Abstract
Hedgehog (Hh) genes play major roles in animal development and studies of their evolution, expression and function point to major differences among chordates. Here we focused on Hh genes in lampreys in order to characterize the evolution of Hh signalling at the emergence of vertebrates. Screening of a cosmid library of the river lamprey Lampetra fluviatilis and searching the preliminary genome assembly of the sea lamprey Petromyzon marinus indicate that lampreys have two Hh genes, named Hha and Hhb. Phylogenetic analyses suggest that Hha and Hhb are lamprey-specific paralogs closely related to Sonic/Indian Hh genes. Expression analysis indicates that Hha and Hhb are expressed in a Sonic Hh-like pattern. The two transcripts are expressed in largely overlapping but not identical domains in the lamprey embryonic brain, including a newly-described expression domain in the nasohypophyseal placode. Global alignments of genomic sequences and local alignment with known gnathostome regulatory motifs show that lamprey Hhs share conserved non-coding elements (CNE) with gnathostome Hhs albeit with sequences that have significantly diverged and dispersed. Functional assays using zebrafish embryos demonstrate gnathostome-like midline enhancer activity for CNEs contained in intron2. We conclude that lamprey Hh genes are gnathostome Shh-like in terms of expression and regulation. In addition, they show some lamprey-specific features, including duplication and structural (but not functional) changes in the intronic/regulatory sequences.
Introduction
Lampreys and hagfish are the only two groups of agnathans (meaning jawless vertebrates) that have survived to date. They belong to a monophyletic group, the cyclostomes, considered as the sister group of extant gnathostomes (or jawed vertebrates) [1]–[3], see also [4]. They are promised to be a first-class model for the study of the evolution of vertebrate developmental mechanisms, owing to this key phylogenetic position and to the special anatomical features they present (reviewed in [5]). The rapidly growing number of reports on various particular genes and gene families in several lamprey species together with the ongoing assembly of genome data for the marine lamprey Petromyzon marinus further give these animals a very crucial status to investigate the origins of the vertebrate genomes, including the genomic and functional outcomes of whole genome duplication events (WGD) [6]–[8].
Hedgehog (Hh) family of intercellular signalling proteins are one of the key mediators of many fundamental processes in embryonic development, and are particularly essential to the development of nervous system in gnathostomes (reviewed in [9], [10]). Hh-related genes have been identified in most of the chordates genomes sequenced so far, such as the cephalochordate Branchiostoma floridae, the urochordate Ciona intestinalis and many vertebrates, although these genomes differ in the number of Hh genes they contain. Amphioxus has only one Hh gene (AmphiHh; [11]) and Ciona intestinalis has two, CiHh1 and CiHh2 [12], which are likely to result from a lineage-specific duplication event. Three Hh genes were identified in tetrapods such as mouse, chick, and human: Desert hedgehog (Dhh), Indian hedgehog (Ihh) and Sonic hedgehog (Shh), with Shh and Ihh appearing more related to each other than to Dhh. Two WGD are most likely at the origin of these paralogous genes. An ancestral Hh gene first duplicated and gave rise to Shh/Ihh and Dhh ancestor genes. An additional duplication event generated Shh, Ihh, Dhh and a fourth gene quickly lost [13], [14] (summarised on Fig. 2, inset). In zebrafish, the Hedgehog family is further enlarged due to a teleost-specific WGD, and there are two Shh (shha and shhb, the latter previously called tiggy-winkle hh), two Ihh (ihha and ihhb), but one Dhh member.
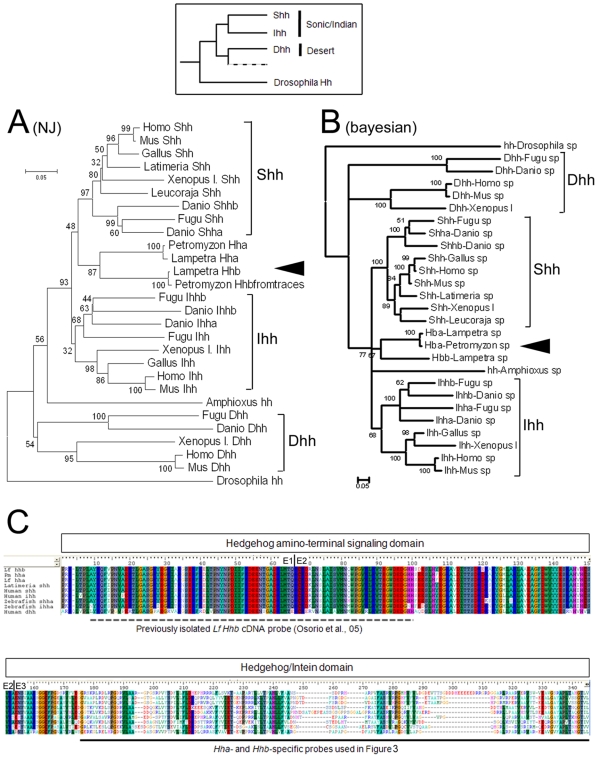
Figure 2. Phylogenetic analysis of lamprey's Hhs.
Inset: a minimal representation of the Hedgehog family in vertebrates, to highlight the classically-described relationships between Sonic, Indian, and Desert groups (see text, Introduction). A and B are Neighbour-Joining (NJ) and Bayesian phylogenetic trees (aligned aminoacids) of 28 (NJ) and 27 (Bayes) Hedgehog family members including the presently found lamprey members (black arrowhead), with the fly and amphioxus Hhs used as out-group. Bootstrap values are given and the 3 orthology groups are indicated on the right (Sonic, Shh; Indian, Ihh; and Desert, Dhh). C is an amino-acid alignment of lamprey Hh proteins with gnathostome family members. The functional domains (HH signal and Hint domain) are indicated, as well as the exon (E) junction positions (e., g., E1/E2). The regions corresponding to Hha- and Hhb-specific in situ hybridization probes used in Figure 3 are also indicated, as well as the previously isolated Lf probe used in [23].
Vertebrate embryos express Shh in key signalling centres from which it is secreted and exerts its so-called morphogen effect: the notochord and the floor plate of the neural tube –together with the zone of polarizing activity (ZPA) of the limb buds [15]. Further functional studies have demonstrated that Shh (and Ihh to a lesser extend) play an essential role in the dorso-ventral and antero-posterior patterning of the neural tube [16], as well as in the definition of anterior-posterior polarity of the limbs. Dhh on the other hand is involved in the development of peripheral nerves (the myelin-forming cells) and is expressed in adult nerves [17].
In a one-day-old zebrafish embryo, Shh is expressed in the notochord and in nested regions of the central nervous system including the floor plate, the zona limitans intrathalamica (zli), the hypothalamus and the retina [18]. Characterisation of regulatory sequences responsible for the spatio-temporal pattern of Shh expression identified at least four enhancer regions, ar-A, ar-B, ar-C, and ar-D (where ar- stands for activating region, see Fig. 1 for a general view). Ar-A, B, and C are intronic, whereas ar-D is located in the 5′ untranslated region. In zebrafish, ar-A and ar-C control notochord expression, whereas ar-D and ar-B are responsible for floor plate expression. Ar-A, ar-B and ar-C are together required to drive the expression in the hypothalamus and tegmentum. Ar-C also mediates expression in the zli [19], [20], which is considered as a secondary forebrain organiser. Several of these enhancers are conserved in sequence with other gnathostomes as demonstrated by phylogenetic footprinting, and they are thus called conserved non-coding elements (CNEs). However, functional conservation of these enhancers is not the rule: for example the ar-C element located in intron2 drives notochord and forebrain expression in zebrafish [19], while its mammalian counterpart (called SFPE2 for Sonic Floor Plate Enhancer2) drives floor plate expression in the mouse [21]. Whether lampreys present such sequence and/or functional conservation in Hh non-coding regions has not been addressed so far.
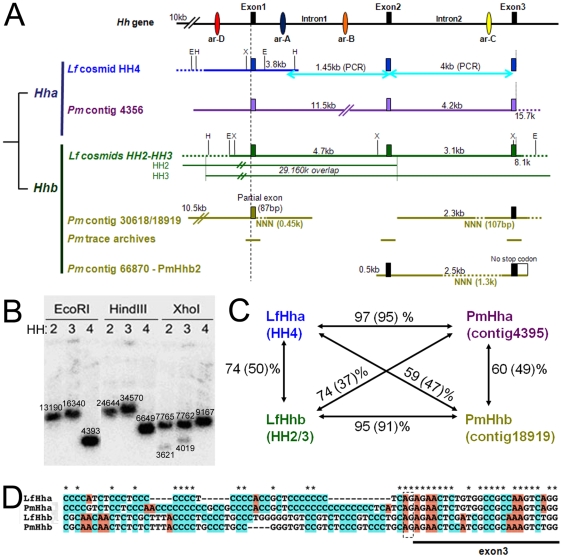
Figure 1. Organisation of Hha and Hhb loci in Lampetra (Lf) and Petromyzon (Pm).
A, Sequence information available after library screening (Lf cosmids), in silico searches (Pm contigs and trace archives) and PCR amplification is schematically depicted. Drawing is not to scale. The top row shows a “generic” Hh gene, with 3 exons (black boxes) and 2 introns containing previously described regulatory elements (coloured ovals, ar-A, B, C, and D; see text). Below, Hha genes are drawn in blue/purple, Hhb genes are drawn in green/kaki. The sizes of introns and the overall length from ATG to Stop codons are indicated. Restriction sites for enzymes EcoRI (E), HindIII (H) and XhoI (X) used for Southern analysis of the cosmids are indicated (restriction sites located on the Lawrist7 vector are not indicated). The trace archives for putative PmHhb exonic sequences have the following IDs (plus and minus are directions for assembly). For exon1: gnl|ti|1427444168+, gnl|ti|1179676842+, gnl|ti|1483498011+, and gnl|ti|1482777522-. For exon2: gnl|ti|1484276315+ and gnl|ti|1482717043+. For exon3: gnl|ti|1470810056+, gnl|ti|1213886743+, gnl|ti|1193744006-, and gnl|ti|1192802884-. B, Southern blot of Lf cosmids HH2, 3, and 4, using the partial previously isolated Lf cDNA encompassing most of exon1 (174 nt) and part of exon2 (100 nt) [23] as a probe (see Figure 2C for probe localisation on cDNA). Restriction enzymes and band sizes (in bp, obtained from in silico restriction digest of the HH cosmid sequences) are indicated. C, Similarity between each coding region and entire locus (in parenthesis) of the lamprey Hh genes. Nucleotide sequences were aligned with the CHAOS/DIALIGN program and the percent identity was calculated using the ClustalX program (ver2.11; [27]). The values show high similarities between the orthologs even when including intronic regions (see the value in the parentheses), while it is much lower between paralogs despite genes of the same species. D, An example of alignment showing the high sequence identity between orthologs and the lower sequence identity between paralogs of lamprey Hh genes. The sequence shown is at the level of the intron2-exon3 junction. The splice acceptor site (AG) is indicated.
Partial Hh cDNA fragments have been isolated for two lamprey species, Lampetra japonica [22] and Lampetra fluviatilis [23]. Lampetra fluviatilis Hh transcripts were detected in the notochord and prechordal plate, the floor plate, the diencephalic zli and a small hypothalamic region, thus strongly resembling the “archetypal” jawed vertebrate Shh pattern [23]. Compared to non-vertebrate chordates, lamprey Hh expression further extends into anterior parts of the neural tube including the forebrain. Unlike gnathostome Shh, however, lamprey Hh expression is absent from the preoptic area and from the ventral telencephalon [5], [23], [24]. These differences in expression further suggest a relationship between Hh expression domains and the development and evolution of the organisation of the central nervous system [5], [25].
Here, we focus on Hh gene(s) in two species, the river lamprey Lampetra fluviatilis (Lf) and the sea lamprey Petromyzon marinus (Pm), to better understand the evolution of the Hh signalling system at the emergence of vertebrates. We provide evidence for the existence of two lamprey Hh genes, we describe their genomic organisation and compare their expression features, and we characterise a functional midline enhancer shared among vertebrates.
Materials and Methods
Ethics statement
Animals were treated according to the French and European regulations for handling of animals in research. SR's authorisation for use of animals in research is number 91–116. This study uses exclusively embryos from aquatic vertebrate (non mammalian) animals and therefore did not require special authorizations.
Screening and sequence analyses (Fig. 1A)
Lampetra library screening
An Lf cosmid genomic library (RZPD library 55) was screened using classical molecular techniques, at low stringency, and using the previously isolated Lf cDNA fragment [23] as a random-primed DNA radioactively labelled probe. Out of the five positive clones identified, four were confirmed by Southern blot and 5′-end sequencing to be Hh positive clones. The full sequence for three of them (HH2, 3, and 4) was obtained after 454 sequencing at the Genoscope Sequencing Centre (Evry, France) and was submitted to GenBank under the accession numbers FP929026 (HH2), FP929027 (HH3) and FP929028 (HH4).
Petromyzon in silico searching
Pm contigs were retrieved in silico after BLAT or BLAST searches on the preliminary assembly of the sea lamprey genome using other vertebrates' Hedgehog genes (mouse and zebrafish) and the Lampetra Hh loci sequences obtained above as queries on the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway?clade=other&org=Lamprey&db=)or the preEnsemble database (http://pre.ensembl.org/Petromyzon_marinus/Info/Index).
Using the LfHhb coding sequence as query, best hit sequences were retrieved by blastn searches on the NCBI Trace Archive for P. marinus whole genome sequence (http://blast.ncbi.nlm.nih.gov/Blast.cgi). “EST” or “other” did not result in obtaining any significantly similar sequences. The retrieved sequence archives were assembled using the CAP3 program (Huang and Madan, 1999) implemented on the website, Mobyle@Pasteur (http://mobyle.pasteur.fr/cgi-bin/portal.py). Each resulting assembled contig was located on Contig30618, Contig31827, and Contig18919. Coding regions were predicted from the contigs with aids of BLASTX. Every putative exons was distinct from those of PmHha located on PmContig4356. Thus, we postulated that these exons belong to a single gene, namely PmHhb, although its continuity is not validated.
PCR amplifications
Intron2 of LfHha was amplified from Lampetra genomic DNA with primers Intron2-for (TGGGTCTACTACGAGTCCAAGG) and PmIn2-rev2 (CTTGGCGGCCACAGAGTT), followed by nested PCR using primers LfHhaI2_fwd (GTACGAATACTGGACTGGGATCG) and LfHhaI2_rev (GCAGTGAGCGGACGTTAGAC), and cloned into pGEM®-T Vector (Promega). A positive clone was sequenced with plasmid walking. To ascertain the continuity of the LfHha locus, partial exon1-exon2-partial exon3 was amplified from Lampetra cDNA obtained previously [23] with primers His_hha_for3 (GTCGCTACGAGGGGAAGAT) and His_hha_rev2 (TTCACGCACGAACACAAAGT), followed by cloning into pGEM®-T Vector and sequencing.
Exons were predicted with BlastX searches and manual editing based on alignment with cDNA sequences and on the canonical splicing rules (see Figure S2B). The nucleotide sequences were aligned with the CHAOS/DIALIGN program and the percent identities between the four identified Hhs were calculated using ClustalX (ver2.11) [26], [27].
Phylogenetic analyses
A multiple sequence alignment was carried out with the ClustalX software [28] and optimized manually using the MUST software [29]. After removing regions of ambiguous homology, an edited alignment of 234 amino acid positions was used in subsequent analyses. We first applied ProtTest [30] to estimate the optimal model of amino acid substitution (LG + Γ + I). Using this model, a maximum likelihood (ML) tree was inferred using PHYML [31]. The robustness of the ML tree was estimated by 100 bootstrap replications. A Bayesian analysis was performed using MrBayes [32] with the optimal closest model implemented in this program (JTT + Γ + I) and the following MCMC parameters: 1,000,000 generations, sampling each 20 generations, 250 “burn-in” trees discarded to reconstruct the consensus tree. Pairwise sequence distances were computed with the JTT + Γ model (alpha parameter, 0.41, estimated with PHYML) and a Neighbor-Joining (NJ) tree was inferred with MEGA software [33]. The robustness of the NJ tree was estimated by 1,000 bootstrap replications.
Accession numbers for Hh proteins included in the phylogenetic tree are as follows: Chicken shh, NP_990152; Human shh, NP_000184;Mouse shh, NP_033196; Xenopus laevis Shh, NP_001081782Leucoraja Shh, ABM66102Zebrafish shhb, NP_571274; Fugu shh, AAT99577; Zebrafish shha, NP_571138; Zebrafish ihhb, NP_571163; Zebrafish ihha, NP_001030165; Fugu Ihh, ENSTRUT00000034629; Fugu Ihhb, ENSTRUT00000031084; Chicken ihh, NP_990288;Xenopus ihh, NP_001079262; Mouse ihh, NP_034674; Human ihh, NP_002172; Fugu Dhh, ENSTRUT00000030985; Xenopus dhh, NP_001079261; Human dhh, NP_066382; Mouse dhh, NP_031883; Zebrafish dhh, NP_001025286; Amphioxus hh, CAA74169; Fruitfly hh, NP_524459.
Sequences alignments for CNEs searching
The global alignment algorithm LAGAN [34] was used to perform the pair-wise alignments for the whole Hh loci, subsequently visualized with VISTA plot [35], [36].
The multiple local alignments of putative ar-C sequences were obtained using the CHAOS/DIALIGN algorithms [26], and then visualized with BioEdit (written by Tom Hall, Ibis Biosciences, Carlsbad, CA, USA).
Whole-mount in situ hybridization
LfHha- and LfHhb-specific probes were designed on the third exon of each paralog, amplified from Lf cDNA with specific primers, and subcloned into pCR-TOPOII (Invitrogen) (see Fig. 2B for exact location of the specific probes). The following primers were used for amplification: HhaP_m_for (5′-GGC GTC GCC GCT CCG CTG CGA-3′), HhaP_m_rev (5′-CGA GCA CCG TGC CGC TCG ACA C-3′), HhbL_f_for (5′-GGC CGA TCC AAG CGG CTC CG-3′) and HhbL_f_rev (5′-CCA GCG TGC CGT GAG CCG T-3′). Digoxigenin-labeled antisense riboprobes against Hha and Hhb mRNAs were synthesised and whole-mount in situ hybridization was performed on Lampetra embryos as previously described [23], [25]. Some embryos were dehydrated with graded ethanol series and cleared with a 1 benzyl alcohol: 2 benzyl benzoate solution for whole-mount observation. Other embryos were dehydrated in ascending ethanol and butanol, embedded in paraffin and sectioned with a Leica microtome at 8 µm. Photographs were taken on a Nikon microscope equipped with a DXM-1200 camera.
DNA constructs
LfHhb intron2 was amplified from the Lf cosmid HH3 using a high-fidelity polymerase, AccuPrime™ Taq DNA Polymerase High Fidelity (cat#12346-086, Invitrogen), and ligated into KpnI and NotI sites of the backbone vector, −0.8Shh:GFP, which contains the minimal promoter of zebrafish shha and a multi-cloning site (MCS) downstream of the GFP reporter [37]. For cloning of a 30 bp fragment of the putative LfHhb C1 element, two complementary oligonucleotides were annealed after heat-induced denaturation, and used as an insert fragment for cloning into the same vector. Each primer sequence is designed to introduce the restriction sites as follows, with the annealing sites and additional restriction sites indicated in capital and small letters, respectively: LfHhb-I2_fwd_NotI (aaagcggccgcACTACGAGTCCAAGGCGCAC) and LfHhb-I2_rev_KpnI (cggggtaccCGGCGATCGAGTTCTCTG) for LfHhb intron2; LfHhb-C1-Fwd (ggccgcAGGGAATTTCGCACCTGAGCAAACGAGGAGggtac) and LfHhb-C1-Rev (cCTCCTCGTTTGCTCAGGTGCGAAATTCCCTgc) for LfHhb putative C1 element.
Microinjection of zebrafish eggs
Fish care and microinjection were carried out as described in [38]. The concentration of DNA constructs was adjusted to 70 ng/µl, and embryos were raised at 28°C. To precisely score the ratio of positive embryos, immuno-staining against GFP was carried out. Briefly, embryos were fixed in 4% PFA, followed by three washes with PBST. They were incubated with rabbit polyclonal anti-GFP at 1/1000 dilution (Molecular Probes, refA6455) at 4°C overnight, followed by washes and incubation in secondary chicken Alexa488 anti-rabbit antibody (Molecular Probes, refA21441) at room temperature for 2 hours. After PBST washes, they were stored in PBS until observation. The GFP fluorescence was scored and photos were taken using a Macrozoom system (Nikon).
Results and Discussion
Two species of lampreys each have two Hedgehog genes
Lampetra fluviatilis
Screening of a river lamprey (Lf) cosmid genomic library generated several positive clones which were subjected to Southern blot and showed two types of restriction profiles (Fig. 1B), raising the possibility of the existence of two distinct Hh genes in Lf genome. Three selected cosmids, HH2, HH3 and HH4, were sequenced and analysis revealed two clearly distinct Lf Hh loci.
Cosmid HH4 (33.4 kb, including 11.4 kb of upstream sequences containing a putative gene, pol polyprotein-like) did not contain the whole Hh locus, spanning the first exon and part of the first intron of an Lf Hh gene corresponding to the Lf cDNA fragment used as a probe [23]. We named this gene LfHha (blue, Fig. 1A). Additional PCR amplifications of the [intron1-exon2] and [exon2-intron2-exon3] regions of LfHha using Lf genomic DNA as template enabled us to obtain an almost complete sequence for LfHha (light blue lines on Fig. 1A). Proof of continuity of the LfHha locus and determination of the intron-exon boundaries was further obtained by PCR amplification on Lf cDNA template of a partial exon1-exon2-partial exon3 sequence (corresponding to amino-acids 38 (R) to 285 (V) of the LfHha protein sequence shown in Fig. 2C).
The other two cosmids, HH2 and HH3 could be assembled into a 48.3 kb contig, with 29.2 kb of overlapping sequence, including 32.4 kb of upstream sequences, 9.2 kb within the Hh locus, and 6.7 kb of downstream sequences. The two cosmids HH2/HH3 encompassed the whole genomic locus of an LfHh which was different from LfHha, as deduced from both sequence comparisons and Southern analysis (Fig. 1B). It was therefore named LfHhb (green, Fig. 1A).
Petromyzon marinus
In silico searches of the preliminary assembled genome of Petromyzon marinus retrieved four hits corresponding to Pm contigs 4356, 30618, 18919 and 66870.
Contig 4356 (23.5 kb) contained the almost full sequence of a Hh gene that was the clear Pm ortholog of the Lf cDNA fragments used as a probe/LfHha and was thus named PmHha (Fig. 1A, purple).
Contig 18919 (19.9 kb long, contains partial intron2 (2.3 kb) with a 107 bp sequencing gap and exon3) and contig 30618 (13.9 kb, contains only part of exon1 (87 bp)) were different in sequence from PmHha (kaki, Fig. 1A). They were however extremely similar to LfHhb, showing for example 92.7% identity with cosmid HH2 in their 5′ overlapping region (10.5 kb) and 97.8% identity in exon1 (partial sequence in contig 30618). Although we cannot prove that these two Pm contigs (30618 and 18919) are indeed from the same Hh locus, it is highly probable, based on the Lf genomic sequences. The existence of this locus is also supported by the finding of PmHhb-related exonic sequences in the trace archives of the Pm genome sequence project (kaki, Fig. 1A and phylogenetic tree in Fig. 2A). Contigs 18919 and 30618 therefore likely correspond to two short pieces of a PmHh locus orthologous to LfHhb, which we thus called PmHhb (kaki, Fig. 1A). Furthermore, BLAT search using Lf cosmids sequences retrieved up- and downstream sequences of the PmHhb genes. For example, the 5′ end of cosmid HH2 retrieved Pm contig18499 (15.0 kb) with 92.8% of similarity in 10.7 kb of overlapping sequence, which may therefore be assigned to be upstream sequence of the PmHhb locus (Figure S1A,B).
Finally, the contig 66870 contained sequences that spanned parts of intron1, exon2, intron2 (with a 1.3 kb gap) and exon3 of a Pm sequence highly similar but not identical to the PmHhb described above (Fig. 1A and Figure S2A). However, a stop codon was not identified in exon3 within the available sequence, and the sequence of the exon3 was highly divergent from the others, suggesting that this locus could be a pseudogene (Figure S2B). We named this putative Hh gene PmHhb2, with the possibility that this sequence may correspond to a Petromyzon-specific duplicate on the way of pseudogenisation.
As the Pm genome is not yet assembled, our in silico search cannot be considered as exhaustive and we cannot rule out the possibility that additional Hh gene(s) remain to be discovered. However, the parallel findings of two Hh sequences in two species of lampreys thought to have diverged 10–40 million years ago [2], using two different experimental approaches (in silico for Pm and in vitro for Lf) strongly support the existence of two Hh family members in the genomes of lampreys.
Lamprey Hh genomic organisation
The overall organisation of lampreys Hha and Hhb are conserved with other species' Hh members, containing three exons and two introns (Fig. 1A). The first intron of PmHha is long (11 kb versus ∼5 kb in other studied species including Lampetra, and 3.2 kb in Fugu). Further sequence analysis detected many repeats corresponding to microsatellites. This phenomenon is coincident with the commonly accepted idea that Pm genome is rich in repeats, hence the difficulties in completing its assembly (University of Washington, http://genome.wustl.edu/genomes/view/petromyzon_marinus/).
Orthology relationships between Hhs in the two lampreys were well supported by cross-comparisons (Fig. 1C,D and Figure S2C). Nucleotide sequences were aligned and the percent identities between the four identified Hhs were calculated. The values presented in Figure 1C show remarkably high similarities between the orthologs, both in coding sequences (>95%) and within intronic regions (>91%, values in parentheses). Identities are much lower between paralogs, being only 60–74% in coding sequences and 50% in non-coding regions, and decrease to even lower values when cross-species/cross-gene comparisons are made (Fig. 1C,D).
Lamprey Hha and Hhb belong to the Sonic/Indian group
Phylogenetic analysis was performed on the predicted amino-acid sequences of the exons. Neighbour-Joining (NJ) and Bayesian methods (together with Maximum Likelihood analysis shown in Figure S3B) all suggest that the two lamprey Hhs tend to cluster together within the Sonic/Indian Hedgehog clade (Fig. 2A and B; Figure S3C for a multiple alignment of the amino-acid sequences). These phylogenies are not very robust but the fact that all three types of analyses gave a similar tree topology favours the hypothesis that lamprey Hh genes indeed belong to the Shh/Ihh family. Of note, teleostean sequences seem to be rather fast-evolving, especially the Ihh and Dhh genes for which the monophyly with other Ihh and Dhh sequences is not well supported (e.g., Fig.2A, bootstrap = 32 and 54, respectively). Removal of these teleostean sequences from the alignment further reinforced the trend of lamprey Hha and Hhb genes to cluster with Shh/Ihh genes (Figure S3A). When included into the alignments, the amino-acid PmHhb sequence translated from trace archives (Fig. 1A, kaki) is the clear ortholog of LfHhb (Fig. 2A), and the partial LfHha sequence is the clear ortholog of PmHha (Fig. 2A and B). These data further support the relationships deduced from the genomic analysis and the existence of two Hhs in the two lamprey species.
This pattern is “classical” for lamprey genes, and has been reported in many studies for other genes/gene families for which orthology relationships are not easily inferred (e.g., for the Otx and other gene families [25], [39], [40]).
At the protein level, lamprey Hha and Hhb are almost identical in their hedgehog amino-terminal signaling domain (only 3 out of 150 amino-acids differ between the two sequences), a domain that is also extremely conserved in Shh and Ihh proteins of other species (Fig. 2C). By contrast, the Hedgehog/Intein domain (encoded by exon 3) is highly divergent between lamprey Hha and Hhb, as well as with Sonic/Indian sequences from other species (Fig. 2C). We took advantage of this feature to design Hha- and Hhb-specific probes (below).
No Dhh in lampreys?
Our searches could not identify a Desert-like Hh gene in lampreys, which means that they either 1) have a Dhh that we have not found yet, 2) once had a Dhh but have lost it, or 3) have no Dhh. As it is clear that the cyclostome genomes have experienced at least one WGD –the event during which Shh/Ihh and Dhh genes arose from the ancestral Hedgehog gene [41], we favour the second possibility for two reasons. First, and as discussed above, we trust that the convergence of data obtained from two independent lamprey species and two independent search methods is a strong support for lampreys having only two Hhs. Then, if the two lamprey Hhs are more related to Shh/Ihh, the common ancestor of cyclostomes and gnathostomes must have had at least two genes, Shh/Ihh and Dhh. It is reasonable to think that Dhh has been lost in cyclostomes after their separation from gnathostomes.
Moreover, reasoning in terms of the function of Dhh and the emergence of novelties in the central nervous system of craniates is also attractive. Functional studies have shown that Dhh plays important roles in the development of peripheral nerves, controlling the formation of myelin nerve sheath [17]. Classical neuroanatomical studies also tell us that cyclostomes (both lampreys and hagfish) lack a myelin sheath around their axons, while all gnathostome brains including those of chondrychtyans like sharks and rays are indeed myelinated [42], making axon myelinisation a gnathostome novelty. A correlation between the emergence of myelin and the maintenance of a Dhh gene in gnathostomes is therefore striking, and myelinisation may be due to the recruitment of Dhh in a new function. A phylogenomics approach including synteny analysis will be helpful on these issues when Pm genome assembly is updated.
LfHha and LfHhb expression patterns
Since the divergence of the sequences of lamprey Hha and Hhb genes suggest an ancient duplication of a Shh/Ihh-like gene, we next asked whether their expression would differ too. Previous analysis using a probe encompassing 276 bp of LfHha in the highly conserved hedgehog amino-terminal signaling domain (Fig. 2C for probe location) had revealed expression in the notochord at early stages and later in the floor plate of the neural tube, the hypothalamus and zli region of the ventral diencephalon, which appeared Shh-like [23]. This short probe was in a region of 93% identity (nucleotide level) between Hha and Hhb.
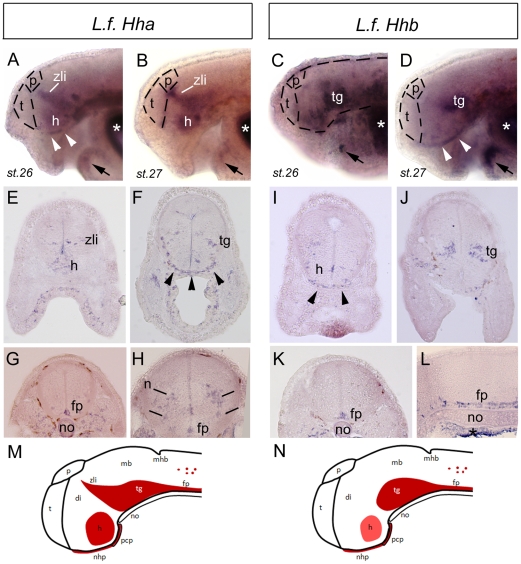
In order to discriminate between LfHha and LfHhb transcripts, we generated paralog-specific probes containing mostly the third exon, which is the most divergent in sequence between LfHha and LfHhb (probe regions indicated on Fig. 2C). In situ hybridization performed on Lampetra embryos at stage 26 and 27, and analysed on whole-mounts (Fig. 3A–D) and on sections (Fig. 3E–L) showed that LfHha and LfHhb are expressed with very similar patterns. Expression was observed in the floor plate, the midbrain tegmentum, the hypothalamus and in scattered cell populations in the posterior neural tube. One important difference between the transcript distributions of the two genes is that LfHha, but not LfHhb, is expressed in the diencephalic zli organiser (summarised on Fig. 3MN). There is no LfHh expression in the basal telencephalon, which is the main difference between the lamprey Hhs and gnathostome Shh. Although we might have expected that LfHhb would be expressed in the telencephalon as a result of sub-functionalisation of the two paralogs in lamprey, it appears that this region of the embryonic forebrain is indeed devoid of Hh signalling. As we have suggested and discussed in previous studies, such differences in midline signalling systems between lampreys and gnathostomes may underlie the major neuro-anatomical differences observed in their telencephalon [23]–[25].
Figure 3. Compared expression patterns for LfHha and LfHhb in Lampetra embryos.
A-D show in toto in situ hybridisation photographs for LfHha (A and B; left) and LfHhb (C and D; right). Stages are indicated. Anterior is to the left and dorsal is to the top. Dotted lines delineate the brain in A, and the telencephalon and pineal gland in A to D. E-L show in situ hybridisation photographs for LfHha (E-H; left) and LfHhb (I-L; right) on transverse sections (except L, saggital section) through the head of stage 27 Lampetra embryos. Sections are organised from the most anterior to the more posterior parts of the embryos. M-N show schematic summary drawings of the expression patterns for LfHha (M; left) and LfHhb (N; right), where Hh expression is red and the intensity of red is related to the expression density observed reproducibly on many sectioned embryos in independent experiments. In all panels, arrowheads indicate expression in a placodal structure identified as the nasohypophyseal placode (nhp) underlying the diencephalon in the continuity of the prechordal plate (pcp), arrows point to lower lip expression, and asterisks indicate background trapping in the branchial arches cavities. Abbreviations are: di, diencephalon; fp, floor plate; h, hypothalamus; p, pineal gland; pcp, prechordal plate; mb, midbrain; mhb, mid-hindbrain boundary; n, unidentified neuronal populations; nhp, nasohypophyseal placode; no, notochord; t, telencephalon; tg, tegmentum; zli, zona limitans intrathalamica.
Importantly, we also observed previously unreported LfHh expression in the distal part of the lower lip and in a thin structure underlying the diencephalon ventrally, that we consider as a placodal structure and identify as the naso-hypophyseal plate/placode (nhp on Fig. 3MN). This finding is particularly interesting to discuss with regards to the developmental evolution of the adenohypophysis in lampreys. Uchida and colleagues [22] had proposed that duplication of Bmp2/4, hypothalamic expression of Fgf8 and involvement of Shh in the hypophyseal placode were developmental novelties acquired in gnathostomes for the control of pituitary organogenesis. However, recent phylogenetic analyses suggest that the lamprey Bmp2/4/16 family has well undergone duplications and secondary losses [43]; Fgf8 expression in the lamprey hypothalamus is now demonstrated [25]; and here we found LfHh expression in the nhp. Thus, it appears that most of the key players involved in pituitary development were already recruited in the common ancestor of all vertebrates, in line with the idea that the lamprey hypothalamo-hypophyseal system displays strong similarities with its gnathostome homolog (e.g., [44]).
Both transcripts in lampreys are Shh-like in terms of their expression patterns. This feature, coupled to the poorly resolved orthology relationships seen in phylogenetic analyses (Fig. 2), leads us to propose that Hha and Hhb are lamprey duplicates belonging to the Shh family. To further test this hypothesis, we next performed a comparative genomic analysis of regulatory elements/CNEs between in lamprey Hhs.
Are there any conserved non-coding elements (CNEs) in lamprey Hh genes?
Given the similarities in Hh/Shh expression patterns between lamprey and gnathostomes, it is challenging to identify functional CNEs shared in their genomes [24], [45]–[47]. The divergence and rapid evolution of teleost (zebrafish and fugu) Hh sequences observed in phylogenetic analyses prompted us to search for another reference genome to perform comparative genomic studies. We chose the coelacanth Latimeria menadoensis, a lobe-finned fish (sarcopterygian), which is considered, after the lungfish, as the closest living relative of tetrapods [14]. The coelacanth genome (as well as its phenotype) indeed seems more stable than any teleost genome, and it was proposed as a reference genome for the search for regulatory elements among gnathostomes using a comparative genomics approach [48]. Here we used the Latimeria Shh sequence (GenBank: FJ603040.1) as the base genome to perform the global alignments of lamprey intronic sequences in search of putative CNEs. Because the lamprey genome sequences are divergent from jawed vertebrate genomes, it was harder to align them with those of other vertebrates, although this type of alignment is reportedly easy to do between jawed vertebrate genomic sequences (e.g., [49], [50] (see also Figure S4). In order to identify putative CNEs, we added the sequences of Latimeria ar-A, ar-B, ar-C and ar-D identified by Hadzhiev, Lang and colleagues [37], [51] to our analysis. A global alignment using the LAGAN algorithm and visualization by VISTA plot easily identifies the three exons of lampreys Hha/Hhb and Latimeria Shh genes (blue peaks, Fig. 4A). Importantly, the CNEs of Latimeria can also be detected as significant conservation peaks in the intronic non-coding region of lamprey Hh genes (pink, Fig. 4A). Namely, ar-A and ar-B in intron1, and ar-C in intron2, can be identified in both PmHha and LfHhb. These findings indicate that the lamprey Hha and Hhb loci share CNEs with gnathostome Shh loci. Of note, when the same in silico analysis was performed using zebrafish Shh sequences as the baseline genome instead of Latimeria, less significant conservation hits were detected in the intronic regions (not shown), strengthening the usefulness of Latimeria as a reference genome.
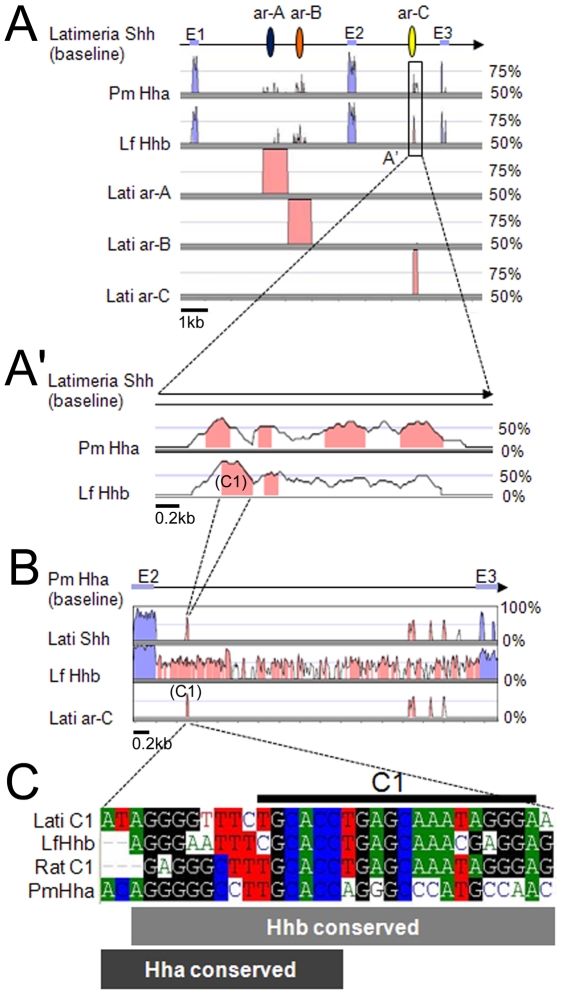
Figure 4. Conserved non coding elements (CNEs) in lamprey's Hha and Hhb.
A is a search of CNEs in PmHha and LfHhb gene sequences, confined with the Latimeria Shh enhancers ar-A, ar-B, and ar-C. Both genes show conserved peaks in the putative enhancer regions. A' is a close-up on ar-C. B is a search of CNEs in intron2 using LfHhb as baseline, showing high conservation between lamprey Hhs, and dispersion of ar-C sub-elements throughout the Latimeria sequence. C is a direct visualization of the underlying nucleotide alignments between coelacanth, lampreys and rat Shh/Hh at the level of C1. Note that the motif conservation extends on the 5′ side in lamprey genes, and is particularly shifted in the case of Hha. Analyses were performed with the global alignment algorithm MLAGAN and visualized with VISTA plot, with Calc Window and Min Cons Width of 20 bp respectively, and 70% Cons Identity. In the baselines, purple boxes indicate the exon positions, while the blue, red, and yellow ovals are the positions of ar-A, ar-B and ar-C, respectively. Conservation peaks representing putative CNEs appear in pink.
Lamprey Hh ar-C enhancer show homologous blocks with other ar-Cs
We next decided to focus on a single CNE, ar-C, whose conservation in sequence but not in function between zebrafish and mammals is quite puzzling [19], [21]. The conservation of ar-C between lampreys and Latimeria is the highest of the 3 enhancers (more than 75% for LfHhb, Fig. 4A and A'). It is also the Hh enhancer that has been analysed in greatest details [19], [20], [37], [52]. Previous functional analyses have subdivided the zebrafish ar-C element into four homology blocks, C1 to C4, which might be targets for transcription factors regulating the differential enhancer activities [37].
As shown on Figure 4A', a magnification of the lampreys ar-C conservation peaks identifies two to four highly conserved blocks (pink on Fig. 4A') for LfHhb and PmHha, respectively. Moreover and strikingly, VISTA visualisation of the global alignment of intron2 using a lamprey locus as baseline instead of the Latimeria locus also identifies four peaks of high conservation which correspond to Latimeria ar-C, but which appear dispersed throughout intron2 (Fig. 4B). This suggested that the Latimeria ar-C conserved elements are subdivided into short elements in lampreys. The most conserved peaks on Figure 4A' and 4B correspond to a C1-like nucleotide sequence, which is unusually located in the 5′ region of the lamprey intron2 (Fig. 4C and see below). These findings prompted us to test the possibility that lamprey ar-C contains the four homology blocks C1 to C4 described in other species, but that these blocks are dispersed or scattered along the entire intron2.
We then used PmHha and LfHhb ar-C sequences and checked for local alignment with Latimeria and rat ar-C sequences. The results show that both lamprey Hhs display homology blocks with other vertebrates in the C1 region, with the LfHhb C1 block displaying very good conservation, while the highly conserved C1 region in PmHha is slightly shifted towards 5′ (Fig. 4C). In addition, PmHha possesses a very short homology fragment within the C2 block, while LfHhb presents a well conserved block at a position corresponding to C3.
Ar-C of gnathostomes is commonly located in the 3′ region of intron2: in zebrafish shha, it is placed at positions 1035–1204 of the 1416 bp long intron2. In lampreys however, the putative ar-C1 region is located at the 5′ side of intron2, namely at positions 378–580 of the 3958 bp PmHha intron2 and at positions 759–959 of the 2589 bp LfHhb intron2. Our analyses demonstrate that putative functional modules can be detected in silico within lamprey CNEs, although they are significantly divergent from other vertebrates. In the following section, we therefore tested whether this conservation of sequence underlies functional conservation of these putative enhancers.
Enhancer activity of lamprey ar-C and ar-C1
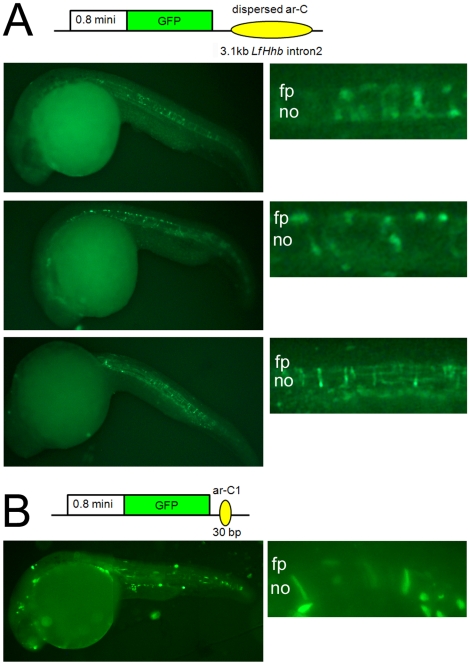
In zebrafish, shha expression at the ventral midline is mediated by ar-C [19], and C1 is crucial for ar-C activity [37]. Here, we have found that lamprey ar-C is dispersed along intron2, and that C1 conservation with gnathostomes is highest for LfHhb (Fig. 4). We therefore tested whether the entire intron2 of LfHhb has any enhancer activity. Injection of a GFP reporter construct in zebrafish embryos demonstrated that LfHhb intron2 drives GFP expression in the notochord and floor plate in 26hpf (hours post-fertilization) embryos (Fig. 5A and Table1). GFP was also observed in other ectopic tissues such as muscles, and considered as non-specific. The GFP reporter could be readily seen after observation in live embryos, and was further amplified for embryo scoring and photographing by immuno-fluorescence staining of the GFP protein (see also below). The backbone vector without any enhancer insertion gave almost no GFP expression, in agreement with previous works (Table 1; see also [20], [37].
Figure 5. Functional analysis of lamprey CNEs in zebrafish embryos.
A, LfHhb intron2 enhancer activity visualized in representative zebrafish embryos at 26hpf, at low (left) and high magnification (right) focusing on the notochord and floor plate. B, LfHhb ar-C1 enhancer activity visualized in two representative embryo at 26hpf, one at low (left) and one at high magnification (right) focusing on the notochord and floor plate. A schematic representation of the reporter constructs is shown in each case. It contains a zebrafish shha -0.8 kb minimal promoter driving GFP expression under control of the 3.1 kb intron2 (shown in A) or the 30 bp LfHhb ar-C1 (shown in B).
Table 1. Quantification of GFP immuno-positive zebrafish embryos after reporter constructs injection at one-cell stage.
| CONSTRUCT | GFP+ [nt/fp] (% of total/of GFP+) | GFP-positive [ectopic] (% of total/of GFP+) | GFP-positive [nt/fp & ectopic] | GFP-negative (including malformed embryos) | total number of injected embryos (n) |
| 0.8Shh:GFP:LfHhb-I2 | 51 (10.1/32.9%) | 104 (20.6/67.1%) | 155 (30.6/−%) | 350 (69.3%) | 505 |
| 0.8Shh:GFP:LfHhb-C1 | 7 (3.7/15.6%) | 38 (20.3/84.4%) | 45 (24.0/−%) | 142 (75.9%) | 187 |
| 0.8Shh:GFP* (*negative control) | 0 (0/0%) | 10 (100/8.1%) | 10 (8.1/−%) | 123 (92.5%) | 133 |
24–26hpf embryos injected with the indicated DNA construct were scored. nt:notochord; fp:floor plate.
Because the ar-C1 motif of ar-C seems to be the primary midline activator while other motifs seem to be repressors (ar-C2 and -C4) or of unknown activity (ar-C3) in zebrafish [37], we next tested the enhancer activity of LfHhb ar-C1 (Fig. 5B). The 30 bp-long LfHhb ar-C1 hardly had any detectable activity when observed in vivo, but clearly showed enhancer activity in the notochord after immuno-fluorescence staining. The global ratio of GFP-expressing embryos was almost the same as with the entire intron2 construct (24% versus 30%, Table 1). However, midline expression, including notochord and floor plate, was found with a ratio that was approximately half that observed with the entire LfHhb intron2 (15% versus 32% of the GFP-positive embryos, respectively, Table1). Furthermore, the relative intensity of GFP expression in the notochord appeared weaker than that driven by intron2. These data suggest that LfHhb C1 “midline” activity is significant, although weaker than that of the complete intron2.
Using a similar approach of phylogenetic footprinting, we have recently found that ar-C is shared and present in chordates Hh genes, but not in other deuterostomes [24]. Ar-C may therefore be a key motif for the chordate lineage, i.e., all the animals which possess a notochord and a floor plate.
General conclusions
Animals share a great number of genes involved in developmental processes, and these are highly conserved in their coding regions, which points to their common and ancient origin, and to the strong selection against changes at amino-acid level in these proteins. Furthermore, CNEs are enriched and clustered around genes involved in developmental regulation. Yet there are few shared CNEs between invertebrates and other deuterostomes and even less related animals. It seems that developmental genes possess different CNEs and are under different regulation mechanisms in distantly-related species [53]–[55]. The comparison of the genome of a chondrichthyan, the elephant shark Callorhinchus milii, with other gnathostome genomes, indicated that the “vertebrate CNEs” had been fixed before the separation of chondrichthyans and other gnathostomes about 450–500 million years ago [56]–[58]. A recent report has identified CNEs in many amphioxus genes, but the single element identified upstream of amphiHh did not show significant enhancer activity in vivo in zebrafish [59]. In lampreys, CNEs were first found in Hox genes [45], [46], [60], and recently confirmed by the findings of McEwen et al.[47] who have surveyed 13 lamprey genes for CNEs, showing that some of them were functional in zebrafish, although they were less numerous than expected. Here, we have performed an in depth analysis of particular developmental genes, the Hh genes. Our findings emphasise the crucial role played by the evolution of developmental gene CNEs for the emergence of the vertebrate body plan. They also confirm that lampreys probably have a genome with all the “vertebrate-like” attributes, although with some degree of divergence. More data from other developmental genes will be necessary to confirm and generalize the case.
It is estimated that Lampetra and Petromyzon diverged from their common ancestor 10-40MYA [2]. Kuraku and Kuratani predicted that the genetic similarity between lamprey species within Petromyzoninae would be too high for phylogenomic footprinting. It is indeed the case. To our knowledge, our paper reports for the first time a genomic comparison between Lf and Pm introns, and it was not effective to detect or predict CNEs (data not shown; see also Fig. 4B). On the contrary, the high similarity in non-coding sequences found between the two species could be used as an additional argument to support the orthology between their respective Hh loci.
The identification of genomic sequences for two Hh genes in two species of lampreys enabled us to present a detailed description and analysis of the Hedgehog “family” (only two members) in an agnathan representative. Our findings that Hha and Hhb have no clear orthology relationship with their gnathostome counterparts (although they are clearly Shh-like in many aspects), that they structurally and functionally possess CNEs shared albeit somewhat divergent with other vertebrates (e.g., in terms of structural dispersion of the conserved motifs), and that their expression patterns have clear lamprey-specific features (an absence of telencephalic expression), suggest that the two lamprey Hh genes have evolved into a quite “lamprey-specific” way after the split between the last common ancestor of cyclostomes and gnathostomes.
Supporting Information
PmContig18499 mapping in the 5'upstream region of the PmHhb locus using Lampetra cosmid sequences.
(0.08 MB DOC)
Detailed sequence comparisons and orthology assessments for lamprey Hh genes.
(0.39 MB DOC)
Additional phylogenetic trees
(2.82 MB DOC)
An example showing difficulty of global alignments of the lamprey genomic sequences with vertebrate sequences.
(0.15 MB DOC)
Acknowledgments
We thank Jean-Stéphane Joly, Maximilian Haeussler and Ashish Maurya for their discussions and scientific enthusiasm, Nadine Peyrieras for free access to her zebrafish colony and embryos, Sylvie Mazan for providing Lampetra genomic DNA sample and coordinating the GIS, and Axel Meyer and Michael Lang for sharing Latimeria sequences before publication.
Petromyzon genomic sequences were produced by the Genome Sequencing Centre at Washington University School of Medicine in St Louis and can be obtained from http://genome.wustl.edu/pub/organism/Other_Vertebrates/Petromyzon_marinus/assembly/Petromyzon_marinus-3.0/ASSEMBLY.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work supported by ANR (Agence Nationale de la Recherche)[MIDLINE] to SR, a GIS (Groupement d'Intérêt Scientifique) Genomique Marine to SR and DC, and by a collaborative student exchange grant between the University of Ottawa (ME) and the CNRS (Centre National de la Recherche Scientifique)(SR). JHX and SK were ANR postdoctoral fellows. JO was supported by a doctoral scholarship from the Foundation for Science and Technology of the Portuguese Ministry of Science and Higher Education, and a short-term scholarship from EMBO (European Molecular Biology Organisation). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kuraku S, Hoshiyama D, Katoh K, Suga H, Miyata T. Monophyly of lampreys and hagfishes supported by nuclear DNA-coded genes. J Mol Evol. 1999;49:729–735. doi: 10.1007/pl00006595. [DOI] [PubMed] [Google Scholar]
- 2.Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zoological Science. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- 3.Mallatt J, Winchell CJ. Ribosomal RNA genes and deuterostome phylogeny revisited: more cyclostomes, elasmobranchs, reptiles, and a brittle star. Mol Phylogenet Evol. 2007;43:1005–1022. doi: 10.1016/j.ympev.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Near TJ. Conflict and resolution between phylogenies inferred from molecular and phenotypic data sets for hagfish, lampreys, and gnathostomes. J Exp Zoolog B Mol Dev Evol. 2009 doi: 10.1002/jez.b.21293. [DOI] [PubMed] [Google Scholar]
- 5.Osorio J, Rétaux S. The lamprey in evolutionary studies. Dev Genes Evol. 2008;218:221–235. doi: 10.1007/s00427-008-0208-1. [DOI] [PubMed] [Google Scholar]
- 6.Donoghue PCJ, Purnell MA. Genome duplication, extinction and vertebrate evolution. Trends in Ecology & Evolution. 2005;20:312–319. doi: 10.1016/j.tree.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Kuraku S, Meyer A, Kuratani S. msn222: Molecular Biology & Evolution; 2008. Timing of genome duplications relative to the origin of the vertebrates: Did cyclostomes diverge before, or after? [DOI] [PubMed] [Google Scholar]
- 8.Ohno S. New York: Springer; 1970. Evolution by gene duplication. [Google Scholar]
- 9.Ingham PW, Placzek M. Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 10.Marti E, Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- 11.Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Development Genes and Evolution. 1999;209:40–47. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- 12.Takatori N, Satou Y, Satoh N. Expression of hedgehog genes in Ciona intestinalis embryos. Mech Dev. 2002;116:235–238. doi: 10.1016/s0925-4773(02)00150-8. [DOI] [PubMed] [Google Scholar]
- 13.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & Development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 14.Zardoya R, Arouheif E, Meyer A. Evolution and orthology of hedgehog genes. Trends in Genetics. 1996;12:496–497. doi: 10.1016/s0168-9525(96)20014-9. [DOI] [PubMed] [Google Scholar]
- 15.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 16.Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, et al. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 17.Parmantier E, Lynn B, Lawson D, Turmaine M, Namini SS, et al. Schwann cell-derived desert hedgehog controls the development of peripheral nerve sheaths. Neuron. 1999;23:713–724. doi: 10.1016/s0896-6273(01)80030-1. [DOI] [PubMed] [Google Scholar]
- 18.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 19.Ertzer R, Muller F, Hadzhiev Y, Rathnam S, Fischer N, et al. Cooperation of sonic hedgehog enhancers in midline expression. Developmental Biology. 2007;301:578–589. doi: 10.1016/j.ydbio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Muller F, Chang B, Albert S, Fischer N, Tora L, et al. Intronic enhancers control expression of zebrafish sonic hedgehog in floor plate and notochord. Development. 1999;126:2103–2116. doi: 10.1242/dev.126.10.2103. [DOI] [PubMed] [Google Scholar]
- 21.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 22.Uchida K, Murakami Y, Kuraku S, Hirano S, Kuratani S. Development of the adenohypophysis in the lamprey: evolution of epigenetic patterning programs in organogenesis. Journal of Experimental Zoology Part B Molecular and Developmental Evolution. 2003;300:32–47. doi: 10.1002/jez.b.44. [DOI] [PubMed] [Google Scholar]
- 23.Osorio J, Mazan S, Rétaux S. Organisation of the lamprey (Lampetra fluviatilis) embryonic brain: Insights from LIM-homeodomain, Pax and hedgehog genes. Developmental Biology. 2005;288:100–112. doi: 10.1016/j.ydbio.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 24.Rétaux S, Kano S. Midline signalling and forebrain evolution in chordates: a focus on the lamprey Hedgehog case. Integrative and Comparative Biology. 2010;50:98–109. doi: 10.1093/icb/icq032. [DOI] [PubMed] [Google Scholar]
- 25.Guérin A, d'Aubenton-Carafa Y, Marrakchi E, Da Silva C, Wincker P, et al. Neurodevelopment genes in lampreys reveal trends for forebrain evolution in craniates. PLoS One. 2009;4:e5374. doi: 10.1371/journal.pone.0005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brudno M, Steinkamp R, Morgenstern B. The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Research. 2004;32:W41–44. doi: 10.1093/nar/gkh361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philippe H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 31.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 32.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 34.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Research. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 37.Hadzhiev Y, Lang M, Ertzer R, Meyer A, Strahle U, et al. Functional diversification of sonic hedgehog paralog enhancers identified by phylogenomic reconstruction. Genome Biology. 2007;8:R106. doi: 10.1186/gb-2007-8-6-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blin M, Norton W, Bally-Cuif L, Vernier P. NR4A2 controls the differentiation of selective dopaminergic nuclei in the zebrafish brain. Mol Cell Neurosci. 2008;39:592–604. doi: 10.1016/j.mcn.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Germot A, Lecointre G, Plouhinec JL, Le Mentec C, Girardot F, et al. Structural evolution of Otx genes in craniates. Mol Biol Evol. 2001;18:1668–1678. doi: 10.1093/oxfordjournals.molbev.a003955. [DOI] [PubMed] [Google Scholar]
- 40.Suda Y, Kurokawa D, Takeuchi M, Kajikawa E, Kuratani S, et al. Evolution of Otx paralogue usages in early patterning of the vertebrate head. Dev Biol. 2009;325:282–295. doi: 10.1016/j.ydbio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Zardoya R, Abouheif E, Meyer A. Evolution and orthology of hedgehog genes. Trends Genet. 1996;12:496–497. doi: 10.1016/s0168-9525(96)20014-9. [DOI] [PubMed] [Google Scholar]
- 42.Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: Both lamprey and hagfish lack myelin. Neuroscience Letters. 1984;48:145–148. doi: 10.1016/0304-3940(84)90010-7. [DOI] [PubMed] [Google Scholar]
- 43.Kuraku S. Impact of hidden paralogy on hagfish and lamprey gene phylogeny. Integrative and Comparative Biology in press; 2010. Paleophylogenomics of the vertebrate ancestor. [DOI] [PubMed] [Google Scholar]
- 44.Sowers SA, Freamat M, Kavanaugh SI. The origins of the vertebrate hypothalamic-pituitary-gonadal (HPG) and hypothalamic-pituitary-thyroid (HPT) endocrine systems: new insights from lampreys. Gen Comp Endocrinol. 2009;161:20–29. doi: 10.1016/j.ygcen.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Irvine SQ, Carr JL, Bailey WJ, Kawasaki K, Shimizu N, et al. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. Journal of Experimental Zoology. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 46.Irvine SQ, Carr JL, Bailey WJ, Kawasaki K, Shimizu N, et al. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J Exp Zool. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 47.McEwen GK, Goode DK, Parker HJ, Woolfe A, Callaway H, et al. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS Genet. 2009;5:e1000762. doi: 10.1371/journal.pgen.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, et al. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemos B, Yunes JA, Vargas FR, Moreira MA, Cardoso AA, et al. Phylogenetic footprinting reveals extensive conservation of Sonic Hedgehog (SHH) regulatory elements. Genomics. 2004;84:511–523. doi: 10.1016/j.ygeno.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biology. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang M, Hadzhiev Y, Siegel N, Parada C, Strahle U, et al. Conservation of Shh cis-regulatory architecture of the coelacanth is consistent with its ancestral phylogenetic position. in preparation. [DOI] [PMC free article] [PubMed]
- 52.Epstein DJ, McMahon AP, Joyner AL. Regionalization of Sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development. 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- 53.Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24:344–352. doi: 10.1016/j.tig.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Vavouri T, Walter K, Gilks W, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biology. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woolfe A, Elgar G. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv Genet. 2008;61:307–338. doi: 10.1016/S0065-2660(07)00012-0. [DOI] [PubMed] [Google Scholar]
- 56.Venkatesh B, Tay A, Dandona N, Patil JG, Brenner S. A compact cartilaginous fish model genome. Current Biology. 2005;15:R82–R83. doi: 10.1016/j.cub.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 57.Woolfe A, Elgar G, Veronica vH, Robert EH. Advances in Genetics: Academic Press; 2008. Chapter 12 Organization of conserved elements near key developmental regulators in vertebrate Genomes. pp. 307–338. [DOI] [PubMed] [Google Scholar]
- 58.Sabarinadh C, Subramanian S, Tripathi A, Mishra RK. Extreme conservation of noncoding DNA near HoxD complex of vertebrates. BMC Genomics. 2004;5:75. doi: 10.1186/1471-2164-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hufton AL, Mathia S, Braun H, Georgi U, Lehrach H, et al. Deeply conserved chordate noncoding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 2009;19:2036–2051. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carr JL, Shashikant CS, Bailey WJ, Ruddle FH. Molecular evolution of Hox gene regulation: cloning and transgenic analysis of the lamprey HoxQ8 gene. J Exp Zool. 1998;280:73–85. doi: 10.1002/(sici)1097-010x(19980101)280:1<73::aid-jez9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PmContig18499 mapping in the 5'upstream region of the PmHhb locus using Lampetra cosmid sequences.
(0.08 MB DOC)
Detailed sequence comparisons and orthology assessments for lamprey Hh genes.
(0.39 MB DOC)
Additional phylogenetic trees
(2.82 MB DOC)
An example showing difficulty of global alignments of the lamprey genomic sequences with vertebrate sequences.
(0.15 MB DOC)