 (Kim et al., 2002), and prostate cancer cells (Moro et al., 2009). Depletion of mtDNA in LNCaP androgen-dependent prostate carcinoma cells induces progression to androgen independence, with increased cell migration and resistance to common chemotherapeutic agents (Higuchi et al., 2006; Moro et al., 2008).
Disruption of mitochondrial membrane potential—through a reduction in mtDNA copy number, drug- and hypoxia-induced mitochondrial dysfunction, or treatment with the mitochondrial ionophore 3-chlorophenylhydrazone (CCCP)—triggers a novel stress-signaling pathway (Biswas et al., 1999; Amuthan et al., 2001, 2002). The signaling onset is marked by an increase in [Ca2+]c and activation of Ca2+-responsive calcineurin (Cn) (Biswas et al., 1999; Amuthan et al., 2001, 2002). Downstream signaling events include activation of the transcription factors nuclear factor of activated T-cells (NFAT), CAAT/enhancer binding protein δ (C/EBPδ), cAMP-responsive element binding protein (CREB), and a novel IκBβ-dependent nuclear factor κB (NFκB) c-Rel/p50 (Amuthan et al., 2001; Arnould et al., 2002; Biswas et al., 2003). A recent study on the characterization of promoter elements of stress target genes, Cathepsin L (CnL), RyR1 (ryanodine receptor 1), and Glut 4 demonstrated the functional importance of these factor binding sites at the 200-1000 nucleotides immediately upstream of transcription start sites (Guha et al., 2009).
Heterogeneous ribonucleoprotein A2 (hnRNPA2), a nuclear RNA-binding protein, which is activated in response to mitochondrial stress, was shown to function as a common transcriptional coactivator for the up-regulation of the mitochondrial stress-target genes Cathepsin L, RyR1, and Glut-4 (Guha et al., 2009). An altered function of hnRNPA2 has been implicated in a variety of cancers including lung, breast, and pancreatic cancers (Garayoa et al., 2003; Patry et al., 2003; Hee et al., 2005; and Yamoka et al., 2006). hnRNPA2 protein is known to associate with a number of oncogenic proteins including TDP-43, TOG2, and SET, although the precise significance of these associations in tumor progression remains unclear. A recent study from our laboratory showed that hnRNPA2 functions as a transcription coactivator by associating with the transcription complexes mainly through protein–protein interactions (Guha et al., 2009). The results of this study suggested that hnRNPA2 plays a key role either in the recruitment of DNA binding factors, NFκB (c-Rel/p50), C/EBPδ, CREB, and NFATc to the promoter sites or in the overall stability of the enhanceosome complex of mitochondrial respiratory stress-responsive genes.
Mitochondrial respiratory stress also increases cellular glucose uptake, dependence on glucose, and glycolysis (Guha et al., 2007) in C2C12 myoblasts, A549 cells, and RAW 264.7 macrophages. These metabolic changes are mediated through Cn-dependent activation of insulin-like growth factor 1R (IGF1R) (Guha et al., 2007). It is now well established that the high metabolic activity of tumor cells is associated with activation of Akt kinase, which is downstream of IGF1R. Akt activation in response to mitochondrial respiratory stress has been reported in different tumor cell systems (Pelicano et al., 2006; Moro et al., 2009).
Here, we show up-regulation of Akt1-gene expression in response to mitochondrial stress, along with a several-fold increase in the level of phosphorylated Akt1 in the nuclear compartment. We also show that Akt activation is crucial for the full stress response, which culminates in altered expression of nuclear gene targets. We show that Akt-mediated phosphorylation of hnRNPA2 is critical for transcriptional modulation and cell survival under mitochondrial respiratory stress. Our results suggest a complex regulatory circuit in which hnRNPA2 and Akt1 function interdependently.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
Murine C2C12 skeletal myoblasts (ATCC, Manassas, VA; CRL1772) were grown in DMEM (Life Technology, Carlsbad, CA) supplemented with 10% fetal bovine serum and 0.1% gentamicin. Depletion of mtDNA was carried out by ethidium bromide treatment (EtBr, 100 ng/ml for 30 passages) as described before (Biswas et al., 1999). Selected clones containing ∼20% mtDNA were grown in the presence of 1 mM sodium pyruvate and 50 μg/ml uridine. We also used 2,3′-dideoxycytidine (ddC; 10 μM, 120 h) in some experiments (as mentioned) for depleting the mtDNA. Reverted cells represent mtDNA-depleted cells (85% depletion) grown subsequently for 30 cycles in the absence of EtBr, for reversing the mtDNA content to 80% of control cells. The mtDNA content was estimated by real-time PCR using a probe for mouse mtDNA (D-loop) and normalized using the nuclear IκBβ, a single-copy gene. The sequences of the primers used are as follows: mtDNA: ACTATCCCTTTCCCCATTTG (forward) and TGTTGGTCATGGGCTGATTA (reverse); and nuclear gene IκBβ: AGCTGGTGTCTGGGGTACAGT (forward) and ATCCTTGGGGAGGCATCTAC (reverse). Aliquots from the same parent stock cultures of control, depleted, and reverted cells were used in all experiments. The relative mtDNA content of each cell groups is shown in Figure S1.
Preparation of Nuclear and Cytosolic Protein Extracts
Nuclear and cytosolic protein fractions were isolated from different cells as described previously (Biswas et al., 1999; Guha et al., 2007).
Immunoblot Analysis
Protein content was measured by the method of Lowry (Lowry et al., 1951). Total cell lysates, cytosolic fractions, and nuclear fractions (30 μg) were solubilized in Laemmli's sample buffer, resolved by electrophoresis on 10 or 12% SDS–polyacrylamide gels, and subjected to immunoblot analysis. The immunoblots were developed using the Super Signal West Femto maximum sensitivity substrate from Pierce Chemical Co. (Rockford, IL). Antibodies to Akt1/2, pAkt (Ser473), and pAkt (Thr 308) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies to p-Serine, p-Thr, and p-Tyr were from Sigma Chemicals (St. Louis, MO). Antibody to nuclear p97 was from Affinity Bioreagents (Golden, CO). Antibodies to DHFR (dihydrofolate reductase), β-actin, and hnRNPA2 were from Santa Cruz Biotechnology (Santa Cruz, CA).
CnAα, IGF1R, and Akt1 mRNA Knockdown by Small Interfering RNA
Predesigned small interfering RNAs (siRNAs) for mouse CnAα (ID no. 292199), IGF1R (ID no. 159115), Akt1 (ID no.162426), and negative controls (scrambled siRNA) were purchased from Ambion (Austin, TX). Control and mtDNA-depleted cells (104 cells/well) were transfected with preannealed double-stranded siRNAs (50 μM stock) at a final concentration of 30 nM by the method of reverse transfection. Transient transfections were carried out in triplicate using siPORT NeoFX reagent (Ambion). RNA was isolated 48 h after transfections using Trizol reagent (Invitrogen, Carlsbad, CA), and the level of silencing of CnAα, IGF1R, and Akt1 mRNA was quantified by real-time PCR, as described later in this section.
Generation of Stable hnRNPA2-silenced Cell Lines
hnRNPA2-silenced stable knockdown of C2C12 cells were described before (Guha et al., 2009). Stable cell lines were generated after transfection (Fugene 6 reagent; Roche Molecular Biochemicals, Indianapolis, IN) of mtDNA-depleted C2C12 cells with hnRNPA2si3-pSilencer2.1neo (or pSilencer2.1neo vector alone) containing a neomycin resistance gene. HnRNPA2-silenced cell lines were generated using G418 (1 mg/ml) as a selection marker.
Cloning and Transfections
Human Akt1 promoter (−900 to +1), a kind gift from J. Q. Chen, was subcloned into the pGL3 mammalian expression vector (Promega, Madison WI). The c-Rel, C/EBPδ, NFAT, and CREB genes were cloned into the pCMV4 expression vector. hnRNPA2 cDNA was subcloned into pCI for transfections of C2C12 cells. The WT, T98A and S219A mutant hnRNPA2 constructs in pcDNA 3.1 (+) hygro plasmid were generated by Gene Pass (Nashville, TN).
Transfections were carried out using Fugene 6 reagent, following the manufacturer's suggested protocol (Guha et al., 2009). Promoter DNA constructs cloned in the pGL3 vector (1 μg) and 0.5 μg of a renilla luciferase construct (Promega) were used as an internal control. Luciferase activity was assayed using the Dual-Luciferase reporter assay system from Promega. Transfections with various cDNAs were carried out using 0.2 μg of cDNA constructs.
mRNA Quantitation by Real-Time PCR
Total RNA was isolated using Trizol reagent as per supplier's protocol (Invitrogen). cDNA was generated from 5 μg RNA using the cDNA Archive kit from Applied Biosystems (Foster City, CA), and 50 ng of this cDNA was used as a template for each reaction. Relative quantification of CnA, IGF1R, Akt1, and Akt2 mRNA by real-time PCR was done using SYBR Green (Applied Biosystems) in an ABI Prism 7300 sequence detection system (Applied Biosystems). Levels of mRNA were normalized to β-actin as an endogenous control.
Immunoprecipitation
To ensure nearly equal amounts of input Akt levels in the control and mtDNA-depleted cell extracts, we used 100 μg of protein from mtDNA-depleted cells and 300 μg protein from control cells. The nuclear and cytosolic fractions were immunoprecipitated overnight at 4°C with hnRNPA2 antibody (20 μg/ml). The immune complexes were collected onto protein A–agarose beads (Sigma Chemicals) and washed extensively. The immunoprecipitates were extracted from the beads with 2× Laemmli buffer devoid of β-mercaptoethanol at 95°C for 5 min for further analysis.
Akt Activity
Akt activity was measured using an Akt/protein kinase B (PKB) kinase activity assay kit (Stressgen Bioreagents, Victoria, BC, Canada). This assay is based on an ELISA that utilizes a synthetic peptide as a substrate for PKB and a polyclonal antibody that recognizes the phosphorylated form of the substrate.
Annexin V Assay
The assay for cells undergoing early apoptosis was performed using the Guava Nexin Kit (Guava Technologies, Hayward, CA) according to the manufacturer's suggested protocol. This method utilizes annexin V-PE to detect phosphatidylserine on the plasma membrane as a marker of apoptotic cells. Cells (106 each) were transfected in triplicate with Akt1 and mock siRNAs, harvested after 48 h, washed with 1 ml 1× Nexin Buffer (Guava Technologies), and resuspended in the same buffer. After labeling with Annexin V-PE, the percentage of apoptotic cells was quantified using the Guava Personal Cytometer (Guava Technologies).
In Vitro Invasion Assay
The in vitro invasion assays were carried out as described previously (Amuthan et al., 2001). The Matrigel invasion chambers were prepared at 1:2 dilution of Matrigel (Becton Dickinson, Belford, MA) as described previously (Guha et al., 2007). Cells from three wells counted in a Guava Personal Cytometer (Guava Technologies) and an equal number of viable cells (1 × 105) were seeded on top of the Matrigel layer. After incubation for 24 h at 37°C, noninvading cells in the Matrigel layer were quantitatively removed, and the microporous membrane containing invaded cells was stained and viewed under an Olympus BX 61 fluorescence microscope (Melville, NY) as described previously (Amuthan et al., 2001; Guha et al., 2007). Each experimental set was carried out in triplicate, and at least six fields were examined within any single experiment for each condition.
In Vitro Kinase Assay
The in vitro kinase assay was carried out according to the methods of Summers and coworkers (Summers and Birnbaum, 1997). 0.1 μg of Recombinant Akt (Millipore, Billerica, MA) was incubated for 30 min at 37°C with purified His-hnRNPA2 (1.0 μg) in a buffer containing 10 mM HEPES (pH 7.4), 1 mM MgCl2, and 1 mM MnCl2 in the presence of 10 μCi [γ-32P]ATP.
Chromatin Immunoprecipitation Analysis
Chromatin immunoprecipitation (ChIP) assays were performed following the protocol of Upstate Biotechnologies (Lake Placid, NY). Cells were fixed by adding 1% formaldehyde to the culture medium and were incubated for 10 min at 37°C. Cells were washed in 1× PBS (containing protease inhibitors), scraped, and pelleted at 2000 × g for 5 min at 4°C. The cell pellet was suspended in SDS lysis buffer (containing 1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) and incubated for 10 min on ice. The cross-linked cell lysates were sonicated using an Ultrasonic processor sonicator (10-s pulses, 20 times, on ice), and the size of sheared DNA was checked on 0.8% agarose gel, such that the DNA fragments were 200-1000 base pairs. The samples were centrifuged for 10 min at 13,000 × g, 4°C. The supernatant was diluted sixfold in a buffer (pH 8.1) containing 167 mM NaCl, 16.7 mM Tris-HCl, 1.2 mM EDTA, 0.01% SDS, and 1.1% Triton X-100; it was then divided into 200-μl aliquots. One aliquot (10%) was retained as an input sample for normalization of the DNA quantity in each sample. Each aliquot was immunoprecipitated overnight at 4°C using antibodies, as mentioned in the figure legends. Preimmune IgG was used as a negative control. The antibody-protein complex was collected by adding protein A–agarose/salmon sperm DNA (50% slurry; Sigma Chemicals) for 1 h at 4°C. The protein A–agarose/antibody/protein complex was washed to remove DNA fragments that were bound nonspecifically. DNA was eluted from the beads, cross-linking was reversed, and after purification the DNA was analyzed by real-time PCR amplification of the mouse Akt1 promoter (−600 to +1). Data were normalized with the corresponding input values.
RESULTS
Increased Expression of Akt1 in Cells Subjected to Mitochondrial Stress
The relative levels of mtDNA and nuclear DNA in control, mtDNA-depleted, and reverted cells are shown in Figure S1. mtDNA content in cells treated with EtBr was reduced by ∼80% relative to that in control cells. We also used 2,3′-ddC to deplete mtDNA by ∼70% compared with control cells (Figure S1). In reverted cells, mtDNA levels were within 20% of control levels. Unless specified, EtBr-induced mtDNA-depleted cells, hereafter referred to as mtDNA-depleted cells, have been used in experiments reported here. The 2,3′-ddC–depleted cells are referred to as ddC mtDNA-depleted cells.
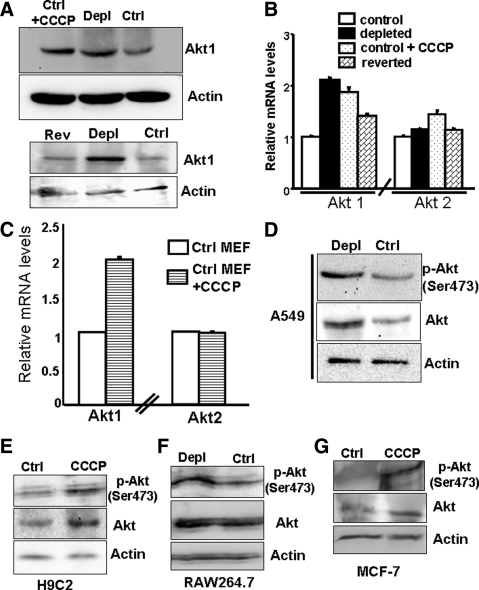
Akt is overexpressed in tumorigenic cells (Plas and Thompson, 2005); therefore we assessed Akt levels after mtDNA depletion. Figure 1A (top) shows the Akt protein level in total lysates from control, mtDNA-depleted cells, and CCCP-treated control cells. Cells subjected to mitochondrial stress via mtDNA depletion or CCCP treatment showed two- to threefold higher Akt protein relative to controls. In reverted cells, the protein level was close to that in control cells (Figure 1A, bottom), indicating the increase in Akt is a direct response to mitochondrial respiratory stress.
Figure 1.
Increased Akt1 protein and mRNA levels in cells subjected to mitochondrial respiratory stress, either by mtDNA depletion or treatment with CCCP. (A) Top, immunoblot analysis of total cell lysate (30 μg protein each) from control, mtDNA-depleted C2C12 cells, and control C2C12 cells treated with CCCP (25 μM, 2 h) using Akt1 antibody (1:1000 dilution). Bottom, immunoblot analysis of total cell lysates (30 μg each) from control, mtDNA-depleted, and reverted C2C12 cells using Akt1 antibody. β-Actin was used as an internal loading control. (B) Real-time PCR analysis of mouse Akt1 and Akt2 mRNA levels in control, mtDNA-depleted, and reverted C2C12 cells and also in control C2C12 cells treated with CCCP (25 μM, 2 h). (C) Real-time PCR analysis of Akt1 and Akt2 mRNA in mouse embryonic fibroblasts (MEFs) treated without and with CCCP (25 μM, 2 h). (D) Immunoblot analysis of total cell extracts (30 μg each) of control and mtDNA-depleted A549 lung carcinoma cells with antibody to Akt and phospho-Ser473 Akt. (E–G) Immunoblot analysis of nuclear extracts from H9C2 cells treated with CCCP, mtDNA-depleted RAW 264.7 cells, and CCCP treated MCF-7 cells with Akt and Ser437 pAkt antibodies. Although not shown, mtDNA contents of A549 and MCF-7 cells were ∼20–25% of control cells. In B and C, mean ± SEM values were derived from three to four separate experiments.
Akt1 mRNA in C2C12 cells was increased twofold in mtDNA-depleted and CCCP-treated cells, compared with control cells (Figure 1B). In reverted cells, Akt1 mRNA was reduced to a level closer to the control cells (1.5-fold above control). The differences in the Akt2 mRNA levels between the cell types were not significant, indicating that Akt2 in these cells is not regulated by mitochondrial stress.
The generality of Akt1 induction in response to mitochondrial stress was further investigated using nontransformed cells (mouse embryonic fibroblasts [MEF] and RAW264.7 macrophages) and transformed cells (A549 lung carcinoma, H9C2 cardiac myoblasts, and MCF-7 breast cancer). As shown in Figure 1C, the level of Akt1 mRNA was increased in MEF cells in response to stress inducer, CCCP. In A549 cells (Figure 1D), H9C2 cells (Figure 1E) and RAW264.7 cells, both the steady-state level of Akt in the total lysate and the level of phosphorylated Akt (Ser 473) were increased in response to mtDNA depletion or CCCP treatment. In the case of MCF-7 cells (Figure 1G), only the level of phosphorylated Akt but not the steady-state Akt increased in response to CCCP treatment. These results show that an increase in steady-state level of Akt under mitochondrial stress is cell specific, although an increase in Ser-473 phosphorylation/activation of Akt1 is a common response to mitochondrial stress in many cell types.
Mitochondrial Stress Results in Activation and Increased Nuclear Localization of Akt
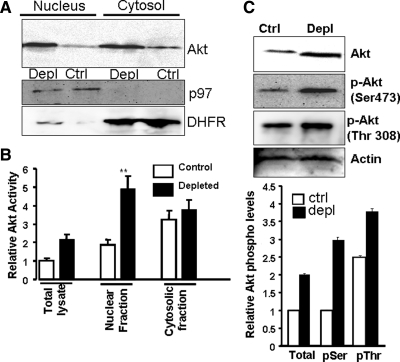
Akt1 exists as a soluble protein in the cytosol and translocates to the nucleus under growth-stimulatory conditions (Trotman et al., 2006). As shown in Figure 2A, steady-state Akt1 levels in the cytosol and nucleus were increased threefold in mtDNA-depleted cells, compared with control cells. 2,3′-ddC–induced mtDNA depletion also caused nuclear accumulation of Akt1 (Figure S1D). Akt activity was increased 2.5-fold in total lysate and nuclear fractions of mtDNA-depleted cells (Figure 2B), but was increased only marginally in the cytosolic fraction.
Figure 2.
Nuclear and cytosolic levels of Akt and phosphorylated Akt in control and mtDNA-depleted C2C12 cells. (A) Immunoblot analysis of nuclear and cytosolic proteins (50 μg) with Akt antibody. The same blot was reprobed with antibody to nuclear protein p97 and cytosolic protein DHFR to determine the extent of cross-contamination and protein-loading levels. (B) Akt activities in the total cell lysate, nuclear, and cytosolic protein fractions were measured using a kit from Assay Design/StressGen (Ann Arbor, MI). Phosphorylation of a synthetic peptide substrate was assayed according to the manufacturer's protocol. (C) Top, immunoblot analysis of nuclear proteins (50 μg protein each) from control and mtDNA-depleted C2C12 cells using antibodies to Akt (top panel), phospho-Ser473 Akt (second panel), phospho-Thr308 Akt (third panel). The level of phosphorylation was quantified by imaging the blots (n = 3) using the Bio-Rad Versa Doc Imaging system (Hercules, CA). Mean ± SEM values were calculated from three separate assays.
Akt activation is dependent on phosphorylation by PDK1 (Ser473) and PDK2 (Thr308) (Chen et al., 2001; Kondapaka et al., 2004). Tyrosine phosphorylation is also important for Akt activation. Nuclear extracts from control and mtDNA-depleted cells were immunoprecipitated using Akt antibody (Figure 2C, top), and the immunoprecipitates were probed with phospho-specific antibodies. Ser473-specific antibody yielded a more intense band with nuclear extract from mtDNA-depleted cells (three- to fourfold higher), compared with control cell extract (Figure 2C). This difference is consistent with the nearly threefold increase of Akt activity in nuclear extract from mtDNA-depleted C2C12 cells (see Figure 2B). Thr308-directed antibody yielded ∼50% higher intensity in extract from mtDNA-depleted cells, relative to that from control cells. The specificity of antibody-based detection of phospho-Akt was confirmed using agents known to inhibit phosphorylation of Akt at Ser473 and Thr308. ML9 markedly reduced staining with anti-Ser473 antibody, and UCNO1 reduced the band intensity with Thr308 antibody (Figure S2). Steady-state protein levels were not affected by these inhibitors.
Akt1 Induction under Mitochondrial Stress Is Dependent on Cn/IGF1 Receptor/Phosphoinositide-3 Kinase Pathway
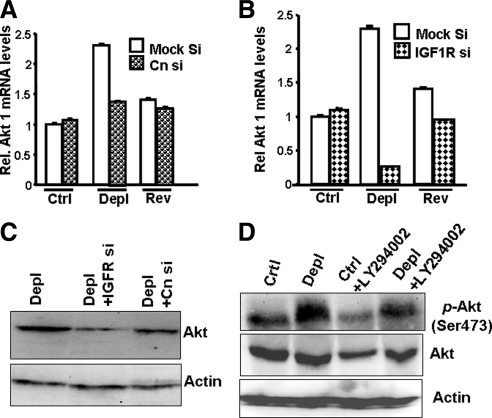
Mitochondrial stress signaling is initiated by Cn activation, which in turn activates IGF1 receptor downstream. We showed previously that inhibiting IGF1R activity makes mtDNA-depleted cells susceptible to apoptosis. We studied the role of IGF1R in the activation of Akt1, which is known to promote cell survival. We used siRNA-targeted silencing of CnAα and IGF1R to investigate the role of Cn/IGF1R in Akt1 induction under mitochondrial stress (Figure 3, A–C). Transient transfection with siRNA for CnAα and IGF1R reduced the respective mRNA levels by 60–80%, relative to levels in mock-transfected (scrambled siRNA) cells (Figure S3, A and B). Akt1 mRNA levels were reduced 40% by CnAα mRNA silencing in mtDNA-depleted cells, but were not changed in control and reverted cells (Figure 3A). IGF1R mRNA silencing caused an 80% reduction in Akt1 mRNA in mtDNA-depleted cells and 10% reduction in reverted cells, but no change in control cells (Figure 3B). Akt1 protein levels were also reduced significantly in mtDNA-depleted cells after either CnAα or IGF1R mRNA silencing (Figure 3C), but neither reduced Akt1 protein in control cells (data not shown).
Figure 3.
The role of calcineurin, IGF1R and PI3-K in the activation of Akt1 in cells subjected to mitochondrial stress. (A) Effects of CnAα mRNA knockdown on the levels of Akt1 mRNA in control, mtDNA-depleted, and reverted C2C12 cells. (B) Effects of IGF1R mRNA knockdown on the levels of Akt1 mRNA in control, mtDNA-depleted, and reverted C 2C12 cells. In both A and B, Akt1 mRNA levels were quantified using real-time PCR analysis. Mean ± SEM values were calculated from three separate estimates. (C) Immunoblot analysis of total cell lysates (50 μg each) from control, mtDNA-depleted C2C12 cells, and control as well as depleted cells expressing siRNAs to CnAα and IGF1R were developed with Akt antibody. A companion blot was probed with actin antibody to assess loading levels. (D) Effects of the PI3-K inhibitor LY294002 (50 μM, 2 h) on Akt and phospho-Ser473 Akt in control and mtDNA-depleted cells. The immunoblot represents the analysis of total lysates (50 μg protein each) developed with the indicated antibodies. A companion blot was probed with actin antibody to assess protein loading.
As shown for other Tyr kinase receptors, IGF1R-dependent Akt activation may involve phosphoinositide-3 kinase (PI3-K) activity. This possibility was tested by treating control and mtDNA-depleted cells with the PI3-K–specific inhibitor LY294002 and measuring the levels of p-Akt (phospho-Ser473) in the nuclear fraction. To compare p-Akt levels, we used equal amounts of input Akt protein from the control and mtDNA-depleted cells. LY294002 treatment resulted in a significant decrease (50%) in p-Akt levels in control and mtDNA-depleted cells, suggesting Akt activation is a PI3-K–dependent process (Figure 3D).
Transcriptional Activation of Akt1 Promoter in Response to Mitochondrial Stress
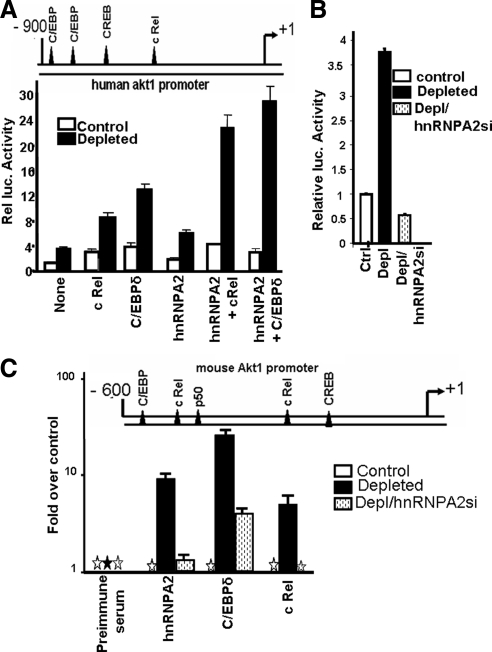
We next examined if the increase in Akt1 mRNA in mtDNA-depleted cells was due to transcriptional activation of the Akt1 promoter, using a human Akt1 promoter DNA-reporter construct (Park et al., 2005). Nucleotide sequence analysis (MatInspector) indicated that the 900-base pairs region of the Akt1 gene upstream of the transcription start site contains consensus sites for binding to the stress-activated transcription factors NFκB c-Rel, C/EBPδ, and CREB (Figure 4A), similar to what has been shown for other stress target genes (Biswas et al., 2005b; Guha et al., 2009). The promoter activity in mtDNA-depleted cells was nearly fourfold higher than in control cells (Figure 4A). Although not shown, 5′ deletion analysis revealed that this minimal promoter region was sufficient to respond to mitochondrial stress–mediated transcription activation. Cotransfection of the promoter-reporter construct with c-Rel, C/EBPδ, and hnRNPA2 cDNAs resulted in 2-, 3-, and 1.5-fold higher promoter driven luciferase activity, respectively, over the basal activity without cDNA transfection. As shown recently for other stress-response genes (Guha et al., 2009), cDNA transfections in control C2C12 cells increased promoter activity only marginally. Cotransfection with hnRNPA2 cDNA alone induced the promoter activity by ∼60%. Cotransfections with hnRNPA2 cDNA and either c-Rel or C/EBPδ cDNAs resulted in a three- to fourfold increase in luciferase activity, suggesting a cooperative interaction between these factors (Figure 4A). Akt1 promoter activity was markedly lower in hnRNPA2-silenced mtDNA-depleted cells (Figure 4B), confirming the role of hnRNPA2 in Akt1 transcription. This is consistent with the reduced nuclear Akt1 protein levels found in hnRNPA2-silenced/mtDNA-depleted cells (Figure S4). In keeping with our observations with other stress responsive target genes (RYR1, CnL, and Glut-4), these results suggest that hnRNPA2 may function as a coactivator of Akt1 transcriptional activity under mitochondrial stress.
Figure 4.
Transcriptional activation of the human Akt1 promoter by mitochondrial stress and promoter occupancy of hnRNPA2 and other transcription factors in mtDNA-depleted C2C12 cells. (A) A cartoon of the human Akt1 promoter, indicating the putative binding sites for stress-activated transcription factors (top). Relative luciferase activity of the Akt1 promoter (cloned in pGL3 basic vector) in control and mtDNA-depleted cells, cotransfected as indicated with various cDNAs. (B) Promoter activity of the human Akt1 promoter in control, mtDNA-depleted and hnRNPA2-silenced/mtDNA-depleted cells. (C) Cartoon of the mouse Akt promoter, indicating the stress-activated factor binding sites on its proximal region (top). ChIP analysis of the mAkt1 promoter region from control, mtDNA-depleted, and mtDNA-depleted/hnRNPA2-silenced cells. Immunoprecipitation of chromatin fragments was carried out with antibodies to hnRNPA2, C/EBPδ, and c Rel proteins as indicated. Real-time PCR analysis was carried out using 10% input DNA for normalization. Values for factor binding relative to the control cell values have been expressed. Preimmune serum was used as a negative antibody control. Mean ± SEM values were calculated from three to four independent assays.
The induction of stress signaling activation of Akt1 is not restricted to EtBr-treated cells because ddC mtDNA-depleted C2C12 cells also showed markedly increased nuclear Akt and also phosphorylated Akt (Figure S7A). Additionally, important marker genes of stress signaling such as RYR1, CnL, IGF1R, hnRNPA2, and Akt are also transcriptionally up-regulated in ddC mtDNA-depleted cells as seen by increase in mRNA levels by 2–3.5-fold (Figure S7B).
Increased Akt1 Promoter Occupancy of Stress-activated Transcription Factors in mtDNA-depleted Cells
The in vivo association of stress activated transcription factors and hnRNPA2 with the Akt1 promoter DNA was investigated by ChIP analysis. The promoter occupancy of these factors in response to mitochondrial respiratory stress was compared in control C2C12 cells, mtDNA-depleted cells, and mtDNA-depleted cells in which hnRNPA2 mRNA was silenced (Figure 4C). As shown in Figure 4C, the footprint of the mouse promoter for stress-specific factor-binding motifs is similar to the human Akt1 promoter, in that the proximal 600-base pair region contains sites for binding to c-Rel, CREB, and C/EBPδ as in the human Akt1 promoter, although the relative order of these factors differs for the two promoters. The mouse promoter also contains a site for CREB immediately upstream of the transcription start site. ChIP analysis in Figure 4 shows that the occupancies of the Akt1 promoter by hnRNPA2, C/EBP δ, c-Rel, and p50 were 10-, 30-, 7- and 6-fold higher, respectively, in mtDNA-depleted C2C12 cells compared with control cells. hnRNPA2 silencing in mtDNA-depleted cells markedly reduced the levels of these factors, confirming that hnRNPA2 is an adaptor protein that facilitates the recruitment of NFκB, c-Rel/p50, and C/EBPδ to the transcription complex.
Interaction of Nuclear Akt1 and hnRNPA2 in mtDNA-depleted Cells
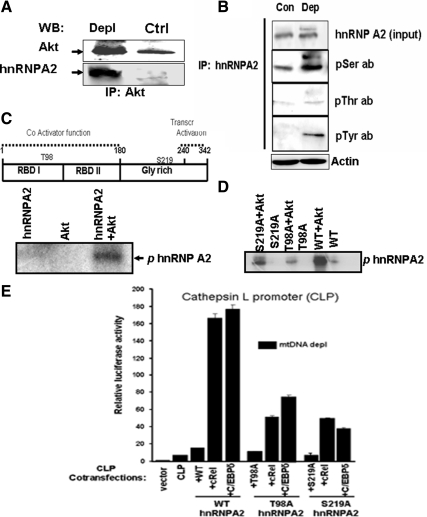
We investigated the physical interaction of Akt1 with hnRNPA2 using an antibody pulldown method, and protein species in the immunoprecipitates were identified by nano-liquid chromatography (LC)/mass spectrometry (MS) analysis. Figure S5A shows the gel pattern of nuclear proteins from control, mtDNA-depleted, and mtDNA-depleted/hnRNPA2 knockdown cells immunoprecipitated by Akt1 antibody. Those protein bands that were more abundantly pulled down from mtDNA-depleted cell extracts compared with control and hnRNPA2 knockdown cell extracts (Figure S5A, arrows) were subjected to LC-MS analysis. Most of the putative Akt1-interacting proteins identified are functionally related to cell morphology, cell survival, resistance to apoptosis, and regulation of oxidative stress (Table 1). Interestingly, the identification of hnRNPA2 as a putative Akt1-interacting protein was most relevant to this article. Notably, low or negligible levels of hnRNPA2 were pulled down from control and mtDNA-depleted/hnRNPA2 knockdown cells, whereas a major population of hnRNPA2 was identified in mtDNA-depleted cells. This was confirmed by immunoblot analysis of protein pulled down by Akt1 antibody. Akt1 antibody pulled down hnRNPA2 in high abundance from mtDNA-depleted cells, but only negligible amounts were precipitated from the control nuclear extract (Figure 5A). Other proteins pulled down from mtDNA-depleted cells (Table 1; e.g., thioredoxin, transgelin, and galectin 3 are involved in tumorigenesis, and some are known substrates of Akt1).
Table 1.
Nuclear proteins associated with Akt in mtDNA-depleted cells
| Proteins | Molecular weight (kDa) |
|---|---|
| Thioredoxin (TRX-1) | 12 |
| Transgelin (SM22, TAGLN) | 22 |
| Myosin | 120 |
| Actin | 43 |
| 14-3-3ϵ | 29 |
| Annexin A2 | 37 |
| HnRNPA2 | 36 |
| HnRNPK | 65 |
| Vimentin | 57 |
| Galectin 3 | 31 |
| Tubulin | 50 |
Distinctive protein bands immunoprecipitated by Akt antibody, as shown in Supplementary Figure S5, were excised and subjected to nano LC-MS/MS analysis.
Figure 5.
hnRNPA2 is a potential substrate for Akt1-mediated phosphorylation. (A) Control and mtDNA-depleted nuclear extracts (150 μg each) were immunoprecipitated using Akt antibody. Immunoprecipitates were probed with Akt antibody (top) and hnRNPA2 antibody (bottom) by immunoblot analysis. (B) Nuclear extracts from control (300 μg protein) and mtDNA-depleted (100 μg protein) C2C12 cells were immunoprecipitated using hnRNPA2 antibody, and the immunoprecipitates were probed with antibodies to hnRNPA2, p-Ser, p-Thr, and p-Tyr antibodies as indicated. Note that the fivefold higher level of nuclear extract from control cells was used to ensure nearly equal input of hnRNPA2 protein. A companion blot was probed with actin antibody for assessing loading level. (C) A cartoon of hnRNPA2 indicating different functional domains has been presented at the top. Putative Akt phosphorylation sites (T98, S219) have been indicated. The bottom panel shows the autoradiogram of phosphorylated recombinant purified hnRNPA2 (1.0 μg), using recombinant Akt1 (0.1 μg) as described in Materials and Methods. Akt alone and hnRNPA2 alone were used as negative controls. (D) Mutational analysis of putative phosphorylation sites of hnRNPA2. Autoradiogram shows the phosphorylation of hnRNPA2 with (0.1 μg of recombinant Akt1) or without Akt1 or purified (1 μg each) recombinant WT, T98A, and S219A mutant forms of hnRNPA2. (E) The functional significance of Akt phosphorylation sites (T98 and S219) of hnRNPA2 was tested by transient transfection CnL promoter in mtDNA-depleted cells. Cells were cotransfected with or without WT and mutant (T98A and S219A) hnRNPA2 cDNAs and c-Rel, C/EBPδ cDNAs as indicated. CLP, cathepsin L promoter. Transfection conditions were as described in Figure 4 and Materials and Methods.
We also observed significantly higher levels of Ser-, Thr-, and Tyr-phosphorylation of nuclear hnRNPA2 in mtDNA-depleted cells (Figure 5B). Sequence analysis (using www.phosphoscan.mit.edu) of hnRNPA2 (Figure 5C, top) revealed two consensus sites for Akt phosphorylation, at Thr98 and Ser219, and four potential Tyr phosphorylation sites at Y123, Y264, Y275, and Y288. We further investigated the role of Akt1 phosphorylation of hnRNPA2 in this study. However, the role of Tyr phosphorylation was not investigated further.
The autoradiogram in Figure 5C of an in vitro kinase assay using recombinant hnRNPA2 shows that hnRNPA2 is phosphorylated in vitro by Akt1 kinase. We investigated the significance of the Ser219 and Thr98 sites for Akt1-dependent phosphorylation using bacterially expressed and purified wild-type (WT) and mutant hnRNPA2 proteins (T98A and S219A). In vitro translation (Figure S6A), as well as the gel pattern of the purified recombinant proteins (Figure S6B), showed no difference in either translation efficiency or purity between WT, T98A, and S219A mutant proteins. Figure 5D shows the gel profiles of purified hnRNPA2 incubated with and without Akt1 in presence of 10 μCi [γ-32P]ATP. Results show that phosphorylation is diminished by ∼90% in the T98A mutant and by almost 70% in the S219A mutant, relative to WT protein. These results suggest that both Thr98 and Ser219 are targets for Akt-mediated phosphorylation and demonstrate that Akt1 phosphorylates hnRNPA2 both under in vitro and in vivo conditions.
The functional significance of Akt1 phosphorylation of hnRNPA2 was further investigated by mutational analysis. Previously we have shown that WT hnRNPA2 with c-Rel or C/EBPδ cooperatively stimulated transcription activation of CnL and RyR1 promoters in mtDNA depleted C2C12 cells (Guha et al., 2009). In here we tested the ability of T98A and S219A mutant hnRNPA2 cDNAs to sustain the c-Rel or C/EBPδ mediated activation of CnL promoter activity in mtDNA-depleted C2C12 cells (Figure 5E). In compliance with previous results (Guha et al., 2009), cotransfection of CnL promoter construct with c-Rel or C/EBPδ cDNAs yielded 16–20-fold higher activity than that of the basal promoter (Figure 5E). Both T98A and S219A mutations had a dominant negative effect because both cDNAs yielded markedly lower activities of 6–7-fold compared with the basal activity. These results confirm that Akt-mediated phosphorylation of hnRNPA2 is critical for its transcription coactivator function.
Functional Significance of Akt-mediated Phosphorylation of hnRNPA2
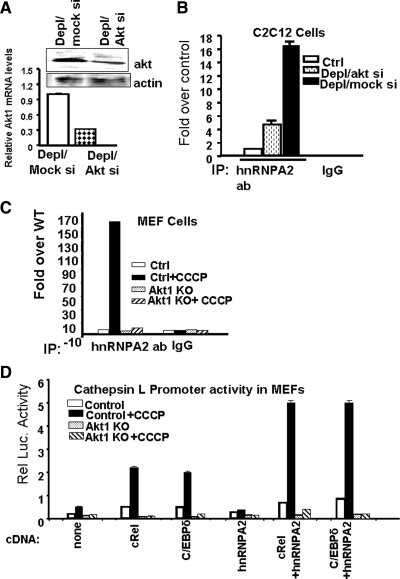
We investigated the role of Akt1 on the transcription-activator function of hnRNPA2, using ChIP analysis of CnL promoter region in C2C12 cells expressing siRNA against Akt1, as well as Akt1 knockout MEFs. Figure 6A shows levels of Akt1 mRNA and protein in Akt1 knockdown/mtDNA-depleted C2C12 cells were reduced by 55–70%, compared with cells expressing scrambled siRNA. ChIP analysis showed hnRNPA2 occupancy of the CnL promoter was reduced by ∼75% in mtDNA-depleted cells, compared with mock siRNA-expressing cells (Figure 6B). The mitochondria-specific ionophore CCCP was used to induce respiratory stress in MEF cells. We showed previously that CCCP induces Ca2+- and Cn-signaling and stress-related transcriptional activation, all of which require the action of hnRNPA2 (Guha et al., 2009). As shown in Figure 6C, CCCP treatment increased the hnRNPA2 occupancy of the CnL promoter by over 100-fold. Akt1-knockout MEFs, however, failed to respond to CCCP-mediated mitochondrial stress, as there was no increase in the association of hnRNPA2 with promoter DNA.
Figure 6.
Effects of Akt1 mRNA silencing on the recruitment of hnRNPA2 to CnL-promoter DNA in vivo. (A) Top, Western blot showing Akt 1 protein levels in mtDNA-depleted C2C12 cells expressing scrambled siRNA (mtDNA-depleted/mock siRNA) and mtDNA-depleted C2C12 cells expressing Akt1 siRNA (mtDNA-depleted/Akt1siRNA). Bottom, real-time PCR analysis of Akt1 mRNA levels in mtDNA-depleted/mock siRNA and mtDNA-depleted/Akt1siRNA cells. (B) ChIP analysis of CnL promoter using hnRNPA2 antibody in control, mtDNA-depleted/mock siRNA, and mtDNA-depleted/Akt1 siRNA cells. Goat IgG was used as a negative antibody control. (C) ChIP analysis of CnL promoter using hnRNPA2 antibodies in control and Akt1 knockout MEFs treated with and without CCCP (25 μM, 5 h). Goat IgG was used as an antibody control. Values have been normalized to 10% of the corresponding input values. (D) Promoter activity of CnL promoter-reporter construct (1.0 μg) in MEFs cotransfected with different cDNAs (0.2 μg) as indicated. Values represent the mean ± SEM of three separate assays. Renilla luciferase activity was used for normalization of transfection efficiency as described in Materials and Methods.
The role of Akt1 as an activator of hnRNPA2 was investigated further by cotransfecting the CnL promoter construct with various cDNAs in control and Akt1-knockout MEFs, with or without CCCP treatment for 5 h. Treatment of control MEFs with CCCP increased the transcriptional activity of the CnL promoter by 2.7-fold, compared with the untreated MEF cells (Figure 6D). Promoter activities in Akt1 knockout cells were markedly lower than in control cells, and in the former case, CCCP treatment had no effect on promoter activity. In control MEF cells treated with CCCP, cotransfection with c-Rel, C/EBPδ, and hnRNPA2 increased promoter activity by 4.4-, 4.0-, and 1.4-fold, respectively, over that in untreated control cells. In Akt1 knockout cells, however, these cDNAs had minimal to marginal effects even after CCCP treatment. Cotransfection with a combination of c-Rel plus hnRNPA2, or C/EBPδ plus hnRNPA2, increased activity more than eightfold in control cells treated with CCCP, compared with untreated control cells. Similar cotransfections in Akt1-knockout cells had no effect on promoter activity, with or without added CCCP. These results support the role of Akt1 in the transcriptional activation of stress-response genes and the promoter occupancy of hnRNPA2.
Akt1 Silencing Results in the Reversal of Invasive Phenotype and Induces Apoptosis in mtDNA-depleted Cells
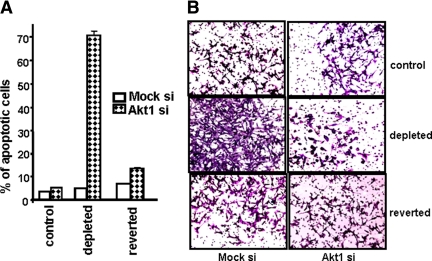
We showed previously that increased invasive behavior and resistance to apoptosis are hallmarks of cells subjected to mitochondrial respiratory stress in different cell types (Amuthan et al., 2001, 2002; Biswas et al., 2005a). Because Akt1 activation is associated with cell tumorigenicity (Semenza et al., 2001; Deberardinis et al., 2008), we tested the effects of Akt1 silencing on apoptosis and in vitro invasiveness in mtDNA-depleted cells. Figure 7A shows the percent apoptosis in control, mtDNA-depleted, and reverted C2C12 cells after Akt1 silencing. All three cell types contain ∼2–5% apoptotic cells when Akt1 expression is unaltered (mock siRNA). Akt1 mRNA silencing increased the apoptotic cell population to ∼70% in mtDNA-depleted cells, but a much smaller effect was seen in control and reverted cells (4–8%).
Figure 7.
Akt1 mRNA silencing in mtDNA-depleted C2C12 cells induces apoptosis and inhibits in vitro invasion. (A) Annexin assay for measuring apoptosis in control, mtDNA-depleted, and reverted cells transfected transiently with mock siRNA (negative control) or Akt1 siRNA. The graph shows the percent apoptotic cells quantified from three independent cell cytometry experiments. (B) Matrigel invasion assay of control, mtDNA-depleted, and reverted C2C12 cells expressing mock or Akt1 siRNA for 24 h.
Figure 7B shows the in vitro Matrigel invasion pattern of control, mtDNA-depleted, and reverted C2C12 cells. As reported previously, mtDNA-depleted cells showed a remarkably higher level of invasiveness compared with control and reverted C2C12 cells. Interestingly, Akt1 mRNA silencing reduced the invasiveness of mtDNA-depleted cells by ∼85%. However, Akt1 mRNA silencing had a marginal effect on control and reverted cells. Because equal numbers of viable cells were used in all three cases, these results show Akt1 plays an important role in the invasive behavior of C2C12 cells subjected to mitochondrial respiratory stress, possibly by regulating the activity of hnRNPA2 and, therefore, the transcriptional activity of stress response genes.
DISCUSSION
The Ser/Thr kinase Akt/PKB is modulated by multiple intra- and extracellular stimuli, including hypoxia, heat shock, oxidative stress, and cytokines (Datta et al., 1997; Semenza et al., 2001; Lawlor and Alessi, 2001; Cho et al., 2001; Deberardinis et al., 2008). Akt is involved in a multitude of cellular processes, including cell survival, cell proliferation, cancer progression, cell metabolism, RNA transport, and transcriptional modulation (Summers and Birnbaum, 1997; Brunet et al., 1999; Datta et al., 1999; Whiteman et al., 2002; Hu et al., 2005; Huang and Chen, 2005; Lee et al., 2008). Akt activation is often coupled to a change in metabolism and a shift to increased glucose uptake and glycolysis. Recent studies have shown that cells subjected to mitochondrial stress develop resistance to apoptosis, which is accompanied by a shift in cellular metabolism (Amuthan et al., 2001; Lee et al., 2004; Meierhofer et al., 2004; Guha et al., 2007; Kulawiec et al., 2009). The metabolic shift to glycolysis involves a switch from insulin- to IGF1 receptor–mediated signaling in C2C12 cells (Guha et al., 2007). We have also shown that IGF1 receptor phosphorylation may be an autocrine process because these cells secrete increased IGF1 (Guha et al., 2007). Preliminary results also suggest that the activation involves IRS1 phosphorylation. In this study, we show the metabolic shift is accompanied by increased nuclear accumulation and activation of Akt under mitochondrial stress.
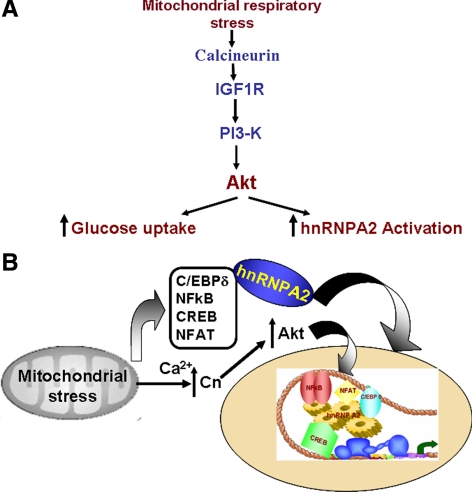
Previous studies showed activation of Akt in mtDNA-depleted tumor cells (Pelicano et al., 2006; Moro et al., 2009), although the precise mechanisms of activation were not elucidated. Using respiration-deficient human leukemia and colon cancer cells, Pelicano et al. (2006) suggested that Akt activation in response to mitochondrial stress may be due to increased mitochondrial NADH accumulation and inactivation of PTEN through a redox regulated mechanism. The nature of this redox regulation remains unclear. More recently, Moro et al. (2009) showed that in mtDNA-depleted prostate epithelial cells, Akt activation was associated with up-regulation p85 and PI3-K. Using mRNA knockdown in mtDNA-depleted C2C12 cells and CCCP treatment of control and Akt1 knockout MEFs, we show that Akt1 activation is critical for the propagation of mitochondrial respiratory stress signaling. Akt1 is activated by multiple pathways. We show here that Akt1 activation in mtDNA-depleted or CCCP-treated C2C12 cells is dependent on activation of Cn, IGF1R, and PI3-K (see Figure 8A), critical hallmarks of stress signaling. Based on our previous results on mechanism of IGF1R activation, AMPK pathway is not likely involved in this activation (Guha et al., 2007). Although not shown, mitochondrial stress induced activation of Akt in A549, H9C2, MEF, and MCF-7 cells also was dependent on IGF1R and PI3-K activation. Here, we extend previous observations, both in terms of the mechanism of stress-induced Akt1 activation and its role in transcriptional activation of nuclear target genes and also identify hnRNPA2 as a novel target for Akt action.
Figure 8.
Proposed mechanism of mitochondrial respiratory stress-induced Akt1 activation and its role in transcriptional activation. (A) A model showing the main mediators of the mitochondrial stress-signaling pathway involving calcineurin. (B) The role of Akt1 in the activation of transcription coactivator functions of hnRNPA2.
It is well recognized that Akt mediates transcription activation directly via phosphorylation and activation of transcription coactivators, such as p300 (Huang and Chen, 2005) and Forkhead transcription factors such as Foxo3a (Brunet et al., 1999). We show here that Akt1 phosphorylates hnRNPA2 in vitro at Thr98 and/or Ser219, and that Akt1 activity in vivo is essential for the recruitment of the transcription coactivator hnRNPA2 to the enhanceosome complex. This possibility is further supported by immunoprecipitation experiments showing that antibody to Akt1 pulled down hnRNPA2 and a number of other nuclear proteins from mtDNA-depleted cells. Negligible or undetectable precipitation of this protein by Akt1 antibody from control and reverted cell extracts suggest the Akt1-hnRNPA2 interaction is predominately a respiratory stress-related process. Some of the other proteins pulled down by Akt1 antibody from mtDNA-depleted cells, such as actin, tubulin, and 14-3-3, are known substrates for Akt1 (Vandermoere et al., 2007). At present the functional significance of the association of Akt1 with other proteins in Table 1 and their relevance to mitochondrial stress remains unclear.
Multiple techniques, including differential mRNA display, cDNA array, and proteomics, have revealed that a wide spectrum of genes encoding mitochondrial respiratory complex proteins, cell surface receptors, and proteins involved in apoptosis, Ca2+ homeostasis, and transcription regulation are either up- or down-regulated in cells lacking or partially depleted of mtDNA (Delsite et al., 2002; Crimi et al., 2005; Jahangir Tafrechi et al., 2005; Biswas et al., 2008; Johnston et al., 2009). Our results on the stress-induced expression of Akt1 mRNA in cells subjected to mitochondrial stress by mtDNA depletion, as well as by CCCP treatment, were surprising in view of studies showing the regulation of human Akt1 gene expression by the Src/Stat3 pathway by Park et al. (2005), who have shown convincingly that Stat3 response elements are localized in the exon 1-intron 1 region of the human Akt1 gene (Park et al., 2005). It is becoming apparent that many genes undergo multimodal regulation under the influences of different transcription activator proteins and coactivators. For example, Glut-4 expression is known to involve cMyc and other factors (Thai et al., 1998), whereas mitochondrial respiratory stress-induced transcriptional up-regulation involves c-Rel/p50, C/EBPδ, CREB, and hnRNPA2 (Guha et al., 2009). Interestingly, respiratory stress-induced transcriptional activation of the human Akt1 promoter involves the same set of transcription factors, and the overall location of putative DNA motifs for these factors within the 900-base pairs region immediately upstream of transcription initiation site resembles those of other stress target genes (Guha et al., 2009). Notably, the footprint of stress response elements for the human Akt1 promoter is conserved in the mouse Akt1 gene.
Our data advances understanding of the process by which mitochondrial respiratory stress signaling modulates the transcription of nuclear genes. This study not only describes the mechanism of stress-induced activation of Akt1, but also defines its role in the phosphorylation and activation of the transcriptional coactivator hnRNPA2. Mutational studies in Figure 5E show that Akt1-mediated phosphorylation is critical for the hnRNPA2 mediated transcription activation of stress target gene. Furthermore, results of ChIP analysis in Figure 6 show that Akt function is critical for the recruitment or association of hnRNPA2 to the enhanceosome complex. Importantly, the activation of Akt1 in response to mitochondrial respiratory stress involves the same four signature factors (C/EBPδ, c-Rel/p50, NFAT, and CREB) and recruitment of hnRNPA2. Previously we have shown that different domains of hnRNPA2 may be involved in the binding or recruitment of different transcription factors (Guha et al., 2009). In view of this mutational analysis, in this study showing the importance of the Akt target sites of hnRNPA2 (T98 and S219) suggests that these two phosphorylation sites may be important for recruiting different transcription factors. The observed 60–70% reduction in activity with either T98A or S219A mutant hnRNPA2 is consistent with our proposed model on the cooperativity between the signature factors (c-Rel/p50, C/EBPδ, CREB and NFAT) in the activation of stress target genes (Guha et al., 2009 and Figure 8).
An intriguing observation is the self-regulatory role of Akt1, in that transcriptional activation of this gene requires Akt1-mediated phosphorylation of hnRNPA2. The general mechanism of stress-induced transcription activation is presented as a model in Figure 8B. We believe the mechanism described here is important in understanding the cellular response to altered mitochondrial function and bioenergetics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. G. Dreyfuss (University of Pennsylvania, Philadelphia, PA) for the hnRNPA2 cDNA, J. Q. Chen (H. Lee Moffitt Cancer Center, Tampa, FL) for the human Akt1 promoter, Shankar Ghosh (Columbia University Medical Center, NY) for cRel cDNA, and Michael Atchison (University of Pennsylvania) for C/EBPδ cDNA. We also thank Satish Srinivasan (University of Pennsylvania) for the mtDNA-depleted RAW264.7 cells and Li Huang for technical help. This research was supported by National Institutes of Health Grants RO1 CA-22762 to N.G.A. and RO1DK-56886 to M.J.B.
Abbreviations used:
- CCCP
carbonylcyanide m-chlorophenylhydrazone
- ChIP
chromatin immunoprecipitation
- Cn
calcineurin
- ddC
dideoxycytidine
- EtBr
ethidium bromide
- hnRNP
heterogeneous ribonucleoprotein
- mtDNA
mitochondrial DNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0192) on August 18, 2010.
REFERENCES
- Amuthan G., Biswas G., Ananadatheerthavarada H. K., Vijayasarathy C., Shephard H. M., Avadhani N. G. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- Amuthan G., Biswas G., Zhang S. Y., Klein-Szanto A., Vijayasarathy C., Avadhani N. G. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould T., Vankoningsloo S., Renard P., Houbion A., Ninane N., Demazy C., Remacle J., Raes M. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock D. F., Herrington J., Goodwin P. C., Park Y. B., Hille B. Mitochondrial participation in the intracellular Ca2+ network. J. Cell Biol. 1997;136:833–844. doi: 10.1083/jcb.136.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Adebanjo O. A., Freedman B. D., Anandatheerthavarada H. K., Vijayasarathy C., Zaidi M., Kotlikoff M., Avadhani N. G. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Anandatheerthavarada H. K., Avadhani N. G. Mechanism of mitochondrial stress-induced resistance to apoptosis in mitochondrial DNA-depleted C2C12 myocytes. Cell Death Differ. 2005a;12:266–278. doi: 10.1038/sj.cdd.4401553. [DOI] [PubMed] [Google Scholar]
- Biswas G., Anandatheerthavarada H. K., Zaidi M., Avadhani N. G. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through Cn-mediated inactivation of IkappaBbeta. J. Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Guha M., Avadhani N. G. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene. 2005b;354:132–139. doi: 10.1016/j.gene.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Tang W., Sondheimer N., Guha M., Bansal S., Avadhani N. G. A distinctive physiological role for Ikappa Bbeta in the propagation of mitochondrial respiratory stress signaling. J. Biol. Chem. 2008;283:12586–12594. doi: 10.1074/jbc.M710481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Chen R., Kim O., Yang J., Sato K., Eisenmann K. M., McCarthy J., Chen H., Qiu Y. Regulation of Akt/PKB activation by tyrosine phosphorylation. J. Biol. Chem. 2001;276:31858–31862. doi: 10.1074/jbc.C100271200. [DOI] [PubMed] [Google Scholar]
- Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B., III, Kaestner K. H., Bartolomei M. S., Shulman G. I., Birnbaum M. J. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- Crimi M., Bordoni A., Menozzi G., Riva L., Fortunato F., Galbiati S., Del B.R., Pozzoli U., Bresolin N., Comi G. P. Skeletal muscle gene expression profiling in mitochondrial disorders. FASEB J. 2005;19:866–868. doi: 10.1096/fj.04-3045fje. [DOI] [PubMed] [Google Scholar]
- Datta S. R., Brunet A., Greenberg M. E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Deberardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Delsite R., Kachhap S., Anbazhagan R., Gabrielson E., Singh K. K. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer. 2002;1:6–16. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R., Moraes C. T. Lack of oxidative phosphorylation and low mitochondrial membrane potential decrease susceptibility to apoptosis and do not modulate the protective effect of Bcl-x(L) in osteosarcoma cells. J. Biol. Chem. 2000;275:7087–7094. doi: 10.1074/jbc.275.10.7087. [DOI] [PubMed] [Google Scholar]
- Garayoa M., Man Y. G., Martínez A., Cuttitta F., Mulshine J. L. Downregulation of hnRNP A2/B1 expression in tumor cells under prolonged hypoxia. Am. J. Respir. Cell. Mol. Biol. 2003;28:80–85. doi: 10.1165/rcmb.4880. [DOI] [PubMed] [Google Scholar]
- Guha M., Pan H., Fang J. K., Avadhani N. G. Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress. Mol. Biol. Cell. 2009;20:4107–4119. doi: 10.1091/mbc.E09-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M., Srinivasan S., Biswas G., Avadhani N. G. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J. Biol. Chem. 2007;282:14536–14546. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Brown M. A., Rothnagel J. A., Saunders N. A., Smith R. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J. Cell Sci. 2005;118:3173–3183. doi: 10.1242/jcs.02448. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Kudo T., Suzuki S., Evans T. T., Sasaki R., Wada Y., Shirakawa T., Sawyer J. R., Gotoh A. Mitochondrial DNA determines androgen dependence in prostate cancer cell lines. Oncogene. 2006;25:1437–1445. doi: 10.1038/sj.onc.1209190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton T. M., Petros J. A., Heddi A., Shoffner J., Kaufman A. E., Graham S. D., Jr, Gramlich T., Wallace D. C. Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosom. Cancer. 1996;15:95–101. doi: 10.1002/(SICI)1098-2264(199602)15:2<95::AID-GCC3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Hu Y., Yao J., Liu Z., Liu X., Fu H., Ye K. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. EMBO J. 2005;24:3543–3554. doi: 10.1038/sj.emboj.7600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. C., Chen C. C. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell. Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahangir Tafrechi R. S., Svensson P. J., Janssen G. M., Szuhai K., Maassen J. A., Raap A. K. Distinct nuclear gene expression profiles in cells with mtDNA depletion and homoplasmic A3243G mutation. Mutat. Res. 2005;578:43–52. doi: 10.1016/j.mrfmmm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Johnston D. S., Su Y. A., Alesci S. Mitochondrial gene profiling: translational perspectives. Pharmacogenomics. 2009;10:1645–1655. doi: 10.2217/pgs.09.112. [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Kim Y. H., Chang I., Kim S., Pak Y. K., Oh B. H., Yagita H., Jung Y. K., Oh Y. J., Lee M. S. Resistance of mitochondrial DNA-deficient cells to TRAIL: role of Bax in TRAIL-induced apoptosis. Oncogene. 2002;21:3139–3148. doi: 10.1038/sj.onc.1205406. [DOI] [PubMed] [Google Scholar]
- Kondapaka S. B., Zarnowski M., Yver D. R., Sausville E. A., Cushman S. W. 7-hydroxystaurosporine (UCN-01) inhibition of Akt Thr308 but not Ser473 phosphorylation: a basis for decreased insulin-stimulated glucose transport. Clin. Cancer Res. 2004;10:7192–7198. doi: 10.1158/1078-0432.CCR-04-0772. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Dallaporta B., Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu. Rev. Physiol. 1998;60:619–642. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- Kulawiec M., Owens K. M., Singh K. K. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol. Ther. 2009;8:1378–1385. doi: 10.4161/cbt.8.14.8751. [DOI] [PubMed] [Google Scholar]
- Lawlor M. A., Alessi D. R. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Kim J. Y., Park S. Y. Resistance of rho(0) cells against apoptosis. Ann. NY Acad. Sci. 2004;1011:146–153. doi: 10.1007/978-3-662-41088-2_15. [DOI] [PubMed] [Google Scholar]
- Lee S. B., Xuan Nguyen T. L., Choi J. W., Lee K. H., Cho S. W., Liu Z., Ye K., Bae S. S., Ahn J. Y. Nuclear Akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc. Natl. Acad. Sci. USA. 2008;105:16584–16589. doi: 10.1073/pnas.0807668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievre A., Chapusot C., Bouvier A. M., Zinzindohoue F., Piard F., Roignot P., Arnould L., Beaune P., Faivre J., Laurent-Puig P. Clinical value of mitochondrial mutations in colorectal cancer. J. Clin. Oncol. 2005;23:3517–3525. doi: 10.1200/JCO.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Linnartz B., Anglmayer R., Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004;64:1966–1971. doi: 10.1158/0008-5472.can-03-2956. [DOI] [PubMed] [Google Scholar]
- Loeb L. A., Wallace D. C., Martin G. M. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc. Natl. Acad. Sci. USA. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mazurek S., Eigenbrodt E. The tumor metabolome. Anticancer Res. 2003;23:1149–1154. [PubMed] [Google Scholar]
- Meierhofer D., Mayr J. A., Foetschl U., Berger A., Fink K., Schmeller N., Hacker G. W., Hauser-Kronberger C., Kofler B., Sperl W. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25:1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- Moro L., Arbini A. A., Marra E., Greco M. Mitochondrial DNA depletion reduces PARP-1 levels and promotes progression of the neoplastic phenotype in prostate carcinoma. Cell Oncol. 2008;30:307–322. doi: 10.3233/CLO-2008-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro L., Arbini A. A., Yao J. L., di Sant'Agnese P. A., Marra E., Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death Differ. 2009;16:571–583. doi: 10.1038/cdd.2008.178. [DOI] [PubMed] [Google Scholar]
- Okochi O., Hibi K., Uemura T., Inoue S., Takeda S., Kaneko T., Nakao A. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin. Cancer Res. 2002;8:2875–2878. [PubMed] [Google Scholar]
- Park J. S., et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum. Mol. Genet. 2009;18:1578–1589. doi: 10.1093/hmg/ddp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim D., Kaneko S., Szewczyk K. M., Nicosia S. V., Yu H., Jove R., Cheng J. Q. Molecular cloning and characterization of the human AKT1 promoter uncovers its up-regulation by the Src/Stat3 pathway. J. Biol. Chem. 2005;280:38932–38941. doi: 10.1074/jbc.M504011200. [DOI] [PubMed] [Google Scholar]
- Parrella P., et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- Patry C., Bouchard L., Labrecque P., Gendron D., Lemieux B., Toutant J., Lapointe E., Wellinger R., Chabot B. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63:7679–7688. [PubMed] [Google Scholar]
- Pelicano H., et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J. Cell Biol. 2006;175:913–923. doi: 10.1083/jcb.200512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros J. A., et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas D. R., Thompson C. B. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- Semenza G. L., et al. ‘The metabolism of tumours’: 70 years later. Novartis Found. Symp. 2001;240:251–260. [PubMed] [Google Scholar]
- Shidara Y., Yamagata K., Kanamori T., Nakano K., Kwong J. Q., Manfredi G., Oda H., Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005b;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- Shidara Y., Yamagata K., Kanamori T., Nakano K., Kwong J. Q., Manfredi G., Oda H., Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005a;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- Singh K. K., Ayyasamy V., Owens K. M., Koul M. S., Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J. Hum. Genet. 2009;54(9):516–524. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers S. A., Birnbaum M. J. A role for the serine/threonine kinase, Akt, in insulin-stimulated glucose uptake. Biochem. Soc. Trans. 1997;25:981–988. doi: 10.1042/bst0250981. [DOI] [PubMed] [Google Scholar]
- Taylor R. W., Turnbull D. M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai M. V., Guruswamy S., Cao K. T., Pessin J. E., Olson A. L. Myocyte enhancer factor 2 (MEF2)-binding site is required for GLUT4 gene expression in transgenic mice. Regulation of MEF2 DNA binding activity in insulin-deficient diabetes. J. Biol. Chem. 1998;273:14285–14292. doi: 10.1074/jbc.273.23.14285. [DOI] [PubMed] [Google Scholar]
- Trotman L. C., Alimonti A., Scaglioni P. P., Koutcher J. A., Cordon-Cardo C., Pandolfi P. P. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermoere F., El Yazidi-Belkoura I., Demont Y., Slomianny C., Antol J., Lemoine J., Hondermarck H. Proteomics exploration reveals that actin is a signaling target of the kinase Akt. Mol. Cell Proteom. 2007;6:114–124. doi: 10.1074/mcp.M600335-MCP200. [DOI] [PubMed] [Google Scholar]
- Whiteman E. L., Cho H., Birnbaum M. J. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- Yamaoka K., Imajoh-Ohmi S., Fukuda H., Akita Y., Kurosawa K., Yamamoto Y., Sanai Y. Identification of phosphoproteins associated with maintenance of transformed state in temperature-sensitive Rous sarcoma-virus infected cells by proteomic analysis. Biochem. Biophys. Res. Commun. 2006;345:1240–1246. doi: 10.1016/j.bbrc.2006.04.183. [DOI] [PubMed] [Google Scholar]
- Yu M., Shi Y., Wei X., Yang Y., Zang F., Niu R. Mitochondrial DNA depletion promotes impaired oxidative status and adaptive resistance to apoptosis in T47D breast cancer cells. Eur. J. Cancer Prev. 2009;18:445–457. doi: 10.1097/CEJ.0b013e32832f9bd6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.