Abstract
Background.
We have recently shown that 12 weeks of progressive aerobic exercise training improves whole-muscle size and function in older women. The purpose of this investigation was to evaluate molecular markers that may be associated with muscle hypertrophy after aerobic training in aging skeletal muscle.
Methods.
Muscle biopsies were obtained before and after 12 weeks of aerobic exercise training on a cycle ergometer in nine older women (70 ± 2 years) to determine basal levels of messenger RNA and protein content of select myogenic, proteolytic, and mitochondrial factors.
Results.
The training program increased (p < .05) aerobic capacity 30 ± 9%, whole-muscle cross-sectional area 11 ± 2%, and whole-muscle force production 29 ± 8%. Basal messenger RNA levels of FOXO3A, myostatin, HSP70, and MRF4 were lower (p < .05) after aerobic training. FOXO3A, FOXO3A phosphorylation, and HSP70 protein content were unaltered after training. Mitochondrial protein COX IV was elevated (p < .05) 33 ± 7% after aerobic training, whereas PGC-1α protein content was 20 ± 5% lower (p < .05).
Conclusions.
These data suggest that reductions in FOXO3A and myostatin messenger RNA are potentially associated with exercise-induced muscle hypertrophy. Additionally, it appears that mitochondrial biogenesis can occur with aerobic training in older women independent of increased PGC-1α protein. Aerobic exercise training alters molecular factors related to the regulation of skeletal muscle, which supports the beneficial role of aerobic training for improving muscle health in older women.
Keywords: FOXO3A, Myostatin, PGC-1α, Aging, Hypertrophy
AGING is associated with significant reductions in muscle mass termed sarcopenia (1). Sarcopenia is clinically relevant as it is associated with elevated risk of cardiac, pulmonary, and metabolic disease processes. Due to these health concerns, the importance of developing interventions that reverse the negative consequences associated with aging is essential.
Aerobic exercise is commonly recommended for older adults due to the cardiovascular and metabolic health benefits (2). Despite the prevalence of aerobic exercise, there has been limited scientific inquiry regarding the effects of aerobic exercise on muscle size. We have recently shown that 12 weeks of progressive aerobic training dramatically increases muscle size in addition to improvements in muscle function and aerobic capacity in older women (3). Although these whole-body adaptations have been established, the underlying molecular mechanisms remain to be elucidated.
The ubiquitin–proteosomal pathway is responsible for the majority of protein degradation within skeletal muscle, and our laboratory has recently shown that older women exhibit greater basal proteolytic gene expression (FOXO3A, MuRF-1, and myostatin) compared with young women (4,5). Additionally, although the influence of aging on mixed skeletal muscle protein breakdown is equivocal (6,7), our laboratory has reported that myofibrillar proteolysis is elevated in older men (8). Due to the elevated myofibrillar proteolysis and related genetic markers in aging skeletal muscle, it is possible that a reduction in these proteolytic mechanisms partially contributes to the improvements in muscle health in response to exercise training.
Thus, the global purpose of this investigation was to examine a cohort of molecular factors within aging skeletal muscle that may be associated with the recently observed enhancements in muscle size and aerobic capacity after an aerobic exercise training program (3). Interestingly, markers associated with muscle oxidative capacity (ie, PGC-1α) and muscle protein preservation (ie, HSP70) have been recently linked to skeletal muscle health in animal models. Mechanistically, these factors appear to decrease protein breakdown through the ubiquitin–proteosomal pathway by inhibiting FOXO signaling (9–11). Although these relationships have not been examined in human skeletal muscle, they represent potential mechanisms for the muscle hypertrophy induced by aerobic training. Therefore, we hypothesized that resting levels of HSP70, PGC-1α, and TFAM (mitochondrial transcription factor A) would be elevated in skeletal muscle of older women after aerobic training and that these muscle oxidative factors will be associated with reductions in proteolytic markers (FOXO3A, MuRF-1, and atrogin-1).
METHODS
Participants
Nine older women (70 ± 2 years) volunteered to participate in this investigation. All participants were cleared to participate by the study physician following a thorough physical examination. Volunteers were excluded based on criteria that have been previously described (3). Written informed consent, approved by the institutional review boards of Ball State University and Ball Memorial Hospital, was obtained from each participant prior to study participation. Data from a subset (n = 7) of these participants regarding whole-muscle and single myofiber contractile physiology have been reported previously (3).
Experimental Design and Methodology
Each participant completed the experimental protocol over a period of approximately 15 weeks consisting of several visits to the laboratory for baseline measurements of aerobic capacity, body composition using dual energy x-ray absorptiometry, whole-muscle cross-sectional area, a muscle biopsy, and 42 exercise training sessions (3). All measurements were repeated after the 12-week training protocol.
Aerobic Exercise Training Protocol
Participants performed 12 weeks of aerobic training on a cycle ergometer (Stairmaster Stratus 3300 CE, Kirkland, WA) as previously described in detail (3). A total of 42 exercise sessions were performed. Duration (20–45 minutes), intensity (60%–80% heart rate reserve), and frequency (three or four sessions per week) of exercise were progressively increased throughout the 12 weeks. The last 5 weeks consisted of four 45-minute sessions at 80% intensity per week.
Aerobic Capacity
Participants performed a physician-supervised graded exercise test for the assessment of VO2max before and after the 12-week aerobic training intervention. The test was performed on an electronically braked cycle ergometer (SensorMedics Ergometrics 800, Yorba Linda, CA) beginning at a very low workload (∼10 W). After a brief warm-up at the initial workload, the workload was progressively increased 10 W every 1-minute stage until exhaustion with a total test time of 10–12 minutes. During the test, participant’s heart rate, blood pressure, rating of perceived exertion, and electrocardiogram were monitored, and ventilation and expired air samples were measured by a metabolic cart (TrueOne 2400 Metabolic System; ParvoMedics, Inc., Sandy, UT) for the determination of VO2.
Whole-Muscle Function
Peak power and peak isometric force of the knee extensor muscle group was assessed before and after the 12-week aerobic training intervention using an inertial ergometer (Inertial Technology, Stockholm, Sweden) connected to a strain gauge load cell and potentiometer interfaced with a personal computer (Gateway E-4200, Irvine, CA). Following multiple orientation sessions with the knee extensor device, participants performed three identical sessions separated by at least 2 days. All tests were bilateral. Prior to any testing, participants performed a 10-minute warm-up on a stationary bicycle at a self-selected intensity followed by small loads on the resistance apparatus. Peak isometric force was assessed at a fixed knee joint angle of 120°. For peak power and peak force, participants completed three submaximal repetitions, followed by three maximal attempts with three minutes rest between sets. The concentric power output was recorded throughout the full range of motion.
Whole-Muscle Size
Before and after the 12-week training intervention, proton magnetic resonance images of the leg were measured using a General Electric Signa 1.5 Tesla imaging system (Milwaukee, WI) at standard settings (TR/TE = 2000/9 milliseconds) as we have previously described (3,12). Images were analyzed in a blinded fashion by the same investigator via NIH Image software (version 1.60). Muscle cross-sectional area was taken as the average of each slice from the first distal image containing the medial rectus femoris and the last proximal image not containing the gluteal muscle. The cross-sectional area (square centimeters) of the whole quadriceps femoris was taken as the sum of the averaged slices for the vastii (vastus lateralis, vastus medialis, and vastus intermedius) and rectus femoris.
Skeletal Muscle Biopsy
Before and after the 12-week aerobic training intervention, a muscle biopsy was obtained from the vastus lateralis of each participant. The posttraining biopsy sample occurred ∼48 hours after the last exercise session. Tissue was obtained following local anesthetic (Lidocaine HCl 1%) using a 5-mm Bergstrom needle with suction. One piece (∼15 mg) was placed in RNAlater and stored at −20°C until RNA extraction, whereas the other pieces were immediately frozen and stored in liquid nitrogen.
Gene Expression
Total RNA extraction and RNA quality check.—
Total RNA was extracted in TRI reagent (Molecular Research Center, Cincinnati, OH). The quality and integrity (RNA Integrity Number of 8.4 ± 0.07) of extracted RNA (158.1 ± 17.17 ng/μL) was evaluated using an RNA 6000 Nano LabChip kit on Agilent 2100 Bioanalyzer (Wilmington, DE).
Reverse transcription and real-time polymerase chain reaction.—
Oligo(dT) primed first-strand cDNA was synthesized (150ng total RNA) using SuperScript II reverse transcription (Invitrogen, Carlsbad, CA). Quantification of messenger RNA (mRNA) transcription (in duplicate) was performed in a 72-well Rotor-Gene 3,000 Centrifugal Real-Time Cycler (Corbett Research, Mortlake, New South Wales, Australia). Housekeeping gene GAPDH was used as a reference gene (5). The validation of GAPDH was performed to ensure that its expression was unaffected by the experimental treatment as we have previously described (13,14). All primers used in this study were mRNA specific (on different exons and/or over an intron) and were designed for SYBR Green chemistry using Vector NTI Advance 9 software (Invitrogen). Primers for MyoD, myogenin, muscle regulatory factor 4 (MRF4), atrogin-1, muscle RING-finger protein-1 (MuRF-1), forkheadbox (FOXO)3A, peroxisome proliferator–activated receptor-gamma coactivator 1-α (PGC-1α), and mitochondrial transcription factor (TFAM) have been reported previously by our laboratory (13–15). The primer sequence for heat-shock protein 70 (HSP70, NM_005345.5) is as follows: forward primer, 5′-CCACGGACAAGAGCACCGGCAA-3′, and reverse primer, 3′-CGCCTCCTGCACCATGCGCTC-5′. A melting curve analysis was generated validating that only one product was present. The details regarding reverse transcription and polymerase chain reaction parameters have been reported previously (5,14).
Relative quantitative polymerase chain reaction data analysis.—
Relative gene expression analysis comparing the expression of a gene of interest in relation to a reference gene is based on the distinct cycle (Ct) differences. The 2−ΔCT method (16) was used in this study as described in detail previously (4,5).
Western Blotting
Muscle samples (∼20 mg) were homogenized using an electrically powered glass pestle in 40 volumes of cold RIPA buffer (Pierce, Rockford, IL) with Halt Protease Inhibitor Cocktail, Halt Phosphatase Inhibitor Cocktail, and 0.5 M EDTA (Pierce). Proteins (20 μg) were separated with a 4%–20% gradient gel (Pierce) using sodium dodecyl sulfate–polyacrylamide gel electrophoresis for 90 minutes at 100 V (Mini Protean 3 system; Bio-Rad Laboratories, Hercules, CA) and then transferred to polyvinylidene flouride membrane (Immobilon-P; Millipore, Bedford, MA) for 2 hours at 200 mA at 4°C. The membrane was blocked with 5% milk for 60 minutes and then incubated with a human primary antibody (PGC-1α #2178S, HSP70 #4872, FOXO3A #9467, phopsho-FOXO3A Serine 253 #9466, or COX IV #4844; Cell Signaling, Danvers, MA) at 4°C overnight. Blots were identified by incubation with horseradish peroxidase–conjugated secondary antibody (#7074, Cell Signaling) and then exposed to an enhanced chemiluminescent substrate (Pierce ECL Western Blotting Substrate or Amersham ECL Plus Western Blotting Detection System, GE, Milwaukee, WI). Digital images were captured using a chemiluminescent imaging system (FluorChem SP; Alpha Innotech, San Leandro, CA). Phosphorylation of FOXO3A was determined by normalizing the expression of the phosphorylated protein to total protein content. Sizes of the immunodetected proteins were confirmed by molecular weight markers (see Blue & Magic Marker; Invitrogen). Equal protein loading was established by results from a bicinchoninic acid protein assay and confirmed by Ponceau S staining. Also, a control sample was included on each blot to serve as an internal standard to verify uniform transfer and protein detection. Both pre- and posttime points for each participant were analyzed on the same blot to control for interassay variability.
Statistics
Pre- and posttraining values for all variables were analyzed using a paired two-tailed Student’s t test. Significance was accepted as p < .05. All values are presented as mean ± SE.
RESULTS
Body Composition
All participants maintained body weight (p > .05) throughout the training program. Although body weight was unchanged, percent body fat decreased (p < .05) while fat-free mass and lean body mass increased (p < .05) after training (Table 1).
Table 1.
Body Composition Before (PRE) and After (POST) 12 Weeks of Aerobic Exercise Training
| PRE | POST | |
| Weight (kg) | 67.2 ± 4.1 | 67.5 ± 3.9 |
| Body fat (%) | 41.4 ± 2.7 | 40.4 ± 2.9* |
| Fat-free mass (kg) | 39.0 ± 1.1 | 39.5 ± 1.0* |
| Lean body mass (kg) | 36.7 ± 1.0 | 37.2 ± 1.0* |
Notes: All values are presented in mean ± SE. *p < .05 vs PRE.
Aerobic Capacity
Absolute VO2max increased (p < .05) 30 ± 9% from 1.07 ± 0.05 to 1.38 ± 0.10 L/min. Additionally, maximum workload during the graded cycle test increased (p < .05) 43% (87 ± 9 to 122 ± 10 W) after aerobic training (Table 2).
Table 2.
Whole-Body Adaptations to 12 Weeks of Aerobic Training in Older Women
| PRE | POST | |
| Graded exercise test | ||
| VO2max (L/min) | 1.07 ± 0.05 | 1.38 ± 0.10* |
| Maximum workload (W) | 87 ± 9 | 122 ± 10* |
| Whole-muscle size | ||
| Cross-sectional area (cm2) | 40 ± 3 | 44 ± 3* |
| Whole-muscle function | ||
| Knee extensor isometric force (Nm) | 247 ± 34 | 301 ± 35* |
| Knee extensor peak power (W) | 273 ± 49 | 328 ± 50* |
Notes: All values are presented in mean ± SE. *p < .05 vs PRE.
Whole-Muscle Size and Function
Cross-sectional area of the quadriceps femoris increased 11 ± 2% (p < .05) in response to 12 weeks of aerobic training (Table 2). Functional capacity of the knee extensor muscles, measured as isometric force and peak power during knee extension, improved (p < .05) 28 ± 8% and 25 ± 4%, respectively.
Basal mRNA Expression
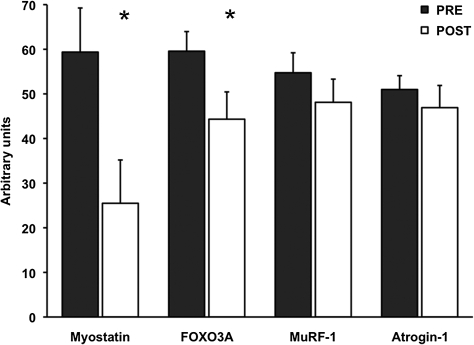
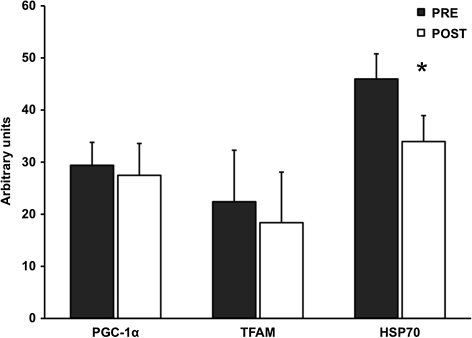
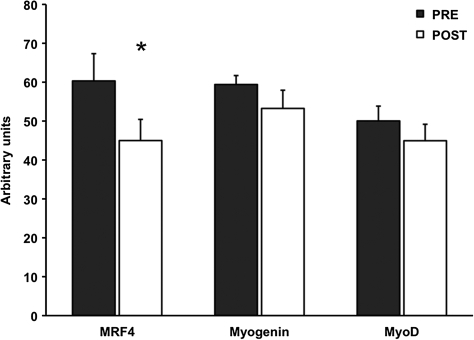
For the proteolytic genes, FOXO3A was reduced 24 ± 9% (p < .05), whereas atrogin-1 and MuRF-1 were unaltered by aerobic training. Myostatin mRNA expression was decreased (p < .05) 49 ± 17% after the aerobic training program (Figure 1). Additionally, HSP70 mRNA was 23 ± 8% lower (p < .05), whereas PGC-1α and TFAM mRNA were not altered after the training intervention (Figure 2). Resting levels of myogenic gene MRF4 was decreased (p < .05) 22 ± 8%, and myogenin trended to decrease (p = .09) 11 ± 6% after training (Figure 3).
Figure 1.
Basal proteolytic mRNA expression before (PRE) and after (POST) 12 weeks of progressive aerobic exercise training. Data are presented in mean ± SE. *p < .05 compared to PRE.
Figure 2.
Basal levels of PGC-1α, TFAM, and HSP70 mRNA before (PRE) and after (POST) 12 weeks of progressive aerobic training. Data are presented in mean ± SE. *p < .05 compared to PRE.
Figure 3.
Myogenic mRNA expression before (PRE) and after (POST) 12 weeks of progressive aerobic exercise training. Data are presented in mean ± SE. *p < .05 compared to PRE.
Protein Expression
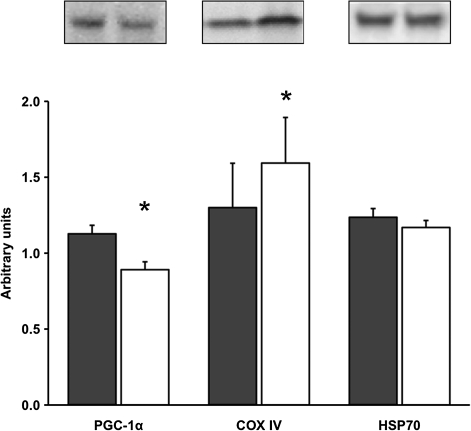
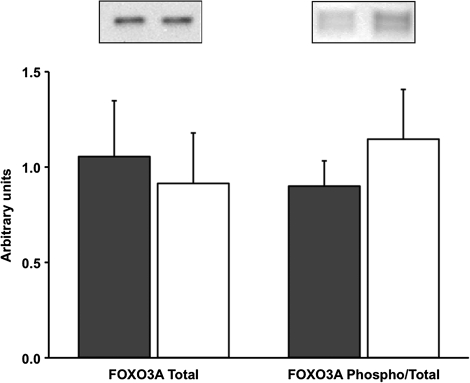
COX IV protein content increased (p < .05) 33 ± 7%, whereas basal levels of PGC-1α protein decreased (p < .05) 20 ± 5% after the aerobic training intervention. There were no changes in the protein content of HSP70, FOXO3A, or FOXO3A phosphorylation after 12 weeks of progressive aerobic training in older women (Figures 4 and 5).
Figure 4.
Basal levels of PGC-1α, COX IV, and HSP70 protein content before (PRE) and after (POST) 12 weeks of progressive aerobic training. A representative Western blot is shown for each respective protein. Data are presented in mean ± SE. *p < .05 compared to PRE.
Figure 5.
Basal levels of FOXO3A total protein content and FOXO3A phosphorylation normalized to total FOXO3A protein before (PRE) and after (POST) 12 weeks of progressive aerobic exercise training. A representative Western blot is shown for each respective protein. Data are presented in mean ± SE.
DISCUSSION
The purpose of this investigation was to elucidate the cellular factors within skeletal muscle that are associated with improvements of skeletal muscle size, function, and aerobic capacity after an aerobic training intervention in older women. The main findings from this investigation were that aerobic training: (a) reduced basal FOXO3A and myostatin mRNA expression, although FOXO3A total protein content and phosphorylation status remained unchanged; (b) reduced basal HSP70 mRNA without a change in HSP70 protein content; and (c) increased mitochondrial protein (COX IV) but reduced PGC-1α protein content. These findings suggest that downregulation of these proteolytic mechanisms may be partially responsible for the training-induced muscle hypertrophy in older women. Furthermore, these data indicate that improvements in whole-body oxidative capacity and muscle mitochondrial content can be enhanced with aerobic training independent of increases in PGC-1α.
Molecular adaptations to aerobic exercise training in older participants are limited in the literature. FOXO3A and myostatin represent attractive targets because they have been implicated in sarcopenia (4,5,17,18). As hypothesized, we observed a decrease in FOXO3A mRNA after training. We also observed a reduction in myostatin mRNA expression, a known potent inhibitor of muscle growth (19). Data from our laboratory suggest that at rest untrained older women display greater expression of these catabolic genes (FOXO3A and myostatin) and exhibit lower muscle mass than their younger counterparts (4,5). Furthermore, it has been purported that elevated levels of myostatin are associated with skeletal muscle atrophy in aging (18) and chronic obstructive pulmonary disease participants (20), whereas a reduction in myostatin mRNA is associated with muscle hypertrophy following resistance training (21). Although the relationship between FOXO3A and myostatin in sarcopenia (4,5,18) and exercise-induced muscle hypertrophy (21) is not clear, their roles in skeletal muscle mass regulation likely target different aspects of protein balance. A decline of FOXO3A mRNA suggests a reduction in muscle protein degradation that is elevated in aging skeletal muscle (8), whereas a reduction of myostatin may indicate an elevation in protein synthesis (22,23). Therefore, it is possible the reduction of myostatin (ie, increase protein synthesis) and FOXO3A (ie, decrease protein breakdown) may act synergistically to create a favorable environment for the muscle hypertrophy induced by aerobic exercise in older women.
Despite reductions in FOXO3A mRNA, we reported no change in FOXO3A total protein content and phosphorylation status. Generally, alterations in mRNA expression are assumed to represent a new demand placed on the cell for the corresponding protein. However, this assumption is not often verified in human skeletal muscle after acute or chronic exercise. To our knowledge, this is the first study to report decreases in the basal levels of FOXO3A mRNA without changes in related protein content after an exercise training program. Urso et al. (24) have also presented discrepancies between mRNA expression and protein content when investigating skeletal muscle in individuals with spinal cord injury. The reason for this disparity between mRNA and protein levels is not readily apparent. We measured total and phospho-FOXO3A in the whole cell lysate, which does not delineate the specific location (ie, cytosol or nucleus) of FOXO3A within the myofiber. Our laboratory has recently shown that resistance training alters the cellular location of FOXO3A (25), which influences its proteolytic activity (26,27). Fujita et al. (28) have shown that aerobic exercise acutely restores the muscle’s anabolic response to insulin through AKT signaling, which may also alter FOXO3A signaling and reduce proteolytic gene expression in the fed state following each exercise session. It should be noted that the posttraining muscle sample was obtained at least 48 hours after the last exercise session in the fasted state to avoid any transient changes of FOXO3A mRNA or signaling from an acute exercise bout as we have previously shown (14,15). For this reason, we are confident that our measure represents a new steady-state level within the muscle.
PGC-1α has also been implicated in reducing muscle protein breakdown by attenuating FOXO3A signaling (11). Therefore, we measured PGC-1α mRNA and protein expression due to its potential role in reducing atrophy through FOXO as well as being a regulator of mitochondrial biogenesis. Interestingly, we observed a 20% reduction in PGC-1α protein, although COX IV was elevated 33 ± 7% after training in our older women, suggesting the training program resulted in mitochondrial biogenesis (29). Acute and chronic aerobic exercise has been shown to increase PGC-1α gene and protein levels in mouse (30,31) and human (32–34) models. PGC-1α is widely believed to be essential for controlling exercise-induced mitochondrial biogenesis (35). However, there are limited data related to PGC-1α protein in older participants. A cross-sectional investigation recently reported that PGC-1α content was higher in skeletal muscle of older aerobically trained than sedentary participants (36). Consequently, we hypothesized that aerobic training would increase PGC-1α in our older women. Our findings may differ from the previous study due to the dissimilarity in study design and participant training history. However, the most unique difference was our stimulus simultaneously induced muscle hypertrophy and increased aerobic capacity. All the above-mentioned differences between investigations could create divergent environments for adaptations to occur.
Although our observed decrease in PGC-1α protein was unexpected, recent research has revealed that PGC-1α protein is not required to initiate adaptations of mitochondrial proteins (37,38). The decrease in PGC-1α protein is uncharacteristic of aerobic exercise in young participants but potentially relates to the effects of the observed muscle hypertrophy, or the elevation of PGC-1α protein expression is not mandatory to induce mitochondrial biogenesis (37). Most importantly, on a global aspect, our participants were able to increase their aerobic capacity by 30 ± 9% without increased PGC-1α mRNA or protein expression.
We also examined HSP70 mRNA and protein content due to its possible role in the preservation of skeletal muscle during atrophic stimuli through FOXO signaling (10,39). Our data do not support this connection in skeletal muscle of older women as we observed a decrease in HSP70 mRNA, although the related protein content of HSP70 was not affected after 12 weeks of aerobic training. HSP70 is responsive to acute aerobic exercise (40); however, it has been reported that aerobic training status may not influence HSP70 protein levels (41). HSP70 is one essential component in remodeling skeletal muscle by repairing and protecting proteins from future insult (42). Thus, a reduction in HSP70 mRNA may reflect a new steady-state level of reduced basal stress in skeletal muscle of older women after aerobic training.
With regard to the myogenic genes, MRF4 decreased and myogenin trended to decrease after aerobic training. Interestingly, basal levels of MyoD, MRF4, and myogenin are elevated in older participants, presumably to offset the increased basal stress that induces muscle loss seen in sarcopenic muscle (5,43). The decrease in myogenic mRNA (MRF4 and myogenin) in our study could potentially be linked to diminished proteolytic machinery. Thus, there may be less need for MRF4 and myogenin to offset elevated proteolysis regulated by catabolic markers (FOXO3A and myostatin).
The aim of this investigation was to examine basal levels of mRNA expression and related protein content of factors associated with whole-muscle adaptations after aerobic exercise training in older women. Consequently, there are inherent limitations when describing skeletal muscle adaptations before and after a 12-week exercise intervention from a relatively small number of older participants (n = 9). Although we based our posttraining biopsy time point on previous investigations (14,25), we acknowledge that acquiring muscle samples 48 hours after training may not have captured alterations in mRNA transcript levels, protein content, or posttranslational modifications that were present at other time points. These limitations reiterate why investigations attempting to connect molecular and whole-muscle adaptations are scarce within the aging literature and warrant additional research.
In conclusion, the current data suggest that aerobic training alters basal levels of growth-related mRNA and proteins within skeletal muscle of older women. More specifically, the decreased mRNA of FOXO3A and myostatin are two factors potentially associated with the exercise-induced muscle hypertrophy. Additionally, these data provide evidence that the enhancement of mitochondrial markers following aerobic training can occur despite decreased PGC-1α protein in older women. These molecular adaptations following aerobic exercise training support the beneficial adaptations seen in the myocellular and whole-muscle environments (3). Therefore, an aerobic training intervention provides an exercise prescription that dynamically combats the debilitating effects of sarcopenia in sedentary older individuals at the molecular, cellular, and whole-muscle levels.
FUNDING
This work was supported by the National Institute of Aging at the National Institutes of Health (AG32127 to M.P.H.).
References
- 1.Doherty TJ. Invited review: aging and sarcopenia. J Appl Physiol. 2003;95(4):1717–1727. doi: 10.1152/japplphysiol.00347.2003. [DOI] [PubMed] [Google Scholar]
- 2.Nelson ME, Rejeski WJ, Blair SN, et al. Physical activity and public health in older adults: recommendation from the American College of Sports Medicine and the American Heart Association. Circulation. 2007;116(9):1094–1105. doi: 10.1161/CIRCULATIONAHA.107.185650. [DOI] [PubMed] [Google Scholar]
- 3.Harber MP, Konopka AR, Douglass MD, et al. Aerobic exercise training improves whole muscle and single myofiber size and function in older women. Am J Physiol Regul Integr Comp Physiol. 2009;297(5):R1452–R1459. doi: 10.1152/ajpregu.00354.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci. 2007;62(12):1407–1412. doi: 10.1093/gerona/62.12.1407. [DOI] [PubMed] [Google Scholar]
- 5.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18-30 yr) and old (80-89 yr) women. J Appl Physiol. 2006;101(1):53–59. doi: 10.1152/japplphysiol.01616.2005. [DOI] [PubMed] [Google Scholar]
- 6.Volpi E, Mittendorfer B, Rasmussen BB, Wolfe RR. The response of muscle protein anabolism to combined hyperaminoacidemia and glucose-induced hyperinsulinemia is impaired in the elderly. J Clin Endocrinol Metab. 2000;85(12):4481–4490. doi: 10.1210/jcem.85.12.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volpi E, Sheffield-Moore M, Rasmussen BB, Wolfe RR. Basal muscle amino acid kinetics and protein synthesis in healthy young and older men. JAMA. 2001;286(10):1206–1212. doi: 10.1001/jama.286.10.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trappe T, Williams R, Carrithers J, et al. Influence of age and resistance exercise on human skeletal muscle proteolysis: a microdialysis approach. J Physiol. 2004;554(Pt 3):803–813. doi: 10.1113/jphysiol.2003.051755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodd S, Hain B, Judge A. Hsp70 prevents disuse muscle atrophy in senescent rats. Biogerontology. 2009;10(5):605–611. doi: 10.1007/s10522-008-9203-1. [DOI] [PubMed] [Google Scholar]
- 10.Senf SM, Dodd SL, McClung JM, Judge AR. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. FASEB J. 2008;22(11):3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandri M, Lin J, Handschin C, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103(44):16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trappe TA, Lindquist DM, Carrithers JA. Muscle-specific atrophy of the quadriceps femoris with aging. J Appl Physiol. 2001;90(6):2070–2074. doi: 10.1152/jappl.2001.90.6.2070. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Creer A, Jemiolo B, Trappe S. Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J Appl Physiol. 2005;98(5):1745–1752. doi: 10.1152/japplphysiol.01185.2004. [DOI] [PubMed] [Google Scholar]
- 14.Louis E, Raue U, Yang Y, Jemiolo B, Trappe S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol. 2007;103(5):1744–1751. doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- 15.Harber MP, Crane JD, Dickinson JM, et al. Protein synthesis and the expression of growth-related genes are altered by running in human vastus lateralis and soleus muscles. Am J Physiol Regul Integr Comp Physiol. 2009;296(3):R708–R714. doi: 10.1152/ajpregu.90906.2008. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab. 2005;288(6):E1110–E1119. doi: 10.1152/ajpendo.00464.2004. [DOI] [PubMed] [Google Scholar]
- 18.Leger B, Derave W, De Bock K, Hespel P, Russell AP. Human sarcopenia reveals an increase in SOCS-3 and myostatin and a reduced efficiency of Akt phosphorylation. Rejuvenation Res. 2008;11(1):163–175B. doi: 10.1089/rej.2007.0588. [DOI] [PubMed] [Google Scholar]
- 19.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 20.Plant PJ, Brooks D, Faughnan M, et al. Cellular markers of muscle atrophy in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2010;42(4):461–471. doi: 10.1165/rcmb.2008-0382OC. [DOI] [PubMed] [Google Scholar]
- 21.Roth SM, Martel GF, Ferrell RE, Metter EJ, Hurley BF, Rogers MA. Myostatin gene expression is reduced in humans with heavy-resistance strength training: a brief communication. Exp Biol Med (Maywood) 2003;228(6):706–709. doi: 10.1177/153537020322800609. [DOI] [PubMed] [Google Scholar]
- 22.Amirouche A, Durieux AC, Banzet S, et al. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology. 2009;150(1):286–294. doi: 10.1210/en.2008-0959. [DOI] [PubMed] [Google Scholar]
- 23.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296(6):C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 24.Urso ML, Chen YW, Scrimgeour AG, Lee PC, Lee KF, Clarkson PM. Alterations in mRNA expression and protein products following spinal cord injury in humans. J Physiol. 2007;579(Pt 3):877–892. doi: 10.1113/jphysiol.2006.118042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson DL, Raue U, Slivka DR, Trappe S. Resistance exercise, skeletal muscle FOXO3A, and 85-year-old women. J Gerontol A Biol Sci Med Sci. 2010;65(4):335–343. doi: 10.1093/gerona/glq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandri M, Sandri C, Gilbert A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stitt TN, Drujan D, Clarke BA, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 28.Fujita S, Rasmussen BB, Cadenas JG, et al. Aerobic exercise overcomes the age-related insulin resistance of muscle protein metabolism by improving endothelial function and Akt/mammalian target of rapamycin signaling. Diabetes. 2007;56(6):1615–1622. doi: 10.2337/db06-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hood DA. Invited review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90(3):1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 30.Baar K, Wende AR, Jones TE, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16(14):1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 31.Terada S, Tabata I. Effects of acute bouts of running and swimming exercise on PGC-1alpha protein expression in rat epitrochlearis and soleus muscle. Am J Physiol Endocrinol Metab. 2004;286(2):E208–E216. doi: 10.1152/ajpendo.00051.2003. [DOI] [PubMed] [Google Scholar]
- 32.Mathai AS, Bonen A, Benton CR, Robinson DL, Graham TE. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. J Appl Physiol. 2008;105(4):1098–1105. doi: 10.1152/japplphysiol.00847.2007. [DOI] [PubMed] [Google Scholar]
- 33.Russell AP, Feilchenfeldt J, Schreiber S, et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52(12):2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 34.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J Physiol. 2003;546(Pt 3):851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 36.Lanza IR, Short DK, Short KR, et al. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57(11):2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. J Biol Chem. 2007;282(1):194–199. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- 38.Leick L, Wojtaszewski JF, Johansen ST, et al. PGC-1alpha is not mandatory for exercise- and training-induced adaptive gene responses in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2008;294(2):E463–E474. doi: 10.1152/ajpendo.00666.2007. [DOI] [PubMed] [Google Scholar]
- 39.Dodd S, Hain B, Judge A. Hsp70 prevents disuse muscle atrophy in senescent rats. Biogerontology. 2009;10(6):605–611. doi: 10.1007/s10522-008-9203-1. [DOI] [PubMed] [Google Scholar]
- 40.Khassaf M, Child RB, McArdle A, Brodie DA, Esanu C, Jackson MJ. Time course of responses of human skeletal muscle to oxidative stress induced by nondamaging exercise. J Appl Physiol. 2001;90(3):1031–1035. doi: 10.1152/jappl.2001.90.3.1031. [DOI] [PubMed] [Google Scholar]
- 41.Morton JP, Maclaren DP, Cable NT, et al. Trained men display increased Basal heat shock protein content of skeletal muscle. Med Sci Sports Exerc. 2008;40(7):1255–1262. doi: 10.1249/MSS.0b013e31816a7171. [DOI] [PubMed] [Google Scholar]
- 42.Morton J. The exercise-induced stress response of skeletal muscle, with specific emphasis on humans. Sports Med. 2009;39(8):643–662. doi: 10.2165/00007256-200939080-00003. [DOI] [PubMed] [Google Scholar]
- 43.Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol. 2005;99(6):2149–2158. doi: 10.1152/japplphysiol.00513.2005. [DOI] [PubMed] [Google Scholar]