Abstract
The regulation of transcription and of many other cellular processes involves large multi-subunit protein complexes. In the context of transcription, it is known that these complexes serve as regulatory platforms that connect activator DNA-binding proteins to a target promoter. However, there is still a lack of understanding regarding the function of these complexes. Why do multi-subunit complexes exist? What is the molecular basis of the function of their constituent subunits, and how are these subunits organized within a complex? What is the reason for physical connections between certain subunits and not others? In this article, I address these issues through a model of network allostery and its application to the eukaryotic RNA polymerase II Mediator transcription complex. The multiple allosteric networks model (MANM) suggests that protein complexes such as Mediator exist not only as physical but also as functional networks of interconnected proteins through which information is transferred from subunit to subunit by the propagation of an allosteric state known as conformational spread. Additionally, there are multiple distinct sub-networks within the Mediator complex that can be defined by their connections to different subunits; these sub-networks have discrete functions that are activated when specific subunits interact with other activator proteins.
Keywords: Mediator, Graph theory, Networks, Transcription
Introduction
An outstanding problem in studies of transcriptional regulation and of many other biological processes is the lack of a complete understanding of multi-subunit protein complexes. Although it is well known that these complexes exist as discrete physical entities, it is not clear how subunits function within a complex. Why do complexes exist? What is the basis of their structure and subunit organization? More specifically, why do certain subunits connect to only a subset of other proteins in the complex? Why are some subunits not essential to the function of a given complex? The answers to these questions probably lie beyond the idea that complexes serve simply to localize several proteins at a point of functional interest. Rather, to accurately interpret data on such complexes — which in some cases approach 2 MDa in size and consist of 20 or more subunits — we need a more comprehensive conceptual framework.
Systems biology and, more specifically, graph theory have been widely applied to help understand biological phenomena (Barabasi and Oltvai, 2004). Graph theory can be used to describe all of the interacting proteins in a cell as networks, similar to the manner in which it can be applied to describe the World Wide Web, an electronic circuit or a genetic regulatory network. Networks consist of nodes that are connected by edges. In a genetic network, the nodes are genes and the connecting edges are regulatory transcription factors (Barabasi and Oltvai, 2004). Spirin and Mirny studied a network of protein interactions in yeast and used their data to construct highly connected graphs consisting of proteins (nodes) and protein-protein interactions (edges) (Spirin and Mirny, 2003). They found that the transcription factor IID (TFIID) complex, the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex, the CCR4-NOT complex and the eukaryotic RNA polymerase II Mediator transcription complex are among protein complexes that have more connections between their constituent subunits than they have with other proteins. These categorizations are based on physical interactions and are in agreement with the biochemical evidence that these factors exist as complexes.
This Hypothesis article takes this interpretation further by hypothesizing what is referred to here as the multiple allosteric networks model (MANM). By using Mediator (Box 1) as an example, the MANM proposes that multi-subunit protein complexes contain multiple regulatory networks and are not simply the sum of several protein-protein interactions. These multiple networks are defined by sets of protein-protein interactions and by the allosteric states that can be adopted by the proteins involved. Furthermore, these networks have diverse outputs that are manifested by differing final conformations of the complex, and these structural outputs are triggered by the binding of different transcriptional activator proteins. The concept that complexes contain multiple allosteric networks offers answers to the questions posed at the beginning of this section.
Box 1. Mediator
The Mediator complex facilitates the interaction of a transcriptional activator to the general transcriptional machinery at the promoter of RNA-polymerase-II-dependent genes (Chadick and Asturias, 2005; Conaway et al., 2005; Kim and Lis, 2005; Kornberg, 2005; Lewis and Reinberg, 2003; Malik and Roeder, 2005). In general, transcriptional activators physically bind to a specific subunit of Mediator and thereby recruit the entire Mediator complex to the target promoter. Subsequently, additional members of the transcriptional machinery are recruited, culminating in the initiation of transcription. Genetic, biochemical and structural data have revealed that the structure of Mediator comprises several modules (head, middle, tail and CDK8) (Chadick and Asturias, 2005; Conaway et al., 2005; Kim and Lis, 2005; Kornberg, 2005; Lewis and Reinberg, 2003; Malik and Roeder, 2005). Furthermore, it has been shown that Mediator can adopt various conformations that are induced by the binding of different transcriptional activators to different Mediator subunits (Taatjes et al., 2002).
Functionally, a multi-subunit protein complex serves to transmit information: it is an information-processing center whose output is in the form of a specific three-dimensional structure. But what are the molecular mechanisms that underpin this information transfer? Allostery has been invoked to explain a variety of protein-protein interactions in signal transduction pathways (Bray and Duke, 2004; Dueber et al., 2004; Duke et al., 2001). Indeed, the propagation of allosteric states (which equate to intra-complex signaling pathways) has previously been termed the conformational state (CS) model (Fig. 1). The CS model extends classic models of allostery to explain conformational changes that occur in large protein complexes such as chemotaxic receptors, flagella and actin filaments. The MANM proposed here combines the CS model with the idea that protein complexes comprise multiple functional networks, and uses principles of the CS model to explain the mechanism of information transmission within the protein complex (Fig. 1). These allosteric states within the Mediator complex are triggered by the binding of external activator proteins to subunits of the Mediator complex. In turn, the allosteric conformational state induced by this activating event induces an allosteric state in a neighboring subunit within the complex. The allosteric state is thus propagated through the complex, from subunit to subunit, resulting in a final conformational state that serves as an output signal. Although I focus here on the Mediator complex (Box 1), it is probable that the MANM can be used as a general model to explain the function of other large protein complexes.
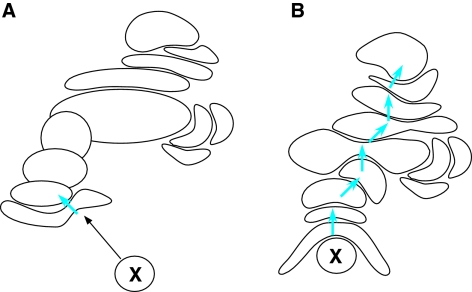
Fig. 1.
Schematic representation of the multiple allosteric network model, illustrating the concept of conformational spread. (A) Activating factor X (either a DNA-binding protein, protein or DNA ligand, or post-translational modification) binds to or modifies a specific binding site on the complex. This binding induces a conformational change (blue arrow) in the subunit bound by X. (B) This, in turn, induces a particular conformational change in a connected protein subunit. The end result is an altered conformation of the complex, which has particular output functions that affect transcription-initiation events at a target promoter. Note that some subunits might not participate in the conformational spread due triggered factor X, but might undergo changes after binding another factor to another subunit, which would induce a second unique conformational state. Examples of different conformational states of Mediator have been published (Naar et al., 2002; Taatjes et al., 2002; Taatjes et al., 2004; Taatjes and Tjian, 2004).
Physical evidence of allosteric pathways in individual proteins
A premise of the MANM is that networks within a protein complex must exist. In fact, there have been several statistical modelings of individual proteins as networks (Amitai et al., 2004; del Sol et al., 2006; del Sol and O'Meara, 2005; Greene and Higman, 2003). Also, for allosteric changes in a protein to occur, residues involved in the allosteric pathway within each protein subunit are predicted to be energetically coupled and evolutionarily co-evolving (Lockless and Ranganathan, 1999). Ranganathan and colleagues exploited this prediction and calculated an evolutionary conservation algorithm to test their idea of functional and evolutionary coupling between any two residues within a protein. These analyses permitted the mapping of physical connections between one domain of a protein and another. They suggested that energetically coupled residues could represent allosteric networks within proteins (Lockless and Ranganathan, 1999). To develop this idea further, they examined three protein families that are known to undergo allosteric changes. They found that certain residues within individual proteins were linked together to form a physical network, and provided a communication link between functional regions in the proteins (Suel et al., 2003). These ideas were examined experimentally in their studies of the liver X receptor (LXR) nuclear receptor. Using their algorithm, they identified statistically coupled residues in LXR and showed by mutagenesis that all of them were functionally involved in ligand binding. However, of these residues, only some were part of the ligand-binding domain, and others were topologically separate from the ligand-binding domain. Therefore, the authors suggested that these residues — only some of which were physically involved in ligand binding — represent an allosteric network. (Shulman et al., 2004). As evolutionarily conserved residues are found in several other ligand-binding proteins (including G-protein-coupled receptors, hemoglobin and serine proteases), allosteric networks probably also exist in other proteins that bind a ligand and that undergo conformational changes after ligand binding (see below). These analyses illustrate that intra-protein allosteric pathways exist and can be functionally identified. Furthermore, it is these principles that suggest that allosteric pathways can also traverse protein subunits in a larger complex.
Interpreting genetic data on Mediator in the context of the MANM
One of the most interesting findings regarding the genetic analysis of Mediator is that the mutation of different Mediator subunits causes disparate effects — that is, some subunits are essential and some are not. It has been shown that the non-essential subunits have activator-specific roles (that is, they interact only with certain activators) whereas the essential subunits have more widespread, pleiotropic functions or serve as scaffolds for the physical assembly of the complex. The different functions of Mediator subunits have been addressed in other articles (Bjorklund and Gustafsson, 2005; Chadick and Asturias, 2005; Conaway et al., 2005; Kim and Lis, 2005; Kornberg, 2005; Malik and Roeder, 2005); however, these previous interpretations do not fully explain how the subunits are organized or offer a framework that can be used to interpret the genetic and structural data on Mediator.
As mentioned above, these data can be interpreted using the MANM by considering Mediator as a set of several functional networks. A property of the MANM (and of networks in general) is that, if genetic experiments indicate that a particular subunit is essential, it probably represents a ‘hub’ — a highly connected protein that is the nexus for all information transfer to and/or from outlying nodes. However, deletions in Mediator-complex proteins that act as peripheral nodes do not disable the entire network and only have effects on the expression of a subset of genes, as reflected by the specific phenotypes of organisms carrying such mutations (Dotson et al., 2000; Jiang et al., 1995; Jiang and Stillman, 1995; Lee et al., 1999; Myers et al., 1998). To further illustrate this point, one can consider the correlation between highly linked proteins and lethality in other types of networks. A study of protein networks in yeast found that 62% of genes encoding highly linked proteins (which amounted to only 0.7% of total interacting proteins) were essential, whereas only 21% of proteins with few links (93% of total interacting proteins) were essential (Jeong et al., 2001).
The fact that the mutation of some Mediator subunits does not disable the function of the complex suggests that, similar to networks such as the Internet, complexes-as-networks are robust. Robust networks are resistant to defects in the majority of their nodes because most nodes connect only to a few other nodes (mainly peripheral nodes). Robustness, then, implies that there is an organization to the network complex, wherein the hub protein is central to the function of the network, much like a server in a computer network. Peripheral nodes have more specialized functions that are not used in every information-processing event, and are probably members of only a subset of the networks contained in the complex.
Genetic data suggests that there are several hubs within the Mediator complex, on the basis of the phenotypic outcome of deleting the genes that encode certain subunits. Five of the eight subunits that make up the head module are essential, and mutations in several of these subunits have general defects in transcription (Myers and Kornberg, 2000). Consistent with its essential functions, the head module contacts RNA polymerase II extensively (Asturias et al., 1999; Davis et al., 2002; Dotson et al., 2000). The middle module connects to the head and has eight subunits, four of which are essential (Myers and Kornberg, 2000); these four subunits are therefore potential hubs. The subunits in the tail module (which connects to the middle module) are a discrete unit that forms extensive connections among its own subunits (Myers and Kornberg, 2000). Curiously, only one of the five tail-module subunits is essential, although several of the subunits interact physically with activators (Myers and Kornberg, 2000). The single essential tail subunit is Rgr1 (known as Med14 in mammals), which seems to act as a connecting node for the tail module because it anchors the tail to the rest of the Mediator complex (Myers and Kornberg, 2000). The capacity of the tail module to interact with activators indicates that the tail is at least one starting point for the allosteric-propagation signal. However, the finding that most subunits in the tail module are not essential suggests that they are peripheral nodes.
A detailed mapping of the subunit interactions that occur within the Mediator complex has been reported for yeast Mediator (Guglielmi et al., 2004). On the basis of these data, putative hubs can be predicted, and the directionality of the allosteric signal can be discerned using genetic data (Fig. 2). For example, only subunits Med17 and Med21 have connections with six other subunits, including each other (Guglielmi et al., 2004). Both are essential for viability in yeast (Myers and Kornberg, 2000). Therefore, it is likely that Med17 (head module) and Med21 (middle module) each act as a hub, or perhaps act together as a double hub, and are central for conformational changes in Mediator.
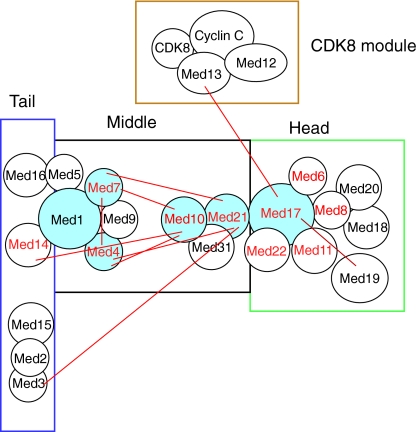
Fig. 2.
Schematic of potential nodes and correlation of nodes to mutant lethality in the Mediator complex. Physical interactions between subunits are indicated by both overlapping circles and/or red lines joining subunits (Guglielmi et al., 2004). The subunit names in red have been shown to be essential in genetic studies (Myers and Kornberg, 2000), and subunits shaded in blue are predicted hubs. Boxes are used to separate subunits that belong to the four different modules of Mediator. Note the high interconnectivity between the hub subunits, indicative of an assortative network. Interaction data for all subunits is not yet available (Guglielmi et al., 2004).
Extending from the Med17 and Med21 hubs are ten connections to other subunits. Med21 is particularly interesting because it is the sole connection between the head module (via Med17) and the rest of the Mediator complex. Therefore, all information transferred to the head module passes through Med21. Furthermore, Med21 directly connects to the head, tail and middle modules, underscoring its position as a hub within the network. Finally, Med21 also connects to three other putative hubs: Med10, Med7 and Med4. Each of these hubs makes a further five connections to other subunits. Additionally, of the Mediator subunits that connect to five or six other subunits, four are essential (Med17, Med21, Med7 and Med4). Of the two Mediator subunits that connect to three other subunits, one of them is essential. Of the 12 subunits that connect to one or two other subunits, five are essential, and four of them are in the crucial head domain (Med22, Med11, Med6 and Med19). Because of the interactions between the head subunits and RNA polymerase II, these latter four subunits might be essential for reasons unrelated to a hub function. In summary, therefore, Mediator contains a distribution of connections per subunit (node) that varies for the different subunits, and a high number of intra-subunit connections appear to correlate to some extent with the essential nature of those subunits.
Why might this distribution of connections between subunits be important? It has been proposed that there are two types of networks: assortative and disassortative (Barabasi and Oltvai, 2004). Assortative networks are defined by a preponderance of direct connections between highly connected nodes or hubs, and seem to be common in non-biological networks (Newman, 2002). Disassortative networks, by contrast, connect hubs to peripheral nodes such that there is a distance between hubs. Assortative networks are more robust to disruption of individual nodes than are disassortative networks (Newman, 2002). As described above, Mediator contains several highly connected subunits (Med1, Med7, Med4, Med10, Med21 and Med17) that all interconnect, suggesting that the complex represents an assortative network.
Also relevant is another property of large protein complexes: coupling energy, which refers to the energy that is required to induce a particular conformational state. In a one-dimensional linear network, coupling energy increases with an increase in the number of subunits that are added. In a two-dimensional system, the coupling energy actually decreases with increasing subunit numbers (Bray and Duke, 2004), and changing allosteric conformations becomes energetically more favorable. Thus, the rapid propagation of allosteric states through a network might be aided by its assortative (i.e. highly connected) nature: the assortative properties reduce the coupling energy, facilitating the allosteric changes required for information transfer.
Finally, there is the important issue of the actual path of the allosteric information within the complex. Genetic analyses of Mediator mutations have been used to discern its internal ‘signaling pathways’. For example, Holstege and colleagues used genetic analysis to show that Med2 and Med18 functions are downstream of signals from the CDK8 module (van de Peppel et al., 2005). This type of analysis, combined with structural and physical interaction data, should allow us to obtain a complete picture of the information transfer within the Mediator complex.
Interpreting structural data in terms of the MANM
Structural studies of Mediator indicate that the complex adopts different conformations after the binding of different activators. Vitamin D receptor (VDR) and thyroid receptor (TR) are both transcriptional activators that bind to the large Med1 subunit of Mediator (Rachez et al., 1999; Ren et al., 2000; Treuter et al., 1999). Cryo-electron microscopy (cryo-EM) at 29-Å resolution shows that structures of Mediator bound to either VDR or TR have similar conformations. These structures differ from that of Mediator associated with the activators VP16 or SREBP, both of which bind to other Mediator subunits (Naar et al., 2002; Taatjes et al., 2002; Taatjes et al., 2004; Taatjes and Tjian, 2004). The finding that the binding of different activators to different Mediator subunits induces markedly different Mediator conformations is the main premise of the MANM: i.e. the binding of different activators to different Mediator subunits activates a different allosteric network of conformational changes. The resolution of these different structures provides support for the idea that different allosteric conformations of Mediator are possible, and that they are induced by the binding of different activators.
Applying the MANM to the function of the CDK8 module
The CDK8 module of Mediator (which contains CDK8, cyclin C, Med12 and Med13) is known to both activate and repress Mediator function (Bjorklund and Gustafsson, 2005). How this occurs is not clear, but several models exist to explain the observation (Bjorklund and Gustafsson, 2005; Malik and Roeder, 2005). It is clear that the repressive functions of the CDK8 module must often be inhibited for transcription to occur (Bjorklund and Gustafsson, 2005; Malik and Roeder, 2005). There is a clear correlation between the presence of the CDK8 module and an inactive promoter, and it has been shown that there is a release of CDK8 from the promoter after induction of transcription (Mo et al., 2004; Pavri et al., 2005). How does the CDK8 module function at the molecular level? It is possible to speculate that the CDK8 module locks the Mediator complex in a ‘negative allosteric state’ such that allosteric conformation propagation does not occur, an active conformational state is not achieved and transcription cannot be activated. Removal of the CDK8 module ‘lock’ allows propagation of the allosteric signal and subsequent transcriptional activation. It is known that certain factors, such as PARP-1, are required for the dissociation of the CDK8 module from the promoter (Pavri et al., 2005). Alternatively, another function of the CDK8 module might be to re-associate with Mediator and ‘reset’ its conformation into an inactive state after the active complex is released from a promoter.
The output signal
What is the output signal of the conformational changes that propagate through Mediator? And why does Mediator adopt different conformations if ultimately these conformations all bind to RNA polymerase II? The answer to these questions requires understanding the mechanisms by which Mediator influences transcription, which are still unclear. Nevertheless, several possibilities can be suggested. First, the end result of the conformational changes might be the targeting and stimulation of the cyclin-dependent kinase 7 (CDK7)-associated functions of the general transcription factor TFIIH; Mediator is known to stimulate these functions (Kim et al., 1994). Second, the various conformational states of Mediator might also influence the functional interactions between Mediator and TFIID that have been shown to occur in vitro (Johnson and Carey, 2003; Johnson et al., 2002). Third, specific conformational changes might define the specificity of different activators for core promoter elements (Butler and Kadonaga, 2001): for example, activator X could induce Mediator conformation Y, which would then force TFIID to adopt a particular conformation Z such that it binds selectively to only certain core promoter elements. Finally, it is possible that RNA polymerase II also undergoes conformational changes in response to conformational changes propagating through the head (and/or middle) modules of Mediator.
Conclusions
There is currently no theoretical framework that can fully explain all aspects of large protein complexes, including their functions, subunit composition and internal organization. The MANM aims to define the Mediator complex and other large complexes in terms of multiple networks that are activated by allosteric states that propagate through a complex. These different networks are defined by the subunits they comprise and by the transcriptional activators that induce that network. The MANM can supply answers to several questions: what is the reason for the internal organization of large protein complexes? How does one explain the distribution of subunits within a complex? Why do some subunits interact with certain subunits and not others?
I have discussed here several lines of experimental evidence in the context of the MANM. Further support for the MANM could be obtained by identifying energetically coupled, co-evolving residues in Mediator subunits and assessing their functional significance by mutagenesis (Shulman et al., 2004). Structural data supports the idea that Mediator can adopt several different conformational states. These types of experiments could be extended to the various reconstituted modules that exist (Koh et al., 1998). If the MANM is correct, one would expect to see different module conformations after activator binding, which should be detectable by cryo-EM. It might even be possible to detect such conformational changes for binary interactions between subunits. The conformational changes required for transcriptional activation to occur might be blocked in vitro by CDK8-module components, if indeed the CDK8 module acts as an allosteric lock, as predicted. Further analysis and modeling of putative Mediator networks using graph theory will aid in understanding the dynamics of the MANM.
The proposed model does not claim that other interpretations are incorrect, but rather that many theories can be brought together under the umbrella of the MANM. Although it remains to be proven, the MANM hypothesis illustrates the potential and application of systems biology to provide a new conceptual framework for understanding transcription biochemistry, and to build bridges between experimental biology and systems biology.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Special thanks to Patrick Trojer, Robert Sims, Michael Hampsey, Joseph Fondell, David Levens, and Dinah Singer for critical reading of the manuscript.
References
- Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., Pietrokovski S. (2004). Network analysis of protein structures identifies functional residues. J. Mol. Biol. 344, 1135-1146 [DOI] [PubMed] [Google Scholar]
- Asturias F. J., Jiang Y. W., Myers L. C., Gustafsson C. M., Kornberg R. D. (1999). Conserved structures of mediator and RNA polymerase II holoenzyme. Science 283, 985-987 [DOI] [PubMed] [Google Scholar]
- Barabasi A. L., Oltvai Z. N. (2004). Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101-113 [DOI] [PubMed] [Google Scholar]
- Bjorklund S., Gustafsson C. M. (2005). The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30, 240-244 [DOI] [PubMed] [Google Scholar]
- Bray D., Duke T. (2004). Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu. Rev. Biophys. Biomol. Struct. 33, 53-73 [DOI] [PubMed] [Google Scholar]
- Butler J. E., Kadonaga J. T. (2001). Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick J. Z., Asturias F. J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30, 264-271 [DOI] [PubMed] [Google Scholar]
- Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W. (2005). The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250-255 [DOI] [PubMed] [Google Scholar]
- Davis J. A., Takagi Y., Kornberg R. D., Asturias F. A. (2002). Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10, 409-415 [DOI] [PubMed] [Google Scholar]
- del Sol A., O'Meara P. (2005). Small-world network approach to identify key residues in protein-protein interaction. Proteins 58, 672-682 [DOI] [PubMed] [Google Scholar]
- del Sol A., Fujihashi H., Amoros D., Nussinov R. (2006). Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol. 2, 2006.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson M. R., Yuan C. X., Roeder R. G., Myers L. C., Gustafsson C. M., Jiang Y. W., Li Y., Kornberg R. D., Asturias F. J. (2000). Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97, 14307-14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber J. E., Yeh B. J., Bhattacharyya R. P., Lim W. A. (2004). Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr. Opin. Struct. Biol. 14, 690-699 [DOI] [PubMed] [Google Scholar]
- Duke T. A., Le Novere N., Bray D. (2001). Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 308, 541-553 [DOI] [PubMed] [Google Scholar]
- Greene L. H., Higman V. A. (2003). Uncovering network systems within protein structures. J. Mol. Biol. 334, 781-791 [DOI] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N. L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H. M., Holstege F. C., Werner M. (2004). A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32, 5379-5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Mason S. P., Barabasi A. L., Oltvai Z. N. (2001). Lethality and centrality in protein networks. Nature 411, 41-42 [DOI] [PubMed] [Google Scholar]
- Jiang Y. W., Stillman D. J. (1995). Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics 140, 103-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. W., Dohrmann P. R., Stillman D. J. (1995). Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 140, 47-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. M., Carey M. (2003). Assembly of a Mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr. Biol. 13, 772-777 [DOI] [PubMed] [Google Scholar]
- Johnson K. M., Wang J., Smallwood A., Arayata C., Carey M. (2002). TFIID and human Mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16, 1852-1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Lis J. T. (2005). Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem. Sci. 30, 245-249 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Bjorklund S., Li Y., Sayre M. H., Kornberg R. D. (1994). A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599-608 [DOI] [PubMed] [Google Scholar]
- Koh S. S., Ansari A. Z., Ptashne M., Young R. A. (1998). An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1, 895-904 [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235-239 [DOI] [PubMed] [Google Scholar]
- Lee Y. C., Park J. M., Min S., Han S. J., Kim Y. J. (1999). An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19, 2967-2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. A., Reinberg D. (2003). The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116, 3667-3675 [DOI] [PubMed] [Google Scholar]
- Lockless S. W., Ranganathan R. (1999). Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295-299 [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R. G. (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30, 256-263 [DOI] [PubMed] [Google Scholar]
- Mo X., Kowenz-Leutz E., Xu H., Leutz A. (2004). Ras induces Mediator complex exchange on C/EBP beta. Mol. Cell 13, 241-250 [DOI] [PubMed] [Google Scholar]
- Myers L. C., Kornberg R. D. (2000). Mediator of transcriptional regulation. Annu. Rev. Biochem. 69, 729-749 [DOI] [PubMed] [Google Scholar]
- Myers L. C., Gustafsson C. M., Bushnell D. A., Lui M., Erdjument-Bromage H., Tempst P., Kornberg R. D. (1998). The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12, 45-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar A. M., Taatjes D. J., Zhai W., Nogales E., Tjian R. (2002). Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16, 1339-1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. E. (2002). Assortative mixing in networks. Phys. Rev. Lett. 89, 208701 [DOI] [PubMed] [Google Scholar]
- Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. (2005). PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18, 83-96 [DOI] [PubMed] [Google Scholar]
- Rachez C., Lemon B. D., Suldan Z., Bromleigh V., Gamble M., Naar A. M., Erdjument-Bromage H., Tempst P., Freedman L. P. (1999). Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398, 824-828 [DOI] [PubMed] [Google Scholar]
- Ren Y., Behre E., Ren Z., Zhang J., Wang Q., Fondell J. D. (2000). Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 20, 5433-5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman A. I., Larson C., Mangelsdorf D. J., Ranganathan R. (2004). Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116, 417-429 [DOI] [PubMed] [Google Scholar]
- Spirin V., Mirny L. A. (2003). Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. USA 100, 12123-12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel G. M., Lockless S. W., Wall M. A., Ranganathan R. (2003). Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10, 59-69 [DOI] [PubMed] [Google Scholar]
- Taatjes D. J., Tjian R. (2004). Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell 14, 675-683 [DOI] [PubMed] [Google Scholar]
- Taatjes D. J., Naar A. M., Andel F., 3rd, Nogales E., Tjian R. (2002). Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058-1062 [DOI] [PubMed] [Google Scholar]
- Taatjes D. J., Schneider-Poetsch T., Tjian R. (2004). Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 11, 664-671 [DOI] [PubMed] [Google Scholar]
- Treuter E., Johansson L., Thomsen J. S., Warnmark A., Leers J., Pelto-Huikko M., Sjoberg M., Wright A. P., Spyrou G., Gustafsson J. A. (1999). Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J. Biol. Chem. 274, 6667-6777 [DOI] [PubMed] [Google Scholar]
- van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T. T., van Leenen D., Holstege F. C. (2005). Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 19, 511-522 [DOI] [PubMed] [Google Scholar]
- Watts D. J., Strogatz S. H. (1998). Collective dynamics of ‘small-world’ networks. Nature 393, 440-442 [DOI] [PubMed] [Google Scholar]