Abstract
Mitochondrial protein translocation machinery in the kinetoplastid parasites, like Trypanosoma brucei, has been characterized poorly. In T. brucei genome data base, one homolog for a protein translocator of mitochondrial inner membrane (Tim) has been found, which is closely related to Tim17 from other species. The T. brucei Tim17 (TbTim17) has a molecular mass 16.2 kDa and it possesses four characteristic transmembrane domains. The protein is localized in the mitochondrial inner membrane. The level of TbTim17 protein is 6–7 fold higher in the procyclic form that has a fully active mitochondrion, than in the mammalian bloodstream form of T. brucei, where many of the mitochondrial activities are suppressed. Knockdown of TbTim17 expression by RNAi caused a cessation of cell growth in the procyclic form and reduced growth rate in the bloodstream form. Depletion of TbTim17 decreased mitochondrial membrane potential more in the procyclic than bloodstream form. However, TbTim17 knockdown reduced the expression level of several nuclear encoded mitochondrial proteins in both the forms. Furthermore, import of presequence containing nuclear encoded mitochondrial proteins was significantly reduced in TbTim17 depleted mitochondria of the procyclic as well as the bloodstream form, confirming that TbTim17 is critical for mitochondrial protein import in both developmental forms. Together, these show that TbTim17 is the translocator of nuclear encoded mitochondrial proteins and its expression is regulated according to mitochondrial activities in T. brucei.
Keywords: Trypanosoma brucei, Tim17, membrane potential, mitochondrial protein import, the bloodstream and procyclic forms
1. Introduction
Mitochondrial biogenesis requires import of up to a thousand nuclear encoded proteins from the cytosol to their proper sub-mitochondrial destination [1,2]. Multi subunit protein translocases of the mitochondrial outer and inner membranes, the TOM and TIM complexes respectively, have been characterized from different systems, including fungi, plants, and animals [1–7]. The TOM complex is needed for the import of virtually all mitochondrial proteins [7]. Proteins destined for the matrix or the mitochondrial inner membrane, are further transported by either one of the two TIM complexes, which are known as TIM23 and TIM22 [2,7–9]. The TIM22 complex mediates import of polytopic membrane proteins with internal targeting signals. On the other hand, the TIM23 complex imports proteins, which contain an N-terminal targeting sequence. These include proteins that are targeted to the matrix and a few inner membrane proteins. The TIM23 core complex contains Tim23, Tim17, and Tim50 [2,7,8,10]. During translocation of the matrix-targeted protein, this core complex associates with the Presequence Activated Motor (PAM) complex consisting of Tim44, mitochondrial Hsp70, Pam16, and Pam18. On the other hand, the core complex associates with Tim21 to form the TIM23-sorting complex (TIM23sort) for translocation of the inner membrane proteins [11]. In fungi, Tim23 forms the import channel [12]. Once the presequence enters into the matrix side, it is cleaved by a Matrix Processing Peptidase (MPP) [13].
Tim17, Tim22, and Tim23 are structurally similar proteins [14, 15]. Each has four characteristic transmembrane domains centrally located in the protein. Tim17 is the most conserved among all Tom and Tim proteins [8]. Recent findings showed that Tim17 is critical for formation of the twin-pore structure of the Tim23 translocase and acts as a voltage sensor for this protein import channel [16,17]. Tim17 also plays role in sorting of inner membrane and matrix localized proteins [18].
S. cerevisiae and N. crassa contain only one copy of each Tim17, Tim22, and Tim23 but Arabidopsis thaliana contains three copies of Tim17 and three copies of Tim23 genes [19]. Humans also contain two copies of the Tim17 gene. The human Tim17 (hTim17a and hTim17b) genes are expressed simultaneously and do not show any tissue specific distribution [8]. In Arabidopsis thaliana, the AtTim17-2 gene is primarily expressed and the other two copies are expressed marginally [19,20].
The haemoflagellated parasitic protozoan Trypanosoma brucei, the causative agent of African trypanosomiasis, possesses a single tubular mitochondrion with many unique characteristics [21]. The organism has a digenetic life cycle and must adapt to the dramatically different environments of an insect’s alimentary gut and the mammalian bloodstream [22,23]. The bloodstream form of this parasite depends on blood glucose as its sole source of energy, and suppresses many of its mitochondrial activities. However, a small number of nuclear encoded mitochondrial enzymes are essential for the survival of this form [24–26]. On the other hand, the procyclic form, which lives in the insect gut, primarily utilizes amino acids as its source of energy and possesses a fully developed mitochondrion [27]. As in other eukaryotes, the majority of the mitochondrial proteins in T. brucei are nuclear encoded and need to be imported into mitochondria. Thus, mitochondrial protein translocation is essential for both life forms of this parasite. Many mitochondrial proteins have been cloned and characterized from T. brucei, however, the mitochondrial protein import machinery has been poorly characterized in these early eukaryotes. Except for a few Tim proteins [28], homologs for any Toms and most of the Tim proteins could not be identified in the T. brucei genome database by homology search, which indicates that the protein translocases are divergent in this organism. Characterization of mitochondrial protein translocator is thus a prerequisite to understand the protein import mechanism in these earliest eukaryotes. Here, we performed the functional characterization of a homologue of Tim17 in T. brucei. The primary sequence of TbTim17 is significantly divergent than Tim17 from other species. Its expression level is developmentally regulated according to mitochondrial activities and it plays a crucial role in the import of nuclear encoded mitochondrial proteins in both forms of the parasite.
2. Materials and methods
2.1. Strains and media
The procyclic form of Trypanosoma brucei 427 double resistant cell line (29–13) expressing the tetracycline repressor gene (TetR) and T7RNA polymerase (T7RNAP) were grown in SDM-79 medium containing 10% fetal bovine serum and appropriate antibiotics (hygromycin; 50 μg/ml; G418; 15 μg/ml) [29]. Bloodstream form cells, single marker 427 (SM427) harboring the TetR and T7RNAP were maintained in HMI-9 media supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals), 10% serum plus (JRH Biosciences) [30] and G418 (2.5 μg/ml). For measurement of cell growth, the procyclic and the bloodstream forms cells were inoculated at a cell density of 2–3 × 106/ml and 2–3 × 105/ml, respectively, in fresh medium containing appropriate antibiotics in the presence and absence of doxycycline. Cells were harvested at different time points of growth (0–5 days) and the number of cells was counted in a Neubauer hemocytometer counter. The cumulative number of cells was plotted versus time of incubation in culture. For large-scale cultivation of the bloodstream form cells, rats (Sprague Dawley) were infected with the parasite (1 × 107 cells/100 g body weight). Rat blood was collected by cardiac puncture when the parasitemia level reached 1 × 109 cells/ml of blood. The bloodstream form cells were separated from blood using DEAE ion exchange resin as described before [31].
2.2. DNA cloning and sequence analysis
The open reading frame (ORF) of the putative TbTim17 (Tb11.01.4870) was PCR amplified from T. brucei genomic DNA using forward (5′ATCTGCAGATGACAACACTTCTCG3′) and reverse (5′GTAAGCTTTTAGCGTTGAGCCAA3′) primers. The PCR product (470 bp) was cloned in TOPO TA cloning vector (Invitrogen) and sequenced. 5′RACE was performed to determine the 5′UTRs of TbTim17 mRNA. First strand synthesis of TbTim17 cDNA was performed using Reverse Transcriptase and oligo dT primer. For 5′RACE the splice leader (SL) primer (5′CTGAATTCGCTATTATTAGAACAGTTTCTG3′) was used as the forward primer and the gene specific primers, Tim17F15 (5′GCCGACTTCTTCCGTTACAGTTTG3′) or Tim17F16 (5′CCACAGACTCGGCTTCAGTTTTG3′) was used as a reverse primer. The PCR product was cloned and sequenced to obtain the 5′UTR of TbTim17. Sequence comparison was performed using ClustalW alignment program [32] in MacVector 7.0. The hydrophobicity profile of predicted Tim17 protein sequences were analyzed by TMpred-Prediction of Transmembrane Regions and Orientation software [33].
2.3. Generation of TbTim17 double stranded RNA expression constructs
To prepare the construct for TbTim17 double stranded RNA expression, the 459 bp fragment of the coding region of TbTim17 was PCR amplified from the TbTim17 cDNA clone using high fidelity Pfu polymerase (Stratagen). Sense and antisense primers containing the proper restriction sites at 5′ ends were designed. The amplified product was cloned into the BamHI/HindIII sites of a tetracycline inducible dual promoter plasmid vector p2T7Ti-177 [34]. From this construct TbTim17 double stranded RNA is produced from two opposing tetracycline regulated T7 promoter/primer and the phleomycin resistant gene is expressed constitutively for selection purposes. The construct for TbTim17 RNAi was verified by sequencing. The purified plasmid DNA was linearized by NotI. The linearized plasmid was used for transfection into procyclic cells (Tb427 29–13) and SM427 bloodstream form cells expressing T7 polymerase and tetracycline repressor proteins according to standard protocols [29]. After transfection, the plasmid was integrated into 177 repeat regions of the minichromosomes in T. brucei.
2.4. RNA isolation, Northern analysis, and Real-Time PCR
RNA was isolated from the bloodstream and procyclic trypanosomes harvested at different time points of growth with or without induction of TbTim17 RNAi, using Trizol reagent (Invitrogen) according to the manufacturer’s protocol and was further precipitated with 2M LiCl. For Northern analysis, RNA was fractionated in formaldehyde-agarose gels (2.0%) and transferred to nitrocellulose membranes [35]. TbTim17 and actin probes were generated using random primer labeling protocol (Invitrogen) of the TbTim17 cDNA clone and the PCR amplified genomic fragment of the T. brucei actin gene. Hybridization was carried out in Rapid-Hyb buffer (Amersham) for 16 hr. The membranes were washed at 65 °C with 0.1 × SSC (150 mM NaCl, 15 mM Na-citrate, pH 7.4) containing 0.1% SDS and exposed to X-ray film [35]. The primers used for the PCR amplification of the T. brucei actin DNA were as follows: forward primer, 5′CAACGTGCTACTGACTG-3′ and reverse primer; 5′-GCACTGTTCGTCATCTC-3′. Quantitation of TbTim17 transcript level in the procyclic and the bloodstream forms of T. brucei was performed by real-time RT-PCR on 50 ng of DNAse treated RNA in a 20 μl reaction containing: primers 900nM each, TbTim17 forward: 5′TGAAGGACAGCACCATTACCCC3′ and reverse: 5′CCGAAAAGGAAACCAAAGTAGGC3′ or actin forward: 5′TGGAAAAGGTTTGGCACCATAC3′ and reverse: 5′CAGAAGAATACAGTGACAACACCGC3′; and iScript One-Step RT-PCR master mix with SYBR Green (Bio-Rad Laboratories). Actin was used as the endogenous control for normalization.
2.5. Generation of TbTim17 antibodies and immunoblot analysis
Polyclonal antibodies against TbTim17 protein were custom synthesized by Bethyl Laboratories, Inc. Montgomery, TX using amino acids 1–17. The synthetic peptide was affinity purified by HPLC and verified by mass-spectrometry. The Tim17 peptide was conjugated to keyhole limpet hemocyanin (KLH) and used to generate specific antibodies in rabbits. Antiserum against TbTim17 was affinity purified using TbTim17 peptide as the ligand. Pre-immune serum collected from the same rabbits was used as control. Total cellular proteins and proteins from isolated mitochondria were analyzed on SDS-PAGE (10 or 15%) as described (36), and transferred to nitrocellulose membrane at 4 °C (100 V with 25 mM Tris-HCl and 192 mM glycine, 20% v/v methanol pH 8.3) [37]. Blots were treated with TbTim17 polyclonal antiserum and antiserum against T. brucei cytochrome C1 (Cyt C1), iron-sulfur protein (ISP) [38], a putative mitochondrial outer membrane protein carnitine palmitoyl transferase (CPT) [39], a cytosolic protein serine/threonine protein phosphatase 5 (TbPP5) [40] and mitochondrial Hsp70 [41] (all at 1 in 1000 dilution in blocking buffer). TAO [42] and KRel1 [43] were detected with the corresponding monoclonal hybridoma supernatants (1 in 50 dilution in 10 mM Tris-HCl, 150 mM NaCl, pH 8.0). Heterologous polyclonal antibodies developed against Neurospora crassa Tom40 [44], were used at 1:1,000 dilution. Monoclonal antibody against T. brucei β-tubulin [45] was used in 1:20,000 dilution in TBST. A peptide antibody was also generated (Bethyl Laboratories, Inc, Montgomery, TX) against T. brucei ATP/ADP carrier protein (AAC) (Tb10.61.1820, amino acid residues from 294 to 307) identified in T. brucei gene database (www.geneDB.org). The antibody was affinity purified using the same peptide as the ligand and verified using recombinant protein as the antigen and the preimmune serum as a control antibody. The purified anti-AAC antibody was used in 1:20,000 dilution for immunoblot analysis of T. brucei mitochondrial proteins. Blots were treated with appropriate secondary antibody and developed using enhanced chemiluminiscence (ECL) detection system (Amersham).
2.6. MitoTracker staining and confocal microscopy
Cells were harvested and suspended in fresh culture medium at a density of 2 × 106/ml. MitoTracker Red CMXRos (Molecular probe) was dissolved in dimethyl sulfoxide at a concentration of 1 mM and added to a final concentration of 0.5 μM for procyclic and 0.05 μM for bloodstream form cells. The mixture was incubated at the normal growth temperature for 10 min. Cells were washed and incubated in fresh culture medium for additional 30 min. Cells were then washed twice with PBS and fixed in 4% paraformaldehyde at 4 °C for 15 min [46]. After another wash in PBS, cell suspension was spread on poly-lysine coated slides. The slides were mounted in Fluoromount G and images were acquired with a Nikon TE2000-U C1 laser scanning confocal microscope outfitted with a 60× 1.4 N.A. Plan Apo oil immersion objective lens. MitoTracker Red was excited with a 578 nm argon laser, and the emitted light was collected at 599 nm. All images were collected with exactly the same detector gain, laser strength, and number of scans in order for the data to be examined for differences in fluorescence intensity. The fluorescence intensity was quantitated using Nikon Elements software program [47]. To nullify the background inhomogeneity, the intensity of the image pixel within the Region of Interest (ROI) is measured by computer software. Here, we used individual cell as ROI and compare the average intensity of ROI among two different samples. The intensity of more than 300 cells from each sample was quantitated.
2.7. Isolation of mitochondria
Mitochondria were isolated from the parasite after lysis via nitrogen cavitation in isotonic buffer as described [44]. Briefly, the cells were washed with a buffer containing 0.15 M NaCl, 20 mM glucose, and 20 mM NaH2PO4, pH 7.4 and resuspended at a density of 1 × 109 cells/ml in SME buffer (0.25 M sucrose, 10 mM MOPS/KOH pH 7.2, 2 mM EDTA, 1 mM PMSF, 0.5 μg/ml leupeptin and 1 μg/ml pepstatin). The cells were equilibrated at 900 psi N2 for 15 min in a nitrogen cavitation bomb (minibomb cell disruption chamber; Kontes, Vineland, NJ). Subsequent release from the bomb resulted in the disruption of more than 90% of cells. The lysate was diluted with one volume of the same buffer and the mitochondrial fraction was isolated by differential centrifugation, as described previously [44]. For isolation of mitochondria from the bloodstream form, TbTim17 RNAi cells were grown in vivo in the rat system as described [25]. To express TbTim17 double stranded RNA, the drinking water for rats were supplemented with 5% sucrose and 250 μg/ml of doxycycline in order to maintain doxycycline concentration in the rat blood. Parasites were separated from the rat blood at the peak parasitemia level after sacrificing the rats and mitochondria were isolated as described in the section 2.1. The isolated mitochondria were stored at a protein concentration of 2–5 mg/ml in MOPS/KOH, buffer containing 50% glycerol at −70°C.
Before using, mitochondria were washed twice with 9 volumes of SME buffer to remove glycerol. For alkali extraction, isolated mitochondria (100 μg) were treated with 100 μl of 100 mM Na2CO3 pH 11.0, for 30 min on ice [44]. The supernatant and pellet fractions were collected after centrifugation and analyzed by SDS-PAGE and immunobloting. Protease digestion of isolated mitochondria was performed by treating mitochondria (50 μg) with various concentration of proteinase K (0–100 μg/ml) in 100 μl reaction volumes of SME for 30 min on ice. After the treatment, proteinase K was inhibited by PMSF (2 mM). Mitochondria were re-isolated and the proteins were analyzed by immunoblot analysis.
2.8. In vitro transcription and translation
The TAO open reading frame (ORF) was PCR amplified using primers 5′AGAAGCTTATGTTTCGTAAC3′ and 5′AAGAATTCTTACTCGTGTTTG3′ containing appropriate restriction site (underlined) at the 5′ ends. TAO cDNA (pTAO25) [42] was used as a template. Similarly, the ORFs for the cytochrome oxidase subunit 4 (COIV) [48] and NADH dehydrogenase subunit k (Ndh K) (49) were also amplified from T. brucei genomic DNA using specific primers. The forward and reverse primers were as follows, COIVFor: 5′AGAAGCTTATGTTTGCTCGCCGCT3′; COIVRev: 5′AAGAATTCCTAAATCTTGTTTGA3′; NdhKFor; 5′AGAAGCTTATGCTTCGTCGCACGT3′; NdhKRev: 5′AAGAATTCTTAATCTCGAACAGAA3′. The PCR product was subcloned in pGEM 4Z vector at the site of HindIII/EcoRI. Radiolabeled precursor protein was synthesized in vitro using a coupled transcription/translation rabbit reticulocyte system (TNT, Promega) according to the manufacturer’s protocol using 35S methionine (DuPont NEN) as the label.
2.9. In vitro import assay
Labeled precursor proteins were used for in vitro import into isolated mitochondria from T. brucei as described [50,51] with a few modifications. Mitochondria (100 μg) were washed with 9 volumes of SME buffer and resuspended in 90 μl of import buffer (0.25 M sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM dithiothreitol, 1.0 mg/ml fatty acid free bovine serum albumin and 10 mM MOPS/KOH at pH 7.2). The mitochondrial suspension was mixed with 10 μl of rabbit reticulocyte translation mixture containing a radiolabeled precursor protein and incubated at room temperature for up to 40 min. Additional ATP was provided at a final concentration of 2 mM and 10 mM creatine phosphate and 0.1 mg/ml creatine phosphokinase were added as an ATP regeneration system. 8 mM potassium ascorbate, 0.2 mM N, N, N′, N′-tetramethylphenylenediamine, and 5 mM NADH were added to energize the mitochondria and provide reducing equivalents. Following import, the mitochondria were treated with proteinase K (50 μg/ml) in ice for 15 min to remove the labeled protein that is bound to the membrane but not imported. PMSF (2.0 mM) was then added to inhibit proteinase K and the mitochondrial fraction was re-isolated by centrifugation. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane as described. After transfer, the blot was dried at 37 °C for 30 min and exposed to an X-ray film (Biomax Film, Kodak) for detection of radioactive proteins.
3. Results
3.1. Cloning and characterization of TbTim17
The recently completed genome sequence of T. brucei revealed the presence of one homolog (Tb11.01.4870) for the Tim17/Tim22/Tim23 family proteins. To characterize the encoded protein in further detail, the complete open reading frame (ORF) of this gene was PCR amplified from T. brucei genomic DNA. The PCR product was cloned in the TOPO TA cloning vector (Invitrogen) and sequenced. The entire coding region is 459 bp long and encodes a protein of molecular mass 16.2 kDa. To determine the 5′ end of the complete cDNA, 5′ RACE analysis was performed. Cloning and sequencing of the 5′ RACE product revealed that the cDNA possesses a 225 base pair 5′ UTR, and the start codon of the ORF is the first methionine, which confirmed that the predicted reading frame was correct.
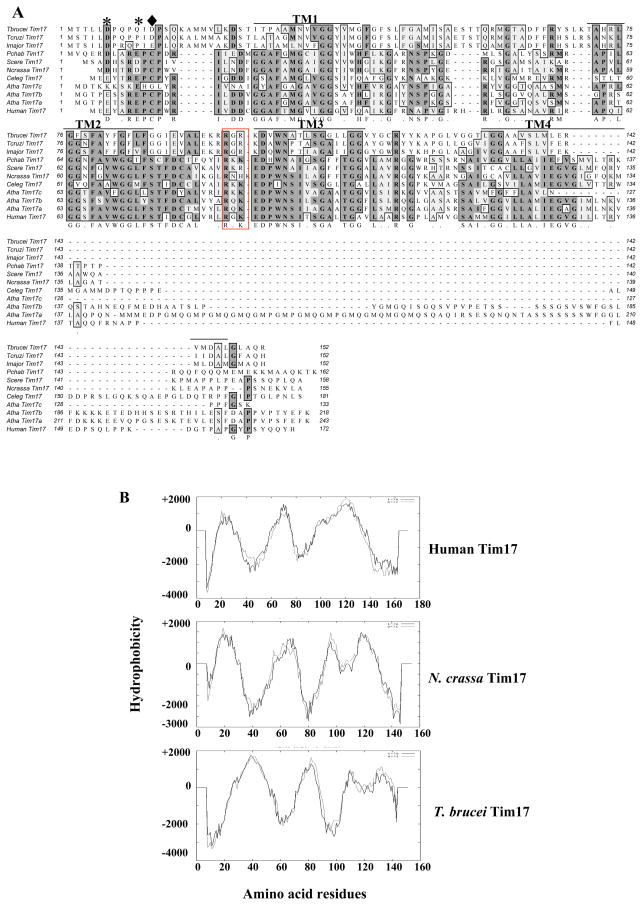
ClustalW alignment of the predicted protein sequence from this ORF with known Tim17 proteins from other species showed a significant amount of homology (Fig. 1A). The overall identity of the putative TbTim17 was 25–26% with Tim17 in other species. We also performed the pair-wise alignments of the T. brucei protein sequence with known Tim17, Tim22, and Tim23 protein sequences from different species (Supplementary Table-1). The putative TbTim17 showed a higher homology with Tim17 proteins than with Tim22 and Tim23. The overall identity of the TbTim17 with Tim22, and Tim23 is about 22%, and 19–20%, respectively suggesting that Tb11.01.4870 encoded protein is the homolog of Tim17. It has been found that Tim17 from human and fungi possess 44% identity and 57% similarity. Thus, T. brucei Tim17 is divergent within the Tim17 family of proteins. Homologues for Tim17 were also identified in the genome databases of two related parasites like Leishmania major and Trypanosoma cruzi. Comparison of Tim17 protein sequences from these three kinetoplastid parasites showed 80% identity among themselves.
Fig. 1.
Analysis of the primary and secondary structure of TbTim17. (A) ClustalW alignment of the predicted protein sequence of TbTim17 with that from other species. Two conserved negatively charged residues at the N-terminal region are shown by astericks (*). In TbTim17, the second charged residue (marked as ◆) was not perfectly aligned to that in other sequences. Four transmembrane regions (TM1-TM4) are underlined. Two conserved positively charged residues in the loop between TM2 and TM3 are in a red box. The accession number of the sequences are Tbrucei; Trypanosoma brucei (EAN 80274), Tcruzi; Trypanosoma cruzi (EAN99897), Lmajor; Leishmania major (CAJ02901), Pchab; Plasmodium chabaudi (CAH84724), Scere; Saccharomyces cerevisiae (CAA54823), Ncrassa; Neurospora crassa (AA072334), Celeg; Caenorhabditis elegans (O44477), Atha a, b, and c; Arabidopsis thaliana (At1g20350, At2g37410, At5g11690, respectively), and Human; Homo sapiens (AAH16817). (B) Hydropathy plots of Tim17 from Human, Neurospora crassa, and Trypanosoma brucei. Protein sequences were analyzed using TMpred software program. TbTim17 possesses 4 transmembrane domains similar to that in other species.
Unlike Tim23 and Tim22, the N-terminal sequence of Tim17 proteins is conserved among different species and contains a motif D/E XXR D/E PCP, in which two negatively charged residues were found to be critical for the function of TIM23 translocase in fungi [16,17]. TbTim17 possesses a similar but not identical motif at the N-terminal region, which consists of two aspartic acid residues but one of these is not at the conserved site (Fig. 1A, marked with * and ◆, respectively).
Hydropathy analysis of the TbTim17 showed that it contains four transmembrane domains (TM1-4) similar to other Tim17 proteins (Fig. 1A & B). The hydrophilic N-terminal region of TbTim17 is about 20 amino acid longer in comparison to that in Human and N. crassa Tim17 (Fig. 1B). Tim17 possesses several other conserved charges in the loops between the membrane spanning domains. In particular, the intermembrane space loop between transmembrane domains TM2 and TM3 contains several positively charged residues, among which two (boxed in red, Fig. 1A) are presumably involved in intragenic ionic interaction with two negative charges in the N-terminal region in fungi Tim17 (17). These two charge residues are conserved in all Tim17 including those of trypanosomatids.
3.2. Expression and Subcellular location of TbTim17
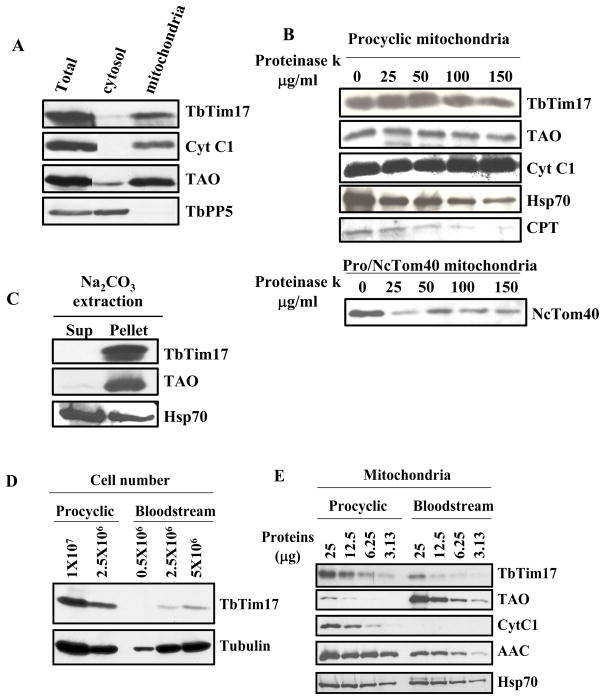
To verify the subcellular location of TbTim17, cell fractionation studies were performed. Differential centrifugation of the cell lysate followed by immunoblot analysis using an affinity purified polyclonal antibodies developed against the N-terminal 17 residues of TbTim17 showed that this protein is present in the crude mitochondrial fraction as are other mitochondrial proteins like Cyt C1 and trypanosome alternative oxidase (TAO). On the other hand, the cytosolic serine/threonine protein phosphatase 5 (TbPP5) [40] is present in the post-mitochondrial supernatant fraction as expected (Fig. 2A). Limited proteolytic digestion of isolated mitochondria with proteinase K (PK) revealed that the level of TbTim17 was not affected even at 100 μg/ml of PK. Similar results were also observed for other mitochondrial inner membrane proteins Cyt C1, and TAO. However, a putative outer membrane protein CPT and an ectopically expressed heterologous mitochondrial outer membrane protein, Tom40 from Neurospora crassa (NcTom40) were degraded at lower concentration of proteinase K (Fig. 2B). It has been previously demonstrated that NcTom40 is targeted to outer mitochondrial membrane while expressed in T. brucei [44]. Alkali extraction of isolated mitochondria of T. brucei followed by immunoblot analysis demonstrated that TbTim17 is present in the alkali-resistant membrane pellet like TAO, whereas Hsp70, a matrix protein is present in the soluble supernatant as expected (Fig. 2C). Thus, TbTim17 is an integral membrane protein and localized in mitochondrial inner membrane.
Fig. 2.
Expression and Subcellular location of TbTim17. (A) Immunoblot analysis of sub cellular fractions using T. brucei Tim17 (TbTim17), Cyt C1 (Cyt C1), serine/threonine protein phosphatase 5 (TbPP5), and trypanosome alternative oxidase (TAO) antibody as probes. Different fractions were total lysate, cytosol, and mitochondria. Ten microgram proteins from each fraction were loaded per lane. (B) Proteinase K digestion of intact mitochondria followed by immunoblot analysis using TbTim17, TAO, Cyt C1, Hsp70, CPT, a putative outer membrane protein, and Neurospora crassa Tom40 (NcTom40) antibodies as probes. Concentration of proteinase K was indicated at the top. After digestion, Mitochondria were re-isolated by centrifugation and electrophorsed as described. (C) Sodium carbonate extraction followed by immunoblot analysis of mitochondrial proteins from T. brucei procyclic form. After Na2CO3 treatment mitochondria were re-isolated by centrifugation. The supernatant (Sup) and the pellet (Pellet) fraction were analyzed using TbTim17, TAO and Hsp70 antibodies as probes. (D) Immunoblot analysis of total cellular proteins from the bloodstream and procyclic form of T. brucei using TbTim17 polyclonal antibody as the probe. Tubulin antibody was used as the loading control. (E) Mitochondrial proteins isolated from the procyclic and bloodstream forms were analyzed by immunobloting using TbTim17, TAO, Cyt C1, AAC, and Hsp70 antibodies as probes. Amount of proteins loaded per lane was indicated on the top.
In fungi, mitochondrial protein translocases are expressed constitutively. However, several reports give evidence that depending upon environmental conditions, protein trafficking in mammalian and plant mitochondria is regulated by altering the expression of the components of translocase proteins [52, 53]. The bloodstream form of T. brucei possesses a rudimentary mitochondrion, which lacks many of the normal activities of typical mitochondria. Upon differentiation to the procyclic form, mitochondrial activities are dramatically increased [21–23, 54]. To assess the expression level of Tim17 in these two developmental forms of T. brucei, we performed western blot analysis using TbTim17 antibody. This antibody specifically recognized a protein of an apparent molecular mass 19 kDa from T. brucei cell extract. The steady state expression level of TbTim17 was 6–7 fold higher in the procyclic than the bloodstream form (Fig. 2D). Analysis of TbTim17 transcript level by real time PCR as described in the materials and methods revealed a two fold difference between the procyclic and the bloodstream forms (not shown). Furthermore, about 3 fold difference in the TbTim17 protein level was also found when purified mitochondria from the procyclic and the bloodstream forms were analyzed (Fig. 2E). As expected, Cyt C1 was not detected and TAO was more abundant in the bloodstream form mitochondria in comparison to that in the procyclic form. The steady state level of AAC was about 3 fold less in the bloodstream form and mHsp70 is constitutively expressed. Thus, in T. brucei, expression level of TbTim17 is developmentally regulated along with mitochondrial activities.
3.3. TbTim17 is essential for cell survival
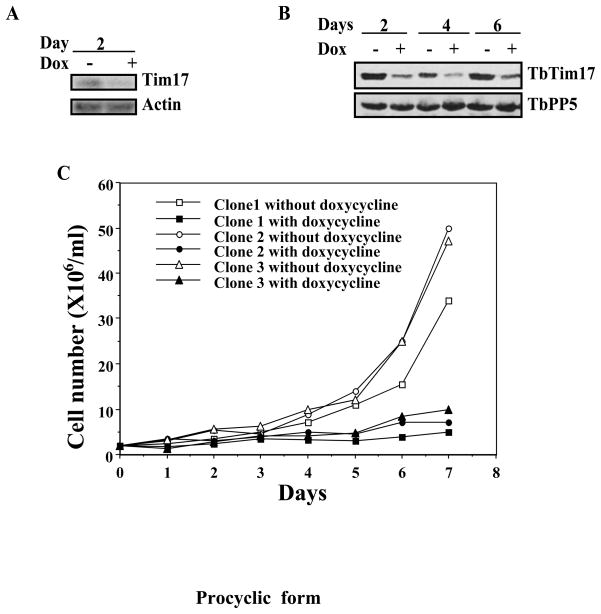
To evaluate the function of TbTim17, RNA interference studies were performed in the T. brucei procyclic and bloodstream forms. Expression of TbTim17 double stranded RNA reduced the steady state level of the specific transcript and protein more than 50% within 2 and 4 days, respectively in the procyclic form (Fig. 3A & B). Measurement of cell numbers at different time points after induction with doxycycline showed that depletion of TbTim17 ceased cell growth within 3 to 4 days in the procyclic form (Fig. 3C). A similar growth pattern was observed for three independent clones of the procyclic TbTim17 RNAi. Thus, as in other organisms, TbTim17 is an essential protein in the T. brucei procyclic form. At day 6 or after, TbTim17 protein level was slightly increased.
Fig. 3.
TbTim17 RNAi in the procyclic form. TbTim17 RNAi cell lines were allowed to grow in the presence and absence of doxycycline (1.0 μg/ml). Cells were harvested at different time points of induction with doxycycline for RNA and protein analysis by Northern (A) and immunoblot (B) analysis, respectively. Actin was used as the control for Northern. TbPP5 antibody was used to show equal loading of the sample for immunoblot analysis. Cell number was counted for three different clones at different time points during induction and plotted versus time (C).
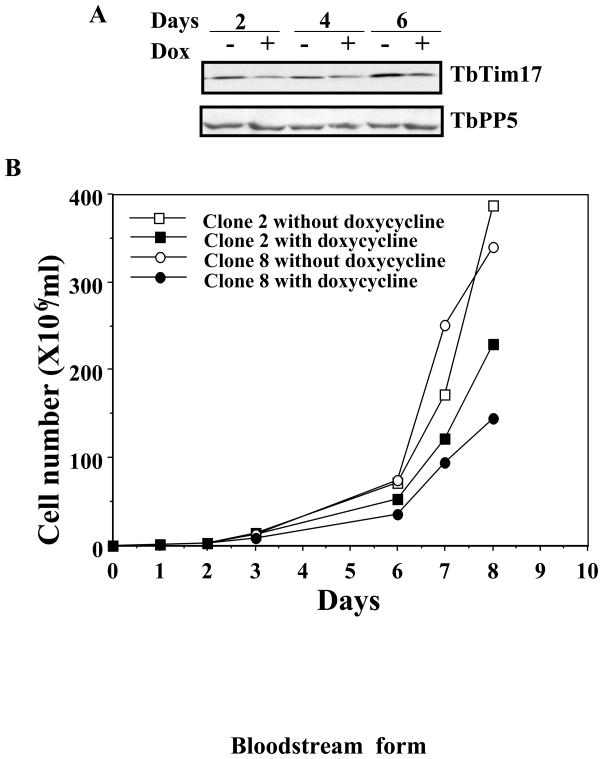
In the bloodstream form, a significant reduction in the level of TbTim17 protein was observed within 2 to 4 days of induction of TbTim17 double stranded RNA (Fig. 4A). However, induction of RNAi reduced the growth of the bloodstream form less dramatically than the procyclic form. The bloodstream form grew at much slower rate but the growth was not ceased due to depletion of TbTim17. A similar growth pattern was observed for two independent clones of the bloodstream form for TbTim17 RNAi. A 30–40% reduction of cumulative cell number was observed at or after day 6 (Fig 4B). Similar to the procyclic form, TbTim17 protein level increased in the bloodstream forms grown for longer time period in the presence of doxycycline due to a failure of RNAi as has been shown for other proteins in T. brucei [55]. Thus, it appears that in comparison to the procyclic form, the requirement of TbTim17 for cell growth is less in the bloodstream form.
Fig. 4.
TbTim17 RNAi in the bloodstream form. The bloodstream form TbTim17 RNAi cell line was allowed to grow in the presence and absence of doxycycline (1.0 μg/ml). Cells were harvested after 2, 4, and 6 days of induction with doxycycline for protein analysis by immunoblot (A) analysis. TbPP5 antibody was used to show equal loading of the sample for immunoblot analysis. Cell number was counted at different time points during induction and plotted versus time (B).
3.4. Depletion of TbTim17 reduced mitochondrial membrane potential
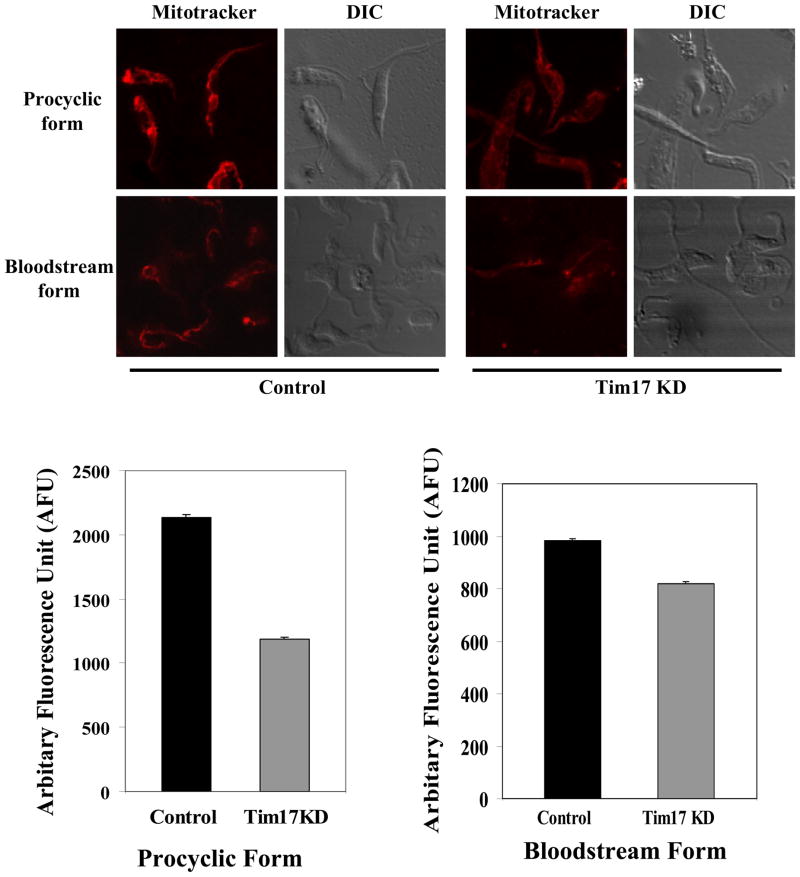
To understand the effect of TbTim17 depletion on mitochondrial structure and membrane potential, we compared MitoTracker stained procyclic and the bloodstream form cells with and without induction of TbTim17 RNAi. Both the bloodstream and the procyclic forms of T. brucei possess mitochondrial membrane potential as found in the confocal images (Fig. 5). The MitoTracker dye was taken up specifically by mitochondria. As expected, in the control procyclic form the mitochondrion is more elaborate and in the bloodstream form it is more rudimentary. When TbTim17 was depleted by RNAi, the uptake of the dye was reduced in both forms. The staining pattern was slightly diffused in TbTim17 depleted procyclic as well as in the bloodstream form cells in comparison to the respective control. This is possibly because the dye from the cytosol was not taken up by mitochondria as efficiently as in the control cells. However, we did not see any changes in the overall structure of mitochondrion in either form. These results showed that TbTim17 is involved to maintain mitochondrial membrane potential. We have quantitated our results by measuring the intensity of 300–400 parasites in each group using computer software. The average drop in fluorescence intensity in the procyclic and the bloodstream forms due to depletion of TbTim17 was 1.8 and 1.2 fold, respectively. Statistical analysis showed that the P value is <0.001 in both cases, indicating that the differences are significant. The average reduction in the fluorescence intensity due to TbTim17 knock down was slightly more in the procyclic form in comparison to the bloodstream form.
Fig. 5.
The effect of Tim17 RNAi on mitochondrial membrane potential in the procyclic form. The procyclic and bloodstream forms TbTim17 RNAi cells were grown for 96 h in the presence and absence of doxycycline. Cells were harvested and incubated with MitoTracker dye as described. Cells were then washed with PBS and fixed with paraformaldehyde. Confocal microscopic images were taken by LSM510 confocal microscope using 100× magnification. The phase contrast (DIC) and MitoTracker stained control and TbTim17 knock-down (Tim17KD) cells were shown. The fluorescence intensity of 300 to 400 parasites in each group was quantitated as described in section 2.6 and the average intensity was plotted for Tim17KD and control cells of both forms. The estimated P values are <0.001.
3.5. Depletion of TbTim17 reduced the steady state level of several nuclear encoded mitochondrial proteins
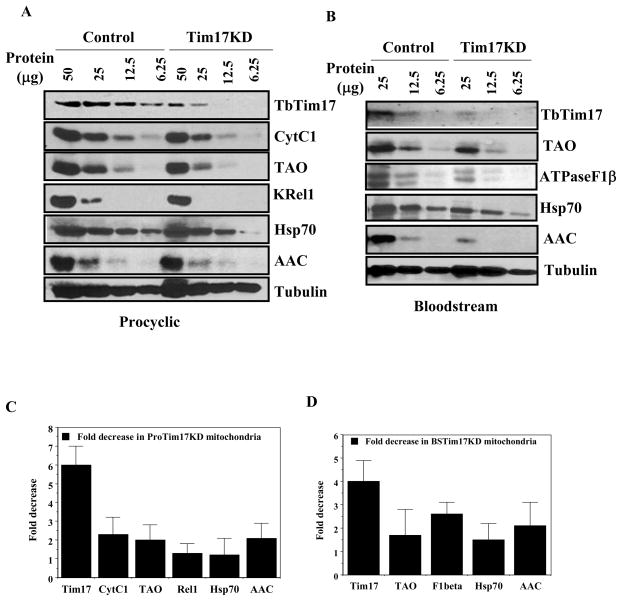
Tim17 is an essential component of the TIM23 complex. In fungi and higher eukaryotes, the TIM23 complex mediates import of precursors for most of the matrix-loalized and some inner-membrane proteins. We monitored the steady state expression level of several such proteins in T. brucei to assess the effect of TbTim17 depletion. A semi-quantitative immunoblot analysis of mitochondrial proteins from Tim17RNAi cells induced for 96 h with doxycycline and that from uninduced cells clearly showed that depletion of TbTim17 was associated with a significant reduction of several mitochondrial proteins destined to the matrix and inner membrane in the procyclic as well as in the bloodstream forms of T. brucei (Fig. 6). In the procyclic form, the level of TbTim17 was reduced about 6–7 fold in the mitochondria isolated from the TbTim17 RNAi cells grown in the presence of doxycycline in comparison to the uninduced control (Fig. 6A & C). Mitochondrial inner membrane localized electron transport proteins such as Cyt C1 and TAO were reduced about 2 fold in the TbTim17 depleted mitochondria in comparison to control. The matrix localized editosome protein KRel1 and mitochondrial Hsp70 were decreased about 1.5 fold due to knock-down of TbTim17. Interestingly, it was observed that T. brucei ATP/ADP carrier protein (AAC) is reduced about 2 fold in TbTim17 depleted mitochondria. Although, it is known in other system that import of AAC occurs via Tim22–54 complex and does not depend on Tim17 [56,57]. Tubulin is not a mitochondrial protein but it is an associated protein found in mitochondrial preparation. As expected, the level of associated tubulin did not alter in TbTim17 depleted mitochondria. We consider this as our loading control.
Fig. 6.
The effect of TbTim17 RNAi on the expression level of various nuclear encoded mitochondrial proteins in T. brucei procyclic and bloodstream forms. (A) The procyclic form of TbTim17 RNAi cells were grown for 96 h in the presence and absence of doxycycline and mitochondria were isolated as described. Mitochondrial proteins (50, 25, 12.5 and 6.25 μg) were analyzed by immunoblot analysis using various antibody probes as follows; T. brucei Tim17 (TbTim17), cytochrome C1 (Cyt C1), Trypanosome alternative oxidase (TAO), Kinetoplast RNA editing ligase (KREL1), heat shock protein 70 (Hsp70), ADP/ATP carrier protein (AAC), and β-tubulin (Tubulin). (B) TbTim17 RNAi bloodstream form cells were grown in vivo in rat model system. To maintain the doxycycline concentration in the rat blood, the drinking water of a group of rat were supplemented with 5% sucrose and 250 μg/ml of doxycycline (Tim17 KD). The control group was supplied with drinking water containing only sucrose (5%). The parasites were harvested from the rat blood at the peak parasitemia level (3–4 days after infection) and mitochondria were isolated as described in the materials and methods. Mitochondrial proteins (25, 12.5 and 6.25 μg) were analyzed by immunoblot analysis using various antibody probes such as T. brucei Tim17 (Tim17), Trypanosome alternative oxidase (TAO), ATPase F1 subunit β (ATPaseF1β), heat shock protein 70 (Hsp70), ADP/ATP carrier protein (AAC), and T. brucei β-tubulin. (C and D) The intensity of the respective protein bands were quantitated using imaging densitometer as described in materials and methods, normalized with the corresponding β-tubulin protein bands and fold decrease in the intensity in TbTim17 knock down cells in comparison to control was plotted for different proteins.
Similar to the mitochondria from the procyclic form, Tim17 protein level was reduced about 4 fold in the mitochondria of the bloodstream form due to expression of TbTim17 double stranded RNA (Fig. 6B & D). TAO, an essential mitochondrial protein in the bloodstream form, is reduced about 1.5 fold. A 2–3 fold reduction on the steady state level of ATPase F1β was observed due to reduction of TbTim17 protein. Similar to the procyclic mitochondria the level of mHsp70 is least affected due to depletion of TbTim17. This is possibly because mHsp70 protein is more stable than other mitochondrial proteins. The steady state level of AAC was also reduced about 2 fold in TbTim17 knock down bloodstream form mitochondria. However, the associated protein tubulin was unaltered in comparison to control. Together, it suggests that TbTim17 is involved in mitochondrial protein biogenesis in both the procyclic and the bloodstream form of T. brucei.
3.5. TbTim17 plays a crucial role in the import of nuclear encoded mitochondrial proteins
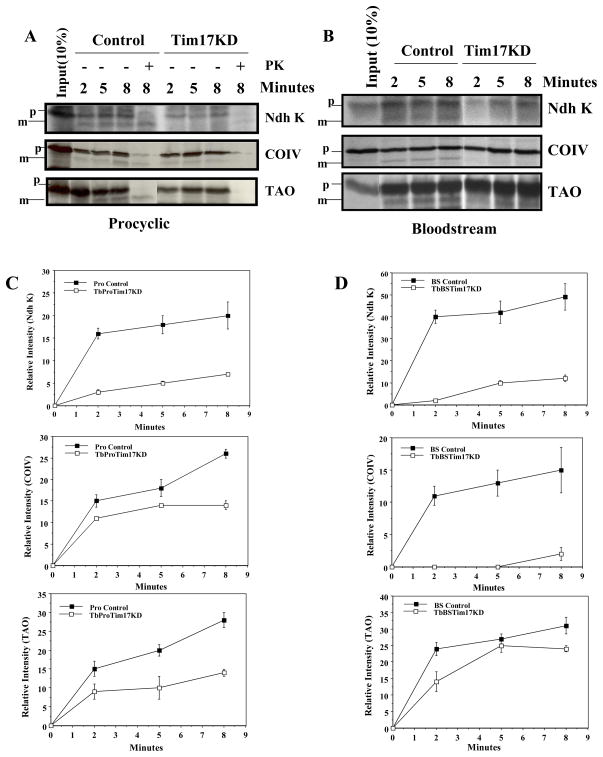
To evaluate whether TbTim17 is involved in mitochondrial protein import, in vitro import assays were performed. We have standardize the in vitro mitochondrial protein import assay system for T. brucei [58] using isolated mitochondria and the radiolabeled precursor proteins following the published protocols [50,51]. Using this system, we found that the precursor proteins are imported and processed inside the isolated mitochondria and the import is inhibited when the membrane potential was disrupted by inhibitors, such as CCCP and valinomycin (58, Supplementary Fig. 1.). To study the effect of TbTim17 knock down on import, we selected three nuclear encoded mitochondrial proteins in T. brucei such as TAO [42], COIV [48], and Ndh K [49]. These proteins were selected because they have relatively longer presequences. TAO and COIV possess the predicted presequence of 22 and 44 amino acids and their expression is upregulated in the bloodstream and the procyclic forms [42, 48, www.genedb.org], respectively. On the other hand, Ndh K is constitutively expressed and has a presequence of 20 amino acids [49]. Results showed that upon incubation with isolated mitochondria from the parental T. brucei procyclic form, the radiolabeled precursor proteins bound to mitochondria and processed to its mature form (Fig. 7A). The mature product was protected from post-import proteinase K digestion but the bound precursor protein was degraded, suggesting that the processed protein is inside the mitochondria. Mature proteins were accumulated in a time-dependent manner. For Ndh K, the import reached almost at the maximum level within 2 mins, which is relatively earlier than the import of other two proteins COIV and TAO (Fig. 7C). In contrast, when we performed the in vitro import reaction of the same three precursor proteins into mitochondria isolated from the TbTim17 knock down procyclic cells, we found that import of all these proteins were significantly reduced. Mitochondria were isolated at day 4 after induction of RNAi. At this time point cell number was reduced about 50% in comparison to untreated control. Depletion of TbTim17 in the procyclic form resulted in a 2.9, 2.0, and 2.5 fold reduction in import of Ndh K, COIV, and TAO, respectively within 8 min (Fig. 7A & C).
Fig. 7.
In vitro import of nuclear encoded mitochondrial proteins in TbTim17 depleted and control mitochondria of the procyclic and bloodstream forms. A) Mitochondria were isolated from TbTim17RNAi procyclic cells grown in the presence and absence of doxycycline for 4 days. In vitro import of radiolabelled proteins (Ndh K, COIV, and TAO) was performed as described in the materials and methods. The precursor (p) and the matured (m) protein bands were visualized by SDS-PAGE and autoradiography. Post-import proteinase k treatment (indicated by +) showed that matured form of the protein was protected from protease digestion as expected. (B) TbTim17 RNAi bloodstream form cells were grown in vivo in rat model system as described in the previous figure for mitochondria isolation. In vitro import of radiolabelled proteins (Ndh K, COIV, and TAO) was performed as described for the procyclic form mitochondria. The precursor (p) and the matured (m) protein bands were visualized by SDS-PAGE and autoradiography. (C and D) The intensity of the matured protein bands was quantitated from three independent experiments by densitometric scanning and plotted vs time of import (2, 5, and 8 mins).
Similar to the procyclic form, we also compared the import of Ndh K, COIV and TAO in the control and TbTim17 knock down bloodstream form mitochondria. These proteins are imported in the bloodstream form mitochondria in vitro and processed to their matured form. During import, the intensity of the mature protein bands at 2 min is closer to that at 8 min in the bloodstream form than in the procyclic form mitochondria, indicating the import of precursor proteins is quicker in the bloodstream in comparison to the procyclic form mitochondria. A similar observation has also been reported previously for import of Ndh K and TAO into the bloodstream and procyclic form mitochondria [50, 58]. Import of Ndh K and COIV into the mitochondria of the bloodstream form was drastically affected due to depletion of TbTim17 (Fig. 7B & D). Within 8 min, the reduction in the cumulative import of Ndh K and COIV was about 4 and 7 fold, respectively. TAO import was also inhibited in TbTim17 reduced bloodstream form mitochondria, particularly at the initial time point. However, very little difference was observed at longer times such as at 5 and 8 min. The Ndh K precursor protein binding was also significantly reduced in the TbTim17 knock down procyclic and bloodstream forms mitochondria. However, the precursor protein binding was not affected during import of COIV and TAO into TbTim17 depleted mitochondria. Overall, these results showed that TbTim17 is crucial for import of nuclear encoded mitochondrial proteins in the procyclic as well as in the bloodstream forms.
4. Discussion
In silico analysis revealed the presence of an ortholog of Tim17 in T. brucei. Tim17 is the most conserved of all Tom and Tim proteins [8]. Here, we found that TbTim17 possesses many characteristics in its primary and predicted secondary structure, which are similar to Tim17 from other species. However, the overall sequence identity of TbTim17 is relatively low. TbTim17 is critical for import of the precursor proteins into mitochondria of both the bloodstream and procyclic forms. While, its expression level is markedly reduced in the bloodstream form in comparison to that in the procyclic form.
Mitochondrial pre-protein translocator Tim17, Tim22, and Tim23 all contain 4 transmembrane regions with the N- and C-terminii on the intermembrane space side of the mitochondrial inner membrane [7,8]. Although the structure of these proteins is very similar, their functions are distinct. The Tim17 and Tim23 are two components of the TIM23 core complex that imports a majority of the mitochondrial proteins (14–20). Tim22 forms a separate complex, which imports the polytopic inner membrane proteins such as mitochondrial metabolite carrier proteins [2–4,9]. Searches in the trypanosomatid genome data bases identified only one homolog of Tim17/Tim22/Tim23 family protein that showed a closer homology to Tim17. Besides Tim17, it showed in a recent report that the homologs for small Tim proteins of mitochondrial intermembrane space are present in T. brucei [28]. These small Tim proteins are needed for import of the polytopic inner membrane proteins through TIM22 complex and also for import of the outer membrane beta-barrel proteins through sorting and assembly (SAM) complex [59]. The presence of small Tims suggests that these two pathways for protein import are possibly present in T. brucei. However, homology searches could not identify any components of the TIM22 complex in these groups of organisms. Therefore, it is not clear at this moment if Tim17 is the only core Tim protein in the kinetoplastid parasites or the divergent homologs of Tim22 and Tim23 are present.
Developmental regulation of mitochondrial activities is a unique phenomenon in T. brucei. The bloodstream forms rely on blood glucose, and thus suppress their mitochondrial activities [21–23]. This form does not have any cytochrome-containing respiratory complexes in mitochondria. On the other hand, the procyclic form possesses a fully functional form of all these complexes and also up-regulates many of the TCA cycle enzymes. All these changes require an increased expression of many nuclear encoded mitochondrial proteins. Thus, the mitochondrion in the procyclic form imports many more proteins in comparison to that in the bloodstream form. For this reason, an increased level of TbTim17 in the procyclic from is properly justifiable. From our analysis we found that the level of TbTim17 transcript and protein both are reduced in the bloodstream form than the procyclic form and the difference in the protein level is more than the level of its transcript. As transcriptional control is absent or rare in trypanosomatids [60], TbTim17 expression is thus regulated at post-transcriptional steps, such as protein and transcript stability and protein synthesis, during differentiation.
In fungi, it has long been known that Tim17 is an essential protein. We have also found that a reduction in the expression level of TbTim17 by RNAi severely inhibit the cell growth in the procyclic form. Thus, TbTim17 is an essential protein in this form. In the bloodstream form, the TbTim17 RNAi reduced growth rate significantly, but did not ceased cell growth like in the procyclic form. This is possible because the bloodstream forms are less dependent on mitochondrial activities. A partial reduction of TbTim17 did not reduce the essential nuclear encoded mitochondrial proteins below their critical level thus showed a reduced effect on cell growth in comparison to the procyclic form. However, a 50% reduction in the cumulative cell number was observed due to 50% reduction of TbTim17 protein level in the bloodstream form, suggesting TbTim17 is an essential protein also in this form.
Recent findings showed that Tim17 is necessary to maintain the structure of the protein import channel and also for gating and sorting of preproteins through this channel [16–18]. Depletion of Tim17 keeps this complex in an open state conformation, which increases the ion permeability across the mitochondrial inner membrane and reduces its membrane potential. A significant reduction in mitochondrial membrane potential was observed in the procyclic form, when TbTim17 was knocked down by RNAi. Thus, it is most likely that similar to fungi, TbTim17 is involved in regulating ion permeability through mitochondrial inner membrane. In fungi, the N-terminal region of Tim17 is found to be critical for this function [16, 17]. A marked difference was observed in the primary sequence of the N-terminal region of TbTim17 in comparison to that in other systems; however, two important positively charged residues are present in this region of TbTim17. Thus, TbTim17 may perform similar function in T. brucei. In the procyclic form, mitochondrial membrane potential is maintained by proton pumping through respiratory complexes like in other systems and thus depends on multiple nuclear encoded proteins. Import of these proteins is likely inhibited due to depletion of TbTim17, which can also be the reason for a reduction of the membrane potential in this form. The effect on membrane potential as a result of TbTim17 RNAi is relatively less in the bloodstream than the procyclic form, which is possibly because TbTim17 protein level was less reduced in the bloodstream than the procyclic form by RNAi.
Tim17 is a core component of the TIM23 complex and is essential for import of a majority of nuclear encoded mitochondrial proteins. Here, we observed that TbTim17 knock down decreased the levels of a number of mitochondrial proteins that are destined to the matrix and inner membrane about 1.5 to 3 fold in both the procyclic and the bloodstream forms. On contrary, there was no effect of TbTim17 depletion on the level of a mitochondrial associated protein such as tubulin, suggesting that TbTim17 is involved in mitochondrial protein biogenesis. A simultaneous reduction of several important mitochondrial proteins could account for a growth defect due to TbTim17 RNAi. The fold reduction on the steady state level of different proteins varied, which may possibly be due to differences in the half-lives of these proteins. Due to RNAi, the level of TbTim17 was reduced about 6–7 fold in the procyclic mitochondria, but in the bloodstream form mitochondria reduction was about 4 to 5 fold. However, the overall reduction of other mitochondrial proteins due to TbTim17 RNAi was very similar in both forms. This is possibly because TbTim17 is expressed more than its requirements in the procyclic form.
Interestingly, we have found that the inner membrane protein AAC, which in other systems, is known to be imported via the TIM22 complex [56, 57] is also reduced about 2 fold in the bloodstream form due to ablation of TbTim17. It may be possible that this reduction is a secondary effect. However, the magnitude of reduction is similar to several other mitochondrial proteins in T. brucei. Thus, further studies are necessary to elucidate if TbTim17 is directly involved for import of AAC.
Finally, we found that the uptake of TAO, COIV, and Ndh K precursor proteins was significantly reduced due to depletion of TbTim17 in mitochondria isolated from both life forms of T. brucei. Thus, TbTim17 is essential for mitochondrial protein import in both forms. Depletion of TbTim17 reduced mitochondrial membrane potential less in the bloodstream form, however, the degree of inhibition of mitochondrial protein import was more in this form than the procyclic form. This indicates that TbTim17 is directly involved for import of precursor proteins. Except for Ndh K, the precursor protein binding was minimally affected in TbTim17 depleted mitochondria suggesting that isolated mitochondria from these cells possesses similar levels of receptors for precursor proteins. Thus, TbTim17 depletion reduced the import efficiency of the preproteins into mitochondria from both forms. TAO and COIV are two stage specific proteins but they are imported under in vitro conditions in both the procyclic and the bloodstream form mitochondria, suggesting that stage specific proteins are capable to be imported into mitochondria in both forms. In contrast to COIV and Ndh K import, TAO import was less affected due to depletion of TbTim17 in the bloodstream form, but not in the procyclic form. Thus, the import of TAO is possibly less dependent on TbTim17 in this form.
Overall, we found that TbTim17 is critical for mitochondrial protein import in both the procyclic and bloodstream forms and essential for cell survival. This is the first characterization of a mitochondrial protein translocator in trypanosomatids parasites. Further characterization of other components of the translocase complex containing TbTim17 is critical to understand the protein import process in T. brucei mitochondria. It may also lead us to identify trypanosome specific essential mitochondrial protein(s).
Supplementary Material
Acknowledgments
We thank George Cross and Elizabeth Wirtz for pLew100 vector, Pro427 (29–13) and SM427 cell lines, Paul Englund for Crithidia Hsp70 antibody, Ken Stuart for anti-KREL1, Steve Hajduk for anti-Cyt C1, Noreen Williams for ATPase F1β, Keith Gull for tubulin antibody and p2T7-177 RNAi vector, Frank Nargang for Neurospora crassa Tom40 antibody. We also thank Susan Opalenik and Latha Raju at the Vanderbilt University Skin Diseases Research Molecular Genetics Core for performing the real-time PCR, an anonymous reviewer for suggestions on MitoTracker staining, and Shvetank Sharma for editing the manuscript. We thank Shawn Goodwin for Confocal Microscopy performed at the Morphology Core Facility in Meharry Medical College, supported by NIH grants U54NS041071-06, RR03032-19, U54CA91408, and U54RR019192-04. The work was supported by Grant 3SO6GM08037-30S1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Neupert W, Herrmann JM. Translocation of Proteins into Mitochondria. Ann Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 2.Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Science. 2003;116:3259–67. doi: 10.1242/jcs.00667. [DOI] [PubMed] [Google Scholar]
- 3.Pfanner N, Geissler A. Versatility of the mitochondrial protein import machinery. Nature molecular cell biology. Nature Mol Cell Biol. 2001;2:339–49. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 4.Lister R, Hulett JM, Lithgow T, Whelan J. Protein import into mitochondria: origin and functions today. Mol Membr Biol. 2005;22:87–100. doi: 10.1080/09687860500041247. [DOI] [PubMed] [Google Scholar]
- 5.Keibler M, Pfaller R, Sollner T, Griffiths G, Horstmann H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertions of precursor proteins. Nature. 1990;348:610–16. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- 6.Rapaport D. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane. J Cell Biol. 2005;171:419–23. doi: 10.1083/jcb.200507147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bomer U, Rassow J, Zufall N, Pfanner N, Meijer M, Maarse M. The preprotein translocase of the inner mitochondrial membrane: evolutionary conservation of targeting and assembly of Tim17. J Mol Biol. 1996;268:389–95. doi: 10.1006/jmbi.1996.0522. [DOI] [PubMed] [Google Scholar]
- 8.Bauer M, Gempel K, Reichert A, Rappold G, Lichtner P, Gerbitz KD, Neupert W, Brunner M, Hofmann S. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J Mol Biol. 1999;289:69–82. doi: 10.1006/jmbi.1999.2751. [DOI] [PubMed] [Google Scholar]
- 9.Koehler CM. Protein translocation pathways of the mitochondrion. FEBS Lett. 2000;476:27–31. doi: 10.1016/s0014-5793(00)01664-1. [DOI] [PubMed] [Google Scholar]
- 10.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–25. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Lann M, Meineeke M, Dudek J, Hutu DP, Lind M, Perchil I, Guiard B, Wagner R, Pfanner N, Rehling P. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- 12.Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. A prsequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struc Biol. 2001;12:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- 13.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 14.Maarse A, Blom J, Keil P, Pfanner N, Meijer M. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 1994;349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- 15.Milisav I, Moro F, Neupert W, Brunner M. Modular structure of the TIM23 preprotein translocase of mitochondria. J Biol Chem. 2001;276:25856–61. doi: 10.1074/jbc.M102132200. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Caballero S, Grigoriev SM, Hermann JM, Campo ML, Kinnally KW. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J Biol Chem. 2007;282:3584–93. doi: 10.1074/jbc.M607551200. [DOI] [PubMed] [Google Scholar]
- 17.Meier S, Neupert W, Herrmann J. Conserved N-terminal negative charge in the Tim17 subunit of TIM23 translocase play a critical role in the import of preproteins into mitochondria. J Biol Chem. 2005;280:7777–85. doi: 10.1074/jbc.M412158200. [DOI] [PubMed] [Google Scholar]
- 18.Chacinska A, Lind M, Frazier A, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer H, Truscott K, Gulard B, Pfanner N, Rehling P. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–29. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Murcha M, Lister R, Ho A, Whelan J. Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol. 2003;131:1731–47. doi: 10.1104/pp.102.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murcha M, Elhafez D, Millar H, Whelan J. The C-terminal region of Tim17 links the outer and inner mitochondrial membrane in Arabidopsis and is essential for protein import. J Biol Chem. 2005;280:16476–83. doi: 10.1074/jbc.M413299200. [DOI] [PubMed] [Google Scholar]
- 21.Schneider A. Unique aspects of mitochondrial biogenesis in trypanosomatids. Inter J Parasitol. 2001;31:1403–15. doi: 10.1016/s0020-7519(01)00296-x. [DOI] [PubMed] [Google Scholar]
- 22.Priest JW, Hajduk SL. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr. 1994;26:179–91. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- 23.Chaudhuri M, Ott RD, Hill GC. Trypanosome alternative oxidase: From molecule to function. Trends in Parasitol. 2006;22:484–91. doi: 10.1016/j.pt.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Schlecker T, Schmidt A, Dirdjaja N, Voncken F, Clayton C, Krauth-Siegel RL. Substrate specificity, localization, and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J Biol Chem. 2005;280:14385–94. doi: 10.1074/jbc.M413338200. [DOI] [PubMed] [Google Scholar]
- 25.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–62. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 26.Helfert S, Estavez AM, Bakker B, Michels P, Clayton C. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem J. 2001;357:117–25. doi: 10.1042/0264-6021:3570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellemond JJ, Baker BM, Tielens AG. Energy metabolism and its compartmentation in Trypanosoma brucei. Adv Microb Physiol. 2005;50:199–226. doi: 10.1016/S0065-2911(05)50005-5. [DOI] [PubMed] [Google Scholar]
- 28.Gentle IE, Perry AJ, Alcock FH, Likic VA, Dolezal P, Ng ET, Purcell AW, McConnville M, Naderer T, Chanez AL, charier F, Aschinger C, Schneider A, Tokatlidis K, Lithgow T. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial inter membrane space. Mol Biol Evol. 2007;24:1149–1160. doi: 10.1093/molbev/msm031. [DOI] [PubMed] [Google Scholar]
- 29.Biebinger S, Wirtz LE, Lorenz P, Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol Biochem Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- 30.Hirumi H, Hirumi K. In vitro cultivation of Trypanosoma congolense bloodstream forms in the absence of feeder cell layers. Parasitol. 1991;112:225–36. doi: 10.1017/s0031182000062533. [DOI] [PubMed] [Google Scholar]
- 31.Chaudhuri M, Sharan R, Hill GC. Trypanosome alternative oxidase is regulated post-transcriptionally at the level of RNA stability. J Eukaryot Microbiol. 2002;49:263–69. doi: 10.1111/j.1550-7408.2002.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 32.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann K, Stoffel W. TMbase A database of membrane protein segments. Biol Chem Hoppe-Seyler. 1993;47:166–70. http://ulrec3.unil.ch/software/TMPRED_form.html.
- 34.Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–16. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E, Maniatis T. Molecular cloning, A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor NY: 1994. [Google Scholar]
- 36.Laemmli UK. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some application. Proc Natl Acad Sci USA. 1979;76:4350–54. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priest JW, Hajduk SL. Developmental regulation of Trypanosoma brucei cytochrome c reductase during bloodstream to procyclic differentiation. Mol Biochem Parasitol. 1994;65:291–304. doi: 10.1016/0166-6851(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 39.Nierlich DP, Kuznair DG, Ghazvini J, Simpson L. Unpublished. [Google Scholar]
- 40.Chaudhuri M. Cloning and characterization of a novel serine/threonine protein phosphatase type 5 from Trypanosoma brucei. Gene. 2001;266:1–13. doi: 10.1016/s0378-1119(01)00367-5. [DOI] [PubMed] [Google Scholar]
- 41.Effron PN, Torri AF, Engman DM, Donelson JE, Englund PT. A mitochondrial heat shock protein from Crithidia fasciculata. Mol Biochem Parasitol. 1993;59:191–200. doi: 10.1016/0166-6851(93)90217-l. [DOI] [PubMed] [Google Scholar]
- 42.Chaudhuri M, Ajayi WU, Hill GC. Biochemical and Molecular properties of the Trypanosoma brucei alternative oxidase. Mol Biochem Parasitol. 1998;95:53–68. doi: 10.1016/s0166-6851(98)00091-7. [DOI] [PubMed] [Google Scholar]
- 43.Panigrahi AK, Schnaufer A, Carmean N, Igo RP, Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, Stuart KD. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:6833–40. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri M, Nargang FE. Import and assembly of Neurospora crassa Tom40 into mitochondria of Trypanosoma brucei in vivo. Curr Genet. 2003;44:85–94. doi: 10.1007/s00294-003-0427-y. [DOI] [PubMed] [Google Scholar]
- 45.MacRae TH, Gull K. Production and characterization of monoclonal antibodies to the mammalian sperm cytoskeleton. Biochem J. 1990;265:87–93. doi: 10.1002/mrd.1080250412. [DOI] [PubMed] [Google Scholar]
- 46.Brown SV, Hosking P, Li J, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Euk Cell. 2006;5:45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen TW, Lin BJ, Brunner E, Schild D. In situ background estimation in quantitative fluorescence Imaging. Biophys J. 2006;90:2534–2547. doi: 10.1529/biophysj.105.070854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maslov DA, Zinkova A, Kyselova I, Lukes J. A putative novel nuclear-encoded subunit of the cytochrome c oxidase complex in trypanosomatids. Mol Biochem Parasitol. 2002;125:113–125. doi: 10.1016/s0166-6851(02)00235-9. [DOI] [PubMed] [Google Scholar]
- 49.Peterson GC, Souza AE, Parsons M. Characterization of a Trypanosoma brucei nuclear gene encoding a protein homologous to a subunit of bovine NADH:ubiquinone oxidoreductase (complex I) Mol Biochem Parasitol. 1993;58:63–70. doi: 10.1016/0166-6851(93)90091-b. [DOI] [PubMed] [Google Scholar]
- 50.Priest JW, Hajduk SL. In vitro import of the Rieske iron-sulfur protein by trypanosome mitochondria. J Biol Chem. 1996;278:15084–94. doi: 10.1074/jbc.271.33.20060. [DOI] [PubMed] [Google Scholar]
- 51.Bertrand KI, Hajduk SL. Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 2000;106:249–60. doi: 10.1016/s0166-6851(99)00218-2. [DOI] [PubMed] [Google Scholar]
- 52.Matsuoka T, Wada J, Hashimoto I, Zhang Y, Eguchi J, Ogawa N, Shikata K, Kanwar YS, Makino H. Gene delivery of Tim44 reduces mitochondrial superoxide production and ameliorates neointimal proliferation of injured carotid artery in diabetic rats. Diabetes. 2005;54:2882–90. doi: 10.2337/diabetes.54.10.2882. [DOI] [PubMed] [Google Scholar]
- 53.Howell KA, Cheng K, Murcha MW, Jenkin LE, Millar AH, Whelan J. Oxygen initiation of respiration and mitochondrial biogenesis in rice. J Biol Chem. 2007;282:15619–15631. doi: 10.1074/jbc.M609866200. [DOI] [PubMed] [Google Scholar]
- 54.Lukes J, Hashimi H, Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–99. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 55.Motyka SA, Englund PT. RNA interference for analysis of gene function in trypanosomatids. Curr Opin Microbiol. 2004;4:563–76. doi: 10.1016/j.mib.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Truscott KN, Wiedemann S, Rehling P, Muller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polcicova K, Kempna P, Sabova L, Gavurmikova G, Polcic P, Kolarov J. The delivery of ADP/ATP carrier protein to mitochondria probed by fusions with green fluorescent protein and beta-galactosidase. FEMS Yeast Res. 2003;4:315–21. doi: 10.1016/S1567-1356(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 58.Williams S, Saha L, Singha UK, Chaudhuri M. Trypanosoma brucei: Differential requirement of membrane potential for import of proteins into mitochondria in two developmental stages. Exp Parasitol. 2007 doi: 10.1016/j.exppara.2007.10.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paschen SA, Neupert W, Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–82. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Teixeira SM, daRocha WD. Control of gene expression and genetic manipulation in the Trypanosomatidae. Genet Mol Res. 2003;2:148–158. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.